Abstract
A novel C25 sterol peroxide, phomasterol A (1), together with two known compounds (2–3), was isolated from the endophytic fungus Phoma sp. EA-122. The structure of phomasterol A (1) was elucidated by MS, 1D, and 2D NMR data analyses. Phomasterol A (1) was evaluated for its inhibitory activities against protein-tyrosine phosphatases MEG2 and PTP1Bc, showing moderate activities with identical IC50 values of 25 μM.
1 Introduction
Endophytic fungi are microorganisms that live in the internal tissues of their host without causing any apparent disease symptoms [1]. They have been proven to be excellent sources of bioactive natural products [2]. The genus Phoma comprises filamentous fungi that inhabit the soil and plant materials, while about 140 species have been defined and recognized. Previous studies within this genus led to the isolation of a variety of novel structures and compounds with interesting bioactivities. Notable examples include epoxyphomalin A and B, prenylated polyketides with potent cytotoxicity [3], while TMC-264 is a novel antiallergic heptaketide [4], and fusidienol A inhibits ras farnesyl-protein transferase [5]. Our investigation of the endophytic fungus Phoma sp. EA-122, which was isolated from the leaves of Eupatorium adenophorum, an invasive plant in Yunnan province, P. R. China, has resulted in the isolation of a novel C25 sterol peroxide, phomasterol A (1), along with two known compounds 3β-hydroxy-(22E, 24R)-ergosta-5,8,22-trien-7-one (2) and (22E,24R)-ergosta-7,22-diene-3β,5α,6β-triol (3) (Figure 1). Herein, we report the isolation and structural elucidation of the new compound, as well as its biological activity.
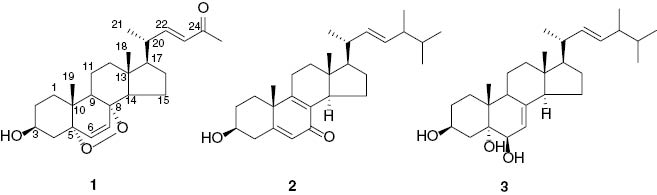
Structures of compounds 1–3.
2 Results and discussion
Phomasterol A (1), exhibited a pseudomolecular ion peak (m/z 423.2503 [M+Na]+, calcd 423.2511), corresponding to the molecular formula C25H36O4, with eight degrees of unsaturation. Its IR data indicated the presence of a hydroxy group (3441 cm–1) and an α, β-unsaturated keto group (1670, 1626 cm–1). The 1H NMR spectrum of compound 1 displayed three singlets for methyls at δH 0.89, 0.91, and 2.24 and a doublet for a methyl at δH 1.12 (3H, d, J=6.6 Hz), four olefinic proton signals at δH 6.02 (1H, d, J=15.9 Hz), 6.28 (1H, d, J=8.5 Hz), 6.54 (1H, d, J=8.5 Hz) and 6.77 (1H, dd, J=15.9, 9.1 Hz). The 13C and DEPT spectra revealed 25 carbon signals, including four methyls, seven methylenes, nine methines (with four olefinic ones) and five quarternary carbons (with one carbonyl and two oxyquarternary carbons). In addition, an α,β-unsaturated ketone moiety was assumed by the diagnostic NMR signals at δC 130.0, 155.8, and 201.9, and the chemical shift values of two oxygenated quaternary carbons at δC 83.4 and 80.6 indicated the presence of an epidioxy group. These observations, in combination with the molecular formula, indicated the presence of an epidioxy group and five rings in 1.
Structural elucidation of 1 was based on COSY and HMBC data (Figure 2). The hydroxy group attached at C-3 was designated by the HMBC correlations from H-3 (δH 3.78 ppm, m) to C-1 and C-5. The angular tcorrelations from Me-18 (δH 0.89 ppm, s) to C-12, C-13, C-14, and C-17, and Me-19 (δH 0.91 ppm, s) to C-1, C-5, C-9, and C-10. The above data suggested an epoxyergostan scaffold similar to that of (22E,24R)-5α,8α-epidioxy-ergosta-6,22-dien-3β-ol (4) [6]. The key differences between the two compounds were that 1 had three carbon atoms <4, and the signals at δH 0.81, 0.82 (each 3H, d, J=6.8 Hz), 1.47 (1H, dq, J=13.5, 6.8 Hz), and at δC 19.7, 20.0, and 33.1 for the isopropyl group were absent in 1. Instead, the corresponding resonances of an acetyl moiety (δH 2.24, δC 26.7, and 201.9) were observed, suggesting an isopropyl moiety was cleaved from the side chain of 4 and the resulting compound was oxygenated at C24 to yield 1, which was further supported by significant HMBC correlations from Me-25 (δH 2.24 ppm, s) to C-23 and C-24.
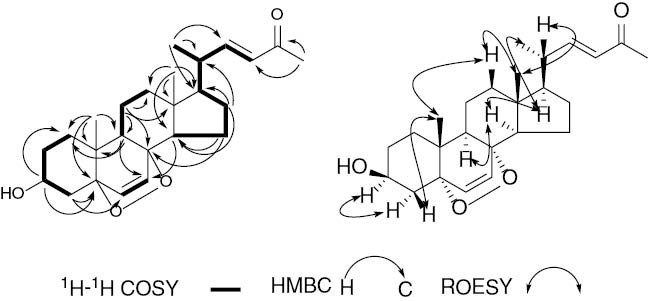
Selected 2D NMR correlations of 1.
The relative configuration of 1 was established by a combination of coupling constant and ROESY experiment. In the 1H NMR spectrum, The E-geometry of the Δ22-double bond was deduced from the large coupling constant observed for H-22 and H-23 (J=15.9 Hz), whereas, the Z-geometry of the Δ6-double bond was deduced from the coupling constants observed for H-6 and H-7 (J=8.5 Hz). In the ROESY spectrum (Figure 2), H-3 showed a correlation with H-4α, while H-4β and H-12β showed correlations with 19-Me, indicating that H-3 was α-oriented, while Me-19 was in the opposite orientation (β). The ROE correlations of Me-18/H-12β, H-20 suggested that Me-18 and H-20 were β-oriented. The ROE correlations of Me-21/H-12α, H-17 and H-17/H-14, H-9 indicated that Me-21, H-17, H-14 and H-9 were α-oriented. Consequently, the structure of 1 was established and named phomasterol A.
The known compounds were identified as 3β-hydroxy-(22E, 24R)-ergosta-5,8,22-trien-7-one (2) [7] and (22E,24R)-ergosta-7,22-diene-3β,5α,6β-triol (3) [8] by comparison with literature data.
There is a structural similarity of 1 to ursolic acid, a triterpene with significant inhibitory activity against protein-tyrosine phosphatases (PTPs). Members of the PTP family are important components of cell signal transduction chains that are involved in the regulation of cell function. Malfunctions of PTPs have been related to human diseases such as cancer, diabetes and immune disorders. As such, PTPs have become a key pharmaceutical target in the treatment of diseases [9]. Phomasterol A (1) was evaluated as an inhibitor of two PTPs: MEG2 and PTP1Bc and was found to exhibit moderate inhibitory activities with identical IC50 values of 25 μM, while the IC50 values of the positive control (ursolic acid) were 1.75 and 2.63 μM, respectively.
3 Experimental
3.1 General experimental procedures
Optical rotations were measured on a Horiba SEAP-300 polarimeter (Horiba, Kyoto, Japan), UV spectra on a Hitachi UV 210A spectrophotometer (Hitachi, Tokyo, Japan), and IR spectra on a Bio-Rad FTS-135 spectrometer (Bio-Rad, Hercules, CA, USA) with KBr pellets; 1D and 2D NMR spectra were recorded on a Bruker Avance III 600 MHz spectrometer (Bruker BioSpin, Rheinstetten, Germany), and ESI-MS and HR-EI-MS on an API QSTAR Pulsar I spectrometer (Applied Biosystems, Warrington, UK). Preparative MPLC was performed on a Büchi apparatus equipped with Büchi fraction collector C-660, Büchi pump module C-605 and manager C-615 (Büchi, Flawil, Switzerland). Column chromatography was performed on silica gel (200–300 mesh; Qingdao Marine Chemical., Qingdao, China) and Sephadex LH-20 (GE Healthcare, Uppsala, Sweden). Fractions were monitored by TLC on silica gel plates with CHCl3–MeOH (95:5, v/v), and spots were visualized by heating the plates after spraying with 10% H2SO4 in EtOH.
3.2 Fungal material
The fungus Phoma sp. EA-122 was isolated from fresh leaves of Eupatorium adenophorum collected in the Kunming Institute of Botany, Kunming, Yunnan province, P. R. China, in July 2012. The fungus was identified by observing the morphological characteristics and analysis of the internal transcribed spacer (ITS) regions. The result from the BLAST search indicated that the sequence was the same (99%) as that for the sequence of Phoma sp. TMS-2011 [10], and the sequence data were deposited in the Genbank (accession no. KM259932). The strain is preserved at the State Key Laboratory of Phytochemistry and Plant Resources in West China, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, P. R. China.
3.3 Fermentation, extraction, and isolation
Phoma sp. EA-122 was cultured on potato dextrose agar medium at 25 °C for 10 days. Agar plugs were inoculated in 500 mL Erlenmeyer flasks, each containing 150 mL of potato dextrose media. Flask cultures were incubated at 28 °C on a rotary shaker at 160 rpm for 2 days as seed culture. Forty 500 mL Erlenmeyer flasks, each containing 150 mL of potato dextrose broth, were individually inoculated with 10 mL of seed culture and were incubated at 25 °C on a rotary shaker at 160 rpm for 15 days.
The filtrate (6 L) of the fermented culture broth was extracted three times with EtOAc (6 L×3) at room temperature, and the organic solvent was evaporated to dryness under reduced pressure to afford a brown crude extract (3.0 g), which was then fractionated by silica gel column chromatography (CC), eluted with a gradient of CHCl3–MeOH (v/v 100:0, 97.5:2.5, 95:5, 9:1, 8:2, and 1:1) to give six fractions. Fraction 3 was eluted with CHCl3/MeOH (95:5), was then purified into five subfractions (3A–3E) by MPLC using MeOH/H2O as eluent. Fraction 3B was then separated by Sephadex LH-20 eluting with MeOH to give 1 (5.2 mg). Fraction 3C was then subjected to Sephadex LH-20 (MeOH) and silica gel CC (petroleum ether–EtoAc, 3:7) to yield 2 (3.2 mg) and 3 (9.0 mg).
Phomasterol A (1). White amorphous solid;
1H and 13C NMR data of 1 in methanol-d4 (δ in ppm).
| No. | δH (J in Hz) | δC, mult |
|---|---|---|
| 1 | 1.73 m | 35.9 t |
| 1.90 m | ||
| 2 | 1.79 m | 30.8 t |
| 1.53 m | ||
| 3 | 3.78 m | 66.9 d |
| 4 | 1.93 m; 1.98 m | 37.7 t |
| 5 | 83.4 s | |
| 6 | 6.28 d (8.5) | 136.9 d |
| 7 | 6.54 d (8.5) | 131.5 d |
| 8 | 80.6 s | |
| 9 | 1.46 m | 52.7 d |
| 10 | 38.1 s | |
| 11 | 1.30 m; 1.57 m | 24.3 t |
| 12 | 2.01 m; 1.32 m | 40.5 t |
| 13 | 46.1 s | |
| 14 | 1.58 m | 52.8 d |
| 15 | 1.56 m | 21.6 t |
| 16 | 1.75 m; 1.41 m | 29.2 t |
| 17 | 1.42 m | 56.3 d |
| 18 | 0.89 s | 13.2 q |
| 19 | 0.91 s | 18.6 q |
| 20 | 2.34 m | 40.9 d |
| 21 | 1.12 d (6.6) | 19.6 q |
| 22 | 6.77 dd (15.9, 9.1) | 155.8 d |
| 23 | 6.02 d (15.9) | 130.0 d |
| 24 | 201.9 s | |
| 25 | 2.24 s | 26.7 q |
3.4 Bioactivity assay
Human MEG2 and PIP1Bc with an N-terminal 6×His-tag were recombinantly expressed in Escherichia coli and purified by Ni-NTA affinity chromatography [11–13]. The enzymatic assay was carried out at room temperature in 96-well plates. The assay mixture (100 μL) containing 100 mM Hepes (pH 6.0), 5 mM DTT 0.015% Brij-35, and PTPase (20 ng PTP1B, 10 ng MEG2, per well) was incubated with various concentrations of the test compounds for 15 min, the reaction was initiated by addition of the substrate p-nitrophenyl phosphate (pNPP, P4744; Sigma-Aldrich, St. Louis, MO, USA) at a final concentration of 2 mM. The activity of PTPase-catalyzed hydrolysis of pNPP was determined by measuring p-nitrophenol absorbance at 405 nm. The IC50 value was determined by non-linear curve fitting of the percentage inhibition versus inhibitor concentration. Ursolic acid (U6753; Sigma) was used as positive control [14]. All assays were carried out in triplicate and the average results are presented.
Acknowledgments
This project was financially supported by National Natural Science Foundation of China (U1132607, 81102346), Natural Science Foundation of Yunnan Province (2011FB099), and West Light Program of CAS (2011312D11016).
References
1. Strobel G, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev 2003;67:491–502.10.1128/MMBR.67.4.491-502.2003Suche in Google Scholar
2. Zhang HW, Song YC, Tan RX. Biology and chemistry of endophytes. Nat Prod Rep 2006;23:753–71.10.1039/b609472bSuche in Google Scholar
3. Mohamed IE, Gross H, Pontius A, Kehraus S, Krick A, Kelter G, et al. Epoxyphomalin A and B, prenylated polyketides with potent cytotoxicity from the marine-derived fungus Phoma sp. Org Lett 2009;11:5014–7.10.1021/ol901996gSuche in Google Scholar
4. Sakurai M, Nishio M, Yamamoto K, Okuda T, Kawano K, Ohnuki T. TMC-264, a novel antiallergic heptaketide produced by the fungus Phoma sp TC 1674. Org Lett 2003;5:1083–5.10.1021/ol034125vSuche in Google Scholar
5. Singh SB, Ball RG, Zink DL, Monaghan RL, Polishook JD, Sanchez M, et al. Fusidienol A: a novel ras farnesyl-protein transferase inhibitor from Phoma sp. J Org Chem 1997;62:7485–8.10.1021/jo9708304Suche in Google Scholar
6. Kobori M, Yoshida M, Ohnishi-Kameyama M, Takei T, Shinmoto H. 5α, 8α-Epidioxy-22E-ergosta-6, 9 (11), 22-trien-3β-ol from an edible mushroom suppresses growth of HL60 leukemia and HT29 colon adenocarcinoma cells. Biol Pharm Bull 2006;29:755–9.10.1248/bpb.29.755Suche in Google Scholar
7. Kawagishi H, Katsumi R, Sazawa T, Mizuno T, Hagiwara T, Nakamura T. Cytotoxic steroids from the mushroom Agaricus blazei. Phytochemistry 1988;27:2777–9.10.1016/0031-9422(88)80662-9Suche in Google Scholar
8. Hu LL, Ma QY, Huang SZ, Dai HF, Guo ZK, Zhao YX. Study on the chemical constituents from Ganoderma tropicum. Zhongguo Yaowu Huaxue Zazhi 2013;23:115–9.Suche in Google Scholar
9. Barr AJ. Protein tyrosine phosphatases as drug targets: strategies and challenges of inhibitor development. Future Med Chem 2010;2:1563–76.10.4155/fmc.10.241Suche in Google Scholar PubMed
10. Shrestha P, Szaro TM, Bruns TD, Taylor JW. Systematic search for cultivatable fungi that best deconstruct cell walls of Miscanthus and sugarcane in the field. Appl Environ Microbiol 2011;77:5490–504.10.1128/AEM.02996-10Suche in Google Scholar PubMed PubMed Central
11. Yang X, Li J, Zhou Y, Shen Q, Chen J, Li J. Discovery of novel inhibitor of human leukocyte common antigen-related phosphatase. Biochim Biophys Acta 2005;1726:34–41.10.1016/j.bbagen.2005.07.001Suche in Google Scholar PubMed
12. Imhof D, Wavreille AS, May A, Zacharias M, Tridandapani S, Pei D. Sequence specificity of SHP-1 and SHP-2 Src homology 2 domains. Critical roles of residues beyond the pY+3 position. J Biol Chem 2006;281:20271–82.10.1074/jbc.M601047200Suche in Google Scholar PubMed
13. Wei M, Wynn R, Hollis G, Liao B, Margulis A, Reid BG, et al. High-throughput determination of mode of inhibition in lead identification and optimization. J Biomol Screen 2007;12:220–8.10.1177/1087057106296679Suche in Google Scholar PubMed
14. Zhang W, Hong D, Zhou Y, Zhang Y, Shen Q, Li JY, et al. Ursolic acid and its derivative inhibit protein tyrosine phosphatase 1B, enhancing insulin receptor phosphorylation and stimulating glucose uptake. Biochim Biophys Acta 2006;1760:1505–12.10.1016/j.bbagen.2006.05.009Suche in Google Scholar PubMed
©2015 by De Gruyter
Artikel in diesem Heft
- Frontmatter
- Essential oil composition, phenolic content, antioxidant, and antimicrobial activity of cultivated Satureja rechingeri Jamzad at different phenological stages
- Characterization of a mineral coating of the plant Dyerophytum indicum
- Semisynthesis and insecticidal activity of arylmethylamine derivatives of the neolignan honokiol against Mythimna separata Walker
- Dynamics of alkylresorcinols during rye caryopsis germination and early seedling growth
- New nitrogenous compounds from a Red Sea sponge from the Gulf of Aqaba
- Synthesis and antiproliferative evaluation of novel 5-(4-methylpiperazin-1-yl)-2-phenyl- 1H-benzimidazole derivatives
- Differential cytotoxic activity of the petroleum ether extract and its furanosesquiterpenoid constituents from Commiphora molmol resin
- A novel C25 sterol peroxide from the endophytic fungus Phoma sp. EA-122
- Bioactive compounds isolated from submerged fermentations of the Chilean fungus Stereum rameale
- Combined Autodock and comparative molecular field analysis study on predicting 5-lipoxygenase inhibitory activity of flavonoids isolated from Spatholobus suberectus Dunn
Artikel in diesem Heft
- Frontmatter
- Essential oil composition, phenolic content, antioxidant, and antimicrobial activity of cultivated Satureja rechingeri Jamzad at different phenological stages
- Characterization of a mineral coating of the plant Dyerophytum indicum
- Semisynthesis and insecticidal activity of arylmethylamine derivatives of the neolignan honokiol against Mythimna separata Walker
- Dynamics of alkylresorcinols during rye caryopsis germination and early seedling growth
- New nitrogenous compounds from a Red Sea sponge from the Gulf of Aqaba
- Synthesis and antiproliferative evaluation of novel 5-(4-methylpiperazin-1-yl)-2-phenyl- 1H-benzimidazole derivatives
- Differential cytotoxic activity of the petroleum ether extract and its furanosesquiterpenoid constituents from Commiphora molmol resin
- A novel C25 sterol peroxide from the endophytic fungus Phoma sp. EA-122
- Bioactive compounds isolated from submerged fermentations of the Chilean fungus Stereum rameale
- Combined Autodock and comparative molecular field analysis study on predicting 5-lipoxygenase inhibitory activity of flavonoids isolated from Spatholobus suberectus Dunn

