The crystal structure of poly[hexaqua-tetrakis(μ4-pyridine-2,4-dicarboxylate-κ5N: O: O′: O″: O‴)-bi(μ2-pyridine-2,4-dicarboxylate-κ3N: O: O′)-digadolinium(III)tricopper (II)], [Gd2Cu3(C7H3NO4)6(H2O)6] n
Abstract
[Gd2Cu3(C7H3NO4)6(H2O)6] n , triclinic, P1̄ (no. 2), a = 9.4244(8) Å, b = 10.6875(9) Å, c = 12.2795(11) Å, α = 86.189(1)°, β = 81.500(1)°, γ = 86.605(1)°, V = 1209.0(2) Å3, Z = 1, Rgt (F) = 0.0337, wRref(F2) = 0.0692, T = 296 K.
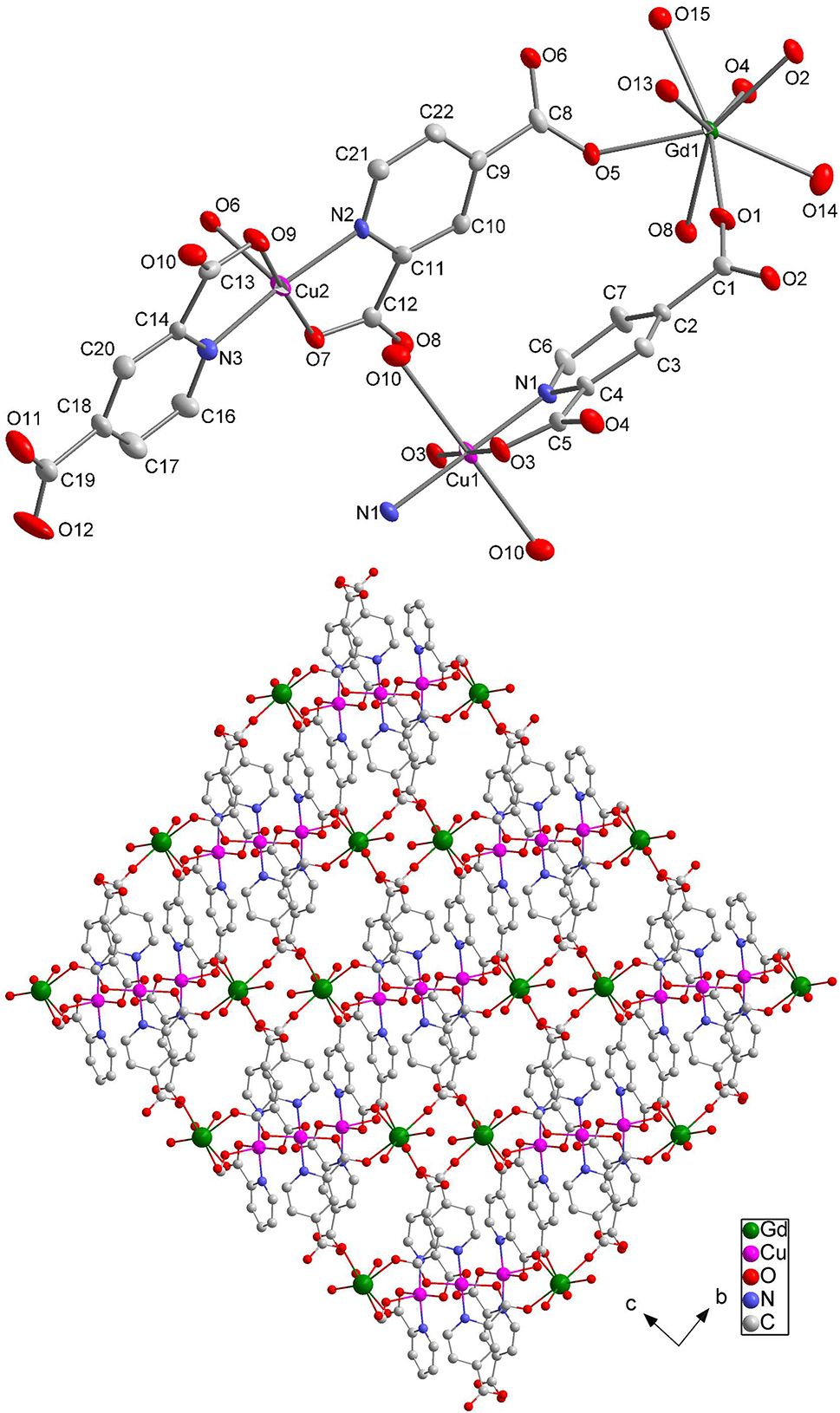
The molecular structure is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Blue block |
| Size: | 0.23 × 0.20 × 0.16 mm |
| Wavelength: | Mo Kα radiation (0.71073 Å) |
| μ: | 4.08 mm−1 |
| Diffractometer, scan mode: | Bruker APEX-II, φ and ω |
| θmax, completeness: | 27.6°, 99 % |
| N(hkl)measured, N(hkl)unique, Rint: | 7,421, 5,318, 0.029 |
| Criterion for Iobs, N(hkl)gt: | Iobs > 2σ(Iobs), 4,450 |
| N(param)refined: | 376 |
| Programs: | Bruker, 1 Diamond, 2 Olex2, 3 SHELX 4 , 5 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | Uiso*/Ueq |
|---|---|---|---|---|
| C1 | 1.0616 (5) | 0.8340 (4) | 0.8846 (4) | 0.0169 (10) |
| C2 | 1.0523 (5) | 0.7575 (4) | 0.7874 (4) | 0.0161 (9) |
| C3 | 1.0375 (5) | 0.6278 (4) | 0.8020 (4) | 0.0155 (9) |
| H3 | 1.038992 | 0.585687 | 0.872476 | 0.019* |
| C4 | 1.0207 (5) | 0.5623 (4) | 0.7121 (4) | 0.0146 (9) |
| C5 | 0.9931 (5) | 0.4254 (4) | 0.7182 (4) | 0.0147 (9) |
| C6 | 1.0428 (5) | 0.7423 (4) | 0.5950 (4) | 0.0185 (10) |
| H6 | 1.048560 | 0.780829 | 0.522583 | 0.022* |
| C7 | 1.0535 (5) | 0.8153 (4) | 0.6824 (4) | 0.0227 (11) |
| H7 | 1.061449 | 0.903480 | 0.670734 | 0.027* |
| C8 | 0.6885 (5) | 1.0867 (4) | 0.6876 (4) | 0.0187 (10) |
| C9 | 0.6523 (5) | 0.9840 (4) | 0.6196 (4) | 0.0164 (9) |
| C10 | 0.7458 (5) | 0.9447 (4) | 0.5287 (4) | 0.0170 (9) |
| H10 | 0.839012 | 0.976574 | 0.511839 | 0.020* |
| C11 | 0.7020 (5) | 0.8593 (4) | 0.4635 (4) | 0.0155 (9) |
| C12 | 0.7822 (5) | 0.8244 (4) | 0.3530 (4) | 0.0171 (10) |
| C13 | 0.3406 (5) | 0.5120 (4) | 0.4349 (4) | 0.0184 (10) |
| C14 | 0.4157 (5) | 0.4787 (4) | 0.3229 (4) | 0.0157 (9) |
| C16 | 0.6056 (5) | 0.5235 (5) | 0.1860 (4) | 0.0283 (12) |
| H16 | 0.683210 | 0.574445 | 0.157160 | 0.034* |
| C17 | 0.5790 (6) | 0.4227 (5) | 0.1275 (4) | 0.0301 (12) |
| H17 | 0.638185 | 0.404394 | 0.060312 | 0.036* |
| C18 | 0.4646 (5) | 0.3493 (4) | 0.1686 (4) | 0.0225 (11) |
| C19 | 0.4366 (5) | 0.2320 (5) | 0.1113 (4) | 0.0243 (11) |
| C20 | 0.3815 (5) | 0.3786 (4) | 0.2677 (4) | 0.0222 (10) |
| H20 | 0.301820 | 0.330209 | 0.297216 | 0.027* |
| C21 | 0.4855 (5) | 0.8381 (4) | 0.5795 (4) | 0.0185 (10) |
| H21 | 0.397604 | 0.797561 | 0.599285 | 0.022* |
| C22 | 0.5195 (5) | 0.9304 (4) | 0.6447 (4) | 0.0196 (10) |
| H22 | 0.452833 | 0.956508 | 0.705679 | 0.024* |
| Cu1 | 1.000000 | 0.500000 | 0.500000 | 0.01613 (17) |
| Cu2 | 0.54274 (6) | 0.68643 (5) | 0.37997 (5) | 0.01944 (13) |
| Gd1 | 0.92937 (2) | 1.15165 (2) | 0.85181 (2) | 0.01283 (7) |
| N1 | 1.0247 (4) | 0.6191 (3) | 0.6102 (3) | 0.0149 (8) |
| N2 | 0.5740 (4) | 0.8056 (3) | 0.4896 (3) | 0.0148 (8) |
| N3 | 0.5256 (4) | 0.5513 (3) | 0.2817 (3) | 0.0186 (8) |
| O1 | 1.0210 (4) | 0.9468 (3) | 0.8770 (3) | 0.0230 (7) |
| O2 | 1.1097 (3) | 0.7799 (3) | 0.9659 (2) | 0.0208 (7) |
| O3 | 0.9657 (3) | 0.3820 (3) | 0.6299 (2) | 0.0203 (7) |
| O4 | 0.9961 (4) | 0.3636 (3) | 0.8073 (2) | 0.0214 (7) |
| O5 | 0.8157 (3) | 1.0860 (3) | 0.7063 (3) | 0.0219 (7) |
| O6 | 0.5895 (4) | 1.1647 (3) | 0.7189 (3) | 0.0279 (8) |
| O7 | 0.7212 (3) | 0.7483 (3) | 0.3010 (2) | 0.0203 (7) |
| O8 | 0.8968 (3) | 0.8744 (3) | 0.3175 (3) | 0.0199 (7) |
| O9 | 0.3810 (3) | 0.6122 (3) | 0.4713 (3) | 0.0260 (8) |
| O10 | 0.2519 (3) | 0.4415 (3) | 0.4858 (3) | 0.0244 (8) |
| O11 | 0.3281 (4) | 0.1750 (3) | 0.1528 (3) | 0.0370 (10) |
| O12 | 0.5225 (4) | 0.2019 (4) | 0.0304 (4) | 0.0525 (13) |
| O13 | 0.7307 (3) | 1.0222 (3) | 0.9481 (3) | 0.0225 (7) |
| H13A | 0.713128 | 0.961298 | 0.913346 | 0.034* |
| H13B | 0.662148 | 1.074808 | 0.962186 | 0.034* |
| O14 | 1.1639 (3) | 1.1693 (3) | 0.9092 (3) | 0.0279 (8) |
| H14A | 1.187917 | 1.099573 | 0.945958 | 0.042* |
| H14B | 1.230317 | 1.202853 | 0.869278 | 0.042* |
| O15 | 0.7121 (3) | 1.2877 (3) | 0.8620 (3) | 0.0248 (8) |
| H15A | 0.669583 | 1.263779 | 0.811506 | 0.037* |
| H15B | 0.656243 | 1.277589 | 0.915976 | 0.037* |
1 Source of materials
Pyridine-2,4-dicarboxylic acid (0.8 mmol 0.134 g), Gd2O3 (0.1 mmol 0.036 g), CuBr2 (0.2 mmol 0.045 g), and 0.4 mol L−1 NaOH solution (1 mL), were dissolved in deionized water (7 mL). The mixture was sealed in a 25 mL Teflon-lined steel autoclave, and heated at 453 K for 8 days, then slowly cooled to room temperature. Blue crystals of title complex could be found in the product.
2 Experimental details
All hydrogen atoms were placed in calculated positions and refined as riding atoms. The values were set to be 1.2 Ueq or 1.5 Ueq of the parent atoms.
3 Comment
Pyridine-2,4-dicarboxylic acid is a very common ligand used to link metal ions in coordination chemistry. Some rare-earth-metal complexes of this ligand have been reported, 6 , 7 , 8 , 9 , 10 , 11 , 12 while heterometallic coordination compounds for this ligand are rarely researched. 13 , 14 A heterometallic complex ([Gd2Cu3(C7H3NO4)6(H2O)6] n ) of pyridine-2,4-dicarboxylic acid has been prepared under hydrothermal reaction conditions. The asymmetric unit of the title complex contains one Gd13+ ion, half Cu12+ ion, one Cu22+ ion, three L2− (H2L = pyridine-2,4-dicarboxylic acid) ligands and three coordinated aqua molecules. Each Gd13+ ion is eight coordinated being bound by five O atoms from five bridging L2− ligands and three O atoms from three water molecules (the mean length of Gd–O = 2.414 Å, the mean value of O–Gd–O = 95.55°), yielding a square anti-prism. Each of Cu1(II) ions is surrounded by two chelating L2− ligands and two oxygen atoms of the other two L2− ligands, forming a slightly distorted octahedron geometry. While each Cu2(II) center is coordinated in a distorted square-pyramidal geometry with two chelating L2− ligands, and one oxygen atom from a terminal carboxyl group of a L2− ligand occupying the basal position. The Cu–O distances are in the range 1.932(3)–2.403(3) Å, and all Cu–N bonds are nearly equivalent spanning a narrow range of 1.966(3)–1.978(3) Å. 13 , 14 In the asymmetric unit, two of the three L2− ligands adopt μ4-bridge modes, while the remaining ligand exhibits μ2-bridged pattern. 15
The Gd1, Cu1 and Cu2 ions are linked by the carboxyl groups of L2− ligands, forming two-dimensional layers parallel to the (1 1 0) plane. Furthermore, the layers are connected together by pyridine rings on the ligands to construct the three-dimensional structure of the title complex.
-
Author contribution: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
-
Research funding: NSF (Natural Science Foundation) of Anhui Province (2108085MB53), the NSF for Distinguished Young Scholars of Anhui University (2022AH020087), University NSF of Anhui Province (2021xsxxkc279) and Program for Innovation Research Team in Huainan Normal University (XJTD202006).
References
1. Bruker. SAINT, APEX3 and SADABS; Bruker AXS Inc.: Madison, WI, USA, 2013.Search in Google Scholar
2. Brandenburg, K. DIAMOND. Visual Crystal Structure Information System. Ver. 4.6.8; Crystal Impact GbR: Bonn, Germany, 2022.Search in Google Scholar
3. Dolomanov, O. V.; Bourhis, L. J.; Gildea, R. J.; Howard, J. A. K.; Puschmann, H. OLEX2: A Complete Structure Solution, Refinement and Analysis Program. J. Appl. Crystallogr. 2009, 42, 339–341; https://doi.org/10.1107/s0021889808042726.Search in Google Scholar
4. Sheldrick, G. M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8; https://doi.org/10.1107/s2053229614024218.Search in Google Scholar
5. Sheldrick, G. M. SHELXTL – Integrated Space-Group and Crystal Structure Determination. Acta Crystallogr. 2015, A71, 3–8.10.1107/S2053273314026370Search in Google Scholar PubMed PubMed Central
6. Wang, C.-C. Crystal Structure of Catena-Tetraaqua-Bis(μ4-Pyridine-3,5-Dicarboxylato)-Μ2-Oxalato-Dicerium(III) Dihydrate, Ce2(H2O)4(C7H3NO4)2(C2O4)·2H2O. Z. Kristallogr. N. Cryst. Struct. 2007, 222, 351–352; https://doi.org/10.1524/ncrs.2007.0148.Search in Google Scholar
7. Wang, X.-N.; Li, F.-F. Crystal Structure of Poly[(5-Carboxy-2,6-Dimethylpyridinium-3-Carboxylato-κO)tris(μ2-2,6-Dimethylpyridinium-3,5-Dicarboxylato-Κ3o: O′: O″)Erbium(III)], C36H33ErN4O16. Z. Kristallogr. N. Cryst. Struct. 2016, 231, 759–761, https://doi.org/10.1515/ncrs-2015-0259.Search in Google Scholar
8. Li, J.; Li, J. A Luminescent Porous Metal-Organic Framework with Lewis Basic Pyridyl Sites as a Fluorescent Chemosensor for TNP Detection. Inorg. Chem. Commun. 2018, 89, 51–54; https://doi.org/10.1016/j.inoche.2018.01.013.Search in Google Scholar
9. Sarkar, S.; Singha, D. K.; Majee, P.; Daga, P.; Mondal, S. K.; Mahata, P. Stabilization of CO2 as Zwitterionic Carbamate within a Coordination Polymer (CP): Synthesis, Structure and Anion Sensing Behaviour of a Tb–CP Composite. CrystEngComm 2022, 24, 5890–5899; https://doi.org/10.1039/d2ce00711h.Search in Google Scholar
10. Lyszczek, R.; Mazur, L. Polynuclear Complexes Constructed by Lanthanides and Pyridine-3,5-Dicarboxylate Ligand: Structures, Thermal and Luminescent Properties. Polyhedron 2012, 41, 7–19; https://doi.org/10.1016/j.poly.2012.04.009.Search in Google Scholar
11. Li, Q.; Qian, J.; Zhou, J.; Du, L.; Zhao, Q. Highly Chemically and Thermally Stable Lanthanide Coordination Polymers for Luminescent Probes and White Light Emitting Diodes. CrystEngComm 2020, 22, 2667–2674; https://doi.org/10.1039/d0ce00228c.Search in Google Scholar
12. Lin, X.-M.; Niu, J.-L.; Wen, P.-X.; Pang, Y.; Hu, L.; Cai, Y.-P. A Polyhedral Metal–Organic Framework Based on Supramolecular Building Blocks: Catalysis and Luminescent Sensing of Solvent Molecules. Cryst. Growth Des. 2016, 16, 4705–4710; https://doi.org/10.1021/acs.cgd.6b00779.Search in Google Scholar
13. Cancino, P.; Santibañez, L.; Fuentealba, P.; Olea, C.; Vega, A.; Spodine, E. Heterometallic CuII/LnIII Polymers Active in the Catalytic Aerobic Oxidation of Cycloalkenes under Solvent-free Conditions. Dalton Trans. 2018, 47, 13360–13367; https://doi.org/10.1039/c8dt01913d.Search in Google Scholar PubMed
14. Liu, J.-H.; Gu, Y.-N.; Chen, Y.; Qi, Y.-J.; Li, X.-X.; Zheng, S.-T. Incorporating Cuprous-Halide Clusters and Lanthanide Clusters to Construct Heterometallic Cluster Organic Frameworks with Luminescence and Gas Adsorption Properties. CrystEngComm 2018, 20, 738–745; https://doi.org/10.1039/c7ce01963g.Search in Google Scholar
15. Liang, Y.; Cao, R.; Hong, M.; Sun, D.; Zhao, Y.; Weng, J.; Wang, R. Syntheses and Characterizations of Two Novel Ln(III)–Cu(II) Coordination Polymers Constructed by Pyridine-2,4-Dicarboxylate Ligand. Inorg. Chem. Commun. 2002, 5, 366–368; https://doi.org/10.1016/s1387-7003(02)00385-4.Search in Google Scholar
© 2024 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Editorial
- Editorial 2024 – New developments and changes of Zeitschrift für Kristallographie – New Crystal Structures
- New Crystal Structures
- Hydrogen bonding and π⋅⋅⋅halogen interactions in the crystal structure of bis(theophyllinium) hexachloridoplatinate(IV) monohydrate
- The crystal structure of 6-amino-2-carboxypyridin-1-ium perchlorate, C6H7ClN2O6
- Crystal structure of poly[(μ4-(3-amino-1H-1,2,4-triazol-1-yl)benzene-1,3-dicarboxylato-κ 4 N:O:O':O')(1-methylpyrroldin-2-one-κ1O)dicopper(II)] – 1-methylpyrroldin-2-one (1/3), C40H48Cu2N12O12
- The crystal structure of 18-crown-6-k6O6(2,4,5-trinitroimidazol-1-ido-k1O)potassium(I)
- Crystal structure of poly[tetraaqua-bis(μ2-5-bromoisophthalato-κ3O,O′:O″)-(μ2-1,5-bis(imidazol-2-methyl)pentane-κ2N:N′)dicadmium(II)] dihydrate
- Crystal structure of (5R,6S,E)-5-acetoxy-2-methyl-6-((2aR,3R,5aS,5bS,11aR,12aS)-2a,5a,8,8-tetramethyl-9-oxotetradecahydro-1H,12H-cyclopenta[a]cyclopropa[e]phenanthren-3-yl)hept-2-enoic acid, C32H48O5
- The crystal structure of poly[diaqua-bis(μ2 -thiocyanato-κ2N:O)cobalt(II) monohydrate
- The crystal structure of 1,3,5-tri(1H-imidazol-1-yl)benzene–2,3,5,6-tetrachlorobenzene-1,4-dicarboxylic acid (1/1)
- Crystal structure of dichlorido-bis(1-[(2-ethyl-benzimidazole-1-yl)methyl]-1H–benzotriazole) cadmium(II), C32H32CdN10OCl2
- The crystal structure of N′-(tert-butyl)-N′-(3,5-dimethylbenzoyl)-3-methoxy-N,2-dimethylbenzohydrazide, C23H30N2O3
- Crystal stucture of 3-benzamido-N-(2-bromo-4-(perfluoropropan-2-yl)-6-(trifluoromethyl)phenyl)-2-fluorobenzamide
- Crystal structure of bis(μ-benzeneselenolato)-(tetracarbonyl)-{μ-[N-(diphenylphosphanyl)-N-(3-ethynylphenyl)-P,P-diphenylphosphinous amide]} diiron, C48H35Fe2NO4P2Se2
- The crystal structure of 2′-(p-tolyl)-4′H-spiro[isochromane-1,1′-naphthalene]-3,4′-dione, C25H18O3
- The crystal structure of poly[hexaqua-tetrakis(μ4-pyridine-2,4-dicarboxylate-κ5N: O: O′: O″: O‴)-bi(μ2-pyridine-2,4-dicarboxylate-κ3N: O: O′)-digadolinium(III)tricopper (II)], [Gd2Cu3(C7H3NO4)6(H2O)6] n
- Crystal structure of poly[bis(4-(4-(pyridin-4-yl)phenyl)pyridin-1-ium-κ1N)-(μ4-benzene-1,2,4,5-tetracarboxylato-κ5O:O′: O″:O‴:O⁗)-(μ2-2,5-dicarboxyterephthalato-κ2O:O′)dizinc(II)], C52H32N4O16Zn2
- The crystal structure of 4-(3-carboxy-1-ethyl-6-fluoro-4-oxo-1,4-dihydroquinolin-7-yl)piperazin-1-ium 2-carboxy-6-nitrobenzoate monohydrate, C24H25FN4O10
- Crystal structure of dichlorido-(1-((3,5-dimethyl-2,3-dihydro-1H-1,2,3-triazol-1-yl)methyl)-1H-benzo[d][1,2,3]triazole-k1N)zinc(II), C22H24ZnN12Cl2
- The crystal structure of (3-chlorothiophene-2-carboxylato-κ2O, O′)-(2,2′-dipyridyl-κ2N,N′)lead(II), C20H12Cl2N2O4S2Pb
- Synthesis and crystal structure of (Z)-4-((1-(3-fluorophenyl)-1H-1,2,3-triazol-4-yl)methylene)-5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one, C19H14FN5O
- The crystal structure of the coordination compound catena-poly[(18-crown-6-ether-κ6O6)(4,5-dinitroimidazolato-κ1O)potassium(I)]
- Crystal structure of 7-(diethylamino)-3-(trifluoroacetyl)-2H-chromen-2-one, C15H14F3NO3
- Crystal structure of dichlorido-1-[(2-ethylimidazole-1-yl)methyl]-1H–benzotriazole κ1N zinc(II), C24H26ZnN10Cl2
- Crystal and molecular structure of 5-bromopyridine-2,3-diamine
- Crystal structure of catena-poly[bis(μ2-1-(3-carboxyphenyl)-5-methyl-4-oxo-1,4-dihydropyridazine-3-carboxylato-k3-O,O′:O″)hexaqua-dicobalt tetrahydrate], C26H36N4O20Co2
- Crystal structure of thiocyanate-κ1N-bis(μ1-2,6-diformyl-4-methylphenol oxime-κ2N,O)-manganese(III) acetonitrile solvate, C21H21MnN6O6S
- The crystal structure of pyrrolidin-1-yl pivalate, C9H13NO4
- The crystal structure of 2,2′-(2,2-diphenylethene-1,1-diyl)bis(1,4-dimethoxybenzene), C30H28O4
- Crystal structure of bis(benzyltrimethylammonium) tetrathiotungstate(VI), {(C6H5CH2)(CH3)3N}2[WS4]
- The crystal structure of ethyl (Z)-2-(ethoxymethylene)-3-oxobutanoate, C9H14O4
- The crystal structure of (E)-6-bromo-3,5-dimethyl-2-(1-phenylprop-1-en-2-yl)-3Himidazo[4,5b]pyridine, C17H16BrN3
- Crystal structure of (3S,3′S,4R,4′S)-3′-(furan-3-yl)-3-hydroxy-4′-methyl-3,5,6′,7′-tetrahydro-1H,3′H-4,5′-spirobi[isobenzofuran]-1,1′(4′H)-dione-methanol (1/1), C21H22O7
- Cocrystal structure of progesterone-isophthalic acid, C25H33O4
- The crystal structure of 3-(6-fluoro-1H-indol-3-yl)-1-methylquinoxalin-2(1H)-one, C17H12FN3O
- Crystal structure of S-(4-carboxybutyl)- l -cysteine
- The cocrystal of 2,2′-(hydrazine-1,1-diyl)bis(1H-imidazole-4,5-dicarbonitrile)– methanol (2/3)
- Crystal structure of (1′R,2′S,4′R,6′S)-4,6-dihydroxy-1′,8′,8′-trimethyl-3-(3-methylbutanoyl)-4′,8′,6′,1′,7,2′-hexahydro-1H-4′,6′-methanoxanthene-8-carbaldehyde, C23H30O5
- Crystal structure of (3,6-di(2-pyridyl)-4-methylphenyl pyridazine-k 2 N,N′)-bis(1-phenyl-pyrazole-κ 2 C,N) iridium(III) hexafluorophosphate, C39H29F6IrN8P
- Crystal structure of 1,5-bis[(E)-1-(2-hydroxyphenyl)ethylidene]thiocarbonohydrazide dimethyl sulfoxide monosolvate, C17H18N4O2S·C2H6OS
- Crystal structure of (S)-4-(2-(4-(2-acetyl-5-chlorophenyl)-3-methoxy-6-oxopyridazin-1(6H)-yl)-3-phenylpropanamido)benzoic acid monohydrate, C29H26ClN3O7
- The crystal structure of 1,3-bis(2,4-dinitro-1H-imidazol-1-yl)propane
- Crystal structure of 4-chlorobenzyl (S)-2-(6-methoxynaphthalen-2-yl)propanoate, C21H19ClO3
- Crystal structure of 1-(5-(benzo[d][1,3]dioxol-5-yl)-4-benzyl-1-(4-bromophenyl)-4,5-dihydro-1H-1,2,4-triazol-3-yl)ethan-1-one, C24H20BrN3O3
- The crystal structure of (Z)-3′-(2-(1-(3,4-dimethyl-phenyl)-3-methyl-5-oxo-1,5-dihydro-4H-pyrazol-4-ylidene)hydrazinyl)-2′-hydroxy-[1,1′-biphenyl]-3-carboxylicacid ─ methanol (1/1), C26H26N4O5
- Crystal structure of (S)-1-phenylpropan-1-aminium (S)-(1-phenylpropyl)carbamate C19H26N2O2
- Synthesis and crystal structure of methyl 2-((5-bromo-4-(4-cyclopropylnaphthalen-1-yl)-4H-1,2,4-triazol-3-yl)thio)acetate, C18H16BrN3O2S
- The crystal structure of trichlorobis(pyridine-2,6-dithio-κS-carbomethylamido)antimony(III), [SbCl3(C9H11N3S2)2]
- Crystal structure of 1,8-dihydroxy-3-{[(triphenylstannyl)oxy]carbonyl} anthracene-9,10-dione, C33H22O6Sn
- The crystal structure of (E)-4-(2-(pyridin-4-ylmethylene)hydrazine-1-carbonyl)pyridin-1-ium-2-olate dihydrate, C12H14N4O4
- The crystal structure of 6-amino-pyridinium-2-carboxylate, C6H6N2O2
- The crystal structure of catena-poly[aqua-nitrato-κ3O,O:O′′-(1,10-phenanthroline-κ2N,N′)sodium(I)], C24H18N6O7Na2
- Retractions
- Retraction of: Crystal structure of bis[diaquaisonicotinatosamarium(III)]-µ-isonicotinato-[diisonicotinatocopper(II)], CuSm2(C6H4NO2)8(H2O)4
- Retraction of: Crystal structure of aqua(2,2′-bipyridine-k 2 N:N′)(nitrato)-(4-aminobenzoato)cadmium(II) nitrate, [Cd(H2O)(NO3)(C10H8N2)(C7H7NO2)][NO3]
Articles in the same Issue
- Frontmatter
- Editorial
- Editorial 2024 – New developments and changes of Zeitschrift für Kristallographie – New Crystal Structures
- New Crystal Structures
- Hydrogen bonding and π⋅⋅⋅halogen interactions in the crystal structure of bis(theophyllinium) hexachloridoplatinate(IV) monohydrate
- The crystal structure of 6-amino-2-carboxypyridin-1-ium perchlorate, C6H7ClN2O6
- Crystal structure of poly[(μ4-(3-amino-1H-1,2,4-triazol-1-yl)benzene-1,3-dicarboxylato-κ 4 N:O:O':O')(1-methylpyrroldin-2-one-κ1O)dicopper(II)] – 1-methylpyrroldin-2-one (1/3), C40H48Cu2N12O12
- The crystal structure of 18-crown-6-k6O6(2,4,5-trinitroimidazol-1-ido-k1O)potassium(I)
- Crystal structure of poly[tetraaqua-bis(μ2-5-bromoisophthalato-κ3O,O′:O″)-(μ2-1,5-bis(imidazol-2-methyl)pentane-κ2N:N′)dicadmium(II)] dihydrate
- Crystal structure of (5R,6S,E)-5-acetoxy-2-methyl-6-((2aR,3R,5aS,5bS,11aR,12aS)-2a,5a,8,8-tetramethyl-9-oxotetradecahydro-1H,12H-cyclopenta[a]cyclopropa[e]phenanthren-3-yl)hept-2-enoic acid, C32H48O5
- The crystal structure of poly[diaqua-bis(μ2 -thiocyanato-κ2N:O)cobalt(II) monohydrate
- The crystal structure of 1,3,5-tri(1H-imidazol-1-yl)benzene–2,3,5,6-tetrachlorobenzene-1,4-dicarboxylic acid (1/1)
- Crystal structure of dichlorido-bis(1-[(2-ethyl-benzimidazole-1-yl)methyl]-1H–benzotriazole) cadmium(II), C32H32CdN10OCl2
- The crystal structure of N′-(tert-butyl)-N′-(3,5-dimethylbenzoyl)-3-methoxy-N,2-dimethylbenzohydrazide, C23H30N2O3
- Crystal stucture of 3-benzamido-N-(2-bromo-4-(perfluoropropan-2-yl)-6-(trifluoromethyl)phenyl)-2-fluorobenzamide
- Crystal structure of bis(μ-benzeneselenolato)-(tetracarbonyl)-{μ-[N-(diphenylphosphanyl)-N-(3-ethynylphenyl)-P,P-diphenylphosphinous amide]} diiron, C48H35Fe2NO4P2Se2
- The crystal structure of 2′-(p-tolyl)-4′H-spiro[isochromane-1,1′-naphthalene]-3,4′-dione, C25H18O3
- The crystal structure of poly[hexaqua-tetrakis(μ4-pyridine-2,4-dicarboxylate-κ5N: O: O′: O″: O‴)-bi(μ2-pyridine-2,4-dicarboxylate-κ3N: O: O′)-digadolinium(III)tricopper (II)], [Gd2Cu3(C7H3NO4)6(H2O)6] n
- Crystal structure of poly[bis(4-(4-(pyridin-4-yl)phenyl)pyridin-1-ium-κ1N)-(μ4-benzene-1,2,4,5-tetracarboxylato-κ5O:O′: O″:O‴:O⁗)-(μ2-2,5-dicarboxyterephthalato-κ2O:O′)dizinc(II)], C52H32N4O16Zn2
- The crystal structure of 4-(3-carboxy-1-ethyl-6-fluoro-4-oxo-1,4-dihydroquinolin-7-yl)piperazin-1-ium 2-carboxy-6-nitrobenzoate monohydrate, C24H25FN4O10
- Crystal structure of dichlorido-(1-((3,5-dimethyl-2,3-dihydro-1H-1,2,3-triazol-1-yl)methyl)-1H-benzo[d][1,2,3]triazole-k1N)zinc(II), C22H24ZnN12Cl2
- The crystal structure of (3-chlorothiophene-2-carboxylato-κ2O, O′)-(2,2′-dipyridyl-κ2N,N′)lead(II), C20H12Cl2N2O4S2Pb
- Synthesis and crystal structure of (Z)-4-((1-(3-fluorophenyl)-1H-1,2,3-triazol-4-yl)methylene)-5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one, C19H14FN5O
- The crystal structure of the coordination compound catena-poly[(18-crown-6-ether-κ6O6)(4,5-dinitroimidazolato-κ1O)potassium(I)]
- Crystal structure of 7-(diethylamino)-3-(trifluoroacetyl)-2H-chromen-2-one, C15H14F3NO3
- Crystal structure of dichlorido-1-[(2-ethylimidazole-1-yl)methyl]-1H–benzotriazole κ1N zinc(II), C24H26ZnN10Cl2
- Crystal and molecular structure of 5-bromopyridine-2,3-diamine
- Crystal structure of catena-poly[bis(μ2-1-(3-carboxyphenyl)-5-methyl-4-oxo-1,4-dihydropyridazine-3-carboxylato-k3-O,O′:O″)hexaqua-dicobalt tetrahydrate], C26H36N4O20Co2
- Crystal structure of thiocyanate-κ1N-bis(μ1-2,6-diformyl-4-methylphenol oxime-κ2N,O)-manganese(III) acetonitrile solvate, C21H21MnN6O6S
- The crystal structure of pyrrolidin-1-yl pivalate, C9H13NO4
- The crystal structure of 2,2′-(2,2-diphenylethene-1,1-diyl)bis(1,4-dimethoxybenzene), C30H28O4
- Crystal structure of bis(benzyltrimethylammonium) tetrathiotungstate(VI), {(C6H5CH2)(CH3)3N}2[WS4]
- The crystal structure of ethyl (Z)-2-(ethoxymethylene)-3-oxobutanoate, C9H14O4
- The crystal structure of (E)-6-bromo-3,5-dimethyl-2-(1-phenylprop-1-en-2-yl)-3Himidazo[4,5b]pyridine, C17H16BrN3
- Crystal structure of (3S,3′S,4R,4′S)-3′-(furan-3-yl)-3-hydroxy-4′-methyl-3,5,6′,7′-tetrahydro-1H,3′H-4,5′-spirobi[isobenzofuran]-1,1′(4′H)-dione-methanol (1/1), C21H22O7
- Cocrystal structure of progesterone-isophthalic acid, C25H33O4
- The crystal structure of 3-(6-fluoro-1H-indol-3-yl)-1-methylquinoxalin-2(1H)-one, C17H12FN3O
- Crystal structure of S-(4-carboxybutyl)- l -cysteine
- The cocrystal of 2,2′-(hydrazine-1,1-diyl)bis(1H-imidazole-4,5-dicarbonitrile)– methanol (2/3)
- Crystal structure of (1′R,2′S,4′R,6′S)-4,6-dihydroxy-1′,8′,8′-trimethyl-3-(3-methylbutanoyl)-4′,8′,6′,1′,7,2′-hexahydro-1H-4′,6′-methanoxanthene-8-carbaldehyde, C23H30O5
- Crystal structure of (3,6-di(2-pyridyl)-4-methylphenyl pyridazine-k 2 N,N′)-bis(1-phenyl-pyrazole-κ 2 C,N) iridium(III) hexafluorophosphate, C39H29F6IrN8P
- Crystal structure of 1,5-bis[(E)-1-(2-hydroxyphenyl)ethylidene]thiocarbonohydrazide dimethyl sulfoxide monosolvate, C17H18N4O2S·C2H6OS
- Crystal structure of (S)-4-(2-(4-(2-acetyl-5-chlorophenyl)-3-methoxy-6-oxopyridazin-1(6H)-yl)-3-phenylpropanamido)benzoic acid monohydrate, C29H26ClN3O7
- The crystal structure of 1,3-bis(2,4-dinitro-1H-imidazol-1-yl)propane
- Crystal structure of 4-chlorobenzyl (S)-2-(6-methoxynaphthalen-2-yl)propanoate, C21H19ClO3
- Crystal structure of 1-(5-(benzo[d][1,3]dioxol-5-yl)-4-benzyl-1-(4-bromophenyl)-4,5-dihydro-1H-1,2,4-triazol-3-yl)ethan-1-one, C24H20BrN3O3
- The crystal structure of (Z)-3′-(2-(1-(3,4-dimethyl-phenyl)-3-methyl-5-oxo-1,5-dihydro-4H-pyrazol-4-ylidene)hydrazinyl)-2′-hydroxy-[1,1′-biphenyl]-3-carboxylicacid ─ methanol (1/1), C26H26N4O5
- Crystal structure of (S)-1-phenylpropan-1-aminium (S)-(1-phenylpropyl)carbamate C19H26N2O2
- Synthesis and crystal structure of methyl 2-((5-bromo-4-(4-cyclopropylnaphthalen-1-yl)-4H-1,2,4-triazol-3-yl)thio)acetate, C18H16BrN3O2S
- The crystal structure of trichlorobis(pyridine-2,6-dithio-κS-carbomethylamido)antimony(III), [SbCl3(C9H11N3S2)2]
- Crystal structure of 1,8-dihydroxy-3-{[(triphenylstannyl)oxy]carbonyl} anthracene-9,10-dione, C33H22O6Sn
- The crystal structure of (E)-4-(2-(pyridin-4-ylmethylene)hydrazine-1-carbonyl)pyridin-1-ium-2-olate dihydrate, C12H14N4O4
- The crystal structure of 6-amino-pyridinium-2-carboxylate, C6H6N2O2
- The crystal structure of catena-poly[aqua-nitrato-κ3O,O:O′′-(1,10-phenanthroline-κ2N,N′)sodium(I)], C24H18N6O7Na2
- Retractions
- Retraction of: Crystal structure of bis[diaquaisonicotinatosamarium(III)]-µ-isonicotinato-[diisonicotinatocopper(II)], CuSm2(C6H4NO2)8(H2O)4
- Retraction of: Crystal structure of aqua(2,2′-bipyridine-k 2 N:N′)(nitrato)-(4-aminobenzoato)cadmium(II) nitrate, [Cd(H2O)(NO3)(C10H8N2)(C7H7NO2)][NO3]

