Abstract
Background
Human serum albumin (HSA) is often selected as a subject of any study because albumin is the most abundant protein in human blood plasma. NMR is recognized as a valuable method to determine the structure of proteins-ligand and protein-drug complexes.
Objective – Aim of the study
In this study, protein drug interactions were investigated using 5-Fluorouracil anti-cancer drug and human serum albumin protein.
Materials and methods
In this context 400 MHz NMR spectrometry was used and NMR relaxation rates in drug-albumin complex were investigated with respect to increase albumin concentration and increase in 5-Fluorouracil (5-FU)-albumin solution temperature.
Results
The results of this study indicated that 5-FU had a weak association with albumin, and it easily dissociated from the protein to which it was attached.
Conclusion
The obtained results also gave us useful information about molecular dynamics of drug-albumin interactions.
Öz
Geçmiş
İnsan serum albümini (HSA) sıklıkla herhangi bir çalışmanın konusu olarak seçilir çünkü albümin, insan kan plazmasında en bol bulunan proteindir. NMR, protein-ligand ve protein-ilaç komplekslerinin yapısını belirlemek için değerli bir yöntem olarak kabul edilmektedir.
Amaç – Aim of the study
Bu çalışmada, 5-Fluorourasil anti-kanser ilacı ve insan serum albümin proteini kullanılarak protein ilaç etkileşimleri araştırılmıştır.
Araç ve Yöntemler
Bu bağlamda, 400 MHz NMR spektrometresi kullanılmış ve ilaç-albümin kompleksi içindeki NMR rölaksasyon (gevşeme) oranları, albümin konsantrasyonunun arttırılması ve 5-Flüoroürasil (5-FU)-albümin çözelti sıcaklığındaki artış açısından incelenmiştir.
Bulgular
Bu çalışmanın sonuçları, 5-FU’nun albümin ile zayıf bir ilişkisinin olduğunu ve ekli olduğu proteinden kolayca ayrıldığını göstermiştir.
Sonuç
Elde edilen sonuçlar bize ilaç-albümin etkileşimlerinin moleküler dinamikleri hakkında da faydalı bilgiler vermiştir.
Introduction
NMR has been a powerful tool for physicochemical analysis over almost 50 years, and NMR Spectroscopy is known as a valuable method for determining the structure of proteins, protein-ligand and protein-drug complexes [1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19]. Since a drug is usually transported as a complex with serum albumins in a circulatory system, the binding affinity of drugs to albumin plays a very important role in the determination of therapeutic dosage of drugs. Such binding affinities can be determined by NMR [20], [21]. During long-term treatment such as chemotherapy, a combination of different drugs is often required [22], [23]. The effect of the secondary drug used in the combination chemotherapy can also be analyzed in terms of NMR parameters such as chemical shifts and the line width of single proton and carbon resonances [24], [25], [26], [27], [28], [29], [30], [31], [32]. Such studies will always be the interest of research due to strong need for new generation drugs.
In the present work, HSA is selected as an object of study because albumin is the most abundant protein in human blood plasma. Albumin also plays an important role while storing and transporting drugs in human blood. 5-Fluorouracil (5-FU) is selected as drug since it is more often used in cancer therapy. To our best knowledge, the interaction between albumin and 5-FU has not been studied by NMR so far.
This work aims to gather information on binding affinity of the drug to albumin and also on the molecular dynamics of the drug-albumin complex. For these reason, the T1 and T2 relaxation times in D2O solutions of albumin, in the presence and absence of 5-FU, were measured by 400 MHz NMR at different temperatures.
Materials and methods
5-FU, HSA, and deuterium oxide (D2O) were obtained from Sigma-Aldrich Chemical Co. Six mixing bowls were prepared and 0.05 M 5-FU was added into each of them. One milliliter D2O is added into each bowl and dissolution of the drug is provided in D2O. Albumin is added to the D2O-5-FU solution in each bowl in the amounts of 0, 0.01, 0.02, 0.03, 0.04, and 0.05 g, respectively, provided that albumin is thoroughly dissolved in the solutions. Similar concentrations of albumin in D2O were also prepared in the absence of 5-FU. After complete dissolution of albumin, six different samples for 5-FU with varying concentrations of albumin were transferred to the NMR tube and measurements were then taken. Similar relaxation measurements were made for pure albumin concentrations (5-FU free samples), too.
To examine the effect of temperature in protein-drug interactions, 0.05 M 5-FU is added into a mixing vessel, 1 mL D2O is added into the vessel, and dissolution of the drug is provided. Then 0.05 g of albumin is added into a D2O-5-FU solution, provided that albumin is thoroughly dissolved in the solutions. Consequently, the resulting solution is transferred to the NMR tube. The 1H NMR spectrum and spin-lattice relaxation time (T1) and spin-spin relaxation time (T2) were measured for this sample at 298, 303, 308, 313, and 318 K.
Relaxation measurements were carried out with a Bruker Avance 400 spectrometer using 5 mm tubes. For proton measurements the probe was tuned at 400 MHz. For water suppression, the presaturation method was used. T1 relaxation times measurements were carried out using Inversion Recovery Technique [the (180-τ-90) pulse sequence.] T2 relaxation times measurements were carried out using CPMG (Carr-Purcell-Meiboom-Gill) technique [the (90-τ-180) pulse sequence]. In order to get the most suitable recovery curve for T1 and decay curves for T2, the more suitable set of τ delay times was chosen for each peak in the observed spectrum. A representative T1 and T2 curves of the HDO solution containing 0.05 M 5-FU+0.01 g albumin are shown in Figure 1.
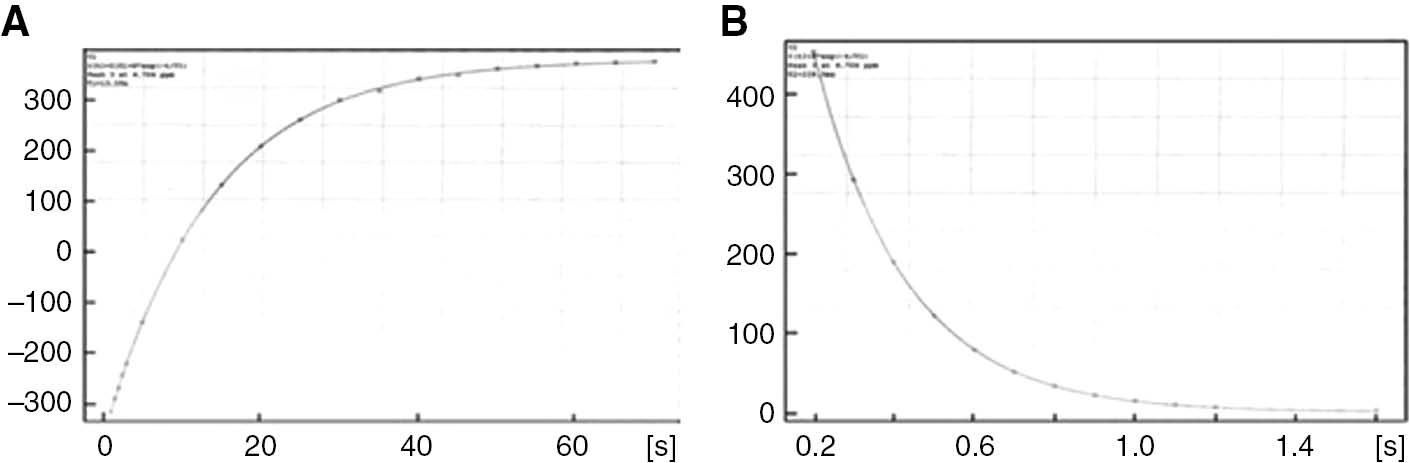
T1 (A) and T2 (B) profile of HDO signal in D20 with 0.05 M 5-FU+0.01 g albumin sample.
Results
Figure 2 shows 400 MHz 1H NMR spectrum of the solution with 0.05 M 5-FU. In this spectrum, the HDO signal was observed at 4.70 ppm, whereas the doublet peak of CH belongs to 5-FU was seen at 7.59 and 7.58 ppm. The CH molecule peak splits into two parts and forms a doublet due to the neighborhood with HN molecule. The NMR spectrum of pure albumin in D20 was not given here for the sake of brevity.
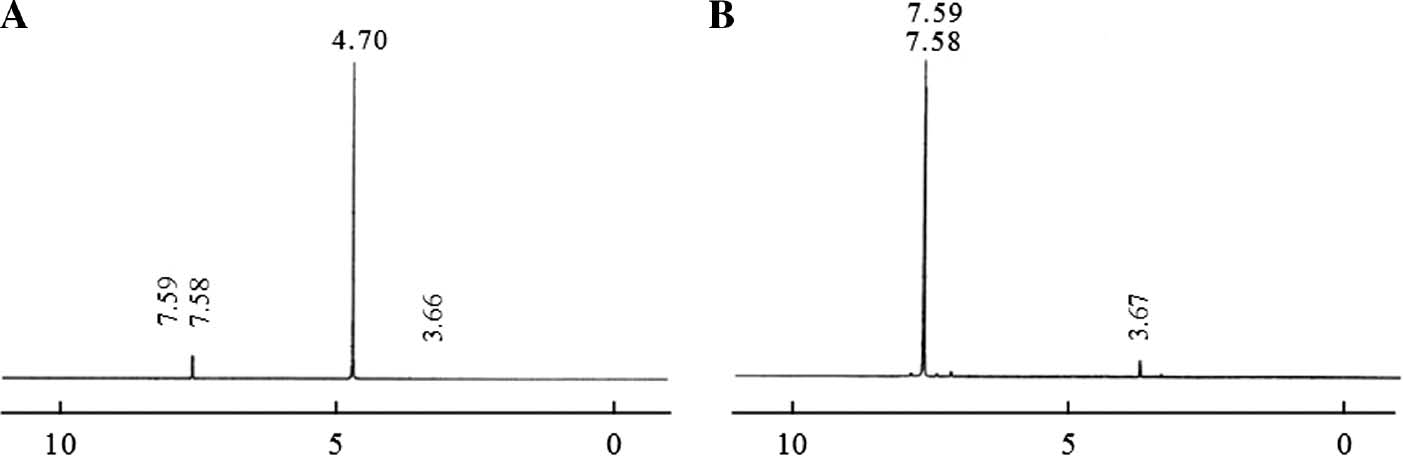
400 MHz 1H NMR spectrum of 5-FU single pulse (A), pre-saturation (B).
The HDO signal in the NMR spectrum, shown in Figure 2, was suppressed using the pre-saturation technique, and in this case the resulting 1H NMR spectrum is shown in Figure 2B. Referring to Figure 2B, the HDO signal is completely eliminated, whereas the intensity of the signal shown in 3.67 ppm for CH was observed to increase. The two spectra shown in Figure 2A, B have been selected in the same vertical axis scale and the corresponding comparisons have been discussed for this case.
The T1 and T2 relaxation rates in the solutions with different albumin contents are shown in Table 1. The relaxation rates of the signal in 3.66 ppm could not be calculated because of T1 and T2 relaxation times measurements did not yield any results. The plots for the 1/T1 and 1/T2 rates of albumin, HDO and the doublet versus albumin concentrations are given in Figure 3A, B and C, respectively. As shown in Figure 3B, C; 1/T1 and 1/T2 rates of HDO and CH signal in the solution with 5-FU increase linearly with the concentration of albumin. The relaxation enhancements for T2 are much higher than those for T1.
The 1/T1 and 1/T2 relaxation rates of each peak in 1H NMR spectrum of six different samples obtained by adding 0, 0.01, 0.02, 0.03, 0.04, and 0.05 g albumin into 1 mL D2O-0.05 M fixed concentration 5-FU at 250 C (298 K) constant temperature.
| Albumin concentration (g/mL) | 1/T1 (s−1) | 1/T2 (s−1) | ||||
|---|---|---|---|---|---|---|
| HDO with albumin | HDO with albumin+5-FU | Doublet (CH) | HDO with albumin | HDQ-ALB+5-FU | Doublet (CH) | |
| 0 | 0.0635 | 0.0563 | 0.0771 | 0.3224 | 0.5353 | 0.1254 |
| 0.01 | 0.0871 | 0.0735 | 0.2142 | 0.8865 | 4.3764 | 1.7870 |
| 0.02 | 0.0986 | 0.0877 | 0.3446 | 1.0995 | 4.7328 | 3.670 |
| 0.03 | 0.1055 | 0.0987 | 0.4384 | 1.1477 | 6.2657 | 5.2970 |
| 0.04 | 0.1242 | 0.1112 | 0.5333 | 1.4524 | 7.1994 | 7.0670 |
| 0.05 | 0.1285 | 0.1279 | 0.5787 | 1.7401 | 8.4317 | 8.7570 |
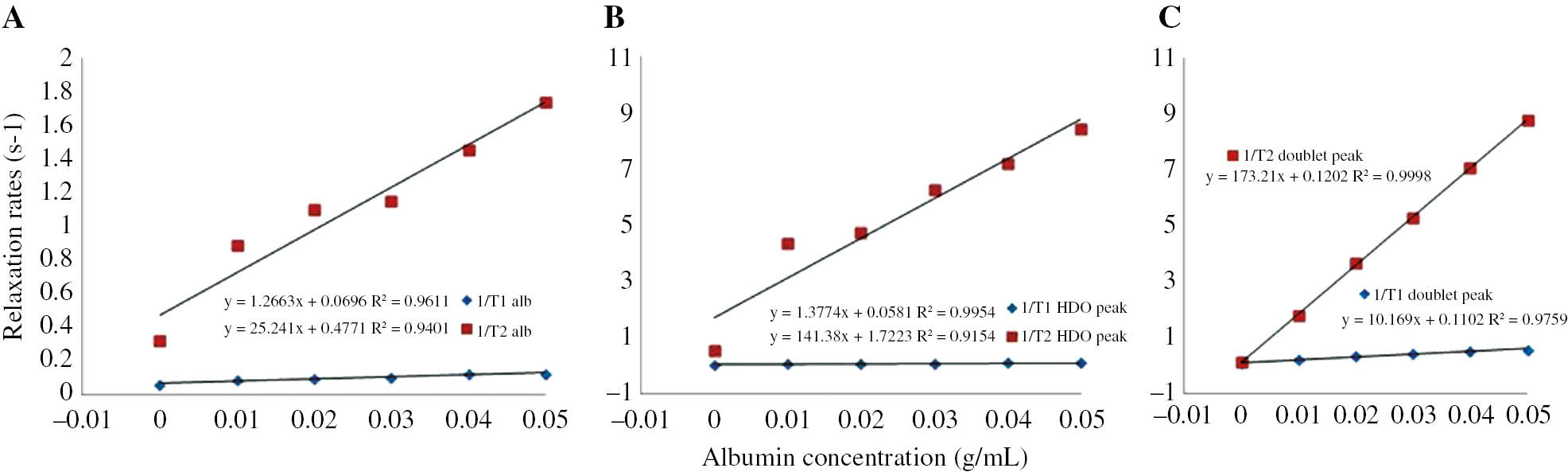
Graph for the relaxation rates of 5-FU peaks versus six different concentrations of albumin.
The relaxation rates in the solution at different temperatures are shown in Table 2, while the temperature dependence of the 1/T1 and 1/T2 for HDO and CH signal are given in Figure 4A and B, respectively. It is seen that 1/T1 values of each peak and 1/T2 values of the HDO increased slightly with temperature but 1/T2 of the CH decreased strongly.
The 1/T1 and 1/T2 relaxation rates of each peak of 1H NMR spectrum for the sample prepared by adding 0.05 g albumin into 1 mL D2O-0.05 M fixed concentration 5-FU at 298, 303, 308, 313, and 318 K.
| Temperature (K) | 1/T1 (s−1) | 1/T2 (s−1) | ||
|---|---|---|---|---|
| HDO+albumin+5-FU | Doublet (CH) | HDO+albumin+5-FU | Doublet (CH) | |
| 298 K | 0.1279 | 0.5787 | 8.432 | 8.756 |
| 303 K | 0.1251 | 0.4847 | 9.009 | 6.784 |
| 308 K | 0.1178 | 0.4353 | 9.107 | 5.171 |
| 313 K | 0.1125 | 0.3886 | 9.208 | 3.825 |
| 318 K | 0.1256 | 0.3859 | 8.865 | 2.940 |
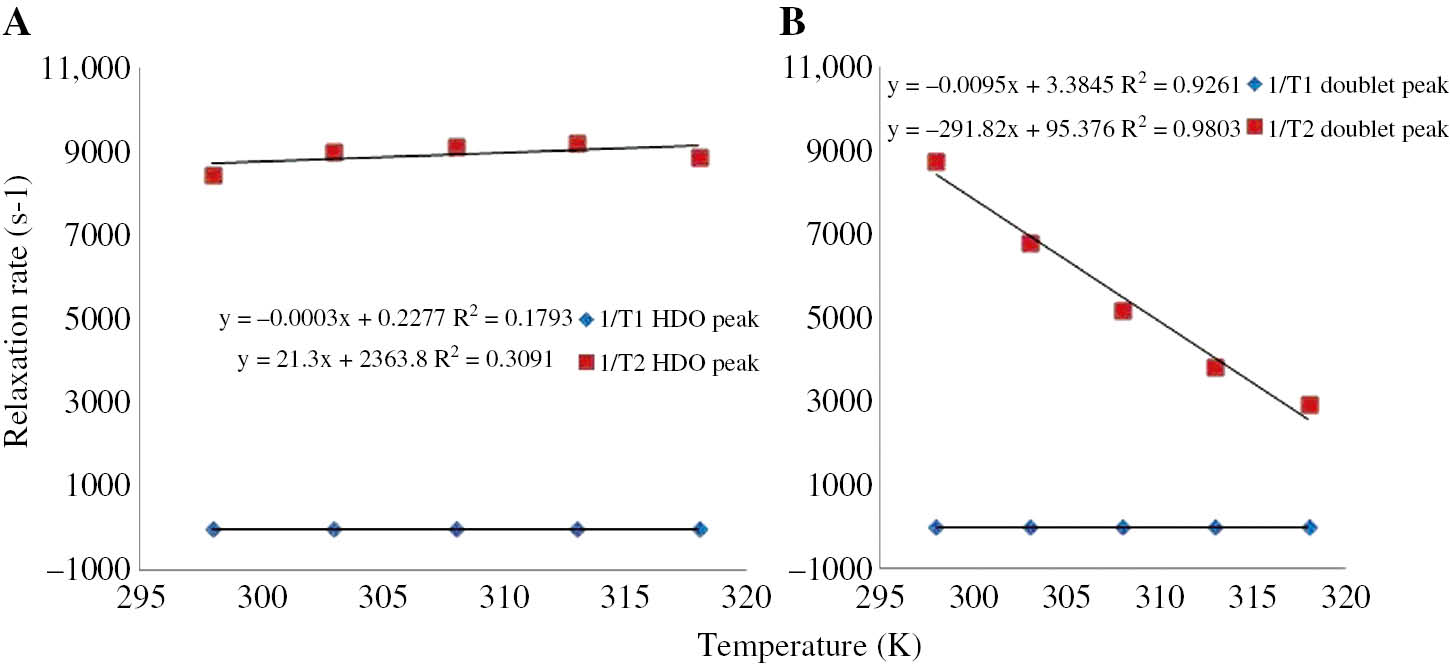
Graph of the relaxation rates of 5-FU peaks versus five different temperatures.
Discussion
Fielding and his collaborators discussed the advantage of using bovine serum albumin (BSA) as a model protein to test NMR techniques for the investigation of protein–ligand interactions. In their work, the binding affinity and stoichiometry of the specific binding site for some drugs such as L-tryptophan, D-tryptophan, naproxen, ibuprofen, salicylic acid, and warfarin have been established. In contrast to other techniques used in structural biology, NMR enables the determination of binding constants, too [33], [34]. Determination of the binding constants or binding affinity is involved in proton T1-relaxation enhancement which is difference of the relaxation rates between drug-bound and -unbound cases [35], [36]. In the present case such difference can be calculated from extraction of values in third column from those of first one in Table 2. The small difference indicates binding of drug to albumin. This is consistent with previous literature given above. On the other hand, crystal structure analyses have showed that the drug binding sites are located in subdomains IIA and IIIA. The IIA subdomain has a large hydrophobic cavity [37], [38]. The present study is also convenient to the presence of drug-binding sites of albumin.
Chemotherapy is a treatment used to treat cancer. Surgical intervention and radiation therapy can kill or damage cancer cells in a particular region, but chemotherapy is used for working on the entire body. Over 100 chemotherapy drugs and their different combinations are used to treat cancer. Chemotherapy drugs can be used individually to treat cancer, but there are also cases of more than one drug being used in combination (called combination therapy). In combination therapy, several drugs with different effects work together to damage or kill large amounts of cancer cells [39], [40]. 5-FU is one of the most commonly used chemotherapy drugs to treat cancer. It has played a major role in the treatment of different types of cancer such as colorectal and breast cancers. As a result of understanding the mechanism of 5-FU action, different methods have been developed to enhance its anticancer activity since the last 50 years. In contrast to all these advances, the clinical use of 5-FU has important limitations due to drug resistance [41], [42]. Many articles contain information related to the chemical structure of 5-FU [43], [44], [45], [46]. The molecular weight of 5-FU is 130.08 g/mol and its chemical formula is C4H3FN2O2. The binding of 5-FU to native and modified HSA was studied by several spectroscopic techniques. In one of these studies Bertucci et al. investigate the binding of 5-FU to native and modified human serum albumin by UV, CD, and 1H and 19F NMR [47]. Dissociation of FU from albumin is also important since albumin is just a carrier to transport drug to cancer tissue. The small relaxation increase in Table 1 indicates that binding is week. As a result, and that 5-FU dissociates from albumin easily. This is also consistent with previous results [48], [49].
On the other hand, the linear relationship between the relaxation rates and albumin concentration, given in Figure 1, indicates the presence of a fast chemical exchange between protons of HDO and albumin. This also consistent with data in Table 2 and Figure 4. However decrease in the 1/T1 of HDO+albumin+5-FU and the relaxation rate of CH versus T indicate dipolar relaxation mechanism. On the other hand, the increase in 1/T2 of HDO+albumin+5-FU versus T indicates a different relaxation mechanism, so called spin rotation mechanism. The spin-rotation theory for the relaxation mechanism of chemical molecules has already been given in several studies. The spin rotation contribution to 1/T2 is given by
where k is the Boltzmann constant, T is the sample temperature, τsr is the correlation time for spin rotation, C is the spin-rotation interaction constant and I is the moment of inertia of the molecule [50]. Linear dependence of 1/T2 on T in Eq. 1 is consistent with T2 data in Table 2 and the plot of 1/T2 in Figure 4A. It is also consistent with T2 data obtained for albumin in D2O.
Conclusions
Our results suggest that 5-FU is weekly associated to albumin, and easily dissociates from protein. The data also suggest that the relaxation mechanisms of CH and T1 mechanism of D2O+albumin+5-FU are dipolar, while 1/T2 mechanism of D2O+albumin+5-FU is spin rotational [50], [51] since probability of bound phase is nearly 1 bound phase dominates the relaxation. Temperature dependence of data suggests that the relaxation of bound phase is caused by spin rotational relaxation mechanism [50].
Acknowledgements
First, I thank my academic advisor Prof. Dr. Ali YILMAZ. This work includes a part of the doctoral thesis who supported Dicle University Scientific Research Project (DÜBAP 08-FF-10).
Conflict of interest: The authors declare that there is no conflict of interests regarding the publication of this article.
References
1. Peters Jr T. Serum albumin. Adv Protein Chem 1985;37:161–245.10.1016/S0065-3233(08)60065-0Search in Google Scholar
2. He XM, Carter DC. Atomic structure and chemistry of human serum albumin. Nature 1992;358:209–15.10.1038/358209a0Search in Google Scholar
3. Ishida S, Kinoshita M, Sakiya Y, Taira Z, Ichikawa T. Glycyrrhizin binding site on human serum albumin. Chem Pharm Bull 1992;40:275–8.10.1248/cpb.40.275Search in Google Scholar
4. Rahman MH, Yamasaki M, Shin YH, Lin CC, Otagiri M. Characterization of high affinity binding sites of non-steroidal anti-inflammatory drugs with respect to site-specific probes on human serum albumin. Biol Pharm Bull 1993;16:1169–74.10.1248/bpb.16.1169Search in Google Scholar
5. Ghuman J, Zunszain PA, Petitpas I, Bhattacharya AA, Otagiri M, Curry S. Structural basis of the drug-binding specificity of human serum albumin. J Mol Biol 2005;353:38–52.10.1016/j.jmb.2005.07.075Search in Google Scholar
6. Sun P, Hoops A, Hartwick RA. Enchanced albumin protein separations and protein-drug binding constant measurements using anti-inflammatory drugs as run buffer additives in affinity capillary electrophoresis. J Chromatogr B Biomed Appl 1994;661:335–40.10.1016/0378-4347(94)00372-6Search in Google Scholar
7. Ascoli G, Bertucci C, Salvadori P. Stereospecific and competitive binding of drugs to human serum albumin: a difference circular dichroism approach. J Pharm Sci 1995;84:737–41.10.1002/jps.2600840615Search in Google Scholar
8. Aubry AF, Markoglou N, Mcgann A. Comparison of drug binding interactions on human, rat and rabbit serum albumin using high-performance displacement chromatography. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol 1995;112:257–66.10.1016/0742-8413(95)02019-5Search in Google Scholar
9. Epps DE, Raub TJ, Kezdy FJ. A general, wide-range spectrofluorometric method for measuring the site-specific affinities of drugs toward human serum albumin. Anal Biochem 1995;227:342–50.10.1006/abio.1995.1290Search in Google Scholar PubMed
10. Castillo M, Smith PC. Disposition and reactivity of ibuprofen and ibufenac acyl glucuronides in vivo in the rhesus monkey and in vitro with human serum albumin. Drug Metab Dispos 1995;23:566–72.Search in Google Scholar
11. Blondeau JP, Robel P. Determination of protein-ligand binding constants at equilibrium in biological samples. Eur J Biochem 1975;55:375–84.10.1111/j.1432-1033.1975.tb02172.xSearch in Google Scholar PubMed
12. Hage DS, Noctor TA, Wainer IW. Characterization of the protein binding of chiral drugs by high-performance affinity chromatography interactions of R- and S-ibuprofen with human serum albumin. J Chromatogr A 1995;693:23–32.10.1016/0021-9673(94)01009-4Search in Google Scholar
13. Hashimoto S, Yabusaki T, Takeuchi H, Harada I. Structure and ligand-binding modes of human serum albumin studied by UV resonance raman spectroscopy. Biospectroscopy 1995;1:375–85.10.1002/bspy.350010603Search in Google Scholar
14. Cheruvallath VK, Riley CM, Narayanan SR, Lindenbaum S, Perrin JH. The effect of octanoic acid on the binding of the enantiomers of ibuprofen and naproxen to human serum albumin: a chromatographic implication. Pharm Res 1996;13:173–8.10.1023/A:1016066325476Search in Google Scholar
15. Andrisano V, Booth TD, Cavrini V, Wainer IW. Enantioselective separation of chiral arylcarboxylic acids on an immobilized human serum albumin chiral stationary phase. Chirality 1997;9:178–83.10.1002/(SICI)1520-636X(1997)9:2<178::AID-CHIR19>3.0.CO;2-KSearch in Google Scholar
16. Cheruvallath VK, Riley CM, Narayanan SR, Lindenbaum S, Perrin JH. A quantitative circular dichroic investigation of the binding of the enantiomers of ibuprofen and naproxen to human serum albumin. J Pharm Biomed Anal 1997;15:1719–24.10.1016/S0731-7085(96)01956-5Search in Google Scholar
17. Itoh T, Saura Y, Tsuda Y, Yamada H. Stereoselectivity and enantiomer-enantiomer interactions in the binding of ibuprofen to human serum albumin. Chirality 1997;9:643–9.10.1002/(SICI)1520-636X(1997)9:7<643::AID-CHIR1>3.0.CO;2-8Search in Google Scholar
18. Barretta GU, Bertucci C, Domenici E, Salvadori P. Conformational and dynamic changes of d- and l-tryptophan due to stereoselective interaction with human serum albumin, as revealed by proton-selective relaxation rate measurements. J Am Chem Soc 1991;113:7017–9.10.1021/ja00018a046Search in Google Scholar
19. Fielding L. NMR methods for the determination of protein–ligand dissociation constants. Prog Magn Reson Spectrosc 2007;51:219–42.10.1016/j.pnmrs.2007.04.001Search in Google Scholar
20. Wood ML, Hardy PA. Proton relaxation enhancement. J Magn Reson Imaging 1993;3:149–56.10.1002/jmri.1880030127Search in Google Scholar
21. Spanoghe M, Lanens D, Dommisse R, Van der Linden A, Alderweireldt F. Proton relaxation enhancement by means of serum albumin and poly-L-lysine labeled with DTPA-Gd3+: relaxivities as a function of molecular weight and conjugation efficiency. Magn Reson Imaging 1992;10:913–7.10.1016/0730-725X(92)90445-6Search in Google Scholar
22. Sułkowska A, Bojko B, Równicka J, Sułkowski W. Competition of drugs to serum albumin in combination therapy. Biopolymers 2004;74:256–62.10.1002/bip.20031Search in Google Scholar
23. Martini S, Bonechi C, Casolaro M, Corbini G, Rossi C. Drug-protein recognition processes investigated by NMR relaxation data. A study on corticosteroid-albumin interactions. Biochem Pharmacol 2006;71:858–64.10.1016/j.bcp.2005.12.010Search in Google Scholar
24. Sułkowska A, Bojko B, Równicka J, Rezner P, Sułkowski W. The competition of drugs to serum albumin in combination chemotherapy: NMR study. J Mol Struct 2005;744747:781–7.10.1016/j.molstruc.2004.11.050Search in Google Scholar
25. Yushmanov VE, Tominaga TT, Borissevitch IE, Imasato H, Tabak M. Binding of manganese and iron tetraphenylporphine sulfonates to albumin is relevant to their contrast properties. Magn Reson Imaging 1996;14:255–61.10.1016/0730-725X(95)02103-ZSearch in Google Scholar
26. Ma Y, Liu M, Mao X, Nicholson JK, Lindon JC. Magn Reson Chem 1999;37:251–330.10.1002/(SICI)1097-458X(199904)37:4<251::AID-MRC406>3.0.CO;2-MSearch in Google Scholar
27. Lucas LH, Yan J, Larive CK, Zartler ER, Shapiro MJ. Transferred nuclear overhauser effect in nuclear magnetic resonance diffusion measurements of ligand-protein binding. Anal Chem 2003;75:627–34.10.1021/ac020563oSearch in Google Scholar
28. Caravan P, Cloutier NJ, Greenfield MT, McDermid SA, Dunham SU, Bulte JW, et al. The interaction of MS-325 with human serum albumin and its effect on proton relaxation rates. J Am Chem Soc 2002;124:3152–62.10.1021/ja017168kSearch in Google Scholar
29. Peng JW, Moore J, Abdul-Manan N. NMR experiments for lead generation in drug discovery. Prog Nucl Magn Reson Spectrosc 2004;44:225–56.10.1016/j.pnmrs.2004.03.001Search in Google Scholar
30. Fielding L, Fletcher D, Rutherford S, Kaur J, Mestres J. Exploring the active site of human factor Xa protein by NMR screening of small molecule probes. Org Biomol Chem 2003;1:4235–41.10.1039/b310265cSearch in Google Scholar
31. Chekin F, Bordbar M, Fathollahi Y, Alizadeh N. The interaction between ketamine and some crown ethers in common organic solvents studied by NMR: the effect of donating atoms and ligand structure. Spectrochimica Acta Part A 2006;63:370–6.10.1016/j.saa.2005.05.022Search in Google Scholar
32. Fielding L, Rutherford S, Fletcher D. Determination of protein-ligand binding affinity by NMR: observations from serum albumin model systems. Magn Reson Chem 2005;43:463–70.10.1002/mrc.1574Search in Google Scholar
33. Clarkson J, Campbell ID. Studies of protein–ligand interactions by NMR. Biochem Soc Trans 2003;31:1006–9.10.1042/bst0311006Search in Google Scholar
34. Fielding L. Determination of Association Constants (Ka) from Solution NMR Data. Tetrahedron 2000;56:6151–70.10.1016/S0040-4020(00)00492-0Search in Google Scholar
35. Lauffer RB. Paramagnetic metal complexes as water proton relaxation agents for NMR imaging: theory and design. Chem Rev 1987;87:901–27.10.1021/cr00081a003Search in Google Scholar
36. Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev 2009;109:4108–39.10.1021/cr900033pSearch in Google Scholar
37. Sulkowska A. Interaction of drugs with bovine and human serum albumin. J Mol Struct 2002;614:227–32.10.1016/S0022-2860(02)00256-9Search in Google Scholar
38. Carter DC, Ho JX. Structure of serum albumin. Adv Protein Chem 1994;45:152–203.10.1016/S0065-3233(08)60640-3Search in Google Scholar
39. American Cancer Society, Last Medical Review 2013.Search in Google Scholar
40. Page R, Takimoto C. In: Principles of chemotherapy. Pazdur R, editor. Cancer management: a multidisciplinary approach: medical, surgical, and radiation oncology, 2004;8:21.Search in Google Scholar
41. Longley DB, Harkin DP, Johnston PG. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 2003;3:330–8.10.1038/nrc1074Search in Google Scholar PubMed
42. Grem JL. 5-Fluorouracil: forty-plus and still ticking. A review of its preclinical and clinical development. Invest New Drugs 2000;18:299–313.10.1023/A:1006416410198Search in Google Scholar
43. Zhang N, Yin Y, Xu S-J, Chen W-S. 5-Fluorouracil: mechanisms of resistance and reversal strategies. Molecules 2008;13:1551–69.10.3390/molecules13081551Search in Google Scholar PubMed PubMed Central
44. Rutman RJ, Cantarow A, Paschkis KE. Studies in 2-acetylaminofluorene carcinogenesis. III. The utilization of uracil-2-C14 by preneoplastic rat liver and rat hepatoma. Cancer Res 1954;14:119–23.Search in Google Scholar
45. Hulme AT, Price SL, Tocher DA. A new polymorph of 5-fluorouracil found following computational crystal structure predictions. J Am Chem Soc 2005;127:1116–7.10.1021/ja044336aSearch in Google Scholar
46. Singh UP, Ghose R, Ghose AK, Sodhi A, Singh SM, Singh RK. The effect of histidine on the structure and antitumor activity of metal-5-halouracil complexes. J Inorg Biochem 1989;37:325–39.10.1016/0162-0134(89)85006-8Search in Google Scholar
47. Bertucci C, Ascoli G, Barretta G, Di Bari L, Salvadori P. The binding of 5-fluorouracil to native and modified human serum albumin: UV, CD, and 1H and 19F NMR investigation. J Pharm Biomed Anal 1995;13:1087–93.10.1016/0731-7085(95)01548-YSearch in Google Scholar
48. Hu Y-J, Liu Y, Shen X-S, Fang X-Y, Qu S-S. J Mol Struct 2005;738:143–7.10.1016/j.molstruc.2004.11.062Search in Google Scholar
49. Pearce S, Dowsett M, McKinna JA. Albumin-bound and non-protein-bound oestradiol and testosterone in postmenopausal breast disease. Eur J Cancer Clin Oncol 1991;27:259–63.10.1016/0277-5379(91)90511-BSearch in Google Scholar
50. Yilmaz A, Zengin B. High-field NMR T2 relaxation mechanism in D2O solutions of albumin. J Appl Spectrosc 2013;80:335–40.10.1007/s10812-013-9769-5Search in Google Scholar
51. Yilmaz A, Zengin B, Ulak FS. NMR proton spin-lattice relaxation mechanism in D2O solutions of albumin determined at 400 MHz. J Appl Spectrosc 2014;81:365–70.10.1007/s10812-014-9938-1Search in Google Scholar
©2019 Walter de Gruyter GmbH, Berlin/Boston
Articles in the same Issue
- Frontmatter
- Research Articles
- Investigating the impact of polysomy 17 in breast cancer patients with HER2 amplification through meta-analysis
- Diagnostic performance of microRNAs in the circulation in differential diagnosis of BPH, chronic prostatitis and prostate cancer
- Enhanced anticancer effect of cetuximab combined with stabilized silver ion solution in EGFR-positive lung cancer cells
- CA125, YKL-40, HE-4 and Mesothelin: a new serum biomarker combination in discrimination of benign and malign epithelial ovarian tumor
- Paricalcitol pretreatment attenuates renal ischemia/reperfusion injury by inhibiting p38 MAPK and activating PI3K/Akt signaling pathways
- Identification of cytoplasmic sialidase NEU2-associated proteins by LC-MS/MS
- Investigation of tyrosinase inhibition by some 1,2,4 triazole derivative compounds: in vitro and in silico mechanisms
- Investigation of alanine, propionylcarnitine (C3) and 3-hydroxyisovalerylcarnitine (C5-OH) levels in patients with partial biotinidase deficiency
- The expression levels of miR-655-3p, miR127-5p, miR-369-3p, miR-544a in gastric cancer
- Evaluation of the JAK2 V617F gene mutation in myeloproliferative neoplasms cases: a one-center study from Eastern Anatolia
- Effects of Rituximab on JAK-STAT and NF-κB signaling pathways in acute lymphoblastic leukemia and chronic lymphocytic leukemia
- Analysis of the effect of DEK overexpression on the survival and proliferation of bone marrow stromal cells
- Serum fetuin-A levels and association with hematological parameters in chronic kidney disease and hemodialysis patients
- Investigation of relaxation times in 5-fluorouracil and human serum albumin mixtures
- Oxydative stress markers and cytokine levels in rosuvastatin-medicated hypercholesterolemia patients
- The protective effects of urapidil on lung tissue after intestinal ischemia-reperfusion injury
- Effects of SR-BI rs5888 and rs4238001 variations on hypertension
- Antioxidant and cytotoxic activity of three Turkish marine-derived fungi
- Is spectrophotometric enzymatic method a cost-effective alternative to indirect Ion Selective Electrode based method to measure electrolytes in small clinical laboratories?
- Plasma presepsin in determining gastric leaks following bariatric surgery
Articles in the same Issue
- Frontmatter
- Research Articles
- Investigating the impact of polysomy 17 in breast cancer patients with HER2 amplification through meta-analysis
- Diagnostic performance of microRNAs in the circulation in differential diagnosis of BPH, chronic prostatitis and prostate cancer
- Enhanced anticancer effect of cetuximab combined with stabilized silver ion solution in EGFR-positive lung cancer cells
- CA125, YKL-40, HE-4 and Mesothelin: a new serum biomarker combination in discrimination of benign and malign epithelial ovarian tumor
- Paricalcitol pretreatment attenuates renal ischemia/reperfusion injury by inhibiting p38 MAPK and activating PI3K/Akt signaling pathways
- Identification of cytoplasmic sialidase NEU2-associated proteins by LC-MS/MS
- Investigation of tyrosinase inhibition by some 1,2,4 triazole derivative compounds: in vitro and in silico mechanisms
- Investigation of alanine, propionylcarnitine (C3) and 3-hydroxyisovalerylcarnitine (C5-OH) levels in patients with partial biotinidase deficiency
- The expression levels of miR-655-3p, miR127-5p, miR-369-3p, miR-544a in gastric cancer
- Evaluation of the JAK2 V617F gene mutation in myeloproliferative neoplasms cases: a one-center study from Eastern Anatolia
- Effects of Rituximab on JAK-STAT and NF-κB signaling pathways in acute lymphoblastic leukemia and chronic lymphocytic leukemia
- Analysis of the effect of DEK overexpression on the survival and proliferation of bone marrow stromal cells
- Serum fetuin-A levels and association with hematological parameters in chronic kidney disease and hemodialysis patients
- Investigation of relaxation times in 5-fluorouracil and human serum albumin mixtures
- Oxydative stress markers and cytokine levels in rosuvastatin-medicated hypercholesterolemia patients
- The protective effects of urapidil on lung tissue after intestinal ischemia-reperfusion injury
- Effects of SR-BI rs5888 and rs4238001 variations on hypertension
- Antioxidant and cytotoxic activity of three Turkish marine-derived fungi
- Is spectrophotometric enzymatic method a cost-effective alternative to indirect Ion Selective Electrode based method to measure electrolytes in small clinical laboratories?
- Plasma presepsin in determining gastric leaks following bariatric surgery

