Abstract
C13H17Cl2NO2, triclinic, P1 (no. 1), a = 7.1486(3) Å, b = 7.8265(3) Å, c = 13.6579(5) Å, α = 81.085(3)°, β = 77.487(4)°, γ = 73.335(4)°, Z = 2, V = 711.13(5) Å3, Rgt(F) = 0.0536, wRref = 0.0878, T = 290 K.
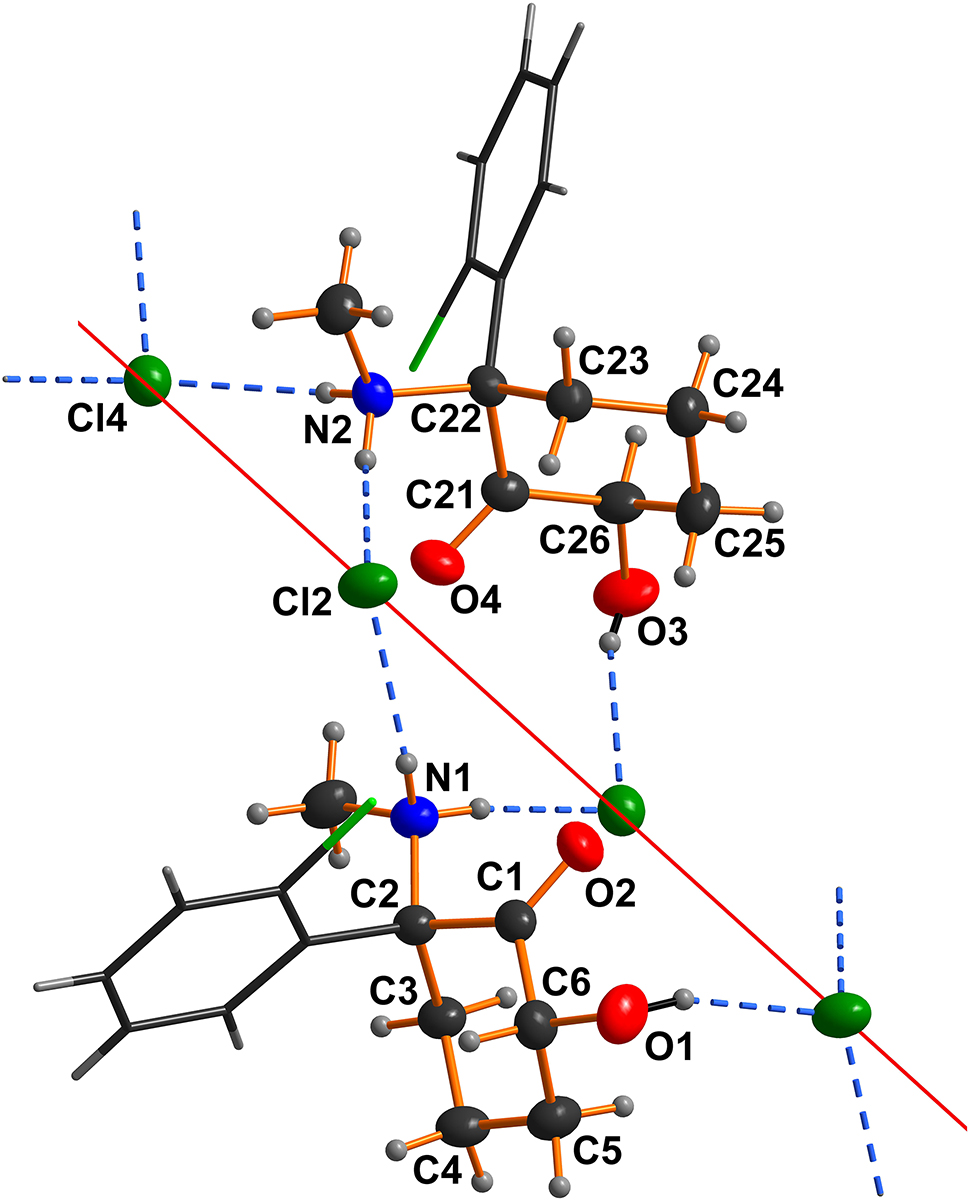
A part of the hydrogen bonded polymeric title structure is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Colorless needle |
| Size: | 0.52 × 0.15 × 0.04 mm |
| Wavelength: | Mo Kα radiation (0.71073 Å) |
| μ: | 0.45 mm−1 |
| Diffractometer, scan mode: | Xcalibur, ω |
| θmax, completeness: | 30.8°, >99% |
| N(hkl)measured, N(hkl)unique, Rint: | 23,627, 8214, 0.056 |
| Criterion for Iobs, N(hkl)gt: | Iobs > 2 σ(Iobs), 5570 |
| N(param)refined: | 335 |
| Programs: | Bruker [1], SHELX [2], [3], Diamond [4] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | Uiso*/Ueq |
|---|---|---|---|---|
| Cl1 | 0.45305 (17) | 0.52741 (15) | 0.19624 (10) | 0.0536 (3) |
| Cl2 | 0.38590 (17) | 0.73388 (14) | 0.41362 (9) | 0.0502 (3) |
| Cl3 | 0.54115 (16) | 0.79443 (17) | 0.83816 (10) | 0.0527 (3) |
| Cl4 | 0.65298 (15) | 1.10762 (13) | 0.60081 (9) | 0.0442 (3) |
| O1 | 0.4597 (5) | 0.0086 (5) | 0.2198 (3) | 0.0531 (10) |
| H1O | 0.418 (9) | −0.048 (8) | 0.278 (5) | 0.09 (2)* |
| O2 | 0.4485 (5) | 0.2071 (4) | 0.3715 (3) | 0.0503 (9) |
| O3 | 0.5377 (6) | 0.3198 (5) | 0.7969 (3) | 0.0589 (11) |
| H3O | 0.578 (7) | 0.287 (7) | 0.744 (4) | 0.053 (18)* |
| O4 | 0.5709 (4) | 0.5911 (4) | 0.6453 (3) | 0.0468 (9) |
| N1 | 0.7089 (5) | 0.3655 (4) | 0.3989 (3) | 0.0379 (9) |
| H11 | 0.606164 | 0.456825 | 0.386775 | 0.045* |
| H12 | 0.666131 | 0.298624 | 0.453246 | 0.045* |
| N2 | 0.3252 (5) | 0.9046 (4) | 0.6130 (3) | 0.0323 (8) |
| H21 | 0.423604 | 0.934784 | 0.630354 | 0.039* |
| H22 | 0.376902 | 0.842764 | 0.559621 | 0.039* |
| C1 | 0.5951 (6) | 0.1825 (5) | 0.3067 (3) | 0.0344 (10) |
| C2 | 0.7736 (6) | 0.2536 (5) | 0.3103 (3) | 0.0319 (9) |
| C3 | 0.9361 (6) | 0.0830 (5) | 0.3364 (3) | 0.0378 (10) |
| H3A | 1.055368 | 0.116487 | 0.338958 | 0.045* |
| H3B | 0.890821 | 0.027556 | 0.402485 | 0.045* |
| C4 | 0.9853 (6) | −0.0523 (6) | 0.2590 (4) | 0.0451 (11) |
| H4A | 1.042621 | −0.000753 | 0.194176 | 0.054* |
| H4B | 1.083658 | −0.158538 | 0.279374 | 0.054* |
| C5 | 0.8035 (7) | −0.1054 (6) | 0.2481 (4) | 0.0505 (13) |
| H5A | 0.759193 | −0.174117 | 0.309727 | 0.061* |
| H5B | 0.839594 | −0.181955 | 0.193792 | 0.061* |
| C6 | 0.6309 (6) | 0.0572 (6) | 0.2259 (4) | 0.0409 (10) |
| H6A | 0.674664 | 0.119639 | 0.160722 | 0.049* |
| C7 | 0.8404 (6) | 0.3655 (5) | 0.2135 (3) | 0.0337 (9) |
| C8 | 1.0412 (7) | 0.3481 (6) | 0.1752 (3) | 0.0447 (11) |
| H8A | 1.134619 | 0.268565 | 0.209835 | 0.054* |
| C9 | 1.1064 (7) | 0.4451 (7) | 0.0874 (4) | 0.0530 (13) |
| H9A | 1.241835 | 0.427834 | 0.063172 | 0.064* |
| C10 | 0.9733 (9) | 0.5658 (7) | 0.0361 (4) | 0.0603 (14) |
| H10A | 1.017392 | 0.632335 | −0.022382 | 0.072* |
| C11 | 0.7724 (8) | 0.5886 (6) | 0.0715 (4) | 0.0501 (12) |
| H11A | 0.680624 | 0.670667 | 0.036937 | 0.060* |
| C12 | 0.7083 (6) | 0.4893 (6) | 0.1584 (3) | 0.0389 (10) |
| C13 | 0.8622 (7) | 0.4391 (6) | 0.4229 (4) | 0.0482 (12) |
| H13A | 0.969936 | 0.342196 | 0.441311 | 0.072* |
| H13B | 0.804504 | 0.511662 | 0.477938 | 0.072* |
| H13C | 0.911149 | 0.511022 | 0.364829 | 0.072* |
| C21 | 0.4192 (6) | 0.6108 (5) | 0.7063 (3) | 0.0343 (10) |
| C22 | 0.2461 (5) | 0.7826 (5) | 0.6987 (3) | 0.0307 (9) |
| C23 | 0.0877 (6) | 0.7155 (5) | 0.6653 (3) | 0.0352 (10) |
| H23A | −0.028084 | 0.815040 | 0.658736 | 0.042* |
| H23B | 0.141021 | 0.670888 | 0.599721 | 0.042* |
| C24 | 0.0255 (6) | 0.5664 (6) | 0.7408 (4) | 0.0437 (11) |
| H24A | −0.036057 | 0.612871 | 0.805249 | 0.052* |
| H24B | −0.071611 | 0.526324 | 0.716886 | 0.052* |
| C25 | 0.2044 (7) | 0.4081 (6) | 0.7543 (4) | 0.0500 (12) |
| H25A | 0.162303 | 0.319156 | 0.805370 | 0.060* |
| H25B | 0.255905 | 0.353335 | 0.691548 | 0.060* |
| C27 | 0.1694 (6) | 0.8801 (5) | 0.7943 (3) | 0.0306 (9) |
| C26 | 0.3676 (6) | 0.4632 (5) | 0.7849 (4) | 0.0433 (11) |
| H26A | 0.314324 | 0.514211 | 0.849528 | 0.052* |
| C28 | −0.0342 (6) | 0.9715 (5) | 0.8182 (3) | 0.0382 (10) |
| H28A | −0.119239 | 0.967991 | 0.776181 | 0.046* |
| C29 | −0.1098 (6) | 1.0649 (6) | 0.9012 (4) | 0.0455 (11) |
| H29A | −0.244206 | 1.124012 | 0.914398 | 0.055* |
| C30 | 0.0120 (7) | 1.0718 (6) | 0.9650 (4) | 0.0488 (12) |
| H30A | −0.039944 | 1.133877 | 1.021792 | 0.059* |
| C31 | 0.2115 (7) | 0.9861 (6) | 0.9442 (3) | 0.0448 (11) |
| H31A | 0.295375 | 0.990535 | 0.986579 | 0.054* |
| C32 | 0.2858 (6) | 0.8937 (5) | 0.8600 (3) | 0.0356 (10) |
| C33 | 0.1791 (6) | 1.0727 (5) | 0.5820 (4) | 0.0429 (11) |
| H33A | 0.080622 | 1.042886 | 0.554803 | 0.064* |
| H33B | 0.246890 | 1.145151 | 0.531613 | 0.064* |
| H33C | 0.115920 | 1.138032 | 0.639500 | 0.064* |
Source of material
The mutated variant I238Q/V286G/L289T/M388A of the cytochrome P450 monooxygenase CYP154E1 from Thermobifida fusca YX was used to catalyze the oxidation of 120 μmol (R)-ketamine hydrochloride to (2R,6R)-hydroxyketamine and -hydroxynorketamine as described by Bokel et al. [5]. The reaction mixture was incubated for 24 h at 25 °C, 250 revolutions per minute. After addition of sodium carbonate, the product mixture was extracted using ethyl acetate. A semi-preparative HPLC was used to purify the (2R,6R)-hydroxy-ketamine as previously described [5]. The solvents (acetonitrile/water) were removed via evaporation and lyophilization. Afterwards, the residue was dissolved in chloroform with hydrochloric acid to form the title compound. The solvent was removed again, and the title compound was dissolved in 2-propanol. Slow evaporation over night at room temperature gave colorless needle crystals.
Experimental details
All hydrogen atoms were added using different riding models depending on the chemical surroundings [3]. The Flack parameter [−0.02(3)] was determined using 1973 quotients ([I+] − [I−])/([I+] + [I−]) using the post-refinement Parsons’ quotients method [3], [6].
Comment
(2R,6R)-hydroxyketamine is one of the minor human metabolites of the anesthetic drug (R,S)-ketamine [7], [8], [9]. Whereas the hepatic metabolism by cytochrome P450s (CYPs) leads to a mixture of several hydroxynorketamines and hydroxyketamines, mutagenesis of the bacterial CYP154E1 allowed the selective synthesis of (2S,6S)- hydroxynorketamine from (S)-ketamine [10] as well as (2R,6R)-hydroxynorketamine from (R)-ketamine [5]. In this study, the crystal structure of one of the side products of the latter reaction is presented.
Molecular description and comparison. The asymmetric unit of the title structure contains two crystallographically independent (2R,6R)-hydroxyketaminium cations (systematic name: (1R,3R)-1-(2-chlorophenyl)-3-hydroxy-N-methyl-2-oxocyclohexan-1-aminium; the protonation at the methylamino group shifts the numbering scheme which includes the chiral centers) and two chloride counter anions (see the figure). In contrast to our previously published structure of (2R,4S)-hydroxyketamine [11], the cyclohexanone ring is in a different chair conformation, which rules out an intramolecular hydrogen bond between the aminium and the hydroxy group. Instead, the conformation of the title cation is very similar to that of the cation seen in the crystal structure of (2R,6R)-hydroxynorketaminium chloride [12]. In general, all molecular geometric parameters are in the expected ranges [11], [12].
Supramolecular aspects. In the title crystal structure each cation donates three classical hydrogen bonds. In detail, each cation donates two NH ··· Cl bonds and one OH ··· Cl hydrogen bond (see the figure). Thus, the (2R,6R)-hydroxyketaminium cation exclusively donates hydrogen bonds. The fact that the carbonyl group is not involved in any classical hydrogen bond is the result of a surplus of hydrogen bond acceptors in this structure. Obviously, the charge-supported hydrogen bonds (NH+···Cl− and NH+···O) are preferred. There are two prominent hydrogen bonding motifs, which should be mentioned and classified using so-called graph set descriptors [13]. There are 12-membered rings consisting of two cation and two chloride anions (graph set descriptor: R24(12); see the figure). Furthermore, a simple chain motif is seen, which is formed by the NH2 groups and the chloride anions (see the figure; graph set descriptor: C12(4)). Consequently, a chain-type polymer is formed, which propagates along the crystallographic b axis (red line in the figure).
A further comparison with the aforementioned crystal structure of (2R,6R)-hydroxynorketaminium chloride [12] shows that NH3 group instead of the H3C–NH2 moiety allows a further hydrogen bonding connection leading to a two-dimensional framework.
Funding source: Federal Ministry of Education and Research
Award Identifier / Grant number: 031A223A
Funding source: Ministry of Innovation, Science and Research of North-Rhine Westphalia
Funding source: German Research Foundation
Award Identifier / Grant number: 162659349
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
Research funding: Federal Ministry of Education and Research, Germany (grant number 031A223A), the Ministry of Innovation, Science and Research of North-Rhine Westphalia and the German Research Foundation (DFG) (Xcalibur diffractometer; INST208/533-1, project no. 162659349).
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
1. Bruker. APEX2, SAINT, SADABS; Bruker AXS Inc.: Madison, Wisconsin, USA, 2012.Search in Google Scholar
2. Sheldrick, G. M. SHELXT – integrated space-group and crystal-structure determination. Acta Crystallogr. 2015, A71, 3–8; https://doi.org/10.1107/s2053273314026370.Search in Google Scholar
3. Sheldrick, G. M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8; https://doi.org/10.1107/s2053229614024218.Search in Google Scholar
4. Brandenburg, K. DIAMOND. Visual Crystal Structure Information System (Ver. 4.6.4). Crystal Impact: Bonn (Germany), 2015.Search in Google Scholar
5. Bokel, A., Hutter, M. C., Urlacher, V. B. Molecular evolution of a cytochrome P450 for the synthesis of potential antidepressant (2R,6R)-hydroxynorketamine. Chem. Commun. 2021, 57, 520–523; https://doi.org/10.1039/d0cc06729f.Search in Google Scholar
6. Parsons, S., Flack, H. D., Wagner, T. Use of intensity quotients and differences in absolute structure refinement. Acta Crystallogr. 2013, B69, 249–259; https://doi.org/10.1107/s2052519213010014.Search in Google Scholar
7. Moaddel, R., Venkata, S. L. V., Tanga, M. J., Bupp, J. E., Green, C. E., Iyer, L., Furimsky, A., Goldberg, M. E., Torjman, M. C., Wainer, I. W. A parallel chiral-achiral liquid chromatographic method for the determination of the stereoisomers of ketamine and ketamine metabolites in the plasma and urine of patients with complex regional pain syndrome. Talanta 2010, 82, 1892–1904; https://doi.org/10.1016/j.talanta.2010.08.005.Search in Google Scholar
8. Desta, Z., Moaddel, R., Ogburn, E. T., Xu, C., Ramamoorthy, A., Venkata, S. L. V., Sanghvi, M., Goldberg, M. E., Torjman, M. C., Wainer, I. W. Stereoselective and regiospecific hydroxylation of ketamine and norketamine. Xenobiotica 2012, 42, 1076–87; https://doi.org/10.3109/00498254.2012.685777.Search in Google Scholar
9. Zanos, P., Moaddel, R., Morris, P. J., Riggs, L. M., Highland, J. N., Georgiou, P., Pereira, E. F. R., Albuquerque, E. X., Thomas, C. J., Zarate, C. A.Jr., Gould, T. D. Ketamine and ketamine metabolite pharmacology: insights into therapeutic mechanisms. Pharmacol. Rev. 2018, 70, 621–660; https://doi.org/10.1124/pr.117.015198.Search in Google Scholar
10. Bokel, A., Rühlmann, A., Hutter, M. C., Urlacher, V. B. Enzyme-mediated two-step regio- and stereoselective synthesis of potential rapid-acting antidepressant (2S,6S)-hydroxynorketamine. ACS Catal. 2020, 10, 4151–4159; https://doi.org/10.1021/acscatal.9b05384.Search in Google Scholar
11. Reiss, G. J., Urlacher, V. B., Luelf, U. J. Enzyme-mediated synthesis and crystal structure of (2R,4S)-hydroxyketamine, C13H16ClNO2. Z. Kristallogr. N. Cryst. Struct. 2020, 235, 1037–1039; https://doi.org/10.1515/ncrs-2020-0157.Search in Google Scholar
12. Morris, P. J., Moaddel, R., Zanos, P., Moore, C. E., Gould, T., Zarate, C. A.Jr., Thomas, C. J. Synthesis and N–Methyl-d- aspartate (NMDA) receptor activity of ketamine metabolites. Org. Lett. 2017, 19, 4572–4575; https://doi.org/10.1021/acs.orglett.7b02177.Search in Google Scholar
13. Grell, J., Bernstein, J., Tinhofer, G. Investigation of hydrogen bond patterns: a review of mathematical tools for the graph set approach. Crystallogr. Rev. 2002, 8, 1–56; https://doi.org/10.1080/08893110211936.Search in Google Scholar
© 2021 U. Joost Luelf et al., published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of poly[(μ2-aqua-tetraaqua-(μ3-glutarato-κ4O,O′:O′:O′′)-(μ5-glutarato-κ6O:O,O′:O′:O′′:O′′′)distrontium(II)], C10H22O13Sr2
- The crystal structure of acetato-κ1O-{(2-(2-(2-aminophenoxy)ethoxy)phenyl)(4-oxo-4-phenylbut-2-en-2-yl)amido-κ2N,N′,O}copper(II), C26H26CuN2O5
- Crystal structure of dimethanolato-k2O:O-bis(1-((2-methyl-1H-benzo[d]imidazol-1-yl)methyl)-1H-benzo[d][1,2,3]triazole-κN)-bis(thiocyanato-κN)dicopper(II), C34H32Cu2N12O2S2
- Crystal structure of poly[diaqua-bis(μ2-3-(pyrimidin-5-yl)benzoato-κ2N:O)cobalt(II)] dihydrate, [Co(C11H11O2N2)2(H2O)2]
- Crystal structure of bis(3,3-dimethyl-1-phenylbut-1-en-2-yl)(trimethylsilyl)amido-k1N)zinc(II), Zn(C15H24NSi)2
- Crystal structure of catena-poly[(μ2-methanolato-κ2O:O)-(μ2-1-((2-methyl-1H-benzo[d]imidazol-1-yl)methyl)-1H-benzo[d][1,2,3]triazole-κ2N:N′)-(thiocyanato-κ1N)copper(II)] 0.25 hydrate, C17H16CuN6OS ⋅ 0.5H2O
- The crystal structure of 2-amino-5-nitroanilinium iodide monohydrate, C6H8IN3O2
- The crystal structure of 3-amino-5-carboxypyridin-1-ium perchlorate monohydrate, C6H9ClN2O7
- Crystal structure of 7-hydroxy-2,4-dimethoxy-9,10-dihydrophenanthrene from Arundina graminifolia, C16H16O3
- Crystal structure of 6,6′-((1E, 1′E)-(((1R, 2R)-1,2-diphenylethane-1,2-diyl) bis(azanylylidene))bis(methanylylidene))bis(2-ethylphenol), C32H32N2O2
- The crystal structure of 2-amino-5-carboxypyridin-1-ium iodide monohydrate, C6H9IN2O3
- The crystal structure of 2-(3,5-difluorophenyl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]diazaborinine, C16H11BF2N2
- Crystal structure of bis{(2-pyridinyl)-1-phenyl-1-isopropylmethanolato-κ2N,O}nickel, C30H32N2NiO2
- Crystal structure of poly[(m3-3-carboxyadamantane-1-carboxylato-κ3O:O′:O″)-(phenanthroline-κ2N,N′)sodium(II)], C24H23N2NaO4
- Crystal structure of 2-phenylethynyl-1,3,6,8-tetramethylBOPHY (BOPHY = bis(difluoroboron)-1,2-bis((1H-pyrrol-2-yl)methylene)hydrazine), C22H20B2F4N4
- Crystal structure of 4-tert-butyl-2-N-(2-pyridylmethyl)aminophenol, C16H20N2O
- The crystal structure of (3Z,3′Z)-4,4′-((1,4-phenylenebis(methylene))bis(azanediyl))bis(pent-3-en-2-one), C18H24N2O2
- Crystal structure of (morpholine-1-carbodithioato-κ2-S,S′)bis(triphenylphosphine-κ-P)gold(I), C41H38AuNOP2S2
- Crystal structure of 1,4-bis(4-bromobenzyl)-4-(4-chlorophenyl)-1,4-dihydropyridine-3-carbonitrile, C26H19Br2ClN2
- The crystal structure of fac-tricarbonyl (N′-benzoyl-N,N-diphenylcarbamimidothioato-κ2S,O)-(pyrazole-κN)rhenium(I) — methanol (1/1) C26H23O4N4SRe
- The crystal structure of Ba2Mn(SeO3)2Cl2 containing 1∞[Mn(SeO3)2Cl2]4− chains
- Crystal structure of 3,3′,3″-((1E,1′E,1″E)-((nitrilotris(ethane-2,1-diyl))tris(azaneylylidene)) tris(methaneylylidene))tris(4-hydroxy-1-naphthaldehyde) monohydrate, C42H36N4O6·H2O
- The crystal structure of 4-(6-acetyl-5-methyl-4,7-dihydrotetrazolo[1,5-a]pyrimidin-7-yl)benzonitrile, C14H12N6O
- Crystal structure of benzo[d][1,3]dioxol-5-yl-2-(6-methoxynaphthalen-2-yl)propanoate, C21H18O5
- The crystal structure of ethyl 5-methyl-7-(4-(phenylthio)phenyl)-4,7-dihydrotetrazolo[1,5-a]pyrimidine-6-carboxylate, C20H19N5O2S
- Crystal structure of N′,N‴-((propane-2,2-diylbis(1H-pyrrole-5,2-diyl))bis(methaneylylidene))-di(isonicotinohydrazide)– water – dimethylformamide (1/4/2), C25H24N8O2·4H2O·2C3H7NO
- Synthesis and crystal structure of 4-(2,4-dinitrophenoxy)benzaldehyde, C13H8N2O6
- The crystal structure of 1-dodecylpyridin-1-ium bromide monohydrate, C17H32BrNO
- Crystal structure of (E)-amino(2-(4-(dimethylamino)benzylidene)hydrazineyl)methaniminium nitrate, C10H16N6O3
- Crystal structure of (E)-(2-((1H-pyrrol-2-yl)methylene)hydrazineyl)(amino)methaniminium nitrate monohydrate, C6H12N6O4
- The crystal structure of hexakis(1-propylimidazole-κ1N)copper(II) dichloride, C36H60Cl2CuN12
- The crystal structure of bis{(μ2-3,3-dimethyl-1-phenylbut-1-en-2-yl)((dimethylamino)dimethylsilyl)amido-κ3N,N′:N′}dilithium, C32H54Li2N4Si2
- The crystal structure of methyl 4-(1H-naphtho[1,8-de][1,3,2]diazaborinin-2(3H)-yl)benzoate, C18H15BN2O2
- Crystal structure of (E)-N-(1-((2-chlorothiazol-5-yl)methyl)pyridin-2(1H)-ylidene)-2,2,2-trifluoroacetamide, C11H7ClF3N3OS
- Crystal structure of N′, N‴-((propane-2,2-diylbis(1H-pyrrole-5,2-diyl))bis (methaneylylidene))di(picolinohydrazide) – water – methanol (1/1/1), C25H24N8O2·H2O·CH3OH
- Crystal structure of 3-(2-chloro-benzyl)-7-[4-(2-chloro-benzyl)-piperazin-1-yl]-5,6,8-trifluoro-3H-quinazolin-4-one, C26H21Cl2F3N4O
- Crystal structure of N1,N2-bis(2-fluorobenzyl)benzene-1,2-diamine,C20H18F2N2
- The crystal structure of 2-(benzo[d][1,3]dioxol-5-yl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]diazaborinine, C17H13BN2O2
- The crystal structure of 6,6′-((1E,1′E)-hydrazine-1,2-diylidenebis(methaneylylidene)) bis(2-bromo-4-nitrophenol) — dimethylsulfoxide (1/2), C14H8Br2N4O6⋅2(C2H6OS)
- Selective biocatalytic synthesis and crystal structure of (2R,6R)-hydroxyketaminium chloride, C13H17Cl2NO2
- Crystal structure of bis{tetraaqua-[μ3-1-(4-carboxylatophenyl)-5-methyl-1H-pyrazole-3-carboxylate-κ4N,O,O′,O″] [μ2-1-methyl-1H-pyrazole-3,5-dicarboxylate-κ3N,O:O]dicobalt(II)} dihydrate, C36H44Co4N8O26
- Crystal structure of diethyl-2,2′-naphthalene-2,3-diylbis(oxy)diacetate, C18H20O6
- Synthesis and crystal structure of poly[(μ3-2-(2-carboxylatophenyl)-1H-benzo[d]imidazole-5-carboxylato-κO,O′:O′;:O″, O″′)-(μ2-1-(4-(1Himidazol-1-yl)phenyl)-1H-imidazole-κ2N:N′)cadmium(II)], C27H18CdN6O4
- The crystal structure of catena-poly[diaqua-bis(μ2-2-((2-(2-phenylacetyl)hydrazineylidene)methyl)benzoato-κ2O:O')zinc(II)], C32H30N4O8Zn
- The crystal structure of 2-(3,4-dimethoxyphenyl)-2,3-dihydro-1H-naphtho [1,8-de][1,3,2]diazaborinine, C18H17BN2O2
- The crystal structure of hexakis(1-ethylimidazole-κ1N)nickel(II) dichloride – 1-ethylimidazole (1/2), C40H64Cl2NiN16
- Crystal structure of diaqua-bis(2,4-dinitrophenolato-κ2O,O′)copper(II) 1.5 hydrate, C12H13CuN4O13.5
- Crystal structure of N′,N‴-((1E,1′E)-((decane-1,10-diylbis(oxy))bis(2,1-phenylene)) bis(methaneylylidene))di(isonicotinohydrazide), C36H40N6O4
- The crystal structure of 2-[(R)-1-(naphthalen-1-yl)ethyl]-2,3,7,7a-tetrahydro-3a,6-epoxyisoindol-1(6H)-one, C19H20NO2
- Synthesis and crystal structure of (1E,2E)-3-(anthracen-9-yl)-1-(4-methoxyphenyl)prop-2-en-1-one oxime, C24H19NO2
- Synthesis and crystal structure of (2E,2′E)-3,3′-(1,3-phenylene)bis(1-(3-bromophenyl)prop-2-en-1-one), C24H16Br2O2
- The crystal structure of catena-poly[bis(µ2-1,2-bis((1H-imidazol-1-yl)methyl)benzene- κ2N:N′)-bis(nitrato-κO)copper(II)], C28H28N10O6Cu
- Synthesis and crystal structure of the novel chiral acetyl-3-thiophene-5-(9-anthryl)-2-pyrazoline, C23H18N2OS
- Crystal structure of (E)-3-(dimethylamino)-1-(thiophen-3-yl)prop-2-en-1-one, C9H11NOS
- Crystal structure of catena-poly[aqua-(4-iodopyridine-2,6-dicarboxylato-κ3N,O,O′)-(μ2-4-amino-4H-1,2,4-triazole-κ2N:N′) copper(II)], C9H8N5O5CuI
- Crystal structure of cyclopropane-1,2,3-triyltris(phenylmethanone), C24H18O3
- Crystal structure of bis(amino(thioureido)methaniminium) terephthalate, C12H18N8O4S2
- A three-dimensional Eu(III) framework in the crystal structure of dimethylaminium poly[dimethylformamide-κ1N)bis(μ4-terephthalato-κ4O:O′:O′′:O′′′)europium(III)] monohydrate, C21H25EuN2O10
- Crystal structure of 2-methoxyphenyl 2-(6-methoxynaphthalen-2-yl)propanoate, C21H20O4
- The crystal structure of Hexakis(diethylamido)dimolybdenum, Mo2(NEt2)6
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of poly[(μ2-aqua-tetraaqua-(μ3-glutarato-κ4O,O′:O′:O′′)-(μ5-glutarato-κ6O:O,O′:O′:O′′:O′′′)distrontium(II)], C10H22O13Sr2
- The crystal structure of acetato-κ1O-{(2-(2-(2-aminophenoxy)ethoxy)phenyl)(4-oxo-4-phenylbut-2-en-2-yl)amido-κ2N,N′,O}copper(II), C26H26CuN2O5
- Crystal structure of dimethanolato-k2O:O-bis(1-((2-methyl-1H-benzo[d]imidazol-1-yl)methyl)-1H-benzo[d][1,2,3]triazole-κN)-bis(thiocyanato-κN)dicopper(II), C34H32Cu2N12O2S2
- Crystal structure of poly[diaqua-bis(μ2-3-(pyrimidin-5-yl)benzoato-κ2N:O)cobalt(II)] dihydrate, [Co(C11H11O2N2)2(H2O)2]
- Crystal structure of bis(3,3-dimethyl-1-phenylbut-1-en-2-yl)(trimethylsilyl)amido-k1N)zinc(II), Zn(C15H24NSi)2
- Crystal structure of catena-poly[(μ2-methanolato-κ2O:O)-(μ2-1-((2-methyl-1H-benzo[d]imidazol-1-yl)methyl)-1H-benzo[d][1,2,3]triazole-κ2N:N′)-(thiocyanato-κ1N)copper(II)] 0.25 hydrate, C17H16CuN6OS ⋅ 0.5H2O
- The crystal structure of 2-amino-5-nitroanilinium iodide monohydrate, C6H8IN3O2
- The crystal structure of 3-amino-5-carboxypyridin-1-ium perchlorate monohydrate, C6H9ClN2O7
- Crystal structure of 7-hydroxy-2,4-dimethoxy-9,10-dihydrophenanthrene from Arundina graminifolia, C16H16O3
- Crystal structure of 6,6′-((1E, 1′E)-(((1R, 2R)-1,2-diphenylethane-1,2-diyl) bis(azanylylidene))bis(methanylylidene))bis(2-ethylphenol), C32H32N2O2
- The crystal structure of 2-amino-5-carboxypyridin-1-ium iodide monohydrate, C6H9IN2O3
- The crystal structure of 2-(3,5-difluorophenyl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]diazaborinine, C16H11BF2N2
- Crystal structure of bis{(2-pyridinyl)-1-phenyl-1-isopropylmethanolato-κ2N,O}nickel, C30H32N2NiO2
- Crystal structure of poly[(m3-3-carboxyadamantane-1-carboxylato-κ3O:O′:O″)-(phenanthroline-κ2N,N′)sodium(II)], C24H23N2NaO4
- Crystal structure of 2-phenylethynyl-1,3,6,8-tetramethylBOPHY (BOPHY = bis(difluoroboron)-1,2-bis((1H-pyrrol-2-yl)methylene)hydrazine), C22H20B2F4N4
- Crystal structure of 4-tert-butyl-2-N-(2-pyridylmethyl)aminophenol, C16H20N2O
- The crystal structure of (3Z,3′Z)-4,4′-((1,4-phenylenebis(methylene))bis(azanediyl))bis(pent-3-en-2-one), C18H24N2O2
- Crystal structure of (morpholine-1-carbodithioato-κ2-S,S′)bis(triphenylphosphine-κ-P)gold(I), C41H38AuNOP2S2
- Crystal structure of 1,4-bis(4-bromobenzyl)-4-(4-chlorophenyl)-1,4-dihydropyridine-3-carbonitrile, C26H19Br2ClN2
- The crystal structure of fac-tricarbonyl (N′-benzoyl-N,N-diphenylcarbamimidothioato-κ2S,O)-(pyrazole-κN)rhenium(I) — methanol (1/1) C26H23O4N4SRe
- The crystal structure of Ba2Mn(SeO3)2Cl2 containing 1∞[Mn(SeO3)2Cl2]4− chains
- Crystal structure of 3,3′,3″-((1E,1′E,1″E)-((nitrilotris(ethane-2,1-diyl))tris(azaneylylidene)) tris(methaneylylidene))tris(4-hydroxy-1-naphthaldehyde) monohydrate, C42H36N4O6·H2O
- The crystal structure of 4-(6-acetyl-5-methyl-4,7-dihydrotetrazolo[1,5-a]pyrimidin-7-yl)benzonitrile, C14H12N6O
- Crystal structure of benzo[d][1,3]dioxol-5-yl-2-(6-methoxynaphthalen-2-yl)propanoate, C21H18O5
- The crystal structure of ethyl 5-methyl-7-(4-(phenylthio)phenyl)-4,7-dihydrotetrazolo[1,5-a]pyrimidine-6-carboxylate, C20H19N5O2S
- Crystal structure of N′,N‴-((propane-2,2-diylbis(1H-pyrrole-5,2-diyl))bis(methaneylylidene))-di(isonicotinohydrazide)– water – dimethylformamide (1/4/2), C25H24N8O2·4H2O·2C3H7NO
- Synthesis and crystal structure of 4-(2,4-dinitrophenoxy)benzaldehyde, C13H8N2O6
- The crystal structure of 1-dodecylpyridin-1-ium bromide monohydrate, C17H32BrNO
- Crystal structure of (E)-amino(2-(4-(dimethylamino)benzylidene)hydrazineyl)methaniminium nitrate, C10H16N6O3
- Crystal structure of (E)-(2-((1H-pyrrol-2-yl)methylene)hydrazineyl)(amino)methaniminium nitrate monohydrate, C6H12N6O4
- The crystal structure of hexakis(1-propylimidazole-κ1N)copper(II) dichloride, C36H60Cl2CuN12
- The crystal structure of bis{(μ2-3,3-dimethyl-1-phenylbut-1-en-2-yl)((dimethylamino)dimethylsilyl)amido-κ3N,N′:N′}dilithium, C32H54Li2N4Si2
- The crystal structure of methyl 4-(1H-naphtho[1,8-de][1,3,2]diazaborinin-2(3H)-yl)benzoate, C18H15BN2O2
- Crystal structure of (E)-N-(1-((2-chlorothiazol-5-yl)methyl)pyridin-2(1H)-ylidene)-2,2,2-trifluoroacetamide, C11H7ClF3N3OS
- Crystal structure of N′, N‴-((propane-2,2-diylbis(1H-pyrrole-5,2-diyl))bis (methaneylylidene))di(picolinohydrazide) – water – methanol (1/1/1), C25H24N8O2·H2O·CH3OH
- Crystal structure of 3-(2-chloro-benzyl)-7-[4-(2-chloro-benzyl)-piperazin-1-yl]-5,6,8-trifluoro-3H-quinazolin-4-one, C26H21Cl2F3N4O
- Crystal structure of N1,N2-bis(2-fluorobenzyl)benzene-1,2-diamine,C20H18F2N2
- The crystal structure of 2-(benzo[d][1,3]dioxol-5-yl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]diazaborinine, C17H13BN2O2
- The crystal structure of 6,6′-((1E,1′E)-hydrazine-1,2-diylidenebis(methaneylylidene)) bis(2-bromo-4-nitrophenol) — dimethylsulfoxide (1/2), C14H8Br2N4O6⋅2(C2H6OS)
- Selective biocatalytic synthesis and crystal structure of (2R,6R)-hydroxyketaminium chloride, C13H17Cl2NO2
- Crystal structure of bis{tetraaqua-[μ3-1-(4-carboxylatophenyl)-5-methyl-1H-pyrazole-3-carboxylate-κ4N,O,O′,O″] [μ2-1-methyl-1H-pyrazole-3,5-dicarboxylate-κ3N,O:O]dicobalt(II)} dihydrate, C36H44Co4N8O26
- Crystal structure of diethyl-2,2′-naphthalene-2,3-diylbis(oxy)diacetate, C18H20O6
- Synthesis and crystal structure of poly[(μ3-2-(2-carboxylatophenyl)-1H-benzo[d]imidazole-5-carboxylato-κO,O′:O′;:O″, O″′)-(μ2-1-(4-(1Himidazol-1-yl)phenyl)-1H-imidazole-κ2N:N′)cadmium(II)], C27H18CdN6O4
- The crystal structure of catena-poly[diaqua-bis(μ2-2-((2-(2-phenylacetyl)hydrazineylidene)methyl)benzoato-κ2O:O')zinc(II)], C32H30N4O8Zn
- The crystal structure of 2-(3,4-dimethoxyphenyl)-2,3-dihydro-1H-naphtho [1,8-de][1,3,2]diazaborinine, C18H17BN2O2
- The crystal structure of hexakis(1-ethylimidazole-κ1N)nickel(II) dichloride – 1-ethylimidazole (1/2), C40H64Cl2NiN16
- Crystal structure of diaqua-bis(2,4-dinitrophenolato-κ2O,O′)copper(II) 1.5 hydrate, C12H13CuN4O13.5
- Crystal structure of N′,N‴-((1E,1′E)-((decane-1,10-diylbis(oxy))bis(2,1-phenylene)) bis(methaneylylidene))di(isonicotinohydrazide), C36H40N6O4
- The crystal structure of 2-[(R)-1-(naphthalen-1-yl)ethyl]-2,3,7,7a-tetrahydro-3a,6-epoxyisoindol-1(6H)-one, C19H20NO2
- Synthesis and crystal structure of (1E,2E)-3-(anthracen-9-yl)-1-(4-methoxyphenyl)prop-2-en-1-one oxime, C24H19NO2
- Synthesis and crystal structure of (2E,2′E)-3,3′-(1,3-phenylene)bis(1-(3-bromophenyl)prop-2-en-1-one), C24H16Br2O2
- The crystal structure of catena-poly[bis(µ2-1,2-bis((1H-imidazol-1-yl)methyl)benzene- κ2N:N′)-bis(nitrato-κO)copper(II)], C28H28N10O6Cu
- Synthesis and crystal structure of the novel chiral acetyl-3-thiophene-5-(9-anthryl)-2-pyrazoline, C23H18N2OS
- Crystal structure of (E)-3-(dimethylamino)-1-(thiophen-3-yl)prop-2-en-1-one, C9H11NOS
- Crystal structure of catena-poly[aqua-(4-iodopyridine-2,6-dicarboxylato-κ3N,O,O′)-(μ2-4-amino-4H-1,2,4-triazole-κ2N:N′) copper(II)], C9H8N5O5CuI
- Crystal structure of cyclopropane-1,2,3-triyltris(phenylmethanone), C24H18O3
- Crystal structure of bis(amino(thioureido)methaniminium) terephthalate, C12H18N8O4S2
- A three-dimensional Eu(III) framework in the crystal structure of dimethylaminium poly[dimethylformamide-κ1N)bis(μ4-terephthalato-κ4O:O′:O′′:O′′′)europium(III)] monohydrate, C21H25EuN2O10
- Crystal structure of 2-methoxyphenyl 2-(6-methoxynaphthalen-2-yl)propanoate, C21H20O4
- The crystal structure of Hexakis(diethylamido)dimolybdenum, Mo2(NEt2)6

