Abstract
C36H60Cl2CuN12, monoclinic, P21/c (no. 14), a = 8.3112(2) Å, b = 15.9513(4) Å, c = 17.1068(4) Å, β = 99.671(2)° V = 2235.69(9) Å3, Z = 2, Rgt(F) = 0.0523, wRref(F2) = 0.1606, T = 298(2) K.
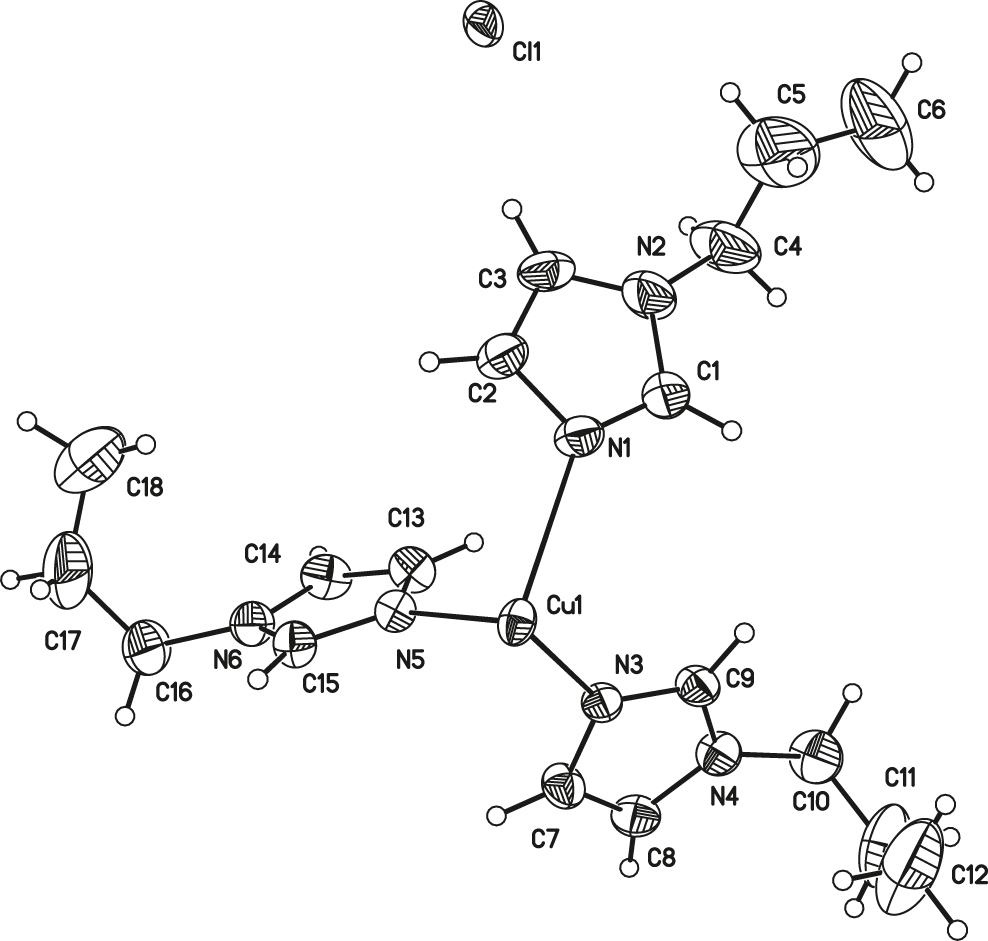
The asymmetric unit of the title crystal structure is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Blue block |
| Size: | 0.19 × 0.18 × 0.16 mm |
| Wavelength: | Cu Kα radiation (1.54178 Å) |
| μ: | 2.09 mm−1 |
| Diffractometer, scan mode: | Bruker SMART APEX II, φ and ω |
| θmax, completeness: | 66.0°, >99% |
| N(hkl)measured, N(hkl)unique, Rint: | 8002, 3880, 0.031 |
| Criterion for Iobs, N(hkl)gt: | Iobs > 2 σ(Iobs), 3310 |
| N(param)refined: | 263 |
| Programs: | Bruker [1], SHELX [2] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | Uiso*/Ueq |
|---|---|---|---|---|
| Cu1 | 1.000000 | 0.500000 | 0.500000 | 0.0424 (3) |
| Cl1 | 0.34311 (8) | 0.23789 (5) | 0.18672 (5) | 0.0611 (3) |
| N1 | 0.7656 (3) | 0.47176 (16) | 0.38734 (15) | 0.0571 (6) |
| N2 | 0.5683 (4) | 0.4642 (2) | 0.2853 (2) | 0.0857 (10) |
| N3 | 1.1108 (2) | 0.58182 (13) | 0.43467 (12) | 0.0431 (5) |
| N4 | 1.1638 (3) | 0.68223 (15) | 0.35540 (14) | 0.0550 (6) |
| N5 | 1.1232 (2) | 0.40701 (13) | 0.45399 (12) | 0.0435 (5) |
| N6 | 1.2850 (3) | 0.30316 (15) | 0.43430 (15) | 0.0563 (6) |
| C1 | 0.6735 (4) | 0.5141 (2) | 0.3341 (2) | 0.0686 (9) |
| H1 | 0.677997 | 0.572144 | 0.329398 | 0.082* |
| C2 | 0.7166 (4) | 0.3898 (2) | 0.3731 (2) | 0.0655 (8) |
| H2 | 0.761207 | 0.344017 | 0.402716 | 0.079* |
| C3 | 0.5970 (4) | 0.3853 (2) | 0.3111 (2) | 0.0687 (9) |
| H3 | 0.543694 | 0.337120 | 0.289833 | 0.082* |
| C4a | 0.4812 (11) | 0.4961 (8) | 0.2043 (6) | 0.117 (3) |
| H4Aa | 0.522219 | 0.550971 | 0.192984 | 0.140* |
| H4Ba | 0.498600 | 0.457833 | 0.162470 | 0.140* |
| C5a | 0.3009 (13) | 0.5010 (10) | 0.2099 (9) | 0.179 (5) |
| H5Aa | 0.287869 | 0.522777 | 0.261381 | 0.215* |
| H5Ba | 0.253229 | 0.445406 | 0.204179 | 0.215* |
| C6a | 0.214 (2) | 0.5579 (15) | 0.1451 (10) | 0.192 (7) |
| H6Aa | 0.099801 | 0.560769 | 0.148930 | 0.288* |
| H6Ba | 0.260183 | 0.613104 | 0.151236 | 0.288* |
| H6Ca | 0.225622 | 0.535911 | 0.094166 | 0.288* |
| C7 | 1.2756 (3) | 0.58484 (19) | 0.43375 (18) | 0.0548 (7) |
| H7 | 1.352791 | 0.549337 | 0.462220 | 0.066* |
| C8 | 1.3086 (3) | 0.6467 (2) | 0.38562 (19) | 0.0576 (7) |
| H8 | 1.410783 | 0.662038 | 0.375137 | 0.069* |
| C9 | 1.0478 (3) | 0.64154 (18) | 0.38679 (16) | 0.0504 (6) |
| H9 | 0.937058 | 0.654276 | 0.375812 | 0.060* |
| C10b | 1.1326 (16) | 0.7546 (3) | 0.3014 (5) | 0.084 (2) |
| H10Ab | 1.017823 | 0.756162 | 0.278271 | 0.101* |
| H10Bb | 1.195080 | 0.748428 | 0.258673 | 0.101* |
| C11b | 1.1789 (12) | 0.8352 (3) | 0.3445 (5) | 0.130 (3) |
| H11Ab | 1.297104 | 0.838427 | 0.354227 | 0.156* |
| H11Bb | 1.139825 | 0.880649 | 0.308553 | 0.156* |
| C12b | 1.1248 (17) | 0.8517 (5) | 0.4179 (5) | 0.194 (5) |
| H12Ab | 1.164468 | 0.905440 | 0.437623 | 0.291* |
| H12Bb | 1.007632 | 0.851524 | 0.409712 | 0.291* |
| H12Cb | 1.165814 | 0.809060 | 0.455648 | 0.291* |
| C13 | 1.1163 (4) | 0.3951 (2) | 0.37366 (16) | 0.0554 (7) |
| H13 | 1.053504 | 0.426649 | 0.334101 | 0.066* |
| C14 | 1.2137 (4) | 0.3311 (2) | 0.36114 (18) | 0.0600 (8) |
| H14 | 1.229418 | 0.309926 | 0.312335 | 0.072* |
| C15 | 1.2274 (3) | 0.35043 (17) | 0.48787 (16) | 0.0514 (6) |
| H15 | 1.257194 | 0.344109 | 0.542421 | 0.062* |
| C16c | 1.4024 (8) | 0.2337 (4) | 0.4495 (8) | 0.084 (2) |
| H16Ac | 1.486449 | 0.248519 | 0.493891 | 0.101* |
| H16Bc | 1.454646 | 0.226089 | 0.403419 | 0.101* |
| C17c | 1.3268 (11) | 0.1542 (4) | 0.4673 (7) | 0.143 (3) |
| H17Ac | 1.402537 | 0.110037 | 0.459102 | 0.172* |
| H17Bc | 1.322256 | 0.154744 | 0.523596 | 0.172* |
| C18c | 1.1680 (12) | 0.1279 (5) | 0.4275 (9) | 0.214 (6) |
| H18Ac | 1.141695 | 0.074445 | 0.447833 | 0.320* |
| H18Bc | 1.087816 | 0.168467 | 0.436658 | 0.320* |
| H18Cc | 1.168747 | 0.123399 | 0.371642 | 0.320* |
| C4′d | 0.4037 (12) | 0.4818 (9) | 0.2431 (7) | 0.117 (3) |
| H4′1d | 0.337798 | 0.506682 | 0.278720 | 0.140* |
| H4′2d | 0.351338 | 0.430512 | 0.221587 | 0.140* |
| C5′d | 0.4211 (19) | 0.5410 (11) | 0.1781 (9) | 0.179 (5) |
| H5′1d | 0.517191 | 0.525766 | 0.156058 | 0.215* |
| H5′2d | 0.438319 | 0.596990 | 0.200086 | 0.215* |
| C6′d | 0.275 (2) | 0.5424 (17) | 0.1120 (10) | 0.192 (7) |
| H6′1d | 0.293794 | 0.581778 | 0.072074 | 0.288* |
| H6′2d | 0.258372 | 0.487558 | 0.088967 | 0.288* |
| H6′3d | 0.179436 | 0.558840 | 0.133030 | 0.288* |
| C10′e | 1.155 (8) | 0.7605 (13) | 0.309 (2) | 0.084 (2) |
| H10Ce | 1.091742 | 0.750621 | 0.256466 | 0.101* |
| H10De | 1.264347 | 0.776844 | 0.301862 | 0.101* |
| C11′e | 1.077 (5) | 0.8314 (12) | 0.349 (3) | 0.130 (3) |
| H11Ce | 1.104128 | 0.823811 | 0.405777 | 0.156* |
| H11De | 0.959854 | 0.827299 | 0.334146 | 0.156* |
| C12′e | 1.130 (6) | 0.9192 (12) | 0.329 (2) | 0.194 (5) |
| H12De | 1.075663 | 0.959902 | 0.356188 | 0.291* |
| H12Ee | 1.245930 | 0.924701 | 0.344188 | 0.291* |
| H12Fe | 1.101397 | 0.928196 | 0.272428 | 0.291* |
| C16′f | 1.378 (4) | 0.2276 (12) | 0.457 (4) | 0.084 (2) |
| H16Cf | 1.410587 | 0.226946 | 0.514340 | 0.101* |
| H16Df | 1.475542 | 0.227769 | 0.433379 | 0.101* |
| C17′f | 1.278 (6) | 0.1485 (11) | 0.431 (4) | 0.143 (3) |
| H17Cf | 1.170614 | 0.152956 | 0.445694 | 0.172* |
| H17Df | 1.265615 | 0.142022 | 0.374213 | 0.172* |
| C18′f | 1.368 (5) | 0.0725 (14) | 0.472 (4) | 0.214 (6) |
| H18Df | 1.306957 | 0.022528 | 0.456631 | 0.320* |
| H18Ef | 1.474257 | 0.068255 | 0.457646 | 0.320* |
| H18Ff | 1.379562 | 0.079153 | 0.528897 | 0.320* |
a Occupancy: 0.524(7), b Occupancy: 0.817(8), c Occupancy: 0.824(10), d Occupancy: 0.476(7), e Occupancy: 0.183(8), f Occupancy: 0.176(10).
Source of material
In a typical experiment, copper chloride (1 mmol) and 1-propylimidazole (10 mmol) were dissolved in H2O (25 mL), and then maintained for 6 h at 80 °C with stirring. After the reaction was completed, the filtrate was left to slowly evaporate at room temperature for about four days, and then the blue rod crystals were filtered off. Yield: 54.5%. Anal. Calcd. for C36H60Cl2CuN12: C, 54.36; H, 7.60; N, 21.13; found: C, 54.39; H 7.52; N 21.18.
Experimental details
Hydrogen atoms were assigned with common isotropic displacement factors Uiso(H) = 1.2 times Ueq (C, imidazole ring and methylene) and Uiso(H) = 1.5 times Ueq(C, methyl carbon). All the H atoms were refined as riding on their parent atom.
Comment
Over the last decades, transition metal complexes have received much attention in the fields of catalysis, magnetism, medicine and material because their bio-compatibility, various coordination modes, and catalytic properties [3], [4], [5]. Among these transition metal complexes, copper complexes with their versatile structures, redox behavior and physicochemical properties have been found to be useful as active agents in chemotherapeutic, catalytic applications. So far, some thrilling work about inorganic-organic hybrid copper complexes have been reported [6], [7], [8]. In addition, imidazoles as multifunctional organic reagents could be used not only as organic base, solvent, but also as N-ligands in the synthesis of transition metal complexes, and some imidazole-based compounds, including some transition metal clusters have been reported [9], [10], [11]. However, despite the progress achieved, newly designed imidazole-based crystalline copper materials are long-sought-after yet still unmet.
Single crystal X-ray diffraction analysis reveals that the asymmetric unit of the title structure consist of half a copper(II) cation, three 1-propylimidazole N-ligands and one free chloride anion. The Cu(II) cation adopted a six-coordinated mode with a twisted octahedral geometry conformation. The bond distances of Cu–N are in the range of 2.034(2) to 2.540(2) Å, which are similar with those of the reported tetrakis (N–methylimidazole–N′)-copper(I) perchlorate complex [12]. Meanwhile, the bond angles of N(5)–Cu(1)–N(3), N(5)–Cu(1)–N(1), and N(3)–Cu(1)–N(1) are 87.22(9), 87.07(8) and 93.27(8)°, respectively. Moreover, the disordered atom sets were (C4, C4′), (C5, C5′), (C6, C6′), (C10, C10′), (C11, C11′), (C12, C12′), (C16, C16′), (C17, C17′), and (C18, C18′), respectively.
Funding source: Liaocheng University
Award Identifier / Grant number: 263222017215 and 263222017214
Funding source: Entrepreneurship Training Program for College Students
Award Identifier / Grant number: CXCY2020Y021, S202010447012X and X202010447012X
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
Research funding: We gratefully acknowledge support by the Research on Experimental Technology of Liaocheng University (263222017215 and 263222017214) and Entrepreneurship Training Program for College Students (CXCY2020Y021, S202010447012X and X202010447012X).
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
1. Bruker. APEX2, SAINT and SADABS; Bruker AXS Inc.: Madison, WI, USA, 2004.Suche in Google Scholar
2. Sheldrick, G. M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8; https://doi.org./10.1107/s2053229614024218.10.1107/S2053229614024218Suche in Google Scholar
3. Yadav, S., Kumar, R., Raj, K. V., Yadav, P., Vanka, K., Sen, S. S. Amidinato germylene-zinc complexes: synthesis, bonding, and reactivity. Chem. Asian J. 2020, 15, 3116–3121; https://doi.org./10.1002/asia.202000807.10.1002/asia.202000807Suche in Google Scholar PubMed
4. Wu, L., Liu, X., Zhang, K., Gao, J., Chen, Y., Tung, C. Monochromophore-based phosphorescence and fluorescence from pure organic assemblies for ratiometric hypoxia detection. Angew. Chem. Int. Ed. 2020, 59, 23456–23460; https://doi.org./10.1002/anie.202007035.10.1002/anie.202007039Suche in Google Scholar PubMed
5. Li, Y., Li, C., Wu, F., Fu, M., Zhang, J., Huang, X. A new Schiff base Ni(II) complex based on 2-hydroxy-1-naphthaldehyde and glycine: synthesis and highly effective construction of C=C bonds. J. Liaocheng Univ. (Nat. Sci. Ed.) 2019, 32, 81–87.Suche in Google Scholar
6. Ohashi, M., Adachi, T., Ishida, N., Kikushima, K., Ogoshi, S. Synthesis and reactivity of fluoroalkyl copper complexes by the oxycupration of tetrafluoroethylene. Angew. Chem. Int. Ed. 2017, 56, 11911–11915; https://doi.org./10.1002/anie.201703923.10.1002/anie.201703923Suche in Google Scholar PubMed
7. Huang, X., Qi, Y., Gu, Y., Gong, S., Shen, G., Li, Q., Li, J. Imidazole-directed fabrication of three polyoxovanadates-based copper frameworks as efficient catalysts for constructing of C–N bonds. Dalton Trans. 2020, 49, 10970–10976; https://doi.org./10.1039/d0dt02162h.10.1039/D0DT02162HSuche in Google Scholar
8. Hayashi, N., Machida, K., Otawara, K., Hasegawa, A., Kosaka, N. Polymer-soluble thermostable phosphate-ester copper complexes for near-infrared absorbing dyes with weak absorbance in the visible region. Opt. Mater. 2018, 77, 111–116; https://doi.org./10.1016/j.optmat.2018.01.020.10.1016/j.optmat.2018.01.020Suche in Google Scholar
9. Gu, X., Lin, J., Zhao, W., Cui, C., Qi, Y., Xue, Z., Huang, X. An organic imidazole functionalized polyoxomolybdenum cluster: synthesis and selectivity switch in the oxidation of sulfides. J. Liaocheng Univ. (Nat. Sci. Ed.) 2021, 34, 77–82.Suche in Google Scholar
10. Chen, B., Huang, X., Wang, B., Lin, Z., Hu, J., Chi, Y., Hu, C. Three new imidazole-functionalized hexanuclear oxidovanadium clusters with exceptional catalytic oxidation properties for alcohols. Chem. Eur J. 2013, 19, 4408–4413; https://doi.org./10.1002/chem.201203854.10.1002/chem.201203854Suche in Google Scholar PubMed
11. Zhao, H., Tao, L., Zhang, F., Zhang, Y., Liu, Y., Xu, H., Diao, G., Ni, L. Transition metal substituted sandwich-type polyoxometalates with a strong metal–C (imidazole) bond as anticancer agents. Chem. Commun. 2019, 55, 1096–1099; https://doi.org./10.1039/c8cc07884j.10.1039/C8CC07884JSuche in Google Scholar PubMed
12. Clegg, W., Acott, S. R., Garner, C. D. Structure of tetrakis(N- methylimidazole-N′) copper(I) perchlorate, [Cu(C4H6N2)4][ClO4]. Acta Crystallogr. 1984, C40, 768–769; https://doi.org./10.1107/s0108270184005667.10.1107/S0108270184005667Suche in Google Scholar
© 2021 Qingpeng He et al., published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Frontmatter
- New Crystal Structures
- Crystal structure of poly[(μ2-aqua-tetraaqua-(μ3-glutarato-κ4O,O′:O′:O′′)-(μ5-glutarato-κ6O:O,O′:O′:O′′:O′′′)distrontium(II)], C10H22O13Sr2
- The crystal structure of acetato-κ1O-{(2-(2-(2-aminophenoxy)ethoxy)phenyl)(4-oxo-4-phenylbut-2-en-2-yl)amido-κ2N,N′,O}copper(II), C26H26CuN2O5
- Crystal structure of dimethanolato-k2O:O-bis(1-((2-methyl-1H-benzo[d]imidazol-1-yl)methyl)-1H-benzo[d][1,2,3]triazole-κN)-bis(thiocyanato-κN)dicopper(II), C34H32Cu2N12O2S2
- Crystal structure of poly[diaqua-bis(μ2-3-(pyrimidin-5-yl)benzoato-κ2N:O)cobalt(II)] dihydrate, [Co(C11H11O2N2)2(H2O)2]
- Crystal structure of bis(3,3-dimethyl-1-phenylbut-1-en-2-yl)(trimethylsilyl)amido-k1N)zinc(II), Zn(C15H24NSi)2
- Crystal structure of catena-poly[(μ2-methanolato-κ2O:O)-(μ2-1-((2-methyl-1H-benzo[d]imidazol-1-yl)methyl)-1H-benzo[d][1,2,3]triazole-κ2N:N′)-(thiocyanato-κ1N)copper(II)] 0.25 hydrate, C17H16CuN6OS ⋅ 0.5H2O
- The crystal structure of 2-amino-5-nitroanilinium iodide monohydrate, C6H8IN3O2
- The crystal structure of 3-amino-5-carboxypyridin-1-ium perchlorate monohydrate, C6H9ClN2O7
- Crystal structure of 7-hydroxy-2,4-dimethoxy-9,10-dihydrophenanthrene from Arundina graminifolia, C16H16O3
- Crystal structure of 6,6′-((1E, 1′E)-(((1R, 2R)-1,2-diphenylethane-1,2-diyl) bis(azanylylidene))bis(methanylylidene))bis(2-ethylphenol), C32H32N2O2
- The crystal structure of 2-amino-5-carboxypyridin-1-ium iodide monohydrate, C6H9IN2O3
- The crystal structure of 2-(3,5-difluorophenyl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]diazaborinine, C16H11BF2N2
- Crystal structure of bis{(2-pyridinyl)-1-phenyl-1-isopropylmethanolato-κ2N,O}nickel, C30H32N2NiO2
- Crystal structure of poly[(m3-3-carboxyadamantane-1-carboxylato-κ3O:O′:O″)-(phenanthroline-κ2N,N′)sodium(II)], C24H23N2NaO4
- Crystal structure of 2-phenylethynyl-1,3,6,8-tetramethylBOPHY (BOPHY = bis(difluoroboron)-1,2-bis((1H-pyrrol-2-yl)methylene)hydrazine), C22H20B2F4N4
- Crystal structure of 4-tert-butyl-2-N-(2-pyridylmethyl)aminophenol, C16H20N2O
- The crystal structure of (3Z,3′Z)-4,4′-((1,4-phenylenebis(methylene))bis(azanediyl))bis(pent-3-en-2-one), C18H24N2O2
- Crystal structure of (morpholine-1-carbodithioato-κ2-S,S′)bis(triphenylphosphine-κ-P)gold(I), C41H38AuNOP2S2
- Crystal structure of 1,4-bis(4-bromobenzyl)-4-(4-chlorophenyl)-1,4-dihydropyridine-3-carbonitrile, C26H19Br2ClN2
- The crystal structure of fac-tricarbonyl (N′-benzoyl-N,N-diphenylcarbamimidothioato-κ2S,O)-(pyrazole-κN)rhenium(I) — methanol (1/1) C26H23O4N4SRe
- The crystal structure of Ba2Mn(SeO3)2Cl2 containing 1∞[Mn(SeO3)2Cl2]4− chains
- Crystal structure of 3,3′,3″-((1E,1′E,1″E)-((nitrilotris(ethane-2,1-diyl))tris(azaneylylidene)) tris(methaneylylidene))tris(4-hydroxy-1-naphthaldehyde) monohydrate, C42H36N4O6·H2O
- The crystal structure of 4-(6-acetyl-5-methyl-4,7-dihydrotetrazolo[1,5-a]pyrimidin-7-yl)benzonitrile, C14H12N6O
- Crystal structure of benzo[d][1,3]dioxol-5-yl-2-(6-methoxynaphthalen-2-yl)propanoate, C21H18O5
- The crystal structure of ethyl 5-methyl-7-(4-(phenylthio)phenyl)-4,7-dihydrotetrazolo[1,5-a]pyrimidine-6-carboxylate, C20H19N5O2S
- Crystal structure of N′,N‴-((propane-2,2-diylbis(1H-pyrrole-5,2-diyl))bis(methaneylylidene))-di(isonicotinohydrazide)– water – dimethylformamide (1/4/2), C25H24N8O2·4H2O·2C3H7NO
- Synthesis and crystal structure of 4-(2,4-dinitrophenoxy)benzaldehyde, C13H8N2O6
- The crystal structure of 1-dodecylpyridin-1-ium bromide monohydrate, C17H32BrNO
- Crystal structure of (E)-amino(2-(4-(dimethylamino)benzylidene)hydrazineyl)methaniminium nitrate, C10H16N6O3
- Crystal structure of (E)-(2-((1H-pyrrol-2-yl)methylene)hydrazineyl)(amino)methaniminium nitrate monohydrate, C6H12N6O4
- The crystal structure of hexakis(1-propylimidazole-κ1N)copper(II) dichloride, C36H60Cl2CuN12
- The crystal structure of bis{(μ2-3,3-dimethyl-1-phenylbut-1-en-2-yl)((dimethylamino)dimethylsilyl)amido-κ3N,N′:N′}dilithium, C32H54Li2N4Si2
- The crystal structure of methyl 4-(1H-naphtho[1,8-de][1,3,2]diazaborinin-2(3H)-yl)benzoate, C18H15BN2O2
- Crystal structure of (E)-N-(1-((2-chlorothiazol-5-yl)methyl)pyridin-2(1H)-ylidene)-2,2,2-trifluoroacetamide, C11H7ClF3N3OS
- Crystal structure of N′, N‴-((propane-2,2-diylbis(1H-pyrrole-5,2-diyl))bis (methaneylylidene))di(picolinohydrazide) – water – methanol (1/1/1), C25H24N8O2·H2O·CH3OH
- Crystal structure of 3-(2-chloro-benzyl)-7-[4-(2-chloro-benzyl)-piperazin-1-yl]-5,6,8-trifluoro-3H-quinazolin-4-one, C26H21Cl2F3N4O
- Crystal structure of N1,N2-bis(2-fluorobenzyl)benzene-1,2-diamine,C20H18F2N2
- The crystal structure of 2-(benzo[d][1,3]dioxol-5-yl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]diazaborinine, C17H13BN2O2
- The crystal structure of 6,6′-((1E,1′E)-hydrazine-1,2-diylidenebis(methaneylylidene)) bis(2-bromo-4-nitrophenol) — dimethylsulfoxide (1/2), C14H8Br2N4O6⋅2(C2H6OS)
- Selective biocatalytic synthesis and crystal structure of (2R,6R)-hydroxyketaminium chloride, C13H17Cl2NO2
- Crystal structure of bis{tetraaqua-[μ3-1-(4-carboxylatophenyl)-5-methyl-1H-pyrazole-3-carboxylate-κ4N,O,O′,O″] [μ2-1-methyl-1H-pyrazole-3,5-dicarboxylate-κ3N,O:O]dicobalt(II)} dihydrate, C36H44Co4N8O26
- Crystal structure of diethyl-2,2′-naphthalene-2,3-diylbis(oxy)diacetate, C18H20O6
- Synthesis and crystal structure of poly[(μ3-2-(2-carboxylatophenyl)-1H-benzo[d]imidazole-5-carboxylato-κO,O′:O′;:O″, O″′)-(μ2-1-(4-(1Himidazol-1-yl)phenyl)-1H-imidazole-κ2N:N′)cadmium(II)], C27H18CdN6O4
- The crystal structure of catena-poly[diaqua-bis(μ2-2-((2-(2-phenylacetyl)hydrazineylidene)methyl)benzoato-κ2O:O')zinc(II)], C32H30N4O8Zn
- The crystal structure of 2-(3,4-dimethoxyphenyl)-2,3-dihydro-1H-naphtho [1,8-de][1,3,2]diazaborinine, C18H17BN2O2
- The crystal structure of hexakis(1-ethylimidazole-κ1N)nickel(II) dichloride – 1-ethylimidazole (1/2), C40H64Cl2NiN16
- Crystal structure of diaqua-bis(2,4-dinitrophenolato-κ2O,O′)copper(II) 1.5 hydrate, C12H13CuN4O13.5
- Crystal structure of N′,N‴-((1E,1′E)-((decane-1,10-diylbis(oxy))bis(2,1-phenylene)) bis(methaneylylidene))di(isonicotinohydrazide), C36H40N6O4
- The crystal structure of 2-[(R)-1-(naphthalen-1-yl)ethyl]-2,3,7,7a-tetrahydro-3a,6-epoxyisoindol-1(6H)-one, C19H20NO2
- Synthesis and crystal structure of (1E,2E)-3-(anthracen-9-yl)-1-(4-methoxyphenyl)prop-2-en-1-one oxime, C24H19NO2
- Synthesis and crystal structure of (2E,2′E)-3,3′-(1,3-phenylene)bis(1-(3-bromophenyl)prop-2-en-1-one), C24H16Br2O2
- The crystal structure of catena-poly[bis(µ2-1,2-bis((1H-imidazol-1-yl)methyl)benzene- κ2N:N′)-bis(nitrato-κO)copper(II)], C28H28N10O6Cu
- Synthesis and crystal structure of the novel chiral acetyl-3-thiophene-5-(9-anthryl)-2-pyrazoline, C23H18N2OS
- Crystal structure of (E)-3-(dimethylamino)-1-(thiophen-3-yl)prop-2-en-1-one, C9H11NOS
- Crystal structure of catena-poly[aqua-(4-iodopyridine-2,6-dicarboxylato-κ3N,O,O′)-(μ2-4-amino-4H-1,2,4-triazole-κ2N:N′) copper(II)], C9H8N5O5CuI
- Crystal structure of cyclopropane-1,2,3-triyltris(phenylmethanone), C24H18O3
- Crystal structure of bis(amino(thioureido)methaniminium) terephthalate, C12H18N8O4S2
- A three-dimensional Eu(III) framework in the crystal structure of dimethylaminium poly[dimethylformamide-κ1N)bis(μ4-terephthalato-κ4O:O′:O′′:O′′′)europium(III)] monohydrate, C21H25EuN2O10
- Crystal structure of 2-methoxyphenyl 2-(6-methoxynaphthalen-2-yl)propanoate, C21H20O4
- The crystal structure of Hexakis(diethylamido)dimolybdenum, Mo2(NEt2)6
Artikel in diesem Heft
- Frontmatter
- New Crystal Structures
- Crystal structure of poly[(μ2-aqua-tetraaqua-(μ3-glutarato-κ4O,O′:O′:O′′)-(μ5-glutarato-κ6O:O,O′:O′:O′′:O′′′)distrontium(II)], C10H22O13Sr2
- The crystal structure of acetato-κ1O-{(2-(2-(2-aminophenoxy)ethoxy)phenyl)(4-oxo-4-phenylbut-2-en-2-yl)amido-κ2N,N′,O}copper(II), C26H26CuN2O5
- Crystal structure of dimethanolato-k2O:O-bis(1-((2-methyl-1H-benzo[d]imidazol-1-yl)methyl)-1H-benzo[d][1,2,3]triazole-κN)-bis(thiocyanato-κN)dicopper(II), C34H32Cu2N12O2S2
- Crystal structure of poly[diaqua-bis(μ2-3-(pyrimidin-5-yl)benzoato-κ2N:O)cobalt(II)] dihydrate, [Co(C11H11O2N2)2(H2O)2]
- Crystal structure of bis(3,3-dimethyl-1-phenylbut-1-en-2-yl)(trimethylsilyl)amido-k1N)zinc(II), Zn(C15H24NSi)2
- Crystal structure of catena-poly[(μ2-methanolato-κ2O:O)-(μ2-1-((2-methyl-1H-benzo[d]imidazol-1-yl)methyl)-1H-benzo[d][1,2,3]triazole-κ2N:N′)-(thiocyanato-κ1N)copper(II)] 0.25 hydrate, C17H16CuN6OS ⋅ 0.5H2O
- The crystal structure of 2-amino-5-nitroanilinium iodide monohydrate, C6H8IN3O2
- The crystal structure of 3-amino-5-carboxypyridin-1-ium perchlorate monohydrate, C6H9ClN2O7
- Crystal structure of 7-hydroxy-2,4-dimethoxy-9,10-dihydrophenanthrene from Arundina graminifolia, C16H16O3
- Crystal structure of 6,6′-((1E, 1′E)-(((1R, 2R)-1,2-diphenylethane-1,2-diyl) bis(azanylylidene))bis(methanylylidene))bis(2-ethylphenol), C32H32N2O2
- The crystal structure of 2-amino-5-carboxypyridin-1-ium iodide monohydrate, C6H9IN2O3
- The crystal structure of 2-(3,5-difluorophenyl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]diazaborinine, C16H11BF2N2
- Crystal structure of bis{(2-pyridinyl)-1-phenyl-1-isopropylmethanolato-κ2N,O}nickel, C30H32N2NiO2
- Crystal structure of poly[(m3-3-carboxyadamantane-1-carboxylato-κ3O:O′:O″)-(phenanthroline-κ2N,N′)sodium(II)], C24H23N2NaO4
- Crystal structure of 2-phenylethynyl-1,3,6,8-tetramethylBOPHY (BOPHY = bis(difluoroboron)-1,2-bis((1H-pyrrol-2-yl)methylene)hydrazine), C22H20B2F4N4
- Crystal structure of 4-tert-butyl-2-N-(2-pyridylmethyl)aminophenol, C16H20N2O
- The crystal structure of (3Z,3′Z)-4,4′-((1,4-phenylenebis(methylene))bis(azanediyl))bis(pent-3-en-2-one), C18H24N2O2
- Crystal structure of (morpholine-1-carbodithioato-κ2-S,S′)bis(triphenylphosphine-κ-P)gold(I), C41H38AuNOP2S2
- Crystal structure of 1,4-bis(4-bromobenzyl)-4-(4-chlorophenyl)-1,4-dihydropyridine-3-carbonitrile, C26H19Br2ClN2
- The crystal structure of fac-tricarbonyl (N′-benzoyl-N,N-diphenylcarbamimidothioato-κ2S,O)-(pyrazole-κN)rhenium(I) — methanol (1/1) C26H23O4N4SRe
- The crystal structure of Ba2Mn(SeO3)2Cl2 containing 1∞[Mn(SeO3)2Cl2]4− chains
- Crystal structure of 3,3′,3″-((1E,1′E,1″E)-((nitrilotris(ethane-2,1-diyl))tris(azaneylylidene)) tris(methaneylylidene))tris(4-hydroxy-1-naphthaldehyde) monohydrate, C42H36N4O6·H2O
- The crystal structure of 4-(6-acetyl-5-methyl-4,7-dihydrotetrazolo[1,5-a]pyrimidin-7-yl)benzonitrile, C14H12N6O
- Crystal structure of benzo[d][1,3]dioxol-5-yl-2-(6-methoxynaphthalen-2-yl)propanoate, C21H18O5
- The crystal structure of ethyl 5-methyl-7-(4-(phenylthio)phenyl)-4,7-dihydrotetrazolo[1,5-a]pyrimidine-6-carboxylate, C20H19N5O2S
- Crystal structure of N′,N‴-((propane-2,2-diylbis(1H-pyrrole-5,2-diyl))bis(methaneylylidene))-di(isonicotinohydrazide)– water – dimethylformamide (1/4/2), C25H24N8O2·4H2O·2C3H7NO
- Synthesis and crystal structure of 4-(2,4-dinitrophenoxy)benzaldehyde, C13H8N2O6
- The crystal structure of 1-dodecylpyridin-1-ium bromide monohydrate, C17H32BrNO
- Crystal structure of (E)-amino(2-(4-(dimethylamino)benzylidene)hydrazineyl)methaniminium nitrate, C10H16N6O3
- Crystal structure of (E)-(2-((1H-pyrrol-2-yl)methylene)hydrazineyl)(amino)methaniminium nitrate monohydrate, C6H12N6O4
- The crystal structure of hexakis(1-propylimidazole-κ1N)copper(II) dichloride, C36H60Cl2CuN12
- The crystal structure of bis{(μ2-3,3-dimethyl-1-phenylbut-1-en-2-yl)((dimethylamino)dimethylsilyl)amido-κ3N,N′:N′}dilithium, C32H54Li2N4Si2
- The crystal structure of methyl 4-(1H-naphtho[1,8-de][1,3,2]diazaborinin-2(3H)-yl)benzoate, C18H15BN2O2
- Crystal structure of (E)-N-(1-((2-chlorothiazol-5-yl)methyl)pyridin-2(1H)-ylidene)-2,2,2-trifluoroacetamide, C11H7ClF3N3OS
- Crystal structure of N′, N‴-((propane-2,2-diylbis(1H-pyrrole-5,2-diyl))bis (methaneylylidene))di(picolinohydrazide) – water – methanol (1/1/1), C25H24N8O2·H2O·CH3OH
- Crystal structure of 3-(2-chloro-benzyl)-7-[4-(2-chloro-benzyl)-piperazin-1-yl]-5,6,8-trifluoro-3H-quinazolin-4-one, C26H21Cl2F3N4O
- Crystal structure of N1,N2-bis(2-fluorobenzyl)benzene-1,2-diamine,C20H18F2N2
- The crystal structure of 2-(benzo[d][1,3]dioxol-5-yl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]diazaborinine, C17H13BN2O2
- The crystal structure of 6,6′-((1E,1′E)-hydrazine-1,2-diylidenebis(methaneylylidene)) bis(2-bromo-4-nitrophenol) — dimethylsulfoxide (1/2), C14H8Br2N4O6⋅2(C2H6OS)
- Selective biocatalytic synthesis and crystal structure of (2R,6R)-hydroxyketaminium chloride, C13H17Cl2NO2
- Crystal structure of bis{tetraaqua-[μ3-1-(4-carboxylatophenyl)-5-methyl-1H-pyrazole-3-carboxylate-κ4N,O,O′,O″] [μ2-1-methyl-1H-pyrazole-3,5-dicarboxylate-κ3N,O:O]dicobalt(II)} dihydrate, C36H44Co4N8O26
- Crystal structure of diethyl-2,2′-naphthalene-2,3-diylbis(oxy)diacetate, C18H20O6
- Synthesis and crystal structure of poly[(μ3-2-(2-carboxylatophenyl)-1H-benzo[d]imidazole-5-carboxylato-κO,O′:O′;:O″, O″′)-(μ2-1-(4-(1Himidazol-1-yl)phenyl)-1H-imidazole-κ2N:N′)cadmium(II)], C27H18CdN6O4
- The crystal structure of catena-poly[diaqua-bis(μ2-2-((2-(2-phenylacetyl)hydrazineylidene)methyl)benzoato-κ2O:O')zinc(II)], C32H30N4O8Zn
- The crystal structure of 2-(3,4-dimethoxyphenyl)-2,3-dihydro-1H-naphtho [1,8-de][1,3,2]diazaborinine, C18H17BN2O2
- The crystal structure of hexakis(1-ethylimidazole-κ1N)nickel(II) dichloride – 1-ethylimidazole (1/2), C40H64Cl2NiN16
- Crystal structure of diaqua-bis(2,4-dinitrophenolato-κ2O,O′)copper(II) 1.5 hydrate, C12H13CuN4O13.5
- Crystal structure of N′,N‴-((1E,1′E)-((decane-1,10-diylbis(oxy))bis(2,1-phenylene)) bis(methaneylylidene))di(isonicotinohydrazide), C36H40N6O4
- The crystal structure of 2-[(R)-1-(naphthalen-1-yl)ethyl]-2,3,7,7a-tetrahydro-3a,6-epoxyisoindol-1(6H)-one, C19H20NO2
- Synthesis and crystal structure of (1E,2E)-3-(anthracen-9-yl)-1-(4-methoxyphenyl)prop-2-en-1-one oxime, C24H19NO2
- Synthesis and crystal structure of (2E,2′E)-3,3′-(1,3-phenylene)bis(1-(3-bromophenyl)prop-2-en-1-one), C24H16Br2O2
- The crystal structure of catena-poly[bis(µ2-1,2-bis((1H-imidazol-1-yl)methyl)benzene- κ2N:N′)-bis(nitrato-κO)copper(II)], C28H28N10O6Cu
- Synthesis and crystal structure of the novel chiral acetyl-3-thiophene-5-(9-anthryl)-2-pyrazoline, C23H18N2OS
- Crystal structure of (E)-3-(dimethylamino)-1-(thiophen-3-yl)prop-2-en-1-one, C9H11NOS
- Crystal structure of catena-poly[aqua-(4-iodopyridine-2,6-dicarboxylato-κ3N,O,O′)-(μ2-4-amino-4H-1,2,4-triazole-κ2N:N′) copper(II)], C9H8N5O5CuI
- Crystal structure of cyclopropane-1,2,3-triyltris(phenylmethanone), C24H18O3
- Crystal structure of bis(amino(thioureido)methaniminium) terephthalate, C12H18N8O4S2
- A three-dimensional Eu(III) framework in the crystal structure of dimethylaminium poly[dimethylformamide-κ1N)bis(μ4-terephthalato-κ4O:O′:O′′:O′′′)europium(III)] monohydrate, C21H25EuN2O10
- Crystal structure of 2-methoxyphenyl 2-(6-methoxynaphthalen-2-yl)propanoate, C21H20O4
- The crystal structure of Hexakis(diethylamido)dimolybdenum, Mo2(NEt2)6

