Abstract
Background
RNAs to be used in transcriptome analysis must be of high quality and pure in order to ensure maximum representation of the expressed genes. RNA isolation is difficult in hazelnut tissues containing large amounts of secondary metabolite, phenolic compounds and the cell wall structure. Commonly used protocols for RNA isolation are those that require a lot of labor and time and also do not allow sufficient RNA isolation when applied to tissues rich in phenolic compounds. This study was aimed to develop an efficient method for isolation of total RNAs from bud of hazelnut to be used in RNA sequencing.
Materials and methods
An optimized new method was successfully applied on three different hazelnuts genotypes (Çakıldak, Palaz, Tombul) and about 25 times higher amount of total RNAs per mg fresh tissues were obtained compared to classical CTAB method. Different methods have been tried for the isolation of RNA from hazelnut tissues and the determination of the quality of the obtained RNAs.
Results
The quality and quantity of isolalated total RNAs were determined by spectrophotometer, electrophoresis and PCR. This success has been caught without any compromise of purity since A260/A280 ratios ranged from 1.90 to 2.04 and A260/A230 ratios were >2.0 in all purified RNAs.
Conclusion
The total RNAs isolated with new protocol was found to be suitable for RNA sequencing and other molecular applications.
Öz
Amaç
Transkriptom analizinde ifade edilen genlerin uygun şekilde temsil edilmesini sağlamak için yüksek kaliteli RNA’nın izolasyonu ve saflaştırılması gerekmektedir. Yüksek miktar sekonder metabolit, fenolik bileşik içeriği ve hücre duvarı yapısından dolayı fındık dokularında RNA izolasyonu zordur. RNA izolasyonu için kullanılan genel protokoller, fazla zaman gerektirmekte ve genellikle fenolik içeriği yüksek dokularda düşük verim sağlamaktadırlar. Bu çalışma, RNA dizilemesinde kullanılacak toplam RNA’ların, fındık tomurcuğundan izole edilmesi için etkili bir yöntem geliştirmeyi amaçlamıştır.
Gereç ve yöntem
Üç farklı fındık genotipinde (Çakıldak, Palaz, Tombul) optimize edilen yeni bir yöntem başarıyla uygulandı ve klasik CTAB yöntemine kıyasla mg taze doku başına yaklaşık 25 kat daha fazla miktarda toplam RNA elde edildi. Fındık dokusundan RNA’yı elde etmek ve bütünlüğünü değerlendirmek için çeşitli yöntemler uygulandı.
Bulgular
RNA kalitesi, spektrofotometre, elektroforez, cDNA sentezi ve PCR ile değerlendirildi. Tüm saflaştırılmış RNA’larda A260 / A280 oranları 1.90 ile 2.04 arasında değiştiği ve A260 / A230 oranları >2.0 olduğundan dolayı, saflıktan ödün verilmeksizin başarılı bir şekilde RNA’lar elde edilmiştir.
Sonuç
Yeni protokolle izole edilen toplam RNA’ların RNA dizilemesi ve diğer moleküler çalışmalar için uygun olduğu bulunmuştur.
Introduction
The genus Corylus consists of a large deciduous shrub and tree species, all of which contain edible nuts, which form an integral part of many temperate forests throughout the Northern Hemisphere. Corylus avellana L. (hazelnut), the most widely studied species of Corylus, ranks sixth in the world after peanuts, cashews, walnuts, chestnuts and almonds. Hazelnut has a long history of use and production, possibly beginning before the Roman period [1], [2]. They have long been known to have both immense dietary and health benefits serving as a good source of dietary fiber, vitamin E, magnesium and B vitamins and antioxidants. Despite its great benefit and long history of utilization, hazelnut cultivation has not shown sufficient improvement compared to other domestic plants.
High-quality and purified total RNA isolation is a crucial prerequisite for downstream molecular biology analyses, such as cDNA library construction, RNA sequencing, Northern hybridization, gene expression and function analyses [3–8]. However, RNA isolation is difficult in plants that contain high levels of endogenous ribonuclease (RNase), polyphenols, polysaccharides, lignins, carbohydrates, etheric oils and the cell wall structure [4, 9–11].
Hazelnut tissue is rich in polysaccharides and polyphenolics that can hinder RNA isolation [12], [13]. Phenolic compounds can be irreversibly oxidized and attached to RNA. When using phenol–chloroform RNA extraction procedure, the presence of high amounts of phenols in the native tissue would lead to a loss of RNA or to formation of insoluble complexes through interactions between organic substances and nucleic acids [14–16]. The presence of polysaccharides in the isolated RNA sample would interfere with solubilization of RNA pellets as because polysaccharides precipitate with RNA to form a colloidal substance, thereby affecting RNA spectroscopic quantification and downstream enzymatic reactions [8], [10], [17]. These compounds form complexes with RNA making cDNA synthesis and RNA sequencing impossible and blocked [18–20].
Different methods are used for RNA isolation [21] specifically modified for each plant species [10], [20], [22]. Due to high phenolic compounds especially in hazelnuts, RNA isolation is very hard and therefore requires either modification of RNA isolation protocols or reproduce new ones. Frequently used commercial RNA isolation kits do not always produce reliable results [22], [23]. Various RNA isolation methods of which (i) phenol-based method such as the use of TRIzol, CTAB and SDS, (ii) column-based method using commercial kit (RNAsy mini kit – Qiagen) (iii) phenol-based & column-based methods [21], [24] have been used to eliminate co-precipitation of phenolic compounds and polysaccharides.
This study describes for the first time a convenient RNA isolation method providing high quality and purified RNA especially for RNA sequencing in hazelnuts buds. In addition, the protocol described herein is applicable in RNA isolation from the other plant species.
Materials and methods
Plant material
Leaf buds from the main stem of 22-year-old C. avellana L. trees growing on a field site in Giresun, Turkey, were collected on February 2015. Leaf buds of three different hazelnut genotypes (Çakıldak, Palaz and Tombul) differing in their leaf bud burst time traits were used for transcriptome sequencing. The hazelnut branch containing leaf buds were stored in a container including liquid nitrogen. The leaf buds were isolated from branch by using sterile scalpel blades and then subsequently stored at −80 °C until they were used for RNA isolation.
RNase removal treatments
Prior to performing RNA isolation, the work environment, mortal, pestle and electrophoresis unit were washed with detergent; rinsed with de-ionized water to remove chloroform and protein residuals and cleaned with RNase-Zap (Zymo Research).
TRIzol protocol
100 mg of hazelnut buds tissue was used to isolate total RNA according to TRIzol manufacturer’s protocol (Thermo Fisher Scientific, Waltham, USA) and re-suspended in a total volume of 50 µl RNase-free water.
CTAB II method
Here, the modified protocol of [25] was used to isolate RNA from hazelnut bud samples. Frozen hazelnut buds (120 mg) were finely crushed with the aid of liquid nitrogen and quickly transferred to 50 mL centrifuge tubes containing 10 volume of extraction buffer (3% CTAB; 100 mM Tris–HCl, pH 8.0; 1.4 M NaCl; 200 mM EDTA, pH 8.0; 2% PVP) preheated to 65 °C. Then, 2% (v/v) β-mercaptoethanol and 80 μg/mL proteinase K were added. The samples were incubated at 65 °C for 30 min and vortexed for at 5 min intervals. An equal volume of chloroform–isoamylalcohol (24:1, v/v) was added to the aqueous phases of the samples, mixed well and centrifuged at 8,000 ×g for 8 min at 15 °C. The centrifugation process was repeated with the same conditions. The resulting supernatants were transferred into new clean tubes, equal volume of phenol–chloroform (24:1, v/v) were added, vortexed and centrifuged at 15 °C at a speed of 11,000 ×g for 20 min. Again, an equal volume of chloroform–isoamylalcohol (24:1, v/v) was added to the aqueous phases, mixed well and centrifuged at 10,000 ×g for 10 min at 15 °C. The supernatants were then collected, one-fourth volume of 10 M LiCl was added to the mixture, stirred and stored at −20 °C for overnight. The following day, the RNAs were recovered by centrifuging at 12,000 ×g for 20 min at 4 °C. The resulting pellets were later dissolved in 4 mL of 0.1 M Tris HCl–EDTA buffer, the aqueous phases were separated with equal amounts of chloroform–isoamylalcohol (24:1, v/v), mixed well and centrifuged at 10,000 ×g for 10 min at 4 °C. The supernatants were then collected and mixed well in a solution containing one-tenth volume of 3 M sodium acetate (pH: 5.2) and 2.5 volume of pre-cooled anhydrous alcohol before incubating for 3 h at −80 °C. Finally, total RNAs were collected by centrifuging at 12,000 ×g for 30 min at 4 °C, washed twice with 500 µL of 75% (v/v) alcohol, air-dried, dissolved in 100 μL RNase-free water and stored at −80 °C.
CTAB buffer + spermidine method
The total RNA isolation method was modified according to [26]. 150 mg of hazelnut buds was crushed to a fine powder in liquid nitrogen and quickly transferred to 50 mL centrifuge tubes containing 900 µL of extraction buffer (2% CTAB; 100 mM Tris–HCl, pH 8.0; 2 M NaCl; 25 mM EDTA, pH 8.0; 2% PVP) preheated to 65 °C. 2% (v/v) β-mercaptoethanol, 80 μg/mL proteinase K and 0.5 g/L spermidine was added. The samples were then incubated at 65 °C for 30 min and vortexed at 5 min intervals. 950 µL of chloroform–isoamylalcohol (24:1, v/v) was added to the aqueous phases of the samples, mixed well and centrifuged at 11,000 ×g for 10 min at 4 °C. The resulting supernatants were transferred into new clean tubes, following the addition of 950 µL of phenol–chloroform (24:1), the samples were vortexed and centrifuged at 4 °C at a speed of 11,000 ×g for 10 min. Another 950 µL of chloroform–isoamylalcohol (24:1, v/v) solution was again added to the aqueous phase, mixed well and centrifuged at 11,000 ×g for 10 min at 4 °C. The supernatants were then collected, following one-third volume of LiCl was added. The mixture is then stirred and stored at −20 °C for overnight. The next day, the RNAs were recovered by centrifuging at 12,000 ×g for 20 min at 4 °C. The pellets were dissolved in 4 mL of 0.1 M Tris·HCl–EDTA buffer, and the aqueous phases were separated with equal amounts of 1 M of chloroform–isoamylalcohol (24:1, v/v) then stirred well and centrifuged at 10,000 ×g for 10 min at 4 °C. After centrifugation, the supernatants were collected to a solution containing one-tenth volume of 3 M sodium acetate (pH: 5.2) and 2.5 volume of pre-cooled anhydrous alcohol, mixed well and incubated for 3 h at −80 °C. Total RNAs were finally collected by centrifuging at 12,000 ×g for 30 min at 4 °C, washed twice with 500 µL of 75% (v/v) alcohol, air-dried, dissolved in 100 μL RNase-free water and stored at −80 °C till further use.
QIAGEN RNeasy plant mini kit
The total RNA extraction was performed according to the manufacturer’s protocol instructions (QIAGEN, Hilden Germany) using 100 mg hazelnut tissue as starting material. Isolated total RNA was eluted with 50 µl RNase-free water.
GeneAll Ribonuclear Plus kit
Total RNA was extracted from the buds of hazelnut samples following the manufacturer’s protocol (Songpa-gu Seoul, Korea) and resuspended in a total volume of 50 µL RNase-free water. 100 mg hazelnut tissue was used in the protocol.
CTAB + QIAGEN RNeasy plant mini kit
The combination of CTAB extraction followed by Qiagen RNA isolation protocol was used in this step. Frozen hazelnut buds (120 mg) were crushed to a fine powder in liquid nitrogen and quickly transferred to 50 mL centrifuge tubes containing 900 µL of extraction buffer (2% CTAB; 100 mM Tris–HCl, pH 8.0; 2 M NaCl; 25 mM EDTA, pH 8.0; 2% PVP) preheated to 65 °C. 2% (v/v) β-mercaptoethanol, 80 μg/mL proteinase K and 0.5 g/L spermidine were added to the samples and incubated at 65 °C for 30 min and vortexed at 5 min intervals. 950 µL of chloroform–isoamylalcohol (24:1, v/v) was added to the aqueous phase of the samples, mixed well before centrifuging at 11,000 ×g for 10 min at 4 °C. The resulting supernatants were transferred into QIAshredder spin columns provided in the QIAGEN kit in accordance with the manufacture’s protocol.
CTAB + GeneAll Ribonuclear Plus kit
A mixture of 4 mL CTAB buffer (2% CTAB; 100 mM Tris–HCl, pH 8.0; 2 M NaCl; 25 mM EDTA, pH: 8.0; 2% PVP) preheated at 65 °C, 2 µL spermidine and 80 µL β-mercaptoethanol were used in this protocol. 300 mg of grounded hazelnut buds was added to 5× CTAB buffer and vortexed for 2 min. The samples were incubated at 65 °C for 20 min. After incubation, the samples were centrifuged at 8,000 ×g for 15 min at 4 °C. The supernatants were put into new centrifuge tubes and equal amounts of chloroform–isoamylalcohol (24:1, v/v) were added, briefly vortexed and centrifuged at 12,000 ×g for 20 min at 4 °C. The supernatants were again transferred to new centrifuge tubes and the same amount of chloroform-isoamylalcohol added, then vortexed and centrifuged at 12,000 ×g for another 20 min at 4 °C. The supernatants were immediately again transferred to new centrifuge tubes, where equivalent amounts of LiCl was added and incubated overnight at −20 °C. The following day, the samples were centrifuged at 14,000 ×g and 4 °C. Without disturbing the pellets, the supernatants were discarded, 800 µL of 70 % cold ethanol added to the pellet and centrifuged at 14,000 ×g for 5 min at 4 °C. The supernatants were discarded and the previous step repeated again. After this step, the supernatants were carefully discarded and the pellets left to dry at room temperature for 5 min. The pellets of RNA were dissolved 100 µL of RNase-free water and then subjected to DNase treatment using the manufacturer’s protocol of the GeneAll Ribonuclear Plus kit.
CTAB + 5′PRIME Phase Lock Gel Heavy (Phenol) column + QIAGEN RNeasy plant mini kit
The buffer used in this protocol was comprised of a solution of 5 mL CTAB and 2.5 µL spermidine (preheated at 65 °C) mixed with 100 µL β-mercaptoethanol. 100 mg of sample powder was added to 950 µL CTAB buffer, vortexed for 2 min and incubated at 65 °C for 15 min. After incubation, 950 µL of cold chloroform isoamyl–alcohol (24:1) was added and the samples were vortexed for 2 min. The mixtures were immediately centrifuged at 12,000 ×g, 20 min and 4 °C. The supernatants were then transferred into the phase lock gel heavy columns (Phase Lock Gel Heavy columns) and equal amounts of cold chloroform isoamyl–alcohol (24:1) were added. The samples were then mixed by inverting the tubes several times and centrifuged at 12,000 ×g for 20 min at 4 °C. After centrifugation, 0.5 volumes of both phenol and cold chloroform isoamyl–alcohol (24:1) were added to the supernatants. Following this step, samples were gently mixed by inversion and centrifuged at 12,000 ×g for 5 min at 4 °C. The same amount of RLT buffer was added to the resulting supernatant and the kit manual was followed. Eluted RNA was treated with DNAse to get rid of any residual DNA.
The concentration of isolated total RNAs was determined by measuring absorbance values from wavelengths at 230, 260 and 280 nm using the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, USA). Purity of the total RNA was estimated by calculating the A260/A230 and A260/A280 ratios to evaluate the levels of protein and polysaccharide/phenolic compound contamination, respectively. The integrity of total RNA was verified by resolving 25 µL of RNA sample on 1% (w/v) agarose gel with ethidium bromide (0.5 mg/mL). Further analysis was performed in an Agilent 2100 Bioanalyzer to calculate RNA integrity number (RIN) values. RINs starting from 1 to 10 were used to assess the distribution and size of the purified RNA molecules.
cDNA synthesis and PCR
First-strand of cDNA was synthesized with total RNAs (2.5 µL) by using cDNA Synthesis kit (Thermo Scientific RevertAid First Strand). The cDNAs were then subjected to PCR to amplify the C. avellana geranylgeranyl diphosphate synthase (GGPPS) (GenBank accession number: EF553534.1) and C. avellana 2S albumin (GenBank accession number: FJ358504.1) genes. The following thermocycling parameters were used during for PCR analyses. These were an initial denaturation at 94 °C for 30 s, followed by 34 cycles of 94 °C denaturation for 15 s, 56 °C annealing for 35 s and 72 °C extension for 30 s, a final extension step at 72 °C for 10 min and an infinite hold at 10 °C. The resulting PCR products were then examined by electrophoresis in 1% agarose gel. The sequences of forward and reverse primers were 5′ TCCATCTCGTCTGTCTCCGT 3′ and 5′ CTCCTAGTACCACAGCCCCT 5′for GGPPS and 5′GCACCACCATAACAACCGTG 3′ and 5′ GGCAAGTCCCTAGCAGTCTC 3′ for 2S albumin genes of C. avellana.
Results
The eight different RNA extraction protocols used in this study were mainly categorized into three subheadings (i) phenol-based methods (the use of Trizol, CTAB II, CTAB & spermidine), (ii) column-based methods (the use of RNesay plant mini kit and GeneAll Ribonuclear Plus kit) and combined phenol & column-based methods (the combinations of CTAB&RNeasy plant mini kit, GeneAll Ribonuclear Plus kit &CTAB and CTAB & Phase Lock Gel Heavy column & RNAeasy plant mini kit ) (Figure 1) to isolate total RNAs from hazelnuts buds. The respective yield and purity of the extracted total RNAs were shown in Table 1.
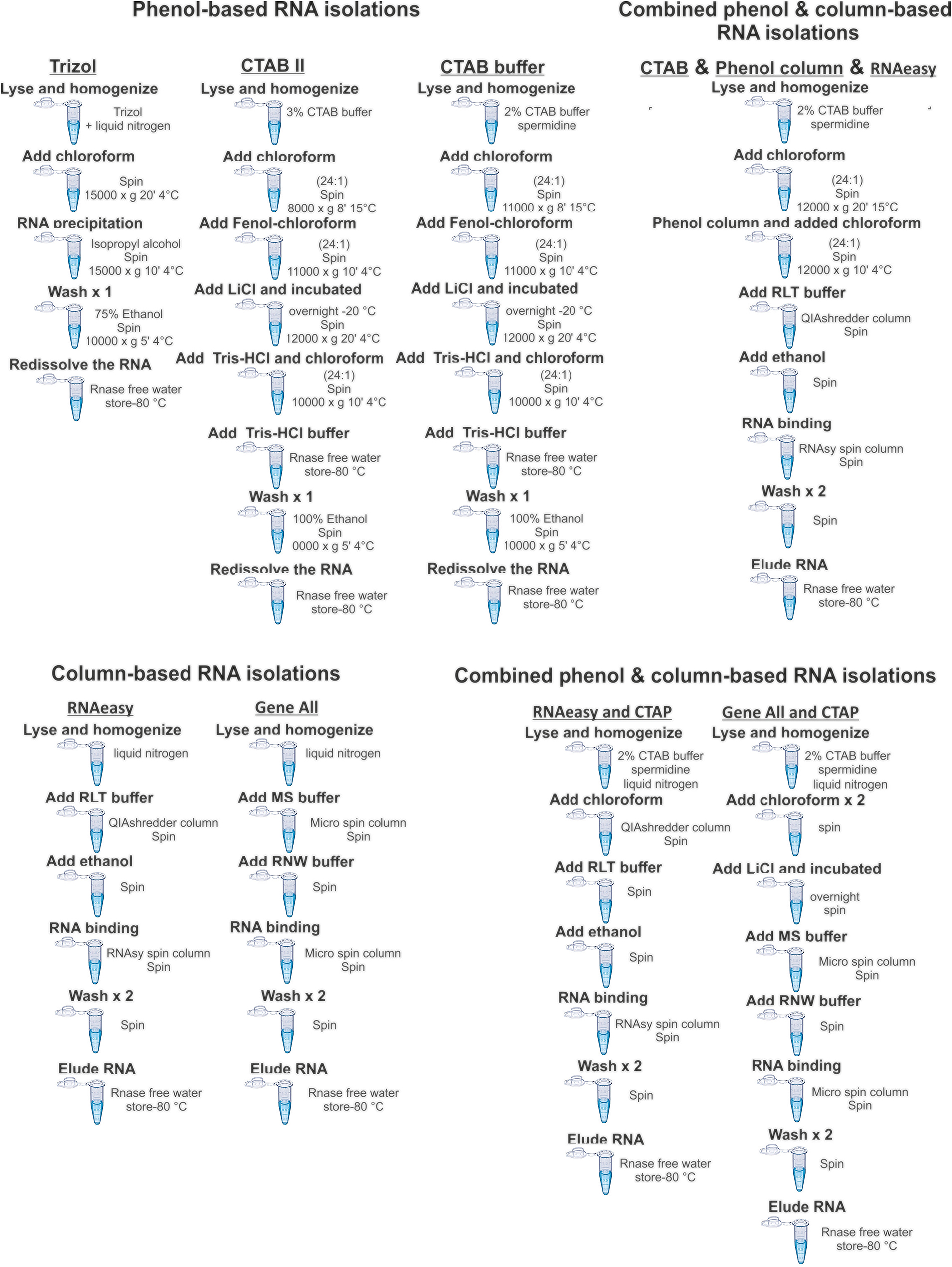
Flow chart showing the tested total RNA isolation methods. The RNAs from hazelnuts buds were isolated by (i) phenol-based method such as Trizol, CTAP and SDS, (ii) column-based method such as commercial kit (RNeasy plant mini kit) (iii) phenol-based & column-based methods.
Yield and absorbance ratio of total RNAs extracted from hazelnut buds with different methods.
| RNA isolation method | NanoDrop measurements | |||
|---|---|---|---|---|
| Yield RNA (ng/µL) | A260/A280 | A260/A280 | ||
| 1 | TRIzol | 2.8 | 1.46 | 0.28 |
| 2 | CTAB II | 22.0 | 1.47 | 0.41 |
| 3 | CTAB Buffer + Spermidine | 209.3 | 1.33 | 0.32 |
| 4 | QIAGEN RNeasy plant kit | 3.1 | 2.53 | 0.36 |
| 5 | GeneAll Riboclear Plus kit | 100.8 | 1.83 | 1.06 |
| 6 | CTAB + QIAGEN RNeasy plant kit | 46.7 | 2.08 | 1.17 |
| 7 | CTAB + GeneAll Ribonuclear Plus kit | 200.0 | 1.85 | 1.06 |
| 8 | CTAB + Phase Lock Gel Heavy column + QIAGEN RNeasy plant mini kit | 492.0 | 2.11 | 2.15 |
The study yielded major differences in terms of RNA quality and purity among the protocols tested. The total RNAs extracted with commercial kits such as Trizol, QIAGEN RNeasy Plant Mini kit and GeneAll Riboclear Plus plant kit were of low quality and quantity (Table 1) and produced indistinct bands. The tested RNA isolation methods using CTAB and CTAB II buffers resulted in total RNAs with low quantity and purity, as well. On the other hand, the combined use of modified CTAB buffer and spermidine protocol proved successful in yielding considerably high quantity RNA but with contaminants such as phenol and protein (Figure 2).
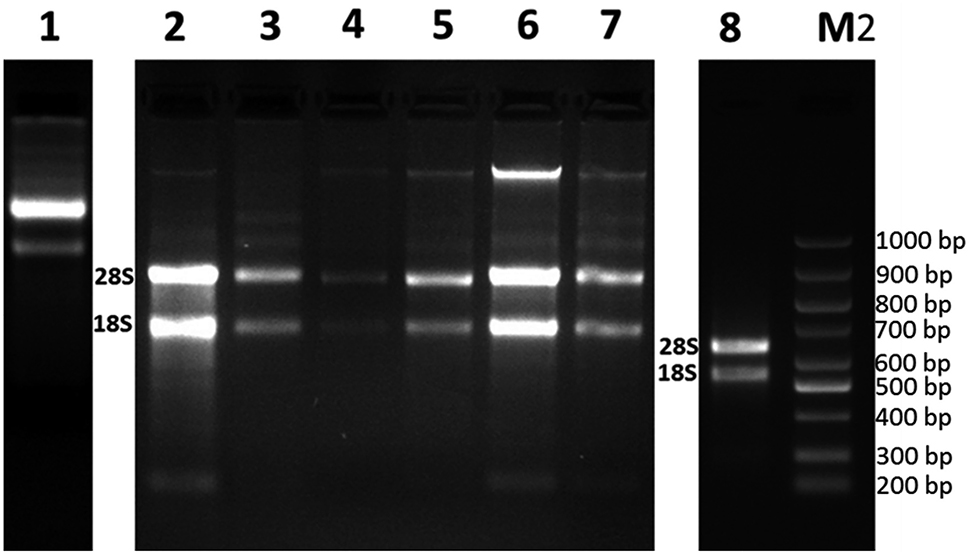
Total RNAs isolated by using different methods. M2: DNA Marker (Bio Basic, 100–1,000 bp). Total RNAs isolated with the use of GeneAll Ribonuclear Plus kit (1), The combined GeneAll Ribonuclear Plus kit and CTAB application (2), Trizol treatment (3), Qiagen RNeasy plant mini kit (4), with the combined use of CTAB treatment and Qiagen RNeasy plant mini kit (5), with the combination of CTAB and spermidine treatments (6), CTAB II protocol (7), with the combined use of CTAB treatment, Phase Lock Gel Heavy column and Qiagen RNeasy plant mini kit (8).
The high quality and pure total RNA was only achieved with the use of combined CTAB & Phase Lock Gel Heavy column & RNeasy plant mini kit methods. The A260/A280 ratio of 2.11 and A260/A230 ratio of 2.15 indicated slight contamination of proteins, polysaccharides and polyphenols (Table 1). The presence of 28S and 18S ribosomal RNA bands obtained through agarose gel electrophoresis confirmed the integrity of isolated total RNAs (Figure 2). In addition, the total RNAs isolated with the combined use of CTAB reagent, Phase Lock Gel Heavy column and Qiagen RNeasy plant mini kit were away from DNA contamination as shown in Figure 2.
The newly developed RNA isolation method in which CTAB, Phase Lock Gel Heavy column and RNeasy plant mini kit protocols were combined, provided high quality and pure total RNA. This procedure was used in RNA isolation from the hazelnut buds belonged to three genotypes (Palaz, Çakıldak and Tombul) and isolated total RNAs were analyzed using Agilent Bioanalyser, NanoDrop and run on agarose gel to check the quality (Figure 3). The yields of Palaz, Çakıldak and Tombul were 3,324, 3,899, 405 ng/µL and the A260/A280 ratios were 2.11, 2.15, 2.18 and A260/A230 ratios were 1.89, 2.08, 2.01, respectively, indicating quality and purity of RNA. The results obtained from both Agilent Bioanalyser and agarose gel electrophoresis confirmed that the respective RNAs were intact and highly pure.
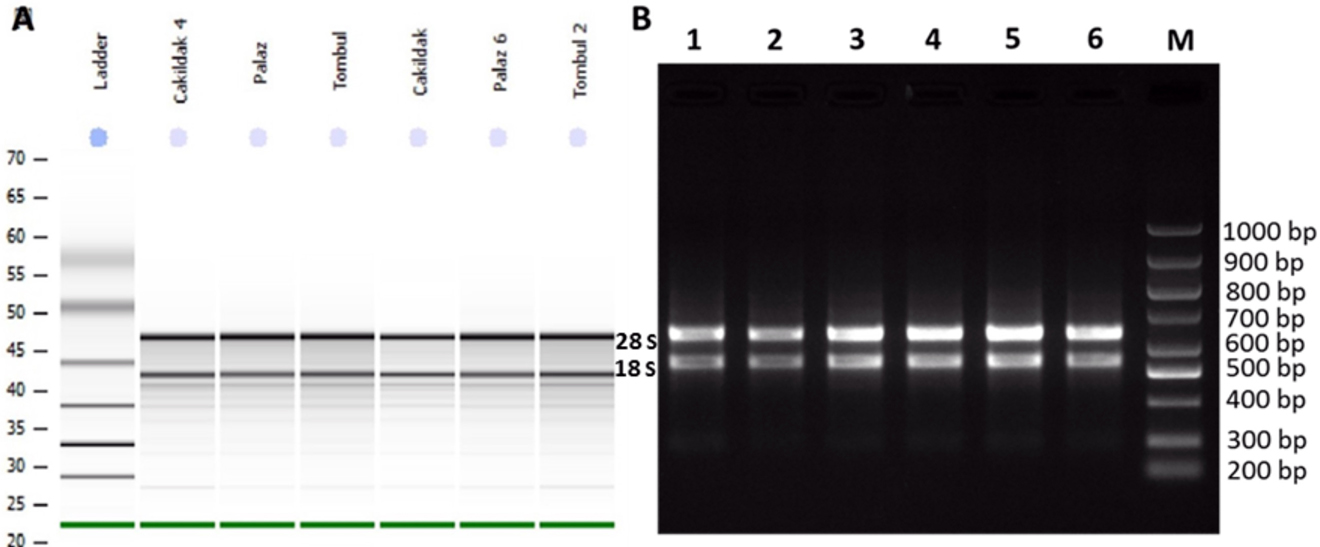
The analysis of RNA quality isolated with the combined CTAB & Phase Lock Gel Heavy column & RNeasy plant mini kit procedure from hazelnut buds of three different genotypes (A) Total RNA samples analyzed in the Agilent 2100 Bioanalyzer with the 6000 Nano LabChip, (B) Agarose gel electrophoresis showing total RNA extracted from different hazelnut varieties. L/M: DNA Ladder/marker (Bio Basic, 100–1,000 bp), the results of 1,4: Çakıldak genotype, 2,5: Palaz genotype, 3,6: Tombul genotype.
Next, the availability of the isolated RNAs for cDNA synthesis was tested by using RNAs as template to amplify the C. avellana GGPPS and 2S albumin genes in each genotype. As shown in Figure 4, the expected amplicons (C. avellana 2S albümin and GGPPS genes are 318 and 657 bp in size, respectively) for both genes were obtained for every genotype showing the success of the isolated RNAs to be used in further molecular processes. Followingly, the RIN values for each RNA sample were also determined to evaluate their potential to be used in RNA sequencing (Figure 5). The electropherograms generated by Bioanalyzer profiling of RNA isolated using the combined CTAB & Phase Lock Gel Heavy column & RNeasy plant mini kit methods exerted the RIN values ranging from 7.30 to 7.90 in Palaz, Çakıldak ve Tombul genotypes. The resultant RIN values are acceptable for performing RNA sequencing. Thus, the RNAs isolated with this new method have passed all the quality parameters and used in RNA sequencing reactions.
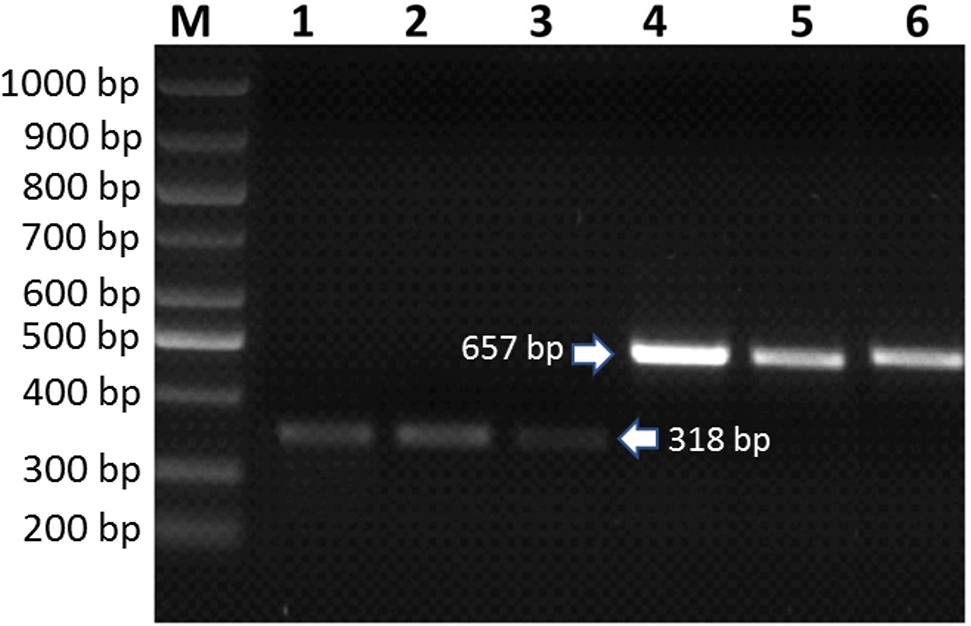
Amplification of Corylus avellana GGPPS and 2S albumin genes by RT-PCR in each genotype using the isolated RNAs as template. M: DNA Ladder (Bio Basic, 100–1,000 bp), Corylus avellana 2S albumin gene amplicon in Çakıldak (1), Tombul (2) and Palaz (3), Corylus avellana GGPPS gene amplicon in Çakıldak (4), Tombul (5) and Palaz (6).
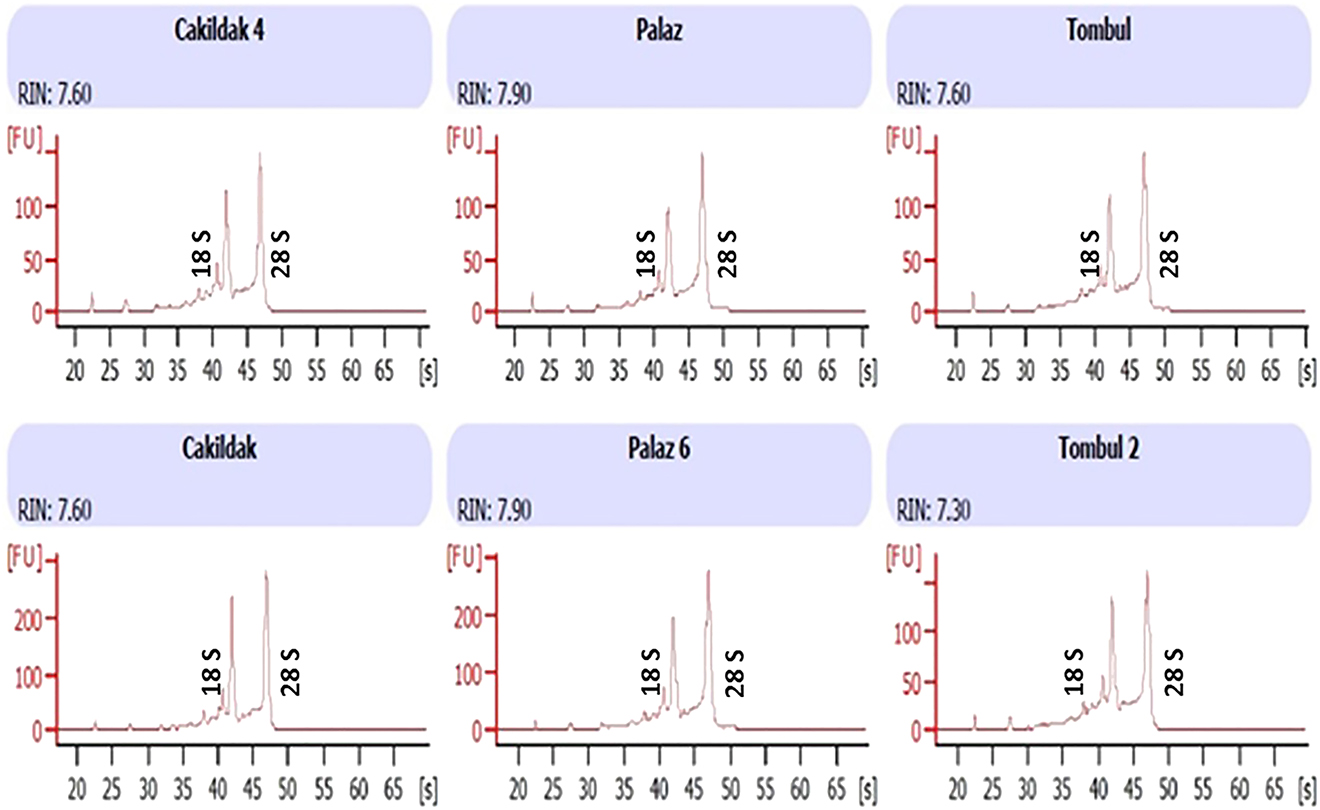
Bioanalyzer results of total RNA extracted using the method described in this study (Combined CTAB & Phase Lock Gel Heavy column & RNeasy plant mini kit method). Extractions were made from different hazelnut genotypes as indicated and run on an Agilent 2100 Bioanalyzer using the Total RNA Pico assay. FU: Fluorescence. S: Time (seconds).
Discussion
Although there are many RNA isolation methods, there is still need for developing new optimized protocols for different plant species and tissues [4]. Phenolic compounds, secondary metabolites and cell structures in hazelnuts are major obstacles encountered during RNA isolation studies and result in problems in making RNA-based studies [27–29]. Therefore, the presence of polysaccharides, proteins and secondary metabolites in the isolated RNA samples prevents downstream processes such as PCR and RNA sequencing. Such kind of problems might be overcomed by making some adjustments in the RNA isolation methods.
Commercially available kits and reagents used for total RNAs isolation offer simple, rapid and non-toxic and provide high quality RNA yield from suitable plant tissues [11]. However, it is very hard to get good yields of RNA and even to extract the RNAs from recalcitrant plant tissues, with the use of such kits or reagents [22]. For instance, different commercial kits were used to isolate RNA from Jatropha curcas and Platycladuc orientalis with high phenolic and polysaccharide content; however, pure RNAs with the high quality were not obtained [22], [23]. In our study, several commercially available kits and reagents, such as Trizol, QIAGEN RNeasy plant mini kit and GeneAll Ribospin Plant Kit used, have also failed to yield good-quality RNA isolation from buds of hazelnuts.
CTAB method should generally be modified, for RNA isolation from plants containing high levels of phenol, polysaccharide and secondary metabolites and having recalcitrant tissues. Using a different modified CTAB method, we isolated high amounts of RNA; however, due to including too much contamination, it was not suitable for a transcriptome study. A modified CTAB method developed for Hippophae sp., a medicine plant sea buckthorn, enabled a suitable RNA isolation for RT-PCR studies [13]. The use of commercial kits together with CTAB method also performed in this study provides isolation of RNAs in higher quality and purity from plants containing high levels of phenol, polysaccharide and secondary metabolites and having recalcitrant tissues. Based on this, before the stage of Phase Lock Gel Heavy column & RNeasy plant mini kit, we used suitable amounts of PVP in order to block the formation of phenolic compounds during tissue pounding and spermidine and β-mercaptoethanol were applied to avoid RNAase activity [20–22]. Plants with high phenol content like hazelnuts require pure RNA for RNA sequencing. Plant RNA samples having RIN values of greater than 6.0, can be considered as intact and can use for highly sensitive downstream applications [30]. Among the tested methods, only the CTAB, Phase Lock Gel Heavy column, Qiagen RNAeasy method combination provided the desired RNA quality and quantity required for RNA sequencing in our study.
Conclusions
The combined use of optimized CTAB, Phase Lock Gel Heavy column, Qiagen RNAeasy methods in the RNA isolation from three different hazelnuts buds (Palaz, Çakıldak, Tombul) gave successful results to provide high purity and quality RNAs for RNA sequencing. The procedure described herein is also advantageous in that it is fast and repeatable. We propose that this efficient procedure will simplify studies on RNA sequencing and transcriptome analysis in hazelnuts and other similar plants with high phenolic content.
Funding source: Türkiye Bilimsel ve Teknolojik Araştırma Kurumu
Award Identifier / Grant number: 114O800
Acknowledgments
This research was supported by a Research Fund of The Scientific and Technological Research Council of Turkey (114O800).
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
Research funding: Türkiye Bilimsel ve Teknolojik Araştırma Kurumu.
Conflict of interest: The authors have no conflict of interest.
References
1. Boccacci, P, Botta, R. Investigating the origin of hazelnut (Corylus avellana L.) cultivars using chloroplast microsatellites. Genet Resour Crop Evol 2009;56:851–9.10.1007/s10722-009-9406-6Search in Google Scholar
2. RosengartenJrF. The book of edible nuts. New York: Dover Publication; 2004:95.Search in Google Scholar
3. Lan, T, Yao, B, Shen, Y, Wang, X. Isolation of high-quality total RNA from lipid-rich seeds. Anal Biochem 2013;438:11–3.10.1016/j.ab.2013.03.012Search in Google Scholar
4. Morante-Carriel, J, Sellés-Marchart, S, Martínez-Márquez, A, Martínez-Esteso, Mj, Luque, I, Bru-Martínez, R. RNA isolation from loquat and other recalcitrant woody plants with high quality and yield. Anal Biochem 2014;452:46–53.10.1016/j.ab.2014.02.010Search in Google Scholar
5. Ghosh, U, Bose, S, Ghosh, T, Bandyopadhyay, Tk, Basu, U. A monophasic solution for isolation of RNA devoid of polymerase chain reaction-detectable genomic DNA contamination. Anal Biochem 2015; 477:50–2.10.1016/j.ab.2015.02.018Search in Google Scholar
6. Körbler, T, Grs̆ković, M, Dominis, M, Antica, MA. Simple method for RNA isolation from formalin-fixed and paraffin-embedded lymphatic tissues. Exp Mol Pathol 2003;74:336–40.10.1016/S0014-4800(03)00024-8Search in Google Scholar
7. Zhang, H, Finiguerra, M, Dam, Hg, Huang, Y, Xu, D, Liu, G, Lin, S. An improved method for achieving high-quality RNA for copepod transcriptomic studies. J Exp Mar Biol Ecol 2013;446:57–66.10.1016/j.jembe.2013.04.021Search in Google Scholar
8. Djami-Tchatchou, A, Straker, C. The isolation of high quality RNA from the fruit of avocado (Persea americana Mill.). S Afr J Bot 2012;78:44–6.10.1016/j.sajb.2011.04.009Search in Google Scholar
9. Dong, JZ, Dunstan, Di. A reliable method for extraction of RNA from various conifer tissues. Plant Cell Rep 1996;15:516–21.10.1007/BF00232985Search in Google Scholar PubMed
10. Santamaría, Me, Toorop, Pe, Rodríguez, R, Cañal, Mj. Dormant and non-dormant Castanea sativa Mill. buds require different polyvinylpyrrolidone concentrations for optimal RNA isolation. Plant Sci 2010;178:55–60.10.1016/j.plantsci.2009.10.002Search in Google Scholar
11. Chang, E, Zhao, Y, Wei, Q, Shi, S, Jiang, Z. Isolation of high-quality RNA from Platycladus orientalis and other Cupressaceae plants. Electron J Biotechn 2016;23:21–7.10.1016/j.ejbt.2016.08.003Search in Google Scholar
12. Sathuvalli, VR, Mehlenbacher, SA. A bacterial artificial chromosome library for ‘Jefferson’hazelnut and identification of clones associated with eastern filbert blight resistance and pollen–stigma incompatibility. Genome 2011;54:862–7.10.1139/g11-048Search in Google Scholar
13. Ghangal, R, Raghuvanshi, S, Sharma, PC. Isolation of good quality RNA from a medicinal plant seabuckthorn, rich in secondary metabolites. Plant Physiol Biochem 2009;47:1113–5.10.1016/j.plaphy.2009.09.004Search in Google Scholar
14. Su, X, Gibor, AA. A method for RNA isolation from marine macro-algae. Anal Biochem 1988;174:650–7.10.1016/0003-2697(88)90068-1Search in Google Scholar
15. Gudenschwager, O, González-Agüero, M, Defilippi, B. A general method for high-quality RNA isolation from metabolite-rich fruits. S Afr J Bot 2012;83:186–92.10.1016/j.sajb.2012.08.004Search in Google Scholar
16. Birtić, S, Kranner, I. Isolation of high-quality RNA from polyphenol-, polysaccharide-and lipid-rich seeds. Phytochem Analysis 2006;17:144–8.10.1002/pca.903Search in Google Scholar PubMed
17. Fang, G, Hammar, S, Grumet, R. A quick and inexpensive method for removing polysaccharides from plant genomic DNA. Biotechniques 1992;13:4–6.Search in Google Scholar
18. Mackenzie, Dj, Mclean, Ma, Mukerji, S, Green, M. Improved RNA extraction from woody plants for the detection of viral pathogens by reverse transcription-polymerase chain reaction. Plant Dis 1997;81:222–6.10.1094/PDIS.1997.81.2.222Search in Google Scholar PubMed
19. Pappi, PG, Chaintoutis, SC, Dovas, CI, Efthimiou, KE, Katis, NI. Development of one-tube real-time qRT-PCR and evaluation of RNA extraction methods for the detection of Eggplant mottled dwarf virus in different species. J Virol Methods 2015;212:59–65.10.1016/j.jviromet.2014.11.001Search in Google Scholar PubMed
20. Ouyang, K, Li, J, Huang, H, Que, Q, Li, P, Chen, X. A simple method for RNA isolation from various tissues of the tree Neolamarckia cadamba. Biotechnol Biotec Eq 2014;28:1008–13.10.1080/13102818.2014.981086Search in Google Scholar PubMed PubMed Central
21. Vicient, Cm, Delseny, M. Isolation of total RNA from Arabidopsis thaliana seeds. Anal Biochem 1999;268:412–3.10.1006/abio.1998.3045Search in Google Scholar PubMed
22. Kumar, GRK, Eswaran, N, Johnson, TS. Isolation of high-quality RNA from various tissues of Jatropha curcas for downstream applications. Anal Biochem 2011;413:63–5.10.1016/j.ab.2011.01.046Search in Google Scholar PubMed
23. Sangha, JS, Gu, K, Kaur, J, Yin, Z. An improved method for RNA isolation and cDNA library construction from immature seeds of Jatropha curcas L. BMC Res Notes 2010;3:1.10.1186/1756-0500-3-126Search in Google Scholar
24. Eldh, M, Lötvall, J, Malmhäll, C, Ekström, K. Importance of RNA isolation methods for analysis of exosomal RNA: evaluation of different methods. Mol Immunol 2012;50:278–86.10.1016/j.molimm.2012.02.001Search in Google Scholar
25. Asif, MH, Dhawan, P, Nath, P. A simple procedure for the isolation of high quality RNA from ripening banana fruit. Plant Mol Biol Rep 2000;18:109–15.10.1007/BF02824018Search in Google Scholar
26. Chang, S, Puryear, J, Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 1993;11:113–6.10.1007/BF02670468Search in Google Scholar
27. Sharma, AD, Gill, PK, Singh, P. RNA isolation from plant tissues rich in polysaccharides. Anal Biochem 2003;314:319–21.10.1016/S0003-2697(02)00689-9Search in Google Scholar
28. Kiefer, E, Heller, W, Ernst, D. A simple and efficient protocol for isolation of functional RNA from plant tissues rich in secondary metabolites. Plant Mol Biol Rep 2000;18:33–9.10.1007/BF02825291Search in Google Scholar
29. Salzman, RA, Fujita, T, Zhu-Salzman, K, Hasegawa, PM, Bressan, RA. An improved RNA isolation method for plant tissues containing high levels of phenolic compounds or carbohydrates. Plant Mol Biol Rep 1999;17:11–7.10.1023/A:1007520314478Search in Google Scholar
30. Nadiya, F, Anjali, N, Gangaprasad, A, Sabu, KK. High-quality RNA extraction from small cardamom tissues rich in polysaccharides and polyphenols. Anal Biochem 2015;485:25–7.10.1016/j.ab.2015.05.017Search in Google Scholar PubMed
© 2020 Walter de Gruyter GmbH, Berlin/Boston
Articles in the same Issue
- Frontmatter
- Research Articles
- Development of new total RNA isolation method for tissues with rich phenolic compounds
- Myofibrillar degeneration with diphtheria toxin
- In vitro and in silico studies on AChE inhibitory effects of a series of donepezil-like arylidene indanones
- In vitro antioxidant, anti-inflammatory and anti-cancer activities of methanolic extract of Asparagus horridus grows in North Cyprus Kuzey Kıbrıs da yetişen Asparagus horridus metanolik ekstraktının in-vitro antioksidan, anti-enflamatuar ve anti-kanser aktivitesi
- Purification and characterization of glucose-6-phosphate dehydrogenase from Eisenia fetida and effects of some pesticides and metal ions
- Nephroprotective effects of eriocitrin via alleviation of oxidative stress and DNA damage against cisplatin-induced renal toxicity
- The impact of orally administered gadolinium orthovanadate GdVO4:Eu3+ nanoparticles on the state of phospholipid bilayer of erythrocytes
- An anxiolytic drug buspirone ameliorates hyperglycemia and endothelial dysfunction in type 2 diabetic rat model
- Effects of mesenchymal stem cell and amnion membrane transfer on prevention of pericardial adhesions
- How potential endocrine disruptor deltamethrin effects antioxidant enzyme levels and total antioxidant status on model organisms
- Antiproliferative effect of rosehip tea phenolics in prostate cancer cell lines
- Investigation of MMP-9 rs3918242 and TIMP-2 rs8179090 polymorphisms in renal cell carcinoma tissues
- Investigation of SR-BI gene rs4238001 and rs5888 polymorphisms prevalence and effects on Turkish patients with metabolic syndrome
- Assessment of the frequency and biochemical parameters of conjunctivitis in COVID-19 and other viral and bacterial conditions
- Short Communication
- Lack of hotspot mutations other than TP53 R249S in aflatoxin B1 associated hepatocellular carcinoma
- Letter to the Editors
- Cornuside, identified in Corni fructus, suppresses melanin biosynthesis in B16/F10 melanoma cells through tyrosinase inhibition
- The extract of male bee and beehive from Bombus terrestris has biological efficacies for promoting skin health
- COVID-19 laboratory biosafety guide
- Retraction note
Articles in the same Issue
- Frontmatter
- Research Articles
- Development of new total RNA isolation method for tissues with rich phenolic compounds
- Myofibrillar degeneration with diphtheria toxin
- In vitro and in silico studies on AChE inhibitory effects of a series of donepezil-like arylidene indanones
- In vitro antioxidant, anti-inflammatory and anti-cancer activities of methanolic extract of Asparagus horridus grows in North Cyprus Kuzey Kıbrıs da yetişen Asparagus horridus metanolik ekstraktının in-vitro antioksidan, anti-enflamatuar ve anti-kanser aktivitesi
- Purification and characterization of glucose-6-phosphate dehydrogenase from Eisenia fetida and effects of some pesticides and metal ions
- Nephroprotective effects of eriocitrin via alleviation of oxidative stress and DNA damage against cisplatin-induced renal toxicity
- The impact of orally administered gadolinium orthovanadate GdVO4:Eu3+ nanoparticles on the state of phospholipid bilayer of erythrocytes
- An anxiolytic drug buspirone ameliorates hyperglycemia and endothelial dysfunction in type 2 diabetic rat model
- Effects of mesenchymal stem cell and amnion membrane transfer on prevention of pericardial adhesions
- How potential endocrine disruptor deltamethrin effects antioxidant enzyme levels and total antioxidant status on model organisms
- Antiproliferative effect of rosehip tea phenolics in prostate cancer cell lines
- Investigation of MMP-9 rs3918242 and TIMP-2 rs8179090 polymorphisms in renal cell carcinoma tissues
- Investigation of SR-BI gene rs4238001 and rs5888 polymorphisms prevalence and effects on Turkish patients with metabolic syndrome
- Assessment of the frequency and biochemical parameters of conjunctivitis in COVID-19 and other viral and bacterial conditions
- Short Communication
- Lack of hotspot mutations other than TP53 R249S in aflatoxin B1 associated hepatocellular carcinoma
- Letter to the Editors
- Cornuside, identified in Corni fructus, suppresses melanin biosynthesis in B16/F10 melanoma cells through tyrosinase inhibition
- The extract of male bee and beehive from Bombus terrestris has biological efficacies for promoting skin health
- COVID-19 laboratory biosafety guide
- Retraction note

