Crystal structure of bis[benzyl(methyl)carbamodithioato-κ 2 S,S′]-di-n-butyltin(IV), C26H38N2S4Sn
-
Nurul Amalina Abd Aziz
, Kok Meng Chan
Abstract
C26H38N2S4Sn, triclinic,
The molecular structure is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
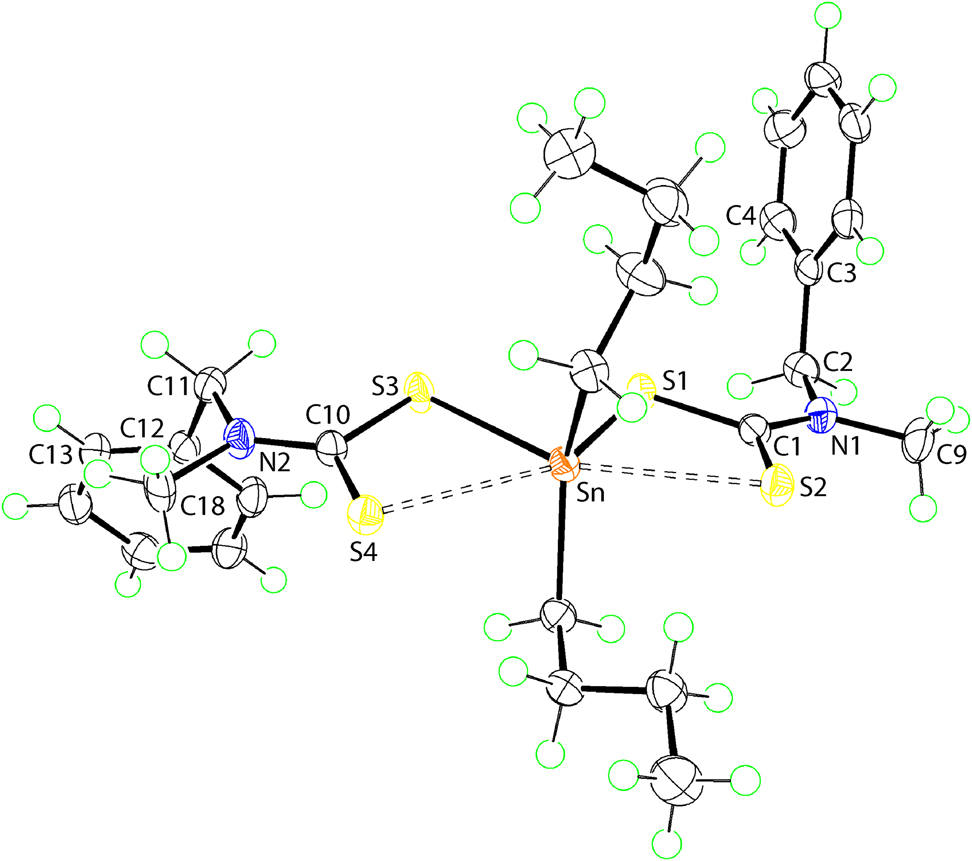
Data collection and handling.
| Crystal: | Colourless prism |
| Size | 0.27 × 0.20 × 0.11 mm |
| Wavelength: | Cu Kα radiation (1.54184 Å) |
| μ: | 10.0 mm−1 |
| Diffractometer, scan mode: | XtaLAB Synergy, ω |
| θ max, completeness: | 75.3°, >99% |
| N(hkl) measured, N(hkl) unique, R int: | 37,119, 5854, 0.033 |
| Criterion for I obs, N(hkl) gt: | I obs > 2 σ(I obs), 5841 |
| N(param)refined: | 302 |
| Programs: | CrysAlis PRO [1], SHELX [2, 3], WinGX/ORTEP [4] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | U iso*/U eq |
|---|---|---|---|---|
| Sn | 0.68916 (2) | 0.59595 (2) | 0.77648 (2) | 0.01437 (5) |
| S1 | 0.59304 (5) | 0.48920 (4) | 0.66375 (3) | 0.01677 (9) |
| S2 | 0.44073 (5) | 0.52679 (4) | 0.86335 (3) | 0.01913 (10) |
| S3 | 0.86956 (5) | 0.58409 (4) | 0.62689 (3) | 0.01739 (9) |
| S4 | 0.89463 (5) | 0.72904 (4) | 0.78246 (3) | 0.01732 (9) |
| N1 | 0.35489 (17) | 0.43578 (14) | 0.71937 (12) | 0.0176 (3) |
| N2 | 1.04222 (17) | 0.71252 (14) | 0.60711 (12) | 0.0174 (3) |
| C1 | 0.4512 (2) | 0.48021 (16) | 0.74830 (14) | 0.0155 (3) |
| C2 | 0.3658 (2) | 0.39303 (17) | 0.62068 (15) | 0.0178 (4) |
| H2A | 0.401166 | 0.444241 | 0.570281 | 0.021* |
| H2B | 0.263747 | 0.403351 | 0.600404 | 0.021* |
| C3 | 0.4714 (2) | 0.26160 (16) | 0.61822 (14) | 0.0163 (3) |
| C4 | 0.5095 (2) | 0.21761 (18) | 0.52511 (15) | 0.0204 (4) |
| H4 | 0.471697 | 0.270418 | 0.464893 | 0.024* |
| C5 | 0.6024 (2) | 0.09730 (19) | 0.51937 (16) | 0.0237 (4) |
| H5 | 0.627313 | 0.068069 | 0.455432 | 0.028* |
| C6 | 0.6589 (2) | 0.01945 (17) | 0.60713 (17) | 0.0222 (4) |
| H6 | 0.722506 | −0.062925 | 0.603351 | 0.027* |
| C7 | 0.6220 (2) | 0.06279 (17) | 0.69997 (16) | 0.0202 (4) |
| H7 | 0.660592 | 0.009995 | 0.760073 | 0.024* |
| C8 | 0.5286 (2) | 0.18340 (17) | 0.70565 (15) | 0.0188 (4) |
| H8 | 0.503768 | 0.212478 | 0.769650 | 0.023* |
| C9 | 0.2402 (2) | 0.4204 (2) | 0.79051 (17) | 0.0240 (4) |
| H9A | 0.168833 | 0.499838 | 0.806301 | 0.036* |
| H9B | 0.289176 | 0.371256 | 0.852715 | 0.036* |
| H9C | 0.185873 | 0.379783 | 0.760100 | 0.036* |
| C10 | 0.9454 (2) | 0.68047 (16) | 0.66750 (14) | 0.0153 (3) |
| C11 | 1.0894 (2) | 0.67291 (17) | 0.50641 (14) | 0.0173 (4) |
| H11A | 1.064418 | 0.602086 | 0.500620 | 0.021* |
| H11B | 1.200276 | 0.646952 | 0.499647 | 0.021* |
| C12 | 1.0145 (2) | 0.77102 (16) | 0.42105 (14) | 0.0163 (3) |
| C13 | 1.0988 (2) | 0.82209 (17) | 0.35781 (15) | 0.0187 (4) |
| H13 | 1.203747 | 0.796966 | 0.369377 | 0.022* |
| C14 | 1.0309 (2) | 0.90965 (18) | 0.27774 (16) | 0.0221 (4) |
| H14 | 1.089805 | 0.943868 | 0.234916 | 0.027* |
| C15 | 0.8780 (2) | 0.94739 (18) | 0.25992 (16) | 0.0218 (4) |
| H15 | 0.831835 | 1.006994 | 0.204929 | 0.026* |
| C16 | 0.7923 (2) | 0.89701 (19) | 0.32351 (16) | 0.0228 (4) |
| H16 | 0.687208 | 0.922744 | 0.312085 | 0.027* |
| C17 | 0.8602 (2) | 0.80941 (18) | 0.40342 (16) | 0.0207 (4) |
| H17 | 0.801235 | 0.775361 | 0.446387 | 0.025* |
| C18 | 1.1072 (2) | 0.7939 (2) | 0.63789 (17) | 0.0255 (4) |
| H18A | 1.170961 | 0.811056 | 0.583192 | 0.038* |
| H18B | 1.168311 | 0.755276 | 0.698550 | 0.038* |
| H18C | 1.025694 | 0.869657 | 0.652497 | 0.038* |
| C19 | 0.8258 (2) | 0.46745 (17) | 0.89439 (14) | 0.0185 (4) |
| H19A | 0.924400 | 0.475071 | 0.895146 | 0.022* |
| H19B | 0.775556 | 0.486449 | 0.960360 | 0.022* |
| C20 | 0.8524 (2) | 0.33720 (18) | 0.88100 (17) | 0.0257 (4) |
| H20A | 0.906075 | 0.317351 | 0.816164 | 0.031* |
| H20B | 0.753651 | 0.330509 | 0.877484 | 0.031* |
| C21 | 0.9450 (2) | 0.24495 (19) | 0.96707 (18) | 0.0280 (4) |
| H21A | 0.897812 | 0.270530 | 1.032446 | 0.034* |
| H21B | 0.941626 | 0.165350 | 0.961319 | 0.034* |
| C22 | 1.1088 (2) | 0.2310 (2) | 0.96693 (17) | 0.0279 (4) |
| H22A | 1.155895 | 0.207226 | 0.901930 | 0.042* |
| H22B | 1.162781 | 0.168659 | 1.021480 | 0.042* |
| H22C | 1.113416 | 0.308125 | 0.977275 | 0.042* |
| C23 | 0.5244 (2) | 0.77921 (16) | 0.75972 (14) | 0.0165 (3) |
| H23A | 0.555598 | 0.825554 | 0.702423 | 0.020* |
| H23B | 0.426504 | 0.778322 | 0.742007 | 0.020* |
| C24 | 0.5010 (2) | 0.84549 (16) | 0.85242 (14) | 0.0175 (4) |
| H24A | 0.437428 | 0.931959 | 0.833227 | 0.021* |
| H24B | 0.600282 | 0.840651 | 0.873559 | 0.021* |
| C25 | 0.4274 (2) | 0.79717 (18) | 0.94263 (15) | 0.0217 (4) |
| H25A | 0.335313 | 0.790351 | 0.919913 | 0.026* |
| H25B | 0.497931 | 0.715202 | 0.969577 | 0.026* |
| C26 | 0.3854 (3) | 0.8784 (2) | 1.02614 (17) | 0.0307 (5) |
| H26A | 0.312985 | 0.958989 | 1.000376 | 0.046* |
| H26B | 0.476366 | 0.884997 | 1.049182 | 0.046* |
| H26C | 0.339849 | 0.843823 | 1.082769 | 0.046* |
Source of material
N–Methylbenzylamine and di-n-butyltin(IV) chloride were purchased from Sigma–Aldrich, carbon disulphide and ammonium solution (25%) were supplied by Merck, and chloroform and ethanol (95%) by System Chemicals.
The title compound was prepared via an in situ method: N-methylbenzylamine (30 mM) was added to ethanol (50 mL) followed by stirring for 1 h under cold temperature (ca 277 K) conditions. The ammonia solution (about 2 mL) was added to the mixture followed by another 30 min stirring. Then, a chilled solution comprising carbon disulphide (30 mM) and ethanol (50 mL) was added drop wise with stirring for 90 min. After that, di-n-butyltin(IV) chloride (15 mM) which had been dissolved in ethanol (50 mL) was added drop wise to the yellow solution with further stirring for 2 h. The white precipitate that formed was filtered, then washed 3 times with cold ethanol to remove unwanted residues. The precipitate was dried in a vacuum desiccator. Yield 75%. Colourless crystals were obtained from the slow evaporation (3–4 days) of its chloroform/ethanol solution (1:2 v/v). Melting point (MPA120 EZ–Melt Automated Melting Point Apparatus): 365.6–368.8 K. A crystal harvested from the mother liquor was transferred directly to the liquid–N2 cold-stream for analysis by X-ray crystallography.
Experimental details
The C-bound H atoms were geometrically placed (C–H = 0.95–0.99 Å) and refined as riding with U iso(H) = 1.2–1.5 U eq(C).
Comment
Organotin dithiocarbamates are well-known to possess several useful properties such as being efficient molecular precursors for the deposition of tin sulphide thin films to biological activity [5], i.e. demonstrating potential as anti-cancer [6] and anti-bacterial agents [7]. It was in the context of biological investigations [8], that the title compound, (n–Bu)2Sn[S2CN(Me)CH2Ph]2, hereafter (I), was investigated crystallographically.
The molecular structure of (I) is shown in the figure (70% displacement ellipsoids). The tin atom in (I) is coordinated by two, asymmetrically bound dithiocarbamate ligands [Sn–S1 = 2.5334(4) and Sn–S3 = 2.5497(4)]. This asymmetry is reflected in the large values of Δ(Sn–S) = [Sn–Slong – Sn–Sshort] = 0.43 and 0.46 Å for the S1- and S3-dithiocarbamate ligands, respectively. The asymmetric mode of coordination exhibited by the dithiocarbamate ligands is seen experimentally in the systematic differences in the associated C–S bond lengths which are rather disparate [C1–S1, S2 = 1.7477(19) & 1.6910(19) Å and 1.7528(19) & 1.6944(19) Å]. The remaining positions in the immediate environment of the tin atom are occupied by the α-carbon atoms of the n-butyl substituents. If a C2S2 donor set is assumed, the range of angles subtended at the tin atom is broad, i.e. 80.289(14)°, for S1–Sn–S3, to 137.83(7)°, for C19–Sn–C23. The distortion from an ideal tetrahedral geometry is also reflected in the value of the geometric parameter τ 4 [9]. This is computed from the equation τ 4 = [360 − (α + β)/141], where α and β are the two widest angles subtended at the tin centre [9]. For an ideal tetrahedral geometry, τ 4 = 1.00 and for an ideal square-planar geometry, τ 4 = 0.00. In (I), τ 4 = 0.80, which is close to τ 4 = 0.85, calculated for a trigonal-pyramidal arrangement. A common description [5] for the molecular geometry observed for (I) is a skewed trapezoidal-bipyramidal geometry with the short trapezoidal edge defined by the more tightly bound S1 and S3 atoms, and the longer edge by the S2 and S4 atoms.
There are three closely related crystal structures to (I) in the literature, namely Ph2Sn[S2CN(Me)CH2C6H4F-4]2, and Me2Sn[S2CN(Me)CH2C6H4Y-4]2, for Y = F & Br [10]. All three molecular structures resemble that observed in (I). Further, it was noted in a recent overview of (n–Bu)2Sn[S2CN(R′)R″]2 molecules [11], that all molecular structures resemble, at least to a first approximation, that noted above for (I).
The most notable aggregate in the crystal is a centrosymmetric dimer within which S3 contacts are apparent [C11–H11a…S3 i : H11a…S3 i = 2.86 Å, C11…S3 i = 3.666(2) Å with angle at H11a = 139° for symmetry operation (i): 2 − x, 1 − y, 1 − z]. The dimeric aggregates form columns along the a-axis. Within columns are reinforcing benzyl–C–H…π(benzyl) contacts [C13–H13…Cg(C12–C16) i : H13…Cg(C12–C16) i = 2.65 Å with angle at H13 = 148° and C16–H16…Cg(C12–C16) ii : H16…Cg(C12–C16) ii = 2.91 Å with angle at H16 = 139° for (ii): 1 − x, 1 − y, 1 − z] implying the C12–C16 benzyl ring forms two such interactions, one to either side of the ring. There are no apparent directional interactions between columns, at least based on standard distance criteria [12].
An analysis of the calculated Hirshfeld surface was conducted [13, 14] in order to probe further the nature of the supramolecular association operating in the crystal of (I). Practically, all surface contacts involve H with by far the greatest contribution due to H…H contacts, i.e. 68.9%, but at separations greater than van der Waals distances. The next most prominent contributions to the Hirshfeld surface come from C…H/H…C [16.9%] and S…H/H…S [13.2%] contacts. The next contribution is 0.6% for N…H/H…N with the balance coming from N…N and S…S, each contributing 0.2%.
Funding source: The Fundamental Research Grants Scheme
Award Identifier / Grant number: (FRGS/1/2021/STG04/UKM/02/5)
Funding source: Crystallographic research at Sunway University is supported by Sunway University Sdn Bhd
Award Identifier / Grant number: (GRTIN–RRO-08–2022 and GRTIN–RRO-56–2022)
-
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Research funding: The Fundamental Research Grants Scheme (FRGS/1/2021/STG04/UKM/02/5) is gratefully acknowledged. Crystallographic research at Sunway University is supported by Sunway University Sdn Bhd (Grant Nos. GRTIN–RRO-08–2022 and GRTIN–RRO-56–2022).
-
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
1. Rigaku Oxford Diffraction. CrysAlisPRO; Rigaku Corporation: Oxford, UK, 2018.Search in Google Scholar
2. Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. 2008, A64, 112–122; https://doi.org/10.1107/s0108767307043930.Search in Google Scholar PubMed
3. Sheldrick, G. M. Crystal structure refinement with SHELX. Acta Crystallogr. 2015, C71, 3–8; https://doi.org/10.1107/s2053229614024218.Search in Google Scholar PubMed PubMed Central
4. Farrugia, L. J. WinGX and ORTEP for Windows: an update. J. Appl. Crystallogr. 2012, 45, 849–854; https://doi.org/10.1107/s0021889812029111.Search in Google Scholar
5. Tiekink, E. R. T. Tin dithiocarbamates: applications and structures. Appl. Organomet. Chem. 2008, 22, 533–550; https://doi.org/10.1002/aoc.1441.Search in Google Scholar
6. Hogarth, G. Metal-dithiocarbamate complexes: chemistry and biological activity. Mini Rev. Med. Chem. 2012, 12, 1202–1215; https://doi.org/10.2174/138955712802762095.Search in Google Scholar PubMed
7. Yeo, C. I., Tiekink, E. R. T., Chew, J. Insights into the antimicrobial potential of dithiocarbamate anions and metal-based species. Inorganics 2021, 9, 48; https://doi.org/10.3390/inorganics9060048.Search in Google Scholar
8. Mohamad, R., Awang, N., Kamaludin, N. F., Abu Bakar, N. F. Synthesis, characterisation and antibacterial activity of new diorganotin(IV) bis(2-methoxyethyl) dithiocarbamate complexes. Res. J. Pharmaceut. Biol. Chem. Sci. 2016, 7, 1269–1274.Search in Google Scholar
9. Yang, L., Powell, D. R., Houser, R. P. Structural variation in copper(I) complexes with pyridylmethylamide ligands: structural analysis with a new four-coordinate geometry index, τ4. Dalton Trans. 2007, 955–964; https://doi.org/10.1039/b617136b.Search in Google Scholar PubMed
10. Khan, N., Farina, Y., Lo, K. M., Rajab, N. F., Awang, N. Syntheses, characterization, X-ray diffraction studies and in vitro antitumor activities of diorganotin(IV) derivatives of bis(p-substituted-N-methylbenzylaminedithiocarbamates). Polyhedron 2015, 85, 754–760; https://doi.org/10.1016/j.poly.2014.08.063.Search in Google Scholar
11. Mohamad, R., Awang, N., Kamaludin, N. F., Jotani, M. M., Tiekink, E. R. T. Di-n-butylbis[N-(2-methoxyethyl)-N-methyldithiocarbamato-κ2S,S′]tin(IV): crystal structure and Hirshfeld surface analysis. Acta Crystallogr. 2017, C73, 260–265; https://doi.org/10.1107/s2056989017001098.Search in Google Scholar PubMed PubMed Central
12. Spek, A. L. checkCIF validation ALERTS: what they mean and how to respond. Acta Crystallogr. 2020, E76, 1–11; https://doi.org/10.1107/s2056989019016244.Search in Google Scholar PubMed PubMed Central
13. Spackman, P. R., Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Jayatilaka, D., Spackman, M. A. CrystalExplorer: a program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011; https://doi.org/10.1107/s1600576721002910.Search in Google Scholar
14. Tan, S. L., Jotani, M. M., Tiekink, E. R. T. Utilizing Hirshfeld surface calculations, non-covalent interaction (NCI) plots and the calculation of interaction energies in the analysis of molecular packing. Acta Crystallogr. 2019, E75, 308–318; https://doi.org/10.1107/s2056989019001129.Search in Google Scholar
© 2022 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of undecacalcium decaarsenide, Ca11As10
- Crystal structure of catena-poly[diiodido-(μ2-1,5-dimethyl-2-phenyl-4-((pyridin-4- ylmethylene)amino)-1,2-dihydro-3H -pyrazol-3-one-κ2 N: O)zinc(II)], C17H16I2N4OZn
- The crystal structure of 5,10,15,20-tetrakis(4-(tert-butyl)phenyl)porphyrin-21,23-diido-κ4 N 4-naphthalocyanido-κ4 N 4-neodymium(IV) - chloroform (1/6) C114H90N12Cl18Nd
- The crystal structure of 1-(4-bromophenyl)-3-(2-chlorobenzyl)urea, C14H12BrClN2O
- Crystal structure of bis[benzyl(methyl)carbamodithioato-κ 2 S,S′]-di-n-butyltin(IV), C26H38N2S4Sn
- Crystal structure of (E)-3-(2-(4-(diethylamino)-2-hydroxystyryl)-3,3-dimethyl-3H-indol-1-ium-1-yl)propane-1-sulfonate – methanol (1/2), C25H32N2O4S⋅2CH3OH
- Synthesis and crystal structure of {(N′,N″-(((ethane-1,2-diylbis(oxy))bis(2,1-phenylene))-bis(methaneylylidene))bis(2-hydroxybenzohydrazonato)-κ6 N 2 O 4}copper(II), C30H24CuN4O6
- The crystal structure of ((E)-2,4-dibromo-6-(((5-(nitro)-2-oxidophenyl)imino)methyl)phenolato-κ 3 N,O,O′)tris(pyridine-κN)manganese(II), C28H21Br2MnN5O4
- Synthesis and crystal structure of 1-((3R,10S,13R,17S)-10,13-dimethyl-3-(p-tolylamino)hexadecahydro-1H-cyclopenta-[a]phenanthren-17-yl)ethan-1-one, C28H41NO
- The crystal structure of 3-nitrobenzene-1,2-dicarboxylic acid—pyrazine-2-carboxamide(1/1), C13H10N4O7
- Crystal structure of poly[tetrakis(μ3-2-aminonicotinato-κ3N,O,O′)-(μ2-oxalato-κ4 O,O′:O″,O′″)-(μ4-oxalato-κ6 N:N′:O,O′:O″,O′″)dicopper(I)-disamarium(III)], [SmCu(C6N2H5O2)2(C2O4)] n
- The crystal structure of 2,3,4-trihydroxybenzoic- acid—pyrazine-2-carboxamide—water (1/1/1), C12H13N3O7
- Crystal structure of N-ethyl-4-[3-(trifluoromethyl)-phenyl]piperazine-1-carbothioamide, C14H18F3N3S
- The crystal structure of 3-anilino-1,4-diphenyl-4H-1,2,4-triazol-1-ium iodide, C20H17N4I
- The crystal structure of (tris(2-benzimidazolylmethyl)amine)-benzoato-copper(II) perchlorate monohydrate, CuC31H28N7O7Cl
- Crystal structure of [2-hydroxy-3-methyl-benzoato-k1 O-triphenyltin(IV)], C26H22O3Sn
- Crystal structure of diaqua-bis(4-(hydroxymethyl)-benzoato-k1 O)zinc(II), C16H18O8Zn
- The crystal structure of dicarbonyl-(N-nitroso-N-oxido-phenylamine-κ 2 O,O)-rhodium(I), C8H5N2O4Rh
- The crystal structure of oxalic acid – 2-ethoxybenzamide (2/1), C20H24N2O8
- The crystal structure of ethyl 7-ethyl-5-methyl-4,7-dihydrotetrazolo[1,5-a]pyrimidine-6-carboxylate, C10H15N5O2
- Crystal structure of poly[(N,N-dimethylacetamide-κO) (μ4-2-nitroisophthalato-κ 4 O:O′:O″:O′″)manganese(II)], C11H10N2O7Mn
- Crystal structure of 14-O-acetyldelcosine, C26H41NO8
- The crystal structure of poly[(1,10-phenanthroline-κ2 N,N′)-(μ 4-2-chlorobenzene-1,3-dicarboxylato-κ5 O:O′:O″:O‴) cadmium(II)] monohydrate, C20H13CdClN2O5
- Crystal structure of propane-1,3-diylbis(diphenylphosphine sulfide) ethanol solvate, C27H26P2S2
- Crystal structure of bis{[(4-diethylamino-2-hydroxy-benzylidene)-hydrazinocarbonylmethyl]-trimethylammonium} tetrabromozincate, C32H54N8O4ZnBr4
- Synthesis and crystal structure of dimethyl 2,2′-(2,5-bis(4-hydroxyphenyl)-2,5-dihydrofuran-3,4-diyl)dibenzoate, C34H30O7
- Synthesis and crystal structure of 2-(2-oxo-2-phenylethyl)-4H-chromen-4-one, C17H12O3
- The crystal structure of tetra(imidazole-κ1 N)zinc(II) μ2-oxido-hexaoxido-divanadium(VI) C12H16N8O6V2Zn
- Crystal structure of S-2-(1-(5-methylpyridin-2-ylamino)octyl)-3-hydroxynaphthalene-1,4-dione, C24H28N2O3
- Crystal structure of bis(μ2-5-chloro-2-oxido-N-(1-oxido-2-oxo-2-phenylethylidene)-benzohydrazonato-κ5 N,O,O′:N′,O′′)-oktakis(pyridine-κ1 N)trinickel(II) – methanol – pyridine (1/1/1) C76H65N13Cl2Ni3O9
- The crystal structure of methyl 3,5-diaminobenzoate, C8H10N2O2
- Crystal structure of 10-(9H-carbazol-9-yl)-5H-dibenzo[a,d][7]annelen-5-one, C27H17NO
- Crystal structure of ethyl 1-(2-hydroxyethyl)-4-((4-methoxyphenyl)amino)-5-oxo-2,5-dihydro-1H-pyrrole-3-carboxylate, C16H20N2O5
- The crystal structure of 1-(4-bromophenyl)-3-cycloheptylurea, C14H19BrN2O
- The crystal structure of 1,4-bis(1,2,3,4,5-pentamethylcyclopenta-2,4-dien-1-yl)-3,6-bis ((4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)methylene)-1,4-dialuminacyclohexane – benzene (1/2), C50H72Al2B2O4
- Crystal structure of bis(μ 3-diphenylphosphinato)-tetrakis(μ 2-diphenylphosphinato)-bis(diphenylphosphinato)-bis(μ 2-hydroxo)dicopper(II)-ditin(IV), C104H100O18P8Cu2Sn2
- Crystal structure of 3-((3,4-dichloroisothiazol-5-yl)methoxy)benzo[d] isothiazole 1,1-dioxide, C11H6Cl2N2O3S2
- Synthesis and crystal structure of 2-(2-(2-fluorophenyl)-2-oxoethyl)-4H-chromen-4-one, C17H11FO3
- The crystal structure of tris(carbonyl)-bis(carbonyl)-[μ-propane-1,2- dithiolato]-(benzyldiphenylphosphine)diiron (Fe—Fe), C27H23Fe2O5PS2
- Crystal structure of 1-(2-(4-chlorophenethyl)-2-hydroxy-3,3-dimethylbutyl)-1H-1,2,4-triazol-4-ium nitrate, C16H23N4O4Cl
- The crystal structure of 3,3′-disulfanediyldi(1H-1,2,4-triazol-5-amine) monohydrate, C4H8N8OS2
- The crystal structure of trans-[bis(4-methylpyridine-κN)bis(quinoline-2-carboxylato- κ 2 N,O)cadmium(II)], C32H26CdN4O4
- The crystal structure of ethyl 2′-hydroxy-4′,6′-dimethoxy-3-(4-methoxynaphthalen-1-yl)-5-oxo-2,3,4,5-tetrahydro-[1,1′-biphenyl]-4-carboxylate, C28H28O7
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of undecacalcium decaarsenide, Ca11As10
- Crystal structure of catena-poly[diiodido-(μ2-1,5-dimethyl-2-phenyl-4-((pyridin-4- ylmethylene)amino)-1,2-dihydro-3H -pyrazol-3-one-κ2 N: O)zinc(II)], C17H16I2N4OZn
- The crystal structure of 5,10,15,20-tetrakis(4-(tert-butyl)phenyl)porphyrin-21,23-diido-κ4 N 4-naphthalocyanido-κ4 N 4-neodymium(IV) - chloroform (1/6) C114H90N12Cl18Nd
- The crystal structure of 1-(4-bromophenyl)-3-(2-chlorobenzyl)urea, C14H12BrClN2O
- Crystal structure of bis[benzyl(methyl)carbamodithioato-κ 2 S,S′]-di-n-butyltin(IV), C26H38N2S4Sn
- Crystal structure of (E)-3-(2-(4-(diethylamino)-2-hydroxystyryl)-3,3-dimethyl-3H-indol-1-ium-1-yl)propane-1-sulfonate – methanol (1/2), C25H32N2O4S⋅2CH3OH
- Synthesis and crystal structure of {(N′,N″-(((ethane-1,2-diylbis(oxy))bis(2,1-phenylene))-bis(methaneylylidene))bis(2-hydroxybenzohydrazonato)-κ6 N 2 O 4}copper(II), C30H24CuN4O6
- The crystal structure of ((E)-2,4-dibromo-6-(((5-(nitro)-2-oxidophenyl)imino)methyl)phenolato-κ 3 N,O,O′)tris(pyridine-κN)manganese(II), C28H21Br2MnN5O4
- Synthesis and crystal structure of 1-((3R,10S,13R,17S)-10,13-dimethyl-3-(p-tolylamino)hexadecahydro-1H-cyclopenta-[a]phenanthren-17-yl)ethan-1-one, C28H41NO
- The crystal structure of 3-nitrobenzene-1,2-dicarboxylic acid—pyrazine-2-carboxamide(1/1), C13H10N4O7
- Crystal structure of poly[tetrakis(μ3-2-aminonicotinato-κ3N,O,O′)-(μ2-oxalato-κ4 O,O′:O″,O′″)-(μ4-oxalato-κ6 N:N′:O,O′:O″,O′″)dicopper(I)-disamarium(III)], [SmCu(C6N2H5O2)2(C2O4)] n
- The crystal structure of 2,3,4-trihydroxybenzoic- acid—pyrazine-2-carboxamide—water (1/1/1), C12H13N3O7
- Crystal structure of N-ethyl-4-[3-(trifluoromethyl)-phenyl]piperazine-1-carbothioamide, C14H18F3N3S
- The crystal structure of 3-anilino-1,4-diphenyl-4H-1,2,4-triazol-1-ium iodide, C20H17N4I
- The crystal structure of (tris(2-benzimidazolylmethyl)amine)-benzoato-copper(II) perchlorate monohydrate, CuC31H28N7O7Cl
- Crystal structure of [2-hydroxy-3-methyl-benzoato-k1 O-triphenyltin(IV)], C26H22O3Sn
- Crystal structure of diaqua-bis(4-(hydroxymethyl)-benzoato-k1 O)zinc(II), C16H18O8Zn
- The crystal structure of dicarbonyl-(N-nitroso-N-oxido-phenylamine-κ 2 O,O)-rhodium(I), C8H5N2O4Rh
- The crystal structure of oxalic acid – 2-ethoxybenzamide (2/1), C20H24N2O8
- The crystal structure of ethyl 7-ethyl-5-methyl-4,7-dihydrotetrazolo[1,5-a]pyrimidine-6-carboxylate, C10H15N5O2
- Crystal structure of poly[(N,N-dimethylacetamide-κO) (μ4-2-nitroisophthalato-κ 4 O:O′:O″:O′″)manganese(II)], C11H10N2O7Mn
- Crystal structure of 14-O-acetyldelcosine, C26H41NO8
- The crystal structure of poly[(1,10-phenanthroline-κ2 N,N′)-(μ 4-2-chlorobenzene-1,3-dicarboxylato-κ5 O:O′:O″:O‴) cadmium(II)] monohydrate, C20H13CdClN2O5
- Crystal structure of propane-1,3-diylbis(diphenylphosphine sulfide) ethanol solvate, C27H26P2S2
- Crystal structure of bis{[(4-diethylamino-2-hydroxy-benzylidene)-hydrazinocarbonylmethyl]-trimethylammonium} tetrabromozincate, C32H54N8O4ZnBr4
- Synthesis and crystal structure of dimethyl 2,2′-(2,5-bis(4-hydroxyphenyl)-2,5-dihydrofuran-3,4-diyl)dibenzoate, C34H30O7
- Synthesis and crystal structure of 2-(2-oxo-2-phenylethyl)-4H-chromen-4-one, C17H12O3
- The crystal structure of tetra(imidazole-κ1 N)zinc(II) μ2-oxido-hexaoxido-divanadium(VI) C12H16N8O6V2Zn
- Crystal structure of S-2-(1-(5-methylpyridin-2-ylamino)octyl)-3-hydroxynaphthalene-1,4-dione, C24H28N2O3
- Crystal structure of bis(μ2-5-chloro-2-oxido-N-(1-oxido-2-oxo-2-phenylethylidene)-benzohydrazonato-κ5 N,O,O′:N′,O′′)-oktakis(pyridine-κ1 N)trinickel(II) – methanol – pyridine (1/1/1) C76H65N13Cl2Ni3O9
- The crystal structure of methyl 3,5-diaminobenzoate, C8H10N2O2
- Crystal structure of 10-(9H-carbazol-9-yl)-5H-dibenzo[a,d][7]annelen-5-one, C27H17NO
- Crystal structure of ethyl 1-(2-hydroxyethyl)-4-((4-methoxyphenyl)amino)-5-oxo-2,5-dihydro-1H-pyrrole-3-carboxylate, C16H20N2O5
- The crystal structure of 1-(4-bromophenyl)-3-cycloheptylurea, C14H19BrN2O
- The crystal structure of 1,4-bis(1,2,3,4,5-pentamethylcyclopenta-2,4-dien-1-yl)-3,6-bis ((4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)methylene)-1,4-dialuminacyclohexane – benzene (1/2), C50H72Al2B2O4
- Crystal structure of bis(μ 3-diphenylphosphinato)-tetrakis(μ 2-diphenylphosphinato)-bis(diphenylphosphinato)-bis(μ 2-hydroxo)dicopper(II)-ditin(IV), C104H100O18P8Cu2Sn2
- Crystal structure of 3-((3,4-dichloroisothiazol-5-yl)methoxy)benzo[d] isothiazole 1,1-dioxide, C11H6Cl2N2O3S2
- Synthesis and crystal structure of 2-(2-(2-fluorophenyl)-2-oxoethyl)-4H-chromen-4-one, C17H11FO3
- The crystal structure of tris(carbonyl)-bis(carbonyl)-[μ-propane-1,2- dithiolato]-(benzyldiphenylphosphine)diiron (Fe—Fe), C27H23Fe2O5PS2
- Crystal structure of 1-(2-(4-chlorophenethyl)-2-hydroxy-3,3-dimethylbutyl)-1H-1,2,4-triazol-4-ium nitrate, C16H23N4O4Cl
- The crystal structure of 3,3′-disulfanediyldi(1H-1,2,4-triazol-5-amine) monohydrate, C4H8N8OS2
- The crystal structure of trans-[bis(4-methylpyridine-κN)bis(quinoline-2-carboxylato- κ 2 N,O)cadmium(II)], C32H26CdN4O4
- The crystal structure of ethyl 2′-hydroxy-4′,6′-dimethoxy-3-(4-methoxynaphthalen-1-yl)-5-oxo-2,3,4,5-tetrahydro-[1,1′-biphenyl]-4-carboxylate, C28H28O7

