Redetermination of the crystal structure of (2E,4Z,13E,15Z)-3,5,14,16-tetramethyl-2,6,13,17-tetraazatricyclo[16.4.0.07,12]docosa-1(22),2,4,7,9,11,13,15,18,20-decaene, C22H24N4
-
Lucky Dey
, Tsugiko Takase
, Edward R. T. Tiekink
and Tapashi Ghosh Roy
Abstract
C22H24N4, triclinic,
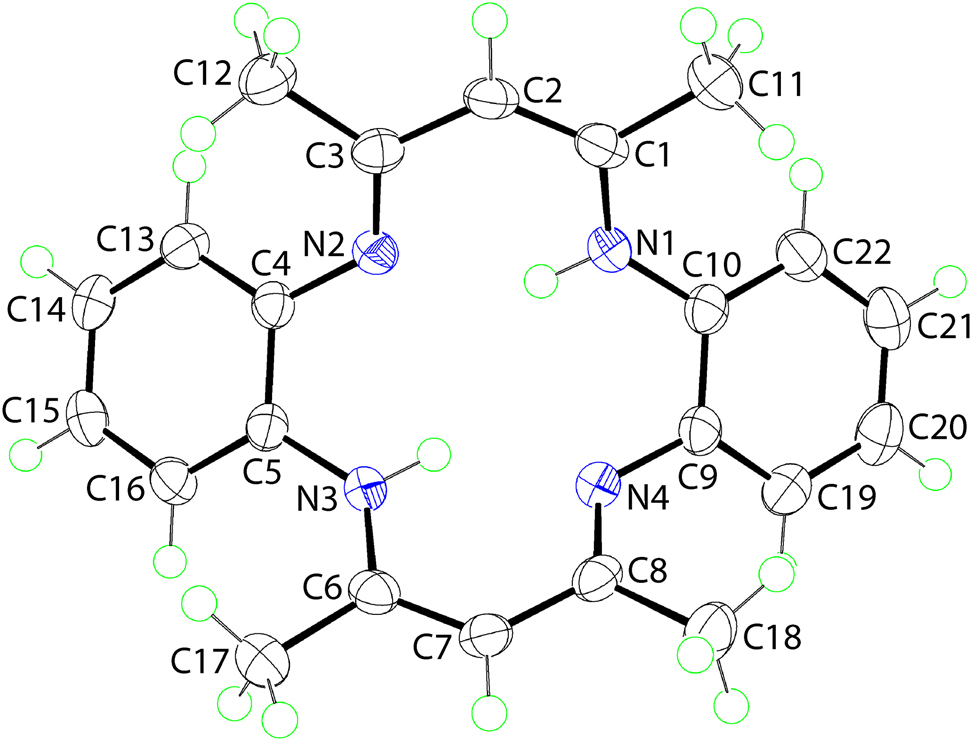
The molecular structure is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Yellow plate |
| Size: | 0.20 × 0.20 × 0.10 mm |
| Wavelength: | Mo Kα radiation (0.71073 Å) |
| μ: | 0.08 mm−1 |
| Diffractometer, scan mode: | Rigaku Saturn724, ω |
| θ max, completeness: | 27.5°, 99% |
| N(hkl)measured, N(hkl)unique, R int: | 9263, 4078, 0.015 |
| Criterion for I obs, N(hkl)gt: | I obs > 2 σ(I obs), 3392 |
| N(param)refined: | 245 |
| Programs: | REQAB [1], CrystalClear [2], SHELX [3, 4], WinGX/ORTEP [5], Diamond [6] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | U iso*/U eq |
|---|---|---|---|---|
| N1 | 0.27109 (12) | 0.10660 (11) | 0.66775 (10) | 0.0213 (2) |
| H1N | 0.3432 (14) | 0.1715 (13) | 0.6454 (13) | 0.026* |
| N2 | 0.44043 (11) | 0.20780 (10) | 0.50519 (9) | 0.0190 (2) |
| N3 | 0.68215 (12) | 0.38152 (11) | 0.69695 (9) | 0.0209 (2) |
| H3N | 0.5912 (12) | 0.3434 (15) | 0.7157 (13) | 0.025* |
| N4 | 0.50957 (11) | 0.28593 (11) | 0.85799 (9) | 0.0209 (2) |
| C1 | 0.21526 (13) | −0.01888 (12) | 0.58099 (11) | 0.0218 (3) |
| C2 | 0.26238 (14) | −0.02912 (12) | 0.46907 (11) | 0.0213 (2) |
| H2 | 0.217027 | −0.118960 | 0.411109 | 0.026* |
| C3 | 0.37297 (13) | 0.08140 (12) | 0.42998 (11) | 0.0199 (2) |
| C4 | 0.54675 (13) | 0.32310 (12) | 0.47286 (11) | 0.0186 (2) |
| C5 | 0.67411 (13) | 0.41255 (12) | 0.57240 (11) | 0.0191 (2) |
| C6 | 0.80638 (13) | 0.38719 (12) | 0.79100 (11) | 0.0204 (2) |
| C7 | 0.78493 (14) | 0.34181 (13) | 0.90335 (11) | 0.0214 (2) |
| H7 | 0.876047 | 0.349610 | 0.966936 | 0.026* |
| C8 | 0.63891 (14) | 0.28408 (12) | 0.93370 (11) | 0.0208 (2) |
| C9 | 0.36310 (14) | 0.23638 (13) | 0.88525 (11) | 0.0208 (2) |
| C10 | 0.23873 (14) | 0.14242 (13) | 0.78643 (11) | 0.0207 (2) |
| C11 | 0.10744 (16) | −0.15005 (14) | 0.61062 (13) | 0.0301 (3) |
| H11A | 0.138388 | −0.152307 | 0.702184 | 0.045* |
| H11B | 0.113235 | −0.237675 | 0.560349 | 0.045* |
| H11C | 0.000499 | −0.145565 | 0.588628 | 0.045* |
| C12 | 0.40622 (15) | 0.03588 (13) | 0.30291 (12) | 0.0252 (3) |
| H12A | 0.328630 | 0.049814 | 0.233011 | 0.038* |
| H12B | 0.401159 | −0.066419 | 0.292176 | 0.038* |
| H12C | 0.510374 | 0.094803 | 0.301296 | 0.038* |
| C13 | 0.52665 (14) | 0.36269 (13) | 0.35216 (11) | 0.0216 (2) |
| H13 | 0.438940 | 0.306934 | 0.285291 | 0.026* |
| C14 | 0.63225 (15) | 0.48175 (13) | 0.32812 (12) | 0.0235 (3) |
| H14 | 0.617882 | 0.505392 | 0.244889 | 0.028* |
| C15 | 0.75840 (14) | 0.56599 (13) | 0.42533 (12) | 0.0235 (3) |
| H15 | 0.831755 | 0.646520 | 0.408565 | 0.028* |
| C16 | 0.77771 (14) | 0.53283 (13) | 0.54722 (12) | 0.0223 (3) |
| H16 | 0.862641 | 0.592897 | 0.614367 | 0.027* |
| C17 | 0.96784 (14) | 0.43626 (15) | 0.77105 (13) | 0.0276 (3) |
| H17A | 0.966101 | 0.397355 | 0.683044 | 0.041* |
| H17B | 1.037648 | 0.401207 | 0.831353 | 0.041* |
| H17C | 1.005088 | 0.542242 | 0.785754 | 0.041* |
| C18 | 0.64604 (16) | 0.21514 (15) | 1.05196 (12) | 0.0288 (3) |
| H18A | 0.683633 | 0.290989 | 1.128847 | 0.043* |
| H18B | 0.717228 | 0.156673 | 1.053412 | 0.043* |
| H18C | 0.541854 | 0.153173 | 1.050106 | 0.043* |
| C19 | 0.33197 (15) | 0.28716 (14) | 0.99972 (12) | 0.0265 (3) |
| H19 | 0.413262 | 0.355817 | 1.064366 | 0.032* |
| C20 | 0.18454 (16) | 0.23921 (15) | 1.02078 (13) | 0.0304 (3) |
| H20 | 0.166081 | 0.272235 | 1.100382 | 0.036* |
| C21 | 0.06437 (15) | 0.14299 (15) | 0.92525 (13) | 0.0286 (3) |
| H21 | −0.036135 | 0.108072 | 0.940215 | 0.034* |
| C22 | 0.08997 (14) | 0.09731 (14) | 0.80777 (12) | 0.0253 (3) |
| H22 | 0.005723 | 0.034890 | 0.741353 | 0.030* |
Source of material
The title macrocycle was isolated after a two-step procedure. Step 1: Preparation of the nickel(II) salt of the macrocycle. A solution of nickel(II) acetate tetrahydrate (2.00 g, 8.03 mmol) in butanol (10 mL) and 1,2-phenylenediamine (1.73 g, 16 mmol) in the same solvent (10 mL) were added to a solution of 2,4-pentanedione (1.7 mL, 16.6 mmol) in butanol (10 mL) in a 100 mL round bottom flask. The mixture was refluxed on a magnetic stirrer for 2–3 h. At this stage, the colour of the mixture was dark-green. After removing the flask from the heat source, the mixture was allowed to cool until just warm to the touch. Then, methanol (30 mL) was added, and the mixture was cooled in an ice-salt bath for at least 15 min to precipitate the purple crystalline product. After that, the mixture was filtered under vacuum and washed with methanol until the washings were colourless to pale-green. The obtained product (1.0 g) was suspended in absolute ethanol (30 mL) in a 100 mL round-bottom flask. A moderate stream of HCl gas was bubbled through the suspension with swirling occasionally. Caution was maintained as the mixture gets quite warm. Once a large quantity of bright-purple precipitate [H4(C22H22N4)][NiCl4], was formed, the mixture was filtered and washed with ethanol followed by diethyl ether. The yield was 95%.
Step 2: Isolation of the free macrocycle [H2(C22H22N4)]. The [H4(C22H22N4)][NiCl4] salt was dissolved in water (10 mL). Some water-insoluble white impurities were present at this stage which were removed by filtration. Solid NH4PF6 (1.0 g) was added to the filtrate which was swirled until a large amount of white precipitate formed. Then, the [H4(C22H22N4)][PF6]2 that had formed was filtered and washed with water until the product was pale-green. In this step, the by-product [NH4]2[NiCl4] was washed away to prevent nickel from re-inserting into the macrocycle upon basification; the large volume of water used resulted in some loss of the desired product. The sticky product [H4(C22H22N4)][PF6]2 that formed was transferred into a 50 mL beaker, rinsing with methanol to maximize the mass of product transferred. Triethylamine was added dropwise to this methanolic suspension with swirling. The bright-yellow free macrocycle [H2(C22H22N4)], crystallised immediately, filtered on a Büchner funnel, washed with methanol and dried in air. The yield was 57%. M. pt. (Microprocessor Melting Point Apparatus, SYSTONIC): 346 K. Elemental analysis (Leco CHNS-932 elemental analyzer) for C22H24N4: C, 76.70; H, 6.97; N, 16.30. Found: C, 76.62; H, 6.99; N, 16.41. IR (Shimadzu IR 20 spectrophotometer, KBr; cm−1): 3255 (w) ν(N–H), 1383 (s) ν(CH3), 1618 (s) and 1551 (s) ν(Ar C=C), 1364 (m) and 1187 (s) ν(Ar CN), 1027 (m) 743 (s) ν(Ar C–H). 1 H NMR (Bruker AVANCE 400 NMR spectrometer, DMSO, ppm): δ 2.16 (s, C(CH3), 12H), 4.91 (s, CH, 2H), 7.02 (m, Aromatic-H, 8H), 12.61 (br, N–H, 2H). 13 C{1 H} NMR (as for 1H NMR) δ: 8.64, 20.911 [C(CH3)], 97.98 [CH], 123.00 [N–C–CH3], 138.54 [Aromatic-C], 158.97 [N–C(Aromatic)]
Experimental details
The C-bound H atoms were geometrically placed (C–H = 0.95–1.00 Å) and refined as riding with U iso(H) = 1.2–1.5U eq(C). The N-bound atoms were located from a Fourier difference map and refined with N–H = 0.88 ± 0.01 Å, and with U iso(H) = 1.2U eq(N). Owing to poor agreement, one reflection, i.e. (2 1 1), was omitted from the final cycles of refinement.
Comment
Macrocyclic chemistry has sustained great fascination to scientists owing to its specific roles in different sectors of contemporary science. Thus, macrocyclic compounds are prominent in coordination chemistry [7], pharmacology [8, 9] as well as in crystal engineering [10]. These compounds also play a remarkable role in medicinal chemistry: as anti-cancer agents [11, 12], radioimmunotherapeutics [13] and as MRI contrast agents [14]. In this connection, researchers continue to report different types of macrocycles and their metal complexes [15], [16], [17], [18], [19], prepared by both the template and non-template methods. In continuation of these studies, herein the synthesis of a nickel(II) complex with a macrocyclic molecule of composition [H2(C22H22N4)] (I), by the template method and the subsequent isolation of the free macrocycle is described. It is noted that the crystal and molecular structures of (I) have been described previously [20]. However, disorder in the positions of the acidic hydrogen atoms precluded a definitive assignment of the putative tautomeric structure. As the authors noted in the Abstract to the paper: “A detailed structural interpretation of the free ligand is complicated by disorder involving degenerate tautomeric structures in the crystal lattice” [20]. The present, low-temperature structure determination of (I) allows a definitive assignment of the tautomeric structure as well as a detailed analysis of the molecular packing. The molecular structure of (I) is shown in figure (70% probably displacement ellipsoids). The molecule lacks symmetry and adopts a saddle-like conformation whereby the phenyl rings lie to one side of the N4-plane, forming a dihedral angle of 47.82(6)°, and with the remaining atoms lying to the other side of the N4 plane; the dihedral angle between the best planes through N1,N2,C1–C3 and N3,N4,C6–C8 atoms = 67.55(4)°. The molecular connectivity and conformation is as reported for the earlier determination [20]. However, the new analysis enables an unambiguous assignment of the tautomeric form of the molecule; no disorder was evident in the positions of the N-bound hydrogen atoms which are located on the N1 and N3 atoms. The imine bonds correspond to C3–N2 [1.3088(15) Å] and C8–N4 [1.3079(16) Å] with the bond lengths significantly shorter than the adjacent C4–N2 [1.4081(15) Å] and C9–N4 [1.4085(16) Å] bonds. The ethylene bonds correspond to C1–C2 [1.3718(18) Å] and C6–C7 [1.3776(17) Å] which are significantly shorter than the C2–C3 [1.4380(17) Å] and C7–C8 [1.4309(18) Å] single bonds. Intramolecular amine N–H⋯N(imine) hydrogen bonds stabilise the observed conformation [N1–H1n⋯N2: H1n⋯N2 = 1.937(14) Å, N1⋯N2 = 2.6856(19) Å with angle at H1n = 142.1(12)° and N3–H3n⋯N4: H3n⋯N4 = 1.926(13) Å, N3⋯N4 = 2.6756(19) Å with angle at H3n = 140.3(12)°]. In the molecular packing, following the distance criteria assumed in PLATON [21], the only directional interactions between molecules of (I) are π–π interactions occurring between centrosymmetrically related (C4,C5,C13–C16) rings [Cg(C4,C5,C13–C16)⋯Cg(C4,C5,C13–C16) i = 3.5776(17) Å, with a slippage value = 1.171 Å for symmetry operation i: 1−x, 1−y, 1−z] to form a dimeric aggregate.
The lack of directional interactions in the crystal is supported by the analysis of the calculated Hirshfeld surfaces and of the full and delineated two-dimensional fingerprint plots. These were calculated with the program Crystal Explorer 17 [22] following literature methods [23]. The surface contacts are dominated by H⋯H contacts, contributing 67.3% of all contacts with significant contributions from C⋯H/H⋯C [22.6%] and N⋯H/H⋯N [6.3%]. The next most significant contributions to the surface contacts are from C⋯C [2.7%] and N⋯C/C⋯N [1.1%].
Funding source: Environmental Radioactivity Research Network Center
Award Identifier / Grant number: I-19-08
Award Identifier / Grant number: I-20-09
Award Identifier / Grant number: I-21-09
Funding source: Grants-in-Aid for Scientific Research
Award Identifier / Grant number: 21K12287
Funding source: Japan Society for the Promotion of Science doi.org/10.13039/501100001691
Funding source: Science and Technology Research Partnership for Sustainable Development doi.org/10.13039/501100009037
Funding source: Japan Science and Technology Agency
Funding source: Japan International Cooperation Agency
Funding source: Sunway University doi.org/10.13039/501100010798
Award Identifier / Grant number: GRTIN-IRG-01-2021
-
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Research funding: This study was supported by the Science and Technology Research Partnership for Sustainable Development (SATREPS) in collaboration with the Japan Science and Technology Agency (JST) and the Japan International Cooperation Agency (JICA) (Grant No. JPMJSA1603), the Environmental Radioactivity Research Network Center at Fukushima University, Japan (Grant Nos I-19-08, I-20-09 and I-21-09), Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) (Grant No. 21K12287) and Sunway University Sdn Bhd (Grant No. GRTIN-IRG-01–2021).
-
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
1. REQAB; Rigaku Corporation, Tokyo, Japan (1998).Search in Google Scholar
2. CrystalClear; Rigaku Corporation, Tokyo, Japan (2010).Search in Google Scholar
3. Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. 2008, A64, 112–122; https://doi.org/10.1107/s0108767307043930.Search in Google Scholar PubMed
4. Sheldrick, G. M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8; https://doi.org/10.1107/s2053229614024218.Search in Google Scholar
5. Farrugia, L. J. WinGX and ORTEP for Windows: an update. J. Appl. Crystallogr. 2012, 45, 849–854; https://doi.org/10.1107/s0021889812029111.Search in Google Scholar
6. Brandenburg, K. DIAMOND; Crystal Impact GbR: Bonn, Germany, 2006.Search in Google Scholar
7. Kolthoff, I. M. Application of macrocyclic compounds in chemical analysis. Anal. Chem. 1979, 51, 1–22; https://doi.org/10.1021/ac50041a001.Search in Google Scholar
8. Chaudhary, A., Bansal, N., Gajraj, A., Singh, R. V. Antifertility, antibacterial, antifungal and percent disease incidence aspects of macrocyclic complexes of manganese(II). J. Inorg. Biochem. 2003, 96, 393–400; https://doi.org/10.1016/s0162-0134(03)00157-0.Search in Google Scholar PubMed
9. Raman, N., Joseph, J., Velan, A. S. K., Pothiraj, C. Antifungal activities of biorelevant complexes of copper(II) with biosensitive macrocyclic ligands. Mycrobiology 2006, 34, 214–218; https://doi.org/10.4489/myco.2006.34.4.214.Search in Google Scholar
10. Blake, A. J., Schröder, M. Thioether macrocycles as spacers for crystal engineering: synthesis and crystal structures of [Ag2([24]aneS8)(CF3SO3)2(MeCN)2]∞ and [Ag([16]aneS4)(BF4)]∞ ([24]aneS8 = 1,4,7,10,13,16,19,22-octathiacyclotetracosane; [16]aneS4 = 1,5,9,13-tetrathiacyclohexadecane). Chem. Commun. 1997, 1943–1944; https://doi.org/10.1039/a704796g.Search in Google Scholar
11. Lamani, D. S., Badiger, S. G., Reddy, K. R. V., Naik, H. S. B. Macrocyclic complexes: synthesis, characterization, antitumor and DNA binding studies. Nucleos Nucleot. Nucleic Acids 2018, 37, 498–517; https://doi.org/10.1080/15257770.2018.1498515.Search in Google Scholar PubMed
12. Ali, S., Singh, V., Jain, P., Tripathi, V. Synthesis, antibacterial, anticancer and molecular docking studies of macrocyclic metal complexes of dihydrazide and diketone. J. Saudi Chem. Soc. 2019, 23, 52–60; https://doi.org/10.1016/j.jscs.2018.04.005.Search in Google Scholar
13. Bernhardt, P. V., Sharpe, P. C. C-substituted macrocycles as candidates for radioimmunotherapy. Inorg. Chem. 2000, 39, 4123–4129; https://doi.org/10.1021/ic000315f.Search in Google Scholar PubMed
14. Xu, K., Xu, N., Zhang, B., Tang, W., Ding, Y., Hu, A. Gadolinium complexes of macrocyclic diethylenetriamine-N-oxide pentaacetic acid-bisamide as highly stable MRI contrast agents with high relaxivity. Dalton Trans. 2020, 49, 8927–8932; https://doi.org/10.1039/d0dt00248h.Search in Google Scholar PubMed
15. Singh, D. P., Malik, V., Kumar, R., Kumar, K. Template synthesis of macrocyclic complexes of Co(II), Ni(II), Cu(II), Zn(II) and Cd(II): spectroscopic, antibacterial and antifungal studies. J. Serb. Chem. Soc. 2010, 75, 763–772; https://doi.org/10.2298/jsc090901050s.Search in Google Scholar
16. Riyadh, M. A., Enaam, I. Y., Hasan, A. H., Mohamad, J. A. Metal complexes of macrocyclic Schiff-base ligand: preparation, characterization, and biological activity. Sci. World J. 2013, 2013, 289805.10.1155/2013/289805Search in Google Scholar PubMed PubMed Central
17. Dey, L., Rabi, S., Palit, D., Hazari, S. K. S., Begum, Z. A., Rahman, I. M. M., Roy, T. G. Syntheses, characterization, and antimicrobial studies of Ni(II), Cu(II), and Co(III) complexes with an N-pendant azamacrocyclic chelator. J. Mol. Struct. 2021, 1240, 130579; https://doi.org/10.1016/j.molstruc.2021.130579.Search in Google Scholar
18. Hood, T. M., Gyton, M. R., Chaplin, A. B. Synthesis and rhodium complexes of macrocyclic PNP and PONOP pincer ligands. Dalton Trans. 2020, 49, 2077–2086; https://doi.org/10.1039/c9dt04474d.Search in Google Scholar PubMed
19. Kedy, S., Almhna, N., Kandil, F. Synthesis and characterization of new macrocyclic Schiff bases by the reaction of: 1,7-bis(6-methoxy-2-formylphenyl)-1,7-dioxaheptane and their use in solvent extraction of metals. Arabian J. Chem. 2015, 8, 93–99; https://doi.org/10.1016/j.arabjc.2011.01.013.Search in Google Scholar
20. Goedken, V. L., Pluth, J. J., Peng, S.-M., Bursten, B. Structure relations between the four-coordinate, S = 1, macrocyclic complex, [Fe(C22H22N4)], and the neutral ligand, C22H24N4. J. Am. Chem. Soc. 1976, 98, 8014–8021; https://doi.org/10.1021/ja00441a023.Search in Google Scholar
21. Spek, A. L. CheckCIF validation ALERTS: what they mean and how to respond. Acta Crystallogr. 2020, E76, 1–11; https://doi.org/10.1107/s2056989019016244.Search in Google Scholar
22. Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D., Spackman, M. A. Crystal Explorer (v17); The University of Western Australia: Australia, 2017.Search in Google Scholar
23. Tan, S. L., Jotani, M. M., Tiekink, E. R. T. Utilizing Hirshfeld surface calculations, non-covalent interaction (NCI) plots and the calculation of interaction energies in the analysis of molecular packing. Acta Crystallogr. 2019, E75, 308–318; https://doi.org/10.1107/s2056989019001129.Search in Google Scholar
© 2021 Lucky Dey et al., published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of [aqua-(4-iodopyridine-2,6-dicarboxylato-κ3 O,N,O′)-(1,10-phenanothroline-κ2 N,N′)copper(II)] dihydrate, C19H16O7N3CuI
- The crystal structure of tetrakis(1-isopropyl-1H-imidazolium) octamolybdate, C24H44Mo8N8O26
- Crystal structure of catena-poly[bis(µ2-3,5-bis(1-imidazolyl)pyridine-κ2 N:N′)-(µ2-3-nitrophthalato-k3 O,O′:O″)cadmium(II)] dihydrate, C30H25N11O8Cd
- The crystal structure of diaqua-bis(2-(3-(1H-pyrazol-4-yl)-1H-1,2,4-triazol-5-yl)pyridine-κ2 N:N′)-bis(3,5-dicarboxybenzoato-κ1 O)cobalt(II), C38H30CoN12O14
- Crystal structure of the nickel(II) complex aqua-(2,6-di(pyrazin-2-yl)-4,4′-bipyridine-κ3 N,N′,N′′)-(phthalato-κ2 O,O′)nickel(II) tetrahydrate, C26H26N6O9Ni
- The crystal structure of 1-[5-(2-fluorophenyl)-1-(pyridine-3-sulfonyl)-1H-pyrrol-3-yl]-N-methylmethanaminium 3-carboxyprop-2-enoate, C21H20FN3O6S
- The crystal structure of 1,2-bis(4-pyridyl)ethane - 4,4-dihydroxydiphenylmethane (1/1), C25H21N2O2
- Crystal structure of bis(2-((E)-5-chloro-2-hydroxybenzylidene)hydrazineyl)methaniminium trifluoroacetate dihydrate, C34H36Cl4N10O12
- Crystal structure of 1-cyclopropyl-7-ethoxy-6,8-difluoro-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid, C15H13F2NO4
- Crystal structure of methyl 3-(1H-naphtho[1,8-de][1,3,2]diazaborinin-2(3H)-yl)benzoate, C18H15BN2O2
- Crystal structure of (E)-N′-(2-chloro-6-hydroxybenzylidene)-2-hydroxybenzohydrazide, C14H11ClN2O3
- Crystal structure of Al-rich fluorophlogopite, K1.0(Mg2.8Al0.2)(Si2.8Al1.2)O10F2
- The crystal structure of 4,5-diiodo-1,3-dimesityl-1H-1,2,3-triazol-3-ium hexafluoridoantimonate(V), C20H22F6I2N3Sb
- Crystal structure of tris(3-iodopyridin-1-ium) catena-poly[(hexachlorido-κ1 Cl)-(μ2-trichlorido-κ2 Cl:Cl)diantimony(III)], C15H15Cl9I3N3Sb2
- Crystal structure of methyl 2-(1H-naphtho[1,8-de][1.3.2]diazaborinin-2(3H-yl)benzoate C18H15BN2O2
- The crystal structure of 1,8-bis(4-methoxybenzoyl)naphthalene-2,7-diyl dibenzoate, C40H28O8
- Crystal structure of 2-bromo-1,3,6,8-tetramethylBOPHY (BOPHY = bis(difluoroboron)-1,2-bis((1H-pyrrol-2-yl)methylene)hydrazine), C14H15B2BrF4N4
- The crystal structure of (E)-3-chloro-2-(2-(2-fluorobenzylidene)hydrazinyl)pyridine, C12H9ClFN3
- Crystal structure of bis(µ2- 4-iodopyridine-2,6-dicarboxylato-κ3O:N:O′)-bis(4-iodopyridine-2,6-dicarboxylato-κ3O:N:O′)-bis(µ2-1-(4-pyridyl)piperazine-κ2N:N′)-hexa-aqua-tetra-copper(II), C46H46Cu4I4N10O22
- Crystal structure of poly[diaqua-(μ2-2,5-dihydroxyterephthalato-κ2O:O′)(μ2-bis(4-pyridylformyl)piperazine-κ2N:N′)cadmium(II)] dihydrate, C24H28CdN4O12
- Crystal structure of poly[aqua-(μ2-1,3-di(1H-imidazol-1-yl)propane-κ2N:N′)-(μ3-2,3,5,6-tetrafluoroterephthalato-κ3O:O′:O′′)cadmium(II)], C17H14N4O5F4Cd
- Crystal structure of 6-(quinolin-8-yl)benzo[a]phenanthridin-5(6H)-one, C26H16N2O
- The crystal structure of aqua-bis(6-chloropicolinato-κ2N,O)copper(II), C12H8Cl2N2O5Cu
- Crystal structure of catena-poly[diaqua-bis(μ2-4,4′-bipyridyl-κ2N:N′) disilver(I)] 4-oxidopyridine-3-sulfonate trihydrate, C25H29Ag2N5O9S
- The crystal structure of 4-(3-bromophenyl)pyrimidin-2-amine, C10H8BrN3
- Crystal structure of 6-oxo-4-phenyl-1-propyl-1,6-dihydropyridine-3-carbonitrile, C15H14N2O
- Crystal structure of 4-(2,2-difluoroethyl)-2,4-dimethyl-6-(trifluoromethyl)isoquinoline-1,3(2H,4H)-dione, C14H12F5NO2
- Crystal structure of dibromido-(1-methyl-1H-imidazole-κ1N)-(3-(3-methyl-1H-imidazol-3-ium-1-yl)propanoato-κ1O)zinc(II), C11H16Br2N4O2Zn
- The crystal structure of 1,1′-(((2 (dimethylamino)ethyl)azanediyl)bis(methylene)) bis(naphthalen-2-olato-κ4 N,N′,O,O′)-(pyridine-2,6-dicarboxylato-N,O,O′)- titanium(IV) ─ dichloromethane (2/1), C33H29N3O6Ti
- The layered crystal structure of bis(theophyllinium) hexachloridostannate (IV), C14H18N8O8SnCl6
- The crystal structre of 3-(1-ethenyl-1H-imidazol-3-ium-3-yl)propane-1-sulfonate, C8H12N2O3S
- Synthesis and crystal structure of di-tert-butyl 1″-acetyl-2,2″,9′-trioxo-4a′,9a′-dihydro-1′H,3′H,9′H-dispiro[indoline-3,2′-xanthene-4′,3″-indoline]-1,3′-dicarboxylate, C39H38N2O9
- The crystal structure of 4-chloro-2-(quinolin-8-yl)isoindoline-1,3-dione, C17H9ClN2O2
- The crystal structure of 1-fluoro-4-(p-tolylethynyl)benzene, C15H11F
- The crystal structure of bis[4-bromo-2-(1H-pyrazol-3-yl) phenolato-κ2N,O] copper(II), C18H12Br2CuN4O2
- The crystal structure of poly[(μ 3-imidazolato-κ 3 N:N:N′)(tetrahydrofuran- κ 1 O)lithium(I)], C7H11LiN2O
- Crystal structure of N′,N′′′-((1E,1′E)-(propane-2,2-diylbis(1H-pyrrole-5,2diyl))bis(methaneylylidene))di(nicotinohydrazide) pentahydrate, C25H24N8O2·5H2O
- Crystal structure of 3-(2-ethoxy-2-oxoethyl)-1-ethyl-1H-imidazol-3-ium hexafluoridophos-phate(V), C9H15F6N2O2P
- Crystal structure of (1,10-phenanthroline-κ2N,N′)-bis(3-thiophenecarboxylato-κ2O,O′)copper(II), C22H14N2O4S2Cu
- The crystal structure of 2-amino-3-carboxypyridin-1-ium iodide hemihydrate, C6H8IN2O2.5
- Crystal structure of (E)-7-methoxy-2-((6-methoxypyridin-2-yl)methylene)-tetralone, C18H17NO3
- The crystal structure of [μ-hydroxido-bis[(5,5′-dimethyl-2,2′-bipyridine-κ2N,N′)-tricarbonylrhenium(I)] bromide hemihydrate, C30H26N4O9Re2Br
- The crystal structure of 2,5-bis(3,5-dimethylphenyl)thiazolo[5,4-d]thiazole, C20H18N2S2
- The crystal structure of 5-benzoyl-1-[(E)-(4-fluorobenzylidene)amino]-4-phenylpyrimidin-2(1H)-one, C24H16FN3O2
- Crystal structure of monocarbonyl(N-nitroso-N-oxido-phenylamine-κ 2 O,O′)(tricyclohexylphosphine-κP)rhodium(I), C25H39N2O3PRh
- Crystal structure of poly[bis[μ3-1,3,5-tris[(1H-imidazol-1-yl)methyl]benzene-κ3N:N′:N″]nickel(II)] hexafluorosilicate, C36H36N12NiSiF6
- The crystal structure of 13-(pyrazole-1-yl-4-carbonitrile)-matrine, C19H25N5O
- Crystal structure of 3,5-bis((E)-4-methoxy-2-(trifluoromethyl)benzylidene)-1-methylpiperidin-4-one, C24H21F6NO3
- The crystal structure of N,N′-(Disulfanediyldi-2,1-phenylene)di(6′-methylpyridine)-2-carboxamide, C26H22N4O2S2
- Crystal structure of (E)-7-fluoro-2-(4-methoxy-2-(trifluoromethyl)benzylidene)-3,4-dihydronaphthalen-1(2H)-one, C19H14F4O2
- Crystal structure of ethyl 1-(4-fluorophenyl)-4-phenyl-1H-pyrrole-3-carboxylate, C19H16FNO2
- The crystal structure of cis-diaqua-bis (N-butyl-N-(pyridin-2-yl)pyridin-2-amine-κ2N,N′)cobalt(II)] dichloride trihydrate, C28H44Cl2N6O5Co
- Crystal structure of (E)-7-methoxy-2-((6-methoxypyridin-3-yl)methylene)-3,4-dihydronaphthalen-1(2H)-one, C18H17NO3
- Crystal structure of (E)-2-((3-fluoropyridin-4-yl)methylene)-7-methoxy-3,4-dihydronaphthalen-1(2H)-one, C17H14FNO2
- The crystal structure of 6-bromohexanoic acid, C6H11BrO2
- The crystal structure of 4-chloro-thiophenol, C6H5ClS
- The crystal structure of 4-bromobenzyl chloride, C7H6BrCl
- The crystal structure of di-tert-butyl dicarbonate, C10H18O5
- The crystal structure of (2-(4-chlorophenyl)-5-methyl-1,3-dioxan-5-yl)methanol, C12H15ClO3
- The crystal structure of the co-crystal: 2-hydroxybenzoic acid – N′-(butan-2-ylidene)pyridine-4-carbohydrazide, C10H13N3O·C7H6O3
- Crystal structure and anti-inflammatory activity of (E)-7-fluoro-2-((5-methoxypyridin-3-yl)methylene)-3,4-dihydronaphthalen-1(2H)-one, C17H14FNO2
- Crystal structure of (E)-7-fluoro-2-((6-methoxypyridin-3-yl)methylene)-3,4-dihydronaphthalen-1(2H)-one, C17H14FNO2
- Crystal structure of 1,1′-(butane-1,4-diyl)bis(3-propyl-1H-imidazol-3-ium) bis(hexafluoridophosphate), C32H56F24N8P4
- The crystal structure of dichlorido-bis(3-methyl-3-imidazolium-1-ylpropionato-κ2)-cadmium(II), C14H20CdCl2N4O4
- Crystal structure of 1-(2-cyanobenzyl)-3-cyano-4-phenyl-4-(2-cyanobenzyl)-1,4-dihydropyridine monohydrate, C56H42N8O
- The crystal structure of 3-(carboxymethyl)-1-ethenyl-1H-imidazol-3-ium chloride, C7H9N2O2Cl
- The crystal structure of adamantylmethoxydiphenylsilane, C23H28OSi
- Redetermination of the crystal structure of (2E,4Z,13E,15Z)-3,5,14,16-tetramethyl-2,6,13,17-tetraazatricyclo[16.4.0.07,12]docosa-1(22),2,4,7,9,11,13,15,18,20-decaene, C22H24N4
- Crystal structure of (E)-7-hydroxy-2-((6-methoxypyridin-2-yl)methylene)-3,4-dihydronaphthalen-1(2H)-one, C17H15NO3
- Crystal structure of catena-poly[diaqua-bis(μ2-1,3-di(1H-imidazol-1-yl)propane-κ2 N:N′)cobalt(II)] dinitrate, C18H28N10O8Co
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of [aqua-(4-iodopyridine-2,6-dicarboxylato-κ3 O,N,O′)-(1,10-phenanothroline-κ2 N,N′)copper(II)] dihydrate, C19H16O7N3CuI
- The crystal structure of tetrakis(1-isopropyl-1H-imidazolium) octamolybdate, C24H44Mo8N8O26
- Crystal structure of catena-poly[bis(µ2-3,5-bis(1-imidazolyl)pyridine-κ2 N:N′)-(µ2-3-nitrophthalato-k3 O,O′:O″)cadmium(II)] dihydrate, C30H25N11O8Cd
- The crystal structure of diaqua-bis(2-(3-(1H-pyrazol-4-yl)-1H-1,2,4-triazol-5-yl)pyridine-κ2 N:N′)-bis(3,5-dicarboxybenzoato-κ1 O)cobalt(II), C38H30CoN12O14
- Crystal structure of the nickel(II) complex aqua-(2,6-di(pyrazin-2-yl)-4,4′-bipyridine-κ3 N,N′,N′′)-(phthalato-κ2 O,O′)nickel(II) tetrahydrate, C26H26N6O9Ni
- The crystal structure of 1-[5-(2-fluorophenyl)-1-(pyridine-3-sulfonyl)-1H-pyrrol-3-yl]-N-methylmethanaminium 3-carboxyprop-2-enoate, C21H20FN3O6S
- The crystal structure of 1,2-bis(4-pyridyl)ethane - 4,4-dihydroxydiphenylmethane (1/1), C25H21N2O2
- Crystal structure of bis(2-((E)-5-chloro-2-hydroxybenzylidene)hydrazineyl)methaniminium trifluoroacetate dihydrate, C34H36Cl4N10O12
- Crystal structure of 1-cyclopropyl-7-ethoxy-6,8-difluoro-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid, C15H13F2NO4
- Crystal structure of methyl 3-(1H-naphtho[1,8-de][1,3,2]diazaborinin-2(3H)-yl)benzoate, C18H15BN2O2
- Crystal structure of (E)-N′-(2-chloro-6-hydroxybenzylidene)-2-hydroxybenzohydrazide, C14H11ClN2O3
- Crystal structure of Al-rich fluorophlogopite, K1.0(Mg2.8Al0.2)(Si2.8Al1.2)O10F2
- The crystal structure of 4,5-diiodo-1,3-dimesityl-1H-1,2,3-triazol-3-ium hexafluoridoantimonate(V), C20H22F6I2N3Sb
- Crystal structure of tris(3-iodopyridin-1-ium) catena-poly[(hexachlorido-κ1 Cl)-(μ2-trichlorido-κ2 Cl:Cl)diantimony(III)], C15H15Cl9I3N3Sb2
- Crystal structure of methyl 2-(1H-naphtho[1,8-de][1.3.2]diazaborinin-2(3H-yl)benzoate C18H15BN2O2
- The crystal structure of 1,8-bis(4-methoxybenzoyl)naphthalene-2,7-diyl dibenzoate, C40H28O8
- Crystal structure of 2-bromo-1,3,6,8-tetramethylBOPHY (BOPHY = bis(difluoroboron)-1,2-bis((1H-pyrrol-2-yl)methylene)hydrazine), C14H15B2BrF4N4
- The crystal structure of (E)-3-chloro-2-(2-(2-fluorobenzylidene)hydrazinyl)pyridine, C12H9ClFN3
- Crystal structure of bis(µ2- 4-iodopyridine-2,6-dicarboxylato-κ3O:N:O′)-bis(4-iodopyridine-2,6-dicarboxylato-κ3O:N:O′)-bis(µ2-1-(4-pyridyl)piperazine-κ2N:N′)-hexa-aqua-tetra-copper(II), C46H46Cu4I4N10O22
- Crystal structure of poly[diaqua-(μ2-2,5-dihydroxyterephthalato-κ2O:O′)(μ2-bis(4-pyridylformyl)piperazine-κ2N:N′)cadmium(II)] dihydrate, C24H28CdN4O12
- Crystal structure of poly[aqua-(μ2-1,3-di(1H-imidazol-1-yl)propane-κ2N:N′)-(μ3-2,3,5,6-tetrafluoroterephthalato-κ3O:O′:O′′)cadmium(II)], C17H14N4O5F4Cd
- Crystal structure of 6-(quinolin-8-yl)benzo[a]phenanthridin-5(6H)-one, C26H16N2O
- The crystal structure of aqua-bis(6-chloropicolinato-κ2N,O)copper(II), C12H8Cl2N2O5Cu
- Crystal structure of catena-poly[diaqua-bis(μ2-4,4′-bipyridyl-κ2N:N′) disilver(I)] 4-oxidopyridine-3-sulfonate trihydrate, C25H29Ag2N5O9S
- The crystal structure of 4-(3-bromophenyl)pyrimidin-2-amine, C10H8BrN3
- Crystal structure of 6-oxo-4-phenyl-1-propyl-1,6-dihydropyridine-3-carbonitrile, C15H14N2O
- Crystal structure of 4-(2,2-difluoroethyl)-2,4-dimethyl-6-(trifluoromethyl)isoquinoline-1,3(2H,4H)-dione, C14H12F5NO2
- Crystal structure of dibromido-(1-methyl-1H-imidazole-κ1N)-(3-(3-methyl-1H-imidazol-3-ium-1-yl)propanoato-κ1O)zinc(II), C11H16Br2N4O2Zn
- The crystal structure of 1,1′-(((2 (dimethylamino)ethyl)azanediyl)bis(methylene)) bis(naphthalen-2-olato-κ4 N,N′,O,O′)-(pyridine-2,6-dicarboxylato-N,O,O′)- titanium(IV) ─ dichloromethane (2/1), C33H29N3O6Ti
- The layered crystal structure of bis(theophyllinium) hexachloridostannate (IV), C14H18N8O8SnCl6
- The crystal structre of 3-(1-ethenyl-1H-imidazol-3-ium-3-yl)propane-1-sulfonate, C8H12N2O3S
- Synthesis and crystal structure of di-tert-butyl 1″-acetyl-2,2″,9′-trioxo-4a′,9a′-dihydro-1′H,3′H,9′H-dispiro[indoline-3,2′-xanthene-4′,3″-indoline]-1,3′-dicarboxylate, C39H38N2O9
- The crystal structure of 4-chloro-2-(quinolin-8-yl)isoindoline-1,3-dione, C17H9ClN2O2
- The crystal structure of 1-fluoro-4-(p-tolylethynyl)benzene, C15H11F
- The crystal structure of bis[4-bromo-2-(1H-pyrazol-3-yl) phenolato-κ2N,O] copper(II), C18H12Br2CuN4O2
- The crystal structure of poly[(μ 3-imidazolato-κ 3 N:N:N′)(tetrahydrofuran- κ 1 O)lithium(I)], C7H11LiN2O
- Crystal structure of N′,N′′′-((1E,1′E)-(propane-2,2-diylbis(1H-pyrrole-5,2diyl))bis(methaneylylidene))di(nicotinohydrazide) pentahydrate, C25H24N8O2·5H2O
- Crystal structure of 3-(2-ethoxy-2-oxoethyl)-1-ethyl-1H-imidazol-3-ium hexafluoridophos-phate(V), C9H15F6N2O2P
- Crystal structure of (1,10-phenanthroline-κ2N,N′)-bis(3-thiophenecarboxylato-κ2O,O′)copper(II), C22H14N2O4S2Cu
- The crystal structure of 2-amino-3-carboxypyridin-1-ium iodide hemihydrate, C6H8IN2O2.5
- Crystal structure of (E)-7-methoxy-2-((6-methoxypyridin-2-yl)methylene)-tetralone, C18H17NO3
- The crystal structure of [μ-hydroxido-bis[(5,5′-dimethyl-2,2′-bipyridine-κ2N,N′)-tricarbonylrhenium(I)] bromide hemihydrate, C30H26N4O9Re2Br
- The crystal structure of 2,5-bis(3,5-dimethylphenyl)thiazolo[5,4-d]thiazole, C20H18N2S2
- The crystal structure of 5-benzoyl-1-[(E)-(4-fluorobenzylidene)amino]-4-phenylpyrimidin-2(1H)-one, C24H16FN3O2
- Crystal structure of monocarbonyl(N-nitroso-N-oxido-phenylamine-κ 2 O,O′)(tricyclohexylphosphine-κP)rhodium(I), C25H39N2O3PRh
- Crystal structure of poly[bis[μ3-1,3,5-tris[(1H-imidazol-1-yl)methyl]benzene-κ3N:N′:N″]nickel(II)] hexafluorosilicate, C36H36N12NiSiF6
- The crystal structure of 13-(pyrazole-1-yl-4-carbonitrile)-matrine, C19H25N5O
- Crystal structure of 3,5-bis((E)-4-methoxy-2-(trifluoromethyl)benzylidene)-1-methylpiperidin-4-one, C24H21F6NO3
- The crystal structure of N,N′-(Disulfanediyldi-2,1-phenylene)di(6′-methylpyridine)-2-carboxamide, C26H22N4O2S2
- Crystal structure of (E)-7-fluoro-2-(4-methoxy-2-(trifluoromethyl)benzylidene)-3,4-dihydronaphthalen-1(2H)-one, C19H14F4O2
- Crystal structure of ethyl 1-(4-fluorophenyl)-4-phenyl-1H-pyrrole-3-carboxylate, C19H16FNO2
- The crystal structure of cis-diaqua-bis (N-butyl-N-(pyridin-2-yl)pyridin-2-amine-κ2N,N′)cobalt(II)] dichloride trihydrate, C28H44Cl2N6O5Co
- Crystal structure of (E)-7-methoxy-2-((6-methoxypyridin-3-yl)methylene)-3,4-dihydronaphthalen-1(2H)-one, C18H17NO3
- Crystal structure of (E)-2-((3-fluoropyridin-4-yl)methylene)-7-methoxy-3,4-dihydronaphthalen-1(2H)-one, C17H14FNO2
- The crystal structure of 6-bromohexanoic acid, C6H11BrO2
- The crystal structure of 4-chloro-thiophenol, C6H5ClS
- The crystal structure of 4-bromobenzyl chloride, C7H6BrCl
- The crystal structure of di-tert-butyl dicarbonate, C10H18O5
- The crystal structure of (2-(4-chlorophenyl)-5-methyl-1,3-dioxan-5-yl)methanol, C12H15ClO3
- The crystal structure of the co-crystal: 2-hydroxybenzoic acid – N′-(butan-2-ylidene)pyridine-4-carbohydrazide, C10H13N3O·C7H6O3
- Crystal structure and anti-inflammatory activity of (E)-7-fluoro-2-((5-methoxypyridin-3-yl)methylene)-3,4-dihydronaphthalen-1(2H)-one, C17H14FNO2
- Crystal structure of (E)-7-fluoro-2-((6-methoxypyridin-3-yl)methylene)-3,4-dihydronaphthalen-1(2H)-one, C17H14FNO2
- Crystal structure of 1,1′-(butane-1,4-diyl)bis(3-propyl-1H-imidazol-3-ium) bis(hexafluoridophosphate), C32H56F24N8P4
- The crystal structure of dichlorido-bis(3-methyl-3-imidazolium-1-ylpropionato-κ2)-cadmium(II), C14H20CdCl2N4O4
- Crystal structure of 1-(2-cyanobenzyl)-3-cyano-4-phenyl-4-(2-cyanobenzyl)-1,4-dihydropyridine monohydrate, C56H42N8O
- The crystal structure of 3-(carboxymethyl)-1-ethenyl-1H-imidazol-3-ium chloride, C7H9N2O2Cl
- The crystal structure of adamantylmethoxydiphenylsilane, C23H28OSi
- Redetermination of the crystal structure of (2E,4Z,13E,15Z)-3,5,14,16-tetramethyl-2,6,13,17-tetraazatricyclo[16.4.0.07,12]docosa-1(22),2,4,7,9,11,13,15,18,20-decaene, C22H24N4
- Crystal structure of (E)-7-hydroxy-2-((6-methoxypyridin-2-yl)methylene)-3,4-dihydronaphthalen-1(2H)-one, C17H15NO3
- Crystal structure of catena-poly[diaqua-bis(μ2-1,3-di(1H-imidazol-1-yl)propane-κ2 N:N′)cobalt(II)] dinitrate, C18H28N10O8Co

