FAIRSpec-ready spectroscopic data collections – advice for researchers, authors, and data managers (IUPAC Technical Report)
-
Mark Archibald
, Ian Bruno
, Stuart Chalk
, Antony N. Davies
, Robert M. Hanson
, Stefan Kuhn
, Robert J. Lancashire
und Henry S. Rzepa
Abstract
In this Technical Report, we introduce the application of FAIR (findable, accessible, interoperable, and reusable) data management in the form of a “FAIRSpec-ready spectroscopic data collection” – that is, a collection of instrument data, chemical structure representations, and related digital items that is ready to be automatically or semi-automatically extracted for metadata that will allow the production of an IUPAC FAIRSpec Finding Aid. Associating this finding aid with the collection produces an IUPAC FAIRSpec Data Collection. The challenge we set for researchers is relatively simple: to maintain their data in a form that allows critical metadata to be extracted in a discipline-specific way, increasing the probability that the data will be findable and reusable both during the research process and after publication. We focus on a few specific suggestions that researchers can use to maximize the “fairness” of their spectroscopic data collection. Most importantly, following these guidelines ensures that instrument datasets are unambiguously associated with the chemical structure. The guidelines promote the inclusion of the instrument dataset itself in the collection and describe ways of organizing the collection such that automated metadata creation is possible. In these guidelines, we emphasize the importance of systematically organizing data throughout the entire research process, not just at the time of publication.
1 Introduction
1.1 The IUPAC “FAIRSpec” project
The IUPAC Project Development of a Standard for FAIR Data Management of Spectroscopic Data 1 was organized in 2019 in order to promote the adoption of FAIR (findable, accessible, interoperable, and reusable) data management principles in chemical sciences throughout the data production, publication, and post-publication process. The overall goals of this ongoing project include:
the development of a clear set of FAIR Data Management Principles specific to chemistry-related spectroscopic data;
the design of a means of describing a spectroscopic data collection that distills critical metadata associated with the digital items in the collection, thus providing a standardized, accessible method of exploring the contents of a data collection without the need to individually access items in the collection (the IUPAC FAIRSpec Finding Aid 2 , 3 );
providing researchers, authors, and data managers with guidelines for the organization of spectra and associated chemical structure information that allows machine-assisted curation of the data, creating the necessary link between chemical structure and spectroscopic data that is often key to its analysis and discussion; and
the specification of a standardized set of metadata keys and values that will allow a broad range of services that can efficiently search for and retrieve spectroscopic datasets of interest.
Previously, we have introduced a set of FAIR management principles relating to spectroscopic data in chemistry. 4 The five main principles and their associated corollaries are shown in Fig. 1.

The five FAIRSpec principles and their corollaries.
In this Technical Report, we focus on the penultimate goal of our project – providing guidelines for the creation of what we are calling FAIRSpec-ready data collections. As summarized in Fig. 2, we propose best practices in relation to the five FAIRSpec principles as applied to developing and managing spectroscopic data collections.

Best practices in spectroscopic data management based on the five FAIRSpec principles.
Of primary relevance to this report is the recognition that context is important . Spectroscopy data objects that are part of a collection generally relate to one or more chemical structures, through diverse relationships, the recognition of which may only develop over time. Further, data must be organized to enable accessibility, and associated metadata must be well designed to facilitate interoperability. It is important to value all data formats, including commonly used proprietary formats as well as recognized standards published by national or international standardization bodies such as IUPAC.
We have chosen to discuss these guidelines prior to describing specifications for the details of our proposed IUPAC FAIRSpec Finding Aid (which will be the focus of a second Technical Report currently in development) because we feel that one of the most important components of the curation necessary to produce an IUPAC FAIRSpec Data Collection is a critical minimal level of curation that can be applied to essentially any working collection of spectroscopic data, whether or not an IUPAC FAIRSpec Finding Aid is ever generated. Effective data management starts immediately after spectroscopic data are generated within a laboratory and continues through publication and beyond. If this minimal curation is done well, over time, many of the goals of FAIR data management can then be accomplished efficiently or even automatically in later stages of the process, however that workflow might ultimately be defined. If done poorly, the effort of data preservation may be painfully time consuming, and meaningful extraction of metadata might even become impossible.
While these guidelines might seem extensive upon first sight, we wish to emphasize that the creation of a FAIRSpec-ready collection can be simplicity itself. Successful efforts can be as involved as implementing a fully “data-aware” laboratory management system or as simple as just maintaining a set of file directories on an instrument, provided appropriate chemical structure representations are added consistently.
1.2 Organization of this report
The guidelines presented here cover four specific areas:
guidance for the generation of structural representations that accompany those datasets (Section 2),
guidance for best practices in the preparation of spectroscopic datasets, particularly in regard to their associated descriptive and relational metadata (Section 3),
guidelines for the organization of digital items in the collections containing spectroscopic data and their associated structural representations (Section 4),
guidance for maximizing the potential for registering metadata with recognized metadata management agencies (Section 5), and
guidance for data and laboratory managers for ensuring that the data collected by their researchers are maintained in a fashion that optimizes the collections’ findability, availability, interoperability, and reusability both within their institution and more generally.
These guidelines are not intended to cover every possible sort of data or structure representation. In several cases involving structures, such as organometallic compounds and polymers, we recognize that there is no perfect solution. As such, these guidelines should be taken as starting points for development of further guidance.
Acronyms and some of the terminology used here are defined more fully in the appendix to this Technical Report.
1.3 Intended audience
The primary intended audience of this Technical Report includes:
practicing researchers who create and work with experimental spectroscopic datasets and are interested in best practices relating to the management of their growing data collections
principal investigators with an interest in maximizing their data’s potential to be found, used, and referenced both internally within their institution and by members of their community
institutional staff responsible for working with researchers to utilize instruments that collect spectroscopic data
librarians working with researchers to develop best practices in relation to data management within their institution
institutional repository managers with responsibilities that include ensuring the highest level of FAIR data management within their institutions
journal editors and publishing house staff tasked with developing guidance for authors and reviewers in the creation and review of electronic supplementary information (ESI) datasets associated with scientific publications involving spectroscopic data
developers of electronic laboratory notebooks (ELNs) and other services associated with the scientific enterprise such as integrated laboratory management platforms and data validation services
funding agencies interested in providing guidance to their grantees in developing practical wide-reaching FAIR data management plans, particularly in the field of chemistry
anyone working outside the context of chemistry-related spectroscopy interested in developing similar guidelines for FAIR data management within their own discipline
1.4 Management of spectroscopic data
Spectroscopic data are key components of many chemistry endeavors both in academia and industry. Spectroscopic analysis forms a principal function in the “proof of structure” in most areas of experimental chemistry, answering questions such as “What have I made?”, “How pure is it?”, and “Why didn’t this reaction work the way I expected it to?” Spectroscopic data provide evidence required for publication of scientific results by journal publishers and is the basis for subsequent experimental replication or extrapolation of results.
Unfortunately, up until very recently, it has been the norm that such data are provided only in packaged document form, vendor-proprietary formats, or in reduced or processed form as a “spectrum”, perhaps even just an image of a spectrum, rather than as the primary or raw instrumental output. The result is that published data are often significantly less useful than they could potentially be.
As primary products of funded research, plans for the maintenance and sharing of spectroscopic data are increasingly becoming a requirement of funding agencies. For example, the U.S. National Science Foundation (NSF) guide to proposal preparation specifies:
Proposals must include a document of no more than two pages uploaded under “Data Management Plan” …. This supplementary document should describe how the proposal will conform to NSF policy on the dissemination and sharing of research results … and may include … the standards to be used for data and metadata format and content. 5
and
Investigators are expected to share with other researchers, at no more than incremental cost and within a reasonable time, the primary data, samples, physical collections, and other supporting materials created or gathered in the course of work under NSF awards. 6
The NSF Directorate for Mathematical and Physical Sciences Division of Chemistry provides more specific guidance to the chemistry community as well. 7
Publishers have also started to at least recommend that authors include their data as ESI for publications, 8 , 9 , 10 though only minimal advice for how to organize those data have been provided.
In strongly regulated industries such as pharmaceutical research and manufacturing, there are long-standing legal requirements to conserve and be able to produce on demand original analytical data such as spectra for auditors. 11 , 12 These data may well be required long after the measuring instruments themselves have been retired.
In both contexts – academic and industrial – this issue is compounded by the international chemistry community’s lack of an agreed-upon, standardized mechanism for organizing and sharing collections of spectroscopic data. Instead, every research group is left to their own devices to save and share data in whatever organizational scheme they choose to implement, often as a single monolithic document supplementing a publication, the ESI. In this form, the data cannot be said to be easily discoverable or findable other than by direct association via the landing page of the journal article reporting the results. The “data” are likely accessible to humans only via copy/paste operations, if these are even permitted by the format. When raw or minimally processed data are provided, they tend to be in the form of an idiosyncratically organized archive file. Such “collections” are not sufficient for optimal broad-context findability or ease of re-use.
To a practicing chemist, it should be obvious that digital entities coming from a laboratory instrument constitute “primary data” (referred to herein as “datasets”). However, the word data as used here is a broader term. For our purposes, data includes processed data such as spectra, as well as derived data and analyses. These might include peak lists or chemical shift splitting, and integration descriptions in nuclear magnetic resonance (NMR) spectroscopy, as well as 1D and 2D spectral assignments in relation to molecular structure. Chemical structure representations such as molfiles (MOL) and structure-data files (SDF) also fall in this category.
1.5 FAIR management of spectroscopic data
Relatively recent discussions of data management have emphasized what are referred to as FAIR data management guidelines. 13 It is important to emphasize that what we are referring to as “FAIR” here is not so much the data themselves as it is the management of that data, and in particular, the production and organization of the metadata associated with experimental data. Metadata are digital items that play the role of documentation for data, resource discovery, and contextualization. Metadata accompanies electronic data in describing the data, making internal relationships among related digital items in a collection, and relating that collection to other work.
Thus, we prefer to refer to “FAIR data management” rather than “FAIR data” itself as a way of emphasizing that we are not just referring to specific file formats (experimental “datasets”) when using these terms, but rather to how those datasets are described and organized as part of data collections. We focus here on the development of a rich metadata context associated with experimental datasets.
It is important to note at the outset that these widely discussed FAIR guidelines were designed not only to facilitate access to such data by humans, but also to allow autonomous discovery and re-use by machines in contexts such as artificial intelligence and machine learning. “FAIR” has been said to also refer to Fully Artificial-Intelligence Ready, and the acronym FAIR2 (“FAIR-squared”) has been suggested for Findable, Accessible, Interoperable, Reusable, and Fully Artificial-Intelligence Ready. 14 Unfortunately, in the chemical spectroscopies, the current forms of ESI as representations of data are rarely if at all conformant with FAIR guidelines or optimal for machine processing.
An important aspect of FAIR data management relates to how a metadata record is stored and shared. There are two main models for the use of metadata. The one most associated with findable, accessible, interoperable, and reusable focuses on the public distribution of data and its associated metadata. The general FAIR guidelines focus on a largely discipline-blind model where a digital object is registered with an appropriate registration agency in exchange for a persistent identifier (PID, normally in the form of a digital object identifier (DOI) from the DOI Foundation, 15 and that record is then aggregated into a metadata store, where it can also be indexed and searched. The metadata records include full file paths (URLs) or database references to the components or collections as held in an appropriate repository. This first model is primarily focused on the post-publication finding and relating of data and their collections on the web.
1.6 Not just for publication: the FAIR data workflow
A more nuanced model for FAIR data management is a private, locally relevant model discussed in detail below. This second discipline-specific model, which we introduce in these guidelines, is one where the metadata are stored on a file system or within a local database where they can be summarized by a local IUPAC FAIRSpec Finding Aid (discussed more specifically in Section 4). This highly structured document describes the accumulating data collection in a machine-readable format. The IUPAC FAIRSpec Finding Aid can then serve as a starting point for both the mostly discipline-blind PID-based model as well as a much more fine-grained private or public discipline-specific purposes.
In the everyday activities of a research laboratory, it is common to acquire numerous spectroscopic data taken by multiple researchers, over multiple timeframes, of samples containing compounds having known or unknown structure. Many researchers already spend significant time curating their data collections, whether that be as complex as a dedicated institutional system or as simple as a hand-written laboratory notebook or spreadsheet. ELNs, when used properly, also serve an important role in the organization of the metadata associated with spectroscopic data. However, with few exceptions 16 such tools (and the metadata they collect) are largely unstandardized, making reusability a problem.
Thus, well before any publication is in sight, there is a need for more standardized workflows for the organization of spectroscopic data collections, particularly in relation to their associated samples and the putative structures of their associated compounds. Every experimental chemist understands the importance of both organizing and communicating such information. The guidelines we present here suggest simple ways that researchers can better organize their data and associated metadata to provide added value throughout the research project lifetime. Indeed, even well past the point of project completion and publication, when digital data collections are stored in and made accessible by repositories, there is a great need for standardization of the metadata associated with spectroscopic data. These guidelines provide the basis for a workflow to produce data collections that are ready to be processed – and, indeed, might be regularly processed privately and locally – to make FAIRSpec-ready collections that are useful throughout the research process.
1.7 Sample-vs. compound-based collections
It is important to understand that the nature of a spectroscopic collection may change over time and be different in different contexts. Specifically, a spectroscopic data collection in its initial context of a laboratory setting tends to be sample oriented. A sample obtained from a reaction or other source is analyzed using an instrument. Data are obtained in the digital form. At this stage, it is not necessarily appropriate to assign any correlation to a chemical structure. However, it is critical that we have a reference to the sample unambiguously associated with the spectral data. Methods of accomplishing this task are discussed in Sections 4.2 and 5.1, below.
As time progresses, in association with internal reports and external publications, the chemical structure is generally associated with spectra. These collections may retain sample-to-spectra associations, but now they add associations referred to here as “compounds”, as discussed in Sections 4.2. These compounds are described digitally as associations made between structure representations and instrument datasets. Section 2 describes in detail our advice for the creation and introduction of structure representations.
1.8 Descriptive and relational metadata
Metadata is central to this discussion. We distinguish two types of metadata – descriptive and relational.
Descriptive metadata in the context of spectroscopy constitute information such as the type of instrument used, the temperature, the solvent, and other details of the analytical methods involved in acquiring and processing the actual “experimental data.” The description also includes declarations of the media types that the data are held in, which will help identify whether it is primary or raw (lossless) data directly captured from an instrument, or whether it has already been processed (possibly with some loss of information) in some form, as for example in a conversion of a time domain or induction form into a frequency domain or spectral form.
Relational metadata include information that ties or associates experimental spectroscopic data to their relevant context or provenance – notebook page and sample references, compound numbers in a publication, researcher and organizational identifiers, proposed chemical structure details and references to both related spectra and other analytical techniques. Relational metadata can also be at the collection level, indicating relationships to other collections such as other datasets or to journal publications. Relational metadata canprovide licensing information together with broader information about sponsoring and research organizations. One of the most important aspects of relational metadata is that it can be registered with agencies such as DataCite 17 specifically for the purposes of improving wide-ranging findability and accessibility. Relational metadata records themselves at the collection level can then be associated with their own individual persistent identifiers.
Relational metadata are dynamic and are likely to increase with time as the project evolves, with the addition of new and related spectra, association with other forms of data such as computational models, and as connections to chemical structure are made or revised. Even after publication, the relationships amongst the items in a collection are likely to change as related publications appear, and additional metadata may be needed to refer to later publications or datasets as well. Descriptive metadata, in contrast, are likely to be more static (and, in many cases, unchangeable for ethical reasons), being produced once and then not changed again.
2 Guidance for digital chemical structure representations in a FAIRSpec-ready data collection
2.1 Digital chemical structure representations
We start with guidelines for creating chemical structure representation specifically in relation to spectroscopic data within a FAIRSpec-ready collection.
There are many ways in which a structure can be represented, ranging from an image or diagram, through electronic formats that capture detailed atomic coordinates and connectivities, to linear representations, chemical names, and other standard identifiers. In practice, such representations perform many functions. Authors use images (or “drawings”), for example, to convey aspects of chemical structure and bonding that relate to their finding. Cheminformaticians use digital representations to reference chemical properties. An important purpose of a digital structure representation in the context of spectroscopic data collections is to provide the critical digital metadata that allow finding and discussing the relationship between structure and spectroscopy. In cases where detailed post-acquisition spectral analysis has been carried out, specific structure representations are necessary to correlate specific atoms and/or functional groups in a compound’s structure with specific signals in the spectroscopic data.
Digital structure representations conveying the chemical structures of compounds associated with spectroscopic data might include one or more of the file types given in Table 1, where pros and cons of each are mentioned. The most useful structure representations in the current context describe the molecule in terms of atoms and bonds with 2D or 3D coordinates (e.g., MDL-MOL 18 , 19 or CDXML 20 ) allowing automated generation of representations that can be included in metadata such as SMILES (simplified molecular input line entry system) 21 , 22 , 23 and InChI (International Chemical Identifier) 24 when appropriate. Although simple images are potentially valuable representations in a variety of contexts, they must not be the sole representation for a compound’s structure, as they cannot generally be reliably converted to any of the other formats.
Common digital chemical structure representations – pros and cons.
| Representation type | Considerations |
|---|---|
| MDL-MOL Version 2000 | Benefits: high interoperability, can contain 2D or 3D coordinates Limitations: limited ability to convey bonding |
| MDL-MOL Version 3000 | Benefits: all the features of Version 2000, with expanded capabilities to describe special bonding types and multi-atom connection bonding (as in ferrocene) Limitations: less widely implemented to date in toolkits and informatics platforms. |
| CDXML | Benefits: well-specified structural format generated and read by popular drawing programs, allows for the expression of “nicknames” such as Ph (phenyl) and TBS (tert-butyldimethylsilyl), highly versatile in expressing nuances of bonding, and may provide warnings of inappropriate bonding or charge states Limitations: generally, less interoperable than MOL (at least at the time of this writing); ambiguity can arise from using bonding and atom elements for nonmolecular depictions, such as titles, labels, and drawn lines; specification is no longer formally maintained, but the last published working specification version is available. 20 |
| CDX | Benefits: well-specified binary equivalent of CDXML, easily converted to CDXML with open-source tools Limitations: binary format lacks the human readability aspects of MOL and CDXML; see note for CDXML in relation to specification |
| SMILES | Benefits: 1D (character string) compact format, generally well-specified standard, allows for stereochemical ambiguity, allows explicit double bond or aromatic bond descriptions, easily interconvertible with 2D or 3D formats within the range of its applicability Limitations: there is no universally accepted “canonical” form; different toolkits and toolkit versions differ in interpretations, particularly of what “aromatic” means and where it is applicable. An IUPAC project to formalize guidelines for reliable use of SMILES is currently in progress. 25 |
| InChI | Benefits: 1D (character string) compact format, canonical, options can include or not include hydrogen and stereochemical “layers”, easily derivable from 2D or 3D formats within the range of its applicability Limitations: depending upon the presence of layers, cannot always be converted unambiguously to 2D or 3D structure representations or to handle tautomeric isomers; does not currently encode bonding details for metal-containing compounds or some advanced stereochemistry features. Projects to address priority limitations are currently in progress. 26 , 27 |
| Image | Benefits: provides a readily interpretable and displayable option for finding aid viewers Limitations: does not generally allow for reliable error-free conversion to any of the other representations; generally, not acceptable as the sole structure representation in a collection |
| Chemical Name | Benefits: particularly the IUPAC preferred name for a compound, when appropriate, is the gold standard for unambiguous description of a chemical entity. Limitations: prone to errors that are not easily discoverable; requires complex processes for converting to other forms of chemical identifiers. |
The key point here is not that one representation is inherently better than another, rather that we value the presence of multiple representations of a structure within a collection. We encourage researchers to provide as many structure representations as they are reasonably able to – 2D drawing file and an image, or a 2D structure as well as a 3D structure. In terms of a collection being FAIRSpec-ready, we do not need all the possible representations, only enough to generate additional ones through automated workflows. For example, a single CDXML representation can be used to generate a SMILES, an InChI, a PNG image, and both 2D- and 3D-molfiles. We do not (and in many cases could not) seek to dictate a single correct method of structural representation; rather we have tried to present here methods most likely to preserve chemical information after machine processing and to highlight common pitfalls where information would be scrambled or lost.
Thus, all chemical structure representations involve priorities and trade-offs. When a chemist draws a chemical structure, the purpose is generally to allow unambiguous communication with other chemists. Additionally, though, the structures we draw could be used also to communicate unambiguously with machines. However, the qualities of a drawn structure that make it unambiguous to a human might introduce ambiguity when interpreted by a machine. The trade-offs and considerations described below are often necessary because the ecosystem of tools to draw, interchange, and process chemical structures electronically lacks the functionality (or broadly agreed upon methods) to handle some of the more nuanced aspects of structural representation. Ultimately, in the context of FAIR data management, the goal is to use a chemical structure to generate the metadata that enables both humans and machines to communicate efficiently and unambiguously with each other. Thus, a focus on unambiguous machine interpretation can lead to slightly different priorities for drawing a structure than chemists might be used to.
2.2 Generating digital chemical structure representations
What follows is a set of suggestions for best practices in producing the chemical structure representations associated with spectroscopic datasets.
Provide multiple representations of the same structure when feasible. The IUPAC FAIRSpec Principles embrace a variety and multitude of structure representations. Many of these structure representations are interconvertible. Thus, if a FAIRSpec-ready data collection includes only a CDXML or MOL representation, that can generally be sufficient to generate all the other representations for inclusion in an IUPAC FAIRSpec Data Collection, such as one or more forms of SMILES or InChI. Nonetheless, the presence of multiple representations for a given structure allows for increased interoperability and improved opportunities for data and metadata validation.
Include only one structure per file. Drawing files containing multiple structures (such as reaction schemes or tables or figures for publication) generally cannot be processed by automated methods. Particularly in relation to spectroscopic collections, correlating specific structures with specific spectra requires that individual structures have their own digital representations (i.e., files). In the same vein, generic labels such as “R” or “X” (“Markush” drawings) should not be used to represent multiple compounds that differ only at specific locations, because doing so does not allow the sort of direct association with spectroscopic data we need in this context.
Produce structure representations free of annotations. In preparation for extraction of metadata from a structure, do not annotate structures with labels or text boxes. Numbers providing compound numbers or adjacent to atoms to identify specific atoms are generally not interpretable by software. Hydrogen bonds and other “weak” bonds, though perhaps important to a discussion, are not advisable for these structure-data associations. Notations such as (R), (S), racemic, scalemic, 93 % ee, 3:1 dr, etc. should not be part of the structure representation itself. If such annotations are important to the discussion, provide a separate representation that includes them. Chemical names should not be included with structures, as this can sometimes result in errors in processing, and can lead to unmanageable image widths. In general, chemical names are not necessary in FAIRSpec-ready collections, as they can be generated from well-made structure or drawing files.
To the extent that it is useful within the given context, we suggest that the place for structure-specific annotation is in metadata, not within the structural representation itself. As such, we describe in Section 5.1 several simple ways of adding annotations in a way that allows them to be associated with structural representations without being hidden within them.
Most importantly, it is advisable to adhere to IUPAC-recommended structure depictions as much as possible. IUPAC recommendations are available for graphical representation of chemical structures 28 and the depiction of stereochemistry. 29 Ongoing IUPAC efforts are expected to expand upon these guidelines specifically in relation to machine-readable formats. 30
Take care when using abbreviations in a structure. Abbreviated atom labels (e.g., ‘OTHP’, meaning “tetrahydropyranyloxy”) may cause problems when converting from a drawing program’s native format to other structure representations, such as MOL, SMILES, and InChI. It is advisable to look for errors in the drawing program. An abbreviated group flagged as a possible error by the program typically means that the drawing program cannot interpret the chemical meaning of the abbreviation. If in doubt, expand all abbreviations, at least temporarily, just to check. Or, if the program allows, check that the calculated molecular formula matches the intended structure. For example, a drawing program may interpret “PMB” as phosphorus-metal-boron, whereas the author intended it to mean para-methoxybenzyl. Expansion of the label or checking the molecular formula would quickly identify this as a problem.
For mixtures of compounds, consider the spectroscopic context. Here we wish to distinguish between structure representation, compound, and sample. For our purposes here, a structure representation is a digital item, a series of bytes, whether that be a SMILES in the form of a string of characters or a MOL file or an image. We contrast that to a compound, which may or may not have an associated chemical structure or spectrum. Thus, we speak of “the structure of a compound” or “this compound’s spectrum.” Essentially, the term compound has an associative nature. It is the connecting link between a spectrum and its associated structure. Chemical samples, on the other hand – the actual starting point for experimental spectroscopy – are generally mixtures of compounds, and the “compounds” themselves might even be mixtures. It is not uncommon, for example, to see in publications the phrase Compound 3c was a 10:1 mixture of diastereomers. Whether or not this is correct usage of the term “compound” is not for us to say. After much discussion within our project group, we have come to the following context-based consensus:
For a single compound for which the NMR spectrum shows (in the author’s opinion) minor impurities or residual solvent, only include the structure of the principal component. In the case of a compound reported with “high enantiomeric excess”, include only the structure of the major enantiomer if, in the given context, the minor enantiomer is nothing more than an undesired impurity.
In contrast, if the context emphasizes the stereochemical nature of the mixture (for example, the analysis is from chromatography that separates and identifies enantiomers, or the NMR spectrum clearly indicates signals from two diastereomers and is used to determine their ratio), multiple structures should be included in individual files. Additional compound association-level descriptive metadata could provide information regarding the nature of the mixture.
For racemates, include only the structure of one of the enantiomers unless the enantiomers are distinguishable by the associated spectroscopic data. Representation of racemates is a special case that is trivially depictable for human consumption but much more difficult to achieve for machine-readability. Current machine-readable formats lack a reliable, consistent way to store the information that the structure is racemic. We note that InChI has a flag for racemates, but it is not part of standard InChI. MOL files may use the “chiral flag” set to zero to indicate a racemic mixture, but this feature has not been implemented reliably in the past. 31
Differentiation between use cases a, b, and c, may well be driven by the environment in which the work was carried out. For example, in pharmaceutical research and manufacturing, it may well be a regulatory requirement that the spectroscopic data is measured specifically to identify and quantify extremely low-concentration compounds within a mixture such as when reporting toxic metabolite levels in a product. Here not only must the compound identification be carried through to the final documentation, it is essential that chiral information, where appropriate, is correctly reported as this can have a direct bearing on the toxicity of the compound identified.
The underlying principle here is that data and metadata should not be mixed in the same representation unless the representations themselves demand it as part of a standard. Providing metadata separate from data makes the metadata itself significantly more findable. The FAIRSpec Finding Aid allows for any amount of additional annotation to be associated with structures as described in Section 5.1.
Do the best you can with compounds that present unsolved challenges for structure description. Few structure representations properly represent coordination and dative bonds or allow for multicenter attachments (as for many inorganic and organometallic compounds). Even when they do, it is not always obvious how to generate the proper representation for machine readability. At the very least, these structure representations should be accompanied by an image representation, as it is quite likely that neither SMILES nor InChI can properly describe them. Other cases exist (such as atropisomerism) where generating suitable machine-readable representations remains an unsolved challenge. Again, the guiding principle here is that the best representation is one that conveys the intention of the creator, whatever that representation might be. To the extent that this can be more than an image, all the better. For example, if a drawing program is used to produce the image, provide both the drawing program’s native representation and the image.
Use standard descriptions for macromolecules, supplemented with images. Macromolecules such as proteins are always best represented by established formats such as Protein Data Bank (PDB), 32 Macromolecular Crystallographic Information File (mmCIF), 33 or BinaryCIF, 34 (the latter two being more extensible and more actively maintained). Additionally, complex biomolecules may be represented in a SMILES-like linear string using the Hierarchical Editing Language for Macromolecules (HELM), 35 a machine-readable linear notation supported by IUPAC and the Pistoia Alliance. 36 Nonetheless, images are welcome additional representations that can be used to supplement and provide meaningful additional interpretation and annotation of these formats. As for organic and inorganic polymers, and network solids, we give no specific guidance other than to provide “appropriate and meaningful” digital representations, whatever that might mean in the context of the collection, even if that is only an image. Additional metadata can be used to be more descriptive.
3 Guidance for instrument dataset representations in a FAIRSpec-ready data collection
In this section, we outline ways in which the instrument dataset itself can be optimized for incorporation into an IUPAC FAIRSpec Data Collection. It is not for these guidelines to elevate one digital representation over another at the instrument level. On this point, we refer to the original recommendations for data representations as given in the FAIR Guiding Principles for scientific data management and stewardship as elaborated by GO FAIR. 37 Specifically:
F2. (Findability) Data are described with rich metadata
R1.3. (Reusability) Data should meet domain-relevant community standards
“Data” in the GO FAIR context means much more than just instrument “datasets”. It includes chemical sample, structure, and analysis representations, as well as all the metadata associated with the collection. Nonetheless, with these goals in mind, specifically in relation to instrument datasets, we suggest:
The original instrument dataset is an important representation. There is no question that the potentially most important digital representation of an instrument dataset is the one that came from the instrument itself. If it is practical to provide this primary data, it should be provided. For NMR, this implies that the free induction decay (FID) is made available.
Multiple representations are valued. If we consider the likely prospects for data reuse, no single data representation will always be the most valuable in all contexts (Table 2). Thus, in the case of NMR spectroscopy, just providing proprietary FID data is not generally as reusable as providing the FID data, a transformed frequency-domain spectrum, and a peak listing.
Generally, do not combine a molecular structure with its associated spectrum within a single representation. While this might seem to be the obvious thing to do, and it can be handled in certain cases during metadata extraction, we suggest that it is bad practice to create “hard-coded” relationships between structure and instrument datasets that may or may not be the final story. The IUPAC FAIRSpec Data Collection separates structures from spectra to associate them in more flexible ways, for example, when the need is for the structure only. Keeping these aspects separate digitally allows for easier later-stage reinterpretation of the data.
Package only one dataset per file. While some digital formats allow combining multiple instrument datasets for the important work of comparative analysis, to be FAIRSpec-ready, just as for structures, there should only be one item per representation. For example, it is not sufficient to have a single file that contains all the spectra in a collection, as each collection may have its own specific associations (all the spectra for this compound, all the spectra of this type, etc.), and the ability to repackage data into (initially unknowable) different collections is an important aspect of the IUPAC FAIRSpec Data Collection. In the case where one file contains both a spectrum and structure representations, it is particularly critical that there be only one spectrum with only one structure (and that both are extractable independently).
Digital dataset representation examples.
| Representation type | Considerations |
|---|---|
| Original instrument data (for example, the NMR FID and associated parameter files) | Benefits: highest integrity with no information loss; high potential for reuse; allows for alternative and/or automated (re)analysis; potential for generation of all other representations; allows possibility of fraud detection; allows the most reliable automated metadata extraction. Limitations: vendor-specific format may require licensing access to a vendor-specific reader; may not express important aspects of the data processing used in the analysis; must contain enough of the key parameters for further processing; may no longer be documented; shortest expected lifetime. |
| Original data exported to an alternative standardized format, such as JCAMP-DX(FID) 38 NMR-Star 39 or nmrML 40 | Benefits: highest possibility of reuse and interoperability; vendor-agnostic open format; allows for automated production of additional representations; recognized by regulators as potentially the longest expected lifetime. Limitations: Depending on implementations potential for loss of data resolution or precision; potential for loss of some metadata fields. |
| Instrument-processed data such as transformed NMR data in the form of a spectrum or spectra | Benefits: most generally and immediately informative to the practicing scientist or educator; can be examined in detail and repurposed. Limitations: does not allow for early-processing adjustments, such as in NMR spectroscopy phasing, line broadening or use of non Fourier-transform methods; may require proprietary software to read; information loss compared to original data; less scope for fraud detection. |
| Third party-processed data | Benefits: concise; convenient if this is the standard process in each laboratory; may include meaningful annotation added by the originator such as integration or peak identification”; may be the only method of getting FAIR metadata to be associated with the data. Limitations: format may require proprietary software to read; possibly limited ability to extract key metadata from proprietary formats; may disallow alternative analysis; may be more subject to fraud or other sorts of spectral editing (removal of solvent peaks, for example) |
| Inline string description (for example, in NMR spectroscopy) a string describing field strength, solvent, chemical shifts, coupling constants, and integration | Benefits: concise; a common requirement for publication as part of the experimental details; distills the essential features of the spectrum. Limitations: minimally informative. |
| Peak listing or peak table | Benefits: easily machine-readable; possibly all that is needed for some forms of reuse. Limitations: minimal semantic information; requires additional context and metadata for interpretation. |
| Image | Benefits: most immediately identifiable and informative to working chemists, educators, and students; excellent for accompanying more robust representations. Limitations: almost certainly reduced resolution; no additional processing possible; susceptible to crude data editing; does not generally allow for conversion to any of the other representations; not acceptable as the sole structure representation in a collection; often cannot represent the details used by automated processing software. |
4 Guidance for the organization of digital items in a FAIRSpec-ready data collection
The essential aspect of a FAIRSpec-ready data collection is that it is organized in a systematic manner that allows (1) machine-based extraction of key metadata and (2) unambiguous association of specific spectra with specific chemical structures (to the extent that such an association is possible). More specifically, the data and associated metadata should, with perhaps a small amount of additional curation, allow for the creation of an IUPAC FAIRSpec Finding Aid. We briefly introduce this metadata document first and then discuss the key elements of a FAIRSpec-ready collection.
4.1 The IUPAC FAIRSpec finding aid
An IUPAC FAIRSpec Finding Aid is a document that describes in detail the contents of a spectroscopic collection. The structure of the document is based on the IUPAC FAIRSpec Metadata Object Model, 41 which describes a small set of abstract objects (for example, “samples”, “structures”, “spectra”, “compounds”, and “analyses”). In addition, the model describes how the various types of digital representations of these abstract objects (CDXML and MOL files, instrumental data sets, PDF reports, etc.) are related and how they are to be described by structured metadata.
A key feature of the IUPAC FAIRSpec Finding Aid is that it is extensible. While it is expected to contain certain metadata in an IUPAC-specified format, the FAIRSpec Metadata Object Model allows for the addition of whatever additional metadata is needed, depending upon the context. Thus, a key feature of a FAIRSpec-ready collection is that it provides for additional (“non-extractable”) metadata in a standardized format. We will see how this additional metadata can be represented in Section 5.
A FAIRSpec-ready data collection provides a means to create such a finding aid and its associated collection and landing page via automation. We want to be able to pass the FAIRSpec-ready data collection to a software tool (perhaps a public or private web site or a local software application), that can read the collection’s digital items, extract the key descriptive and relational metadata, and create what we are calling an IUPAC FAIRSpec Finding Aid. In the process, the extractor may generate additional representations, such as images of structures, predicted spectra, or peak listings. The extractor would only be limited by its sophistication, but it could only work if the FAIRSpec-ready collection is properly organized. This section describes how this organization might be achieved.
The outline of an IUPAC FAIRSpec Finding Aid serialized as JavaScript object notation (JSON) is shown in Fig. 3. The opening view of a web-based landing page that interprets the JSON is shown in Fig. 4, 42 which was created for a recent publication in inorganic chemistry. 43 Without going into extensive detail here, the essence of an IUPAC FAIRSpec Finding Aid is that it consists of the following parts:
A mandatory header section that
identifies the document as an IUPAC FAIRSpec Finding Aid
identifies related work, such as publications or related data collections
identifies the target repositories and pointers to local or remote data items
specifies licensing and other access-related aspects of the collection
An optional section listing individual samples, each with its own set of representations, key metadata, and identifier
An optional section listing individual structures, each with its own set of representations, key metadata, and identifier
An optional section listing individual experimentally or computationally derived datasets, each with its own set of representations, key metadata, and identifier. (The specification allows for predicted, simulated, and experimental spectra to be represented, as long as they are identified as such in their associated metadata.)
An optional compounds section making one-to-one, one-to-many, or many-to-one associations among items on two or more of the above lists. For example, a collection of sample-structure, sample-spectra, or structure-spectrum relationships.
An optional section listing individual structure-spectral analyses, each with its own set of specific representations, relating specific spectra and their related structures
Additional custom sections as needed
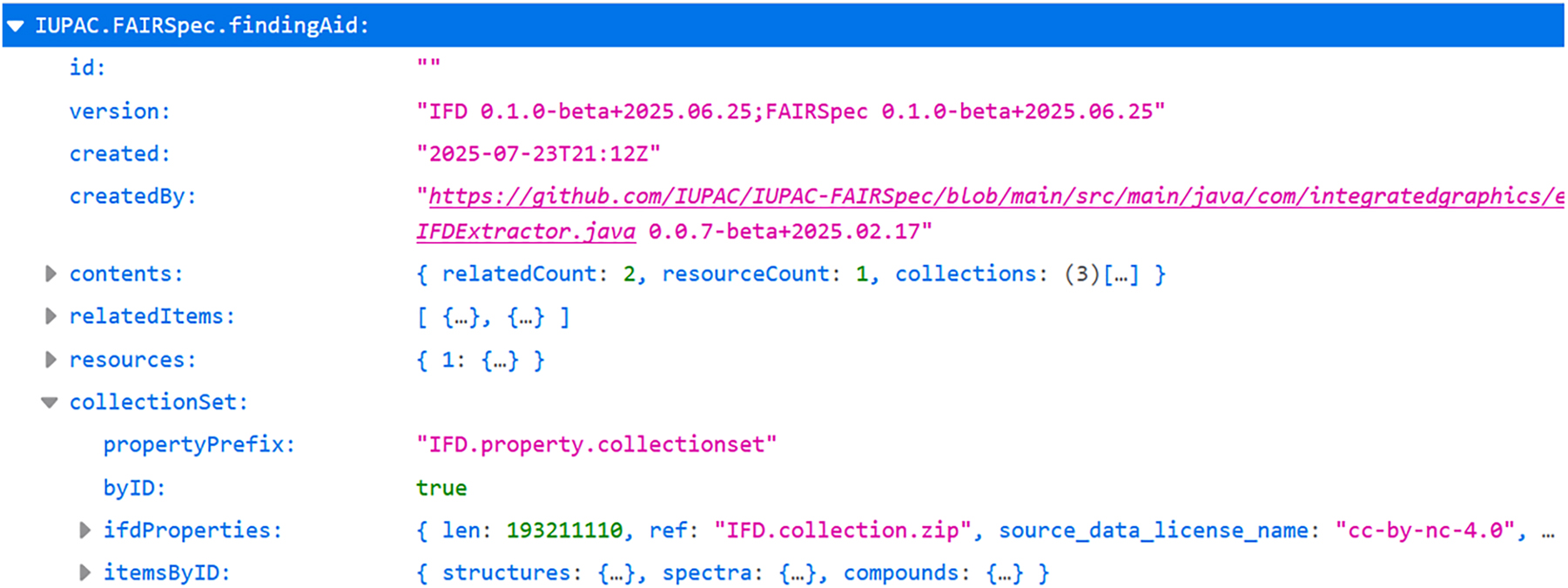
The top-level view of an IUPAC FAIRSpec Finding Aid as JSON as displayed in a web browser. The document was created by an automated process that extracted metadata from a single ZIP file associated with a publication and contains compound-based structure-spectra associations.

The top of a web page designed to interpret IUPAC FAIRSpec Finding Aids. In this case, the web page reads the IUPAC FAIRSpec Finding Aid shown in Fig. 3. Clicking the search link allows text, substructure, and property searching of the collection in relation to compounds, structures, and spectra.
Fig. 5 and 6 illustrate the sort of interpretation that a relatively simple web-based landing page might make of structure and spectrum entries, respectively.
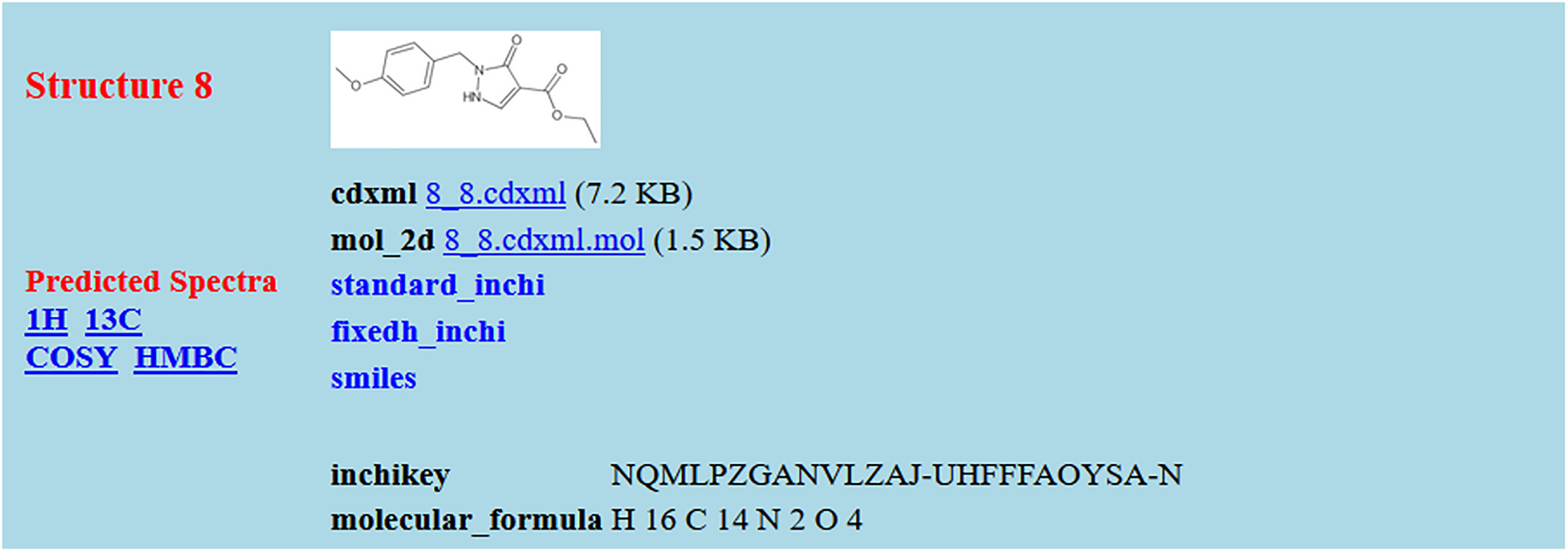
An example of a structure entry based on the presence of a single CDXML file in the FAIRSpec-ready collection. The automated extractor has added a PNG image, a MOL file, standard and fixed-H InChIs, InChIKey, SMILES, molecular formula, and links to interactive predicted spectra.
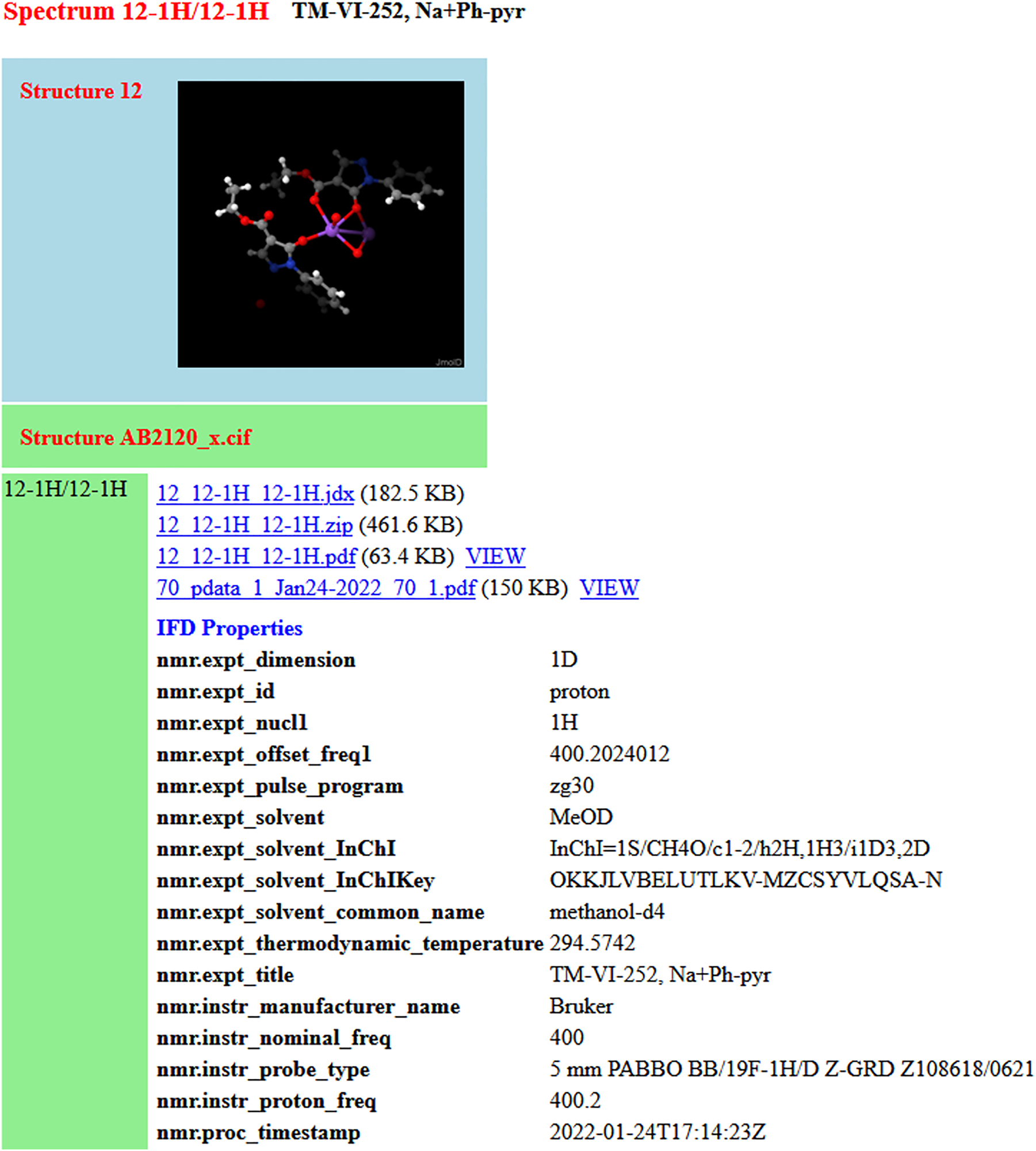
A data entry including an x-ray structure determination and an NMR spectrum with several representations. IFD Properties are extracted from the FAIRSpec-ready collection automatically in the process of creating the IUPAC FAIRSpec Finding Aid. PDF representations are viewable within the web browser. (Interactive spectral representations might also be available via a web browser, but that feature was not implemented in this particular case.)
In principle, except for the header, any combination of the additional sections is possible. Thus, a FAIRSpec-ready collection could be as simple as a single structure representation and a single spectrum or as complicated as the full set of structures, spectra, and analyses associated with a project or publication.
4.2 The FAIRSpec-ready collection
The automated process of adding an IUPAC FAIRSpec Finding Aid to a FAIRSpec-ready collection creates an IUPAC FAIRSpec Data Collection, which can then be searched and interpreted as shown above. The goal, then, is to create well-characterized FAIRSpec-ready collections, whether they be sample- or compound-based collections for use within a research group during the course of a project, or “final” collections associated with publications. The guidelines presented here for such FAIRSpec-ready collections have the following features:
The most important characteristic of a FAIRSpec-ready data collection is that it is organized systematically and consistently. In principle, any systematic organization that conveys appropriate relationships can be processed to become an IUPAC FAIRSpec Data Collection with an associated IUPAC FAIRSpec Finding Aid. Nonetheless, the organizational principles described herein specifically illustrate a small number of suitable FAIRSpec-ready data collection organizations.
Organization will depend upon context. The FAIRSpec-ready Data Collection created on the day a dataset is generated will likely look quite different from one that ultimately is used in creating the IUPAC FAIRSpec Data Collection associated with a publication. At the beginning of an endeavor, for example, there is typically just a physical sample. A spectrum is taken. There may be the expectation of a certain structure, but perhaps not. If nothing else, it is hoped that the spectrum will at least support a hypothesis relating to chemical structure. It may be the case that the structure is truly unknown, and it is only after the spectrum is analyzed that it is “known”. (More precisely, only with the help of spectroscopy can the structure be hypothesized.) Thus, an initial collection may be just one or more spectra and their associated sample identifier. One or more (plausible) structures are added a week later. More spectra are taken. More samples are created and analyzed. Everything is sample-based at first, but then, later, the key organizing principle shifts to chemical structure – chemical “compounds”. Ultimately, upon publication, the key organizing principle will be “compound number in the article,” and only selected spectra will be included. Importantly, the underlying data have not changed. (We hope!) Only the organization has changed. And, importantly, the FAIRSpec-ready collection has probably not changed at all, except for some key metadata.
Extractor utilities may impose their own conventions. Individual tools developed to extract metadata from a data collection to create an IUPAC FAIRSpec Data Collection and its associated IUPAC FAIRSpec Finding Aid may develop their own specific requirements that go beyond what is suggested here. However, an FAIRSpec-ready data collection must follow the conventions described here. Metadata tools that refer to themselves as “FAIRSpec-ready extractors” may enforce occurrences of “should” in what follows as “must” but may not impose additional restrictions that contradict these conventions. For example, an extractor might allow Unicode characters outside the specified range for identifiers, but if it does, it must change those characters to the allowed set for identifiers described for IUPAC FAIRSpec Data Collections.
Unique identifiers should be used for samples, structures, and datasets. One characteristic of the IUPAC FAIRSpec Finding Aid is that it involves “pointing” to digital representations. This is analogous to file names in a file system – and, for that matter, may be exactly that. For example, a compound might have a name “C3” as its identifier; an NMR dataset might be referred to as C3_H1_NMR. It is not important that these identifiers be semantic – that is, that they convey meaning, such as this example does – and, in fact, it is often desirable that they not be descriptive. They simply need to be unique. The following guidelines for identifiers should be followed:
Characters used throughout the metadata should be represented in the UTF-8 character set. This is the standard encoding on the web and in many software applications, allowing for the full range of international language characters.
Characters specifically in relation to identifiers should be limited to 7-bit ASCII characters. IUPAC FAIRSpec Finding Aids utilize string-based identifiers for cross-referencing digital objects. For flexibility of processing, identifiers should only utilize simple alphanumeric [A-Za-z0-9] ASCII characters along with a limited set of punctuation, namely “+-’.,_()[]”. Single spaces are allowed only if not leading or trailing; multiple sequential spaces characters, tab, and new-line characters are not allowed within identifiers.
The organization of files should reflect the types of spectroscopy used. Subdirectories such as “NMR”, “IR” (infrared), “UVVIS” (ultraviolet–visible spectroscopy), and “HRMS” (high-resolution mass spectrometry) can assist in both human and machine readability. These specific examples of analytical techniques are not meant to be exclusive. The IUPAC FAIRSpec Finding Aid in principle can describe any spectroscopic (or non-spectroscopic) dataset. The key for FAIRSpec-ready collections is that whatever techniques are described, they are described consistently within the collection. We focus primarily on NMR spectroscopy in this report simply because we have had more experience with that in our working group to date.
Initially, data should be organized by unique sample identifiers. If a collection is being created at a stage in the research endeavor when the compound’s structure has not been identified, the appropriate association is to a specific sample. This might be a unique identifier automatically created by an ELN or some encoding used by a research group internally. For example, “RMH-III-23.rf-0.65” might be sufficient immediately after chromatographic isolation, with no structure representations provided.
Thus, we might have a sample-based data collection organized by instrumental method and sample ID:
NMR/
RMH-IV-13b/
RMH-IV-13c/
RMH-IV-13d/
IR/
HSR-II-112/
HSR-II-113/
HSR-II-114/
or, alternatively, the other way around:
RMH-IV-13b/
NMR/
IR/
RMH-IV-13c/
NMR/
IR/
RMH-IV-13d/
NMR/
IR/
HSR-II-112/
NMR/
IR/
HSR-II-113/
NMR/
IR/
HSR-II-114/
NMR/
IR/
etc.,
where unique laboratory sample identifiers are used instead of compound identifiers, because at the stage in the process, that is all that is available. Whatever system is used, it must be systematic, with unique identifiers (think “file paths”) for each item in the collection.
Alternatively, if an instrument vendor or ELN provides a means of associating a sample ID with a spectrum, this should be used, as it will provide a well-characterized method of automatically associating a sample ID with spectral data. The common method of working a sample ID into a title field is not advised, because, though it might be easily human readable, co-opting a title field to indicate something as important as a sample ID is fundamentally unsound.
At the point of publication, some representative subset of the sample-based FAIRSpec-ready collection will be used to make a publication-oriented compound-based FAIRSpec-ready collection. This allows for the natural relationship of “structure of a compound”, and “spectrum of a compound”, with a direct relationship to typical publications in the field. For example:
1a/
NMR/
1H-NMR/
13C-NMR/
2a/
NMR/
1H-NMR/
13C-NMR/
Where now “1a” and “2a” are references to compounds described in a specific publication. Here we have unique identifiers of the form 2a/NMR/1H-NMR/xxxx. The file system-like hierarchy provides unique identifiers for all representations. Note that the file paths shown in this example are themselves semantic. The file path itself is conveying the relationship between a structure and spectrum that will become codified in the IUPAC FAIRSpec Finding Aid. This is not absolutely necessary, but it is advisable both in terms of human readability and ease of machine processing. Only selected representative spectra will be included, often from more than one sample of a given compound.
Organizing by compound or sample identifier could be facilitated by ELNs that allow such modes. We feel that the benefits of doing so, including the ELN-based creation of the IUPAC FAIRSpec Finding Aid based on chemical compounds, would be considerable. This principle could also be included in the specifications for future designs of “IUPAC FAIRSpec-Compliant” ELNs.
The organization of files should minimize the duplication of information. Data collections are often used in multiple contexts. Ongoing research collections are generally sample-based and created long before any knowledge of final publication compound numbers. Data published for one manuscript, with a specific set of compound numbers, might also be referred to in another manuscript, with different compound numbers. If maximum flexibility is desired, paths such as “3a/3a-NMR/3a-1HNMR” should be avoided, using simply “3a/NMR/1HNMR” instead. In this way, if “3a” is not the final compound number in the publication, or the data are copied to a collection for a different publication, there is only one directory to rename.
Associating a FAIRSpec-ready collection with chemical structure is a straightforward process. At whatever point in a research program the identity of an isolated or reference compound is established, the association of spectrum with a sample can remain, but an additional association with a chemical structure becomes appropriate. This association can be simplicity itself. All that is needed is a well-crafted (see Section 2) CDXML or MOL representation. The next several points discuss simple methods to accomplish this association.
Structure representations can be added along the path to a dataset. One of the simplest ways to associate structures with spectra is to place a structure representation within the directory path leading to the spectral data. Thus, using the example given above, we might have:
RMH-IV-13b/
structure.cdxml
NMR/
IR/
UVVIS/
HRMS/
RMH-IV-13c/
structure.cdxml
NMR/
IR/
UVVIS/
RMH-IV-13d/
structure.cdxml
… etc.
This is enough to make the key association between the structure and spectrum in simple cases and has the advantage that changes in structure assignment are trivial to implement. Just replace the file.
Structure representations can be integrated into datasets if desired. Some vendors allow one or more structure files to be added to an instrument dataset. For example:
RMH-IV-13b/
NMR/
NMR-2024.04.15a/
structure.mol
NMR-2024.04.15b/
structure.mol
…etc.
This can work, but it suffers from the issue that there is potential for considerable unnecessary duplication. Should the structural hypothesis be modified, then we have the error-prone challenge of modifying all of the structural representations within this directory path as well. Nonetheless, this is a standard way to introduce structures into datasets that is recognized also by some vendors within their own software. Thus, a structure introduced for display in the vendor’s system can be effortlessly introduced also into an IUPAC FAIRSpec Finding Aid.
Structure representations can be placed in a parallel set of directories. Perhaps the cleanest way to associate structures with spectra is to keep them separate but to provide associated identifiers. This could be compound based, as in:
data/
RH0013/
NMR/
IR/
RH0014/
NMR/
IR/
… …
structures/
RH0013/
structure.cdxml
RH0014/
structure.cdxml
…
This method has the advantage that there is no duplication. It is particularly useful for compound-based collections. Unique local lab-based compound identifiers (e.g., RH0013) make the metadata connection between structures and spectra. A modification of a structure will be registered for all its associated spectra automatically. At some later date, a correlation can be made between the lab-based compound identifiers and the compound numbering used in a specific publication.
Compound associations that are mixtures preferably should identify as a mixture using a “+” sign and include separately identified structure representations, one for each component of the mixture, within a “structures” subdirectory. Thus:
3c+3c’/
structures/
3c.cdxml
3c’.cdxml
NMR/
…
This is enough to make it clear that the multiple CDXML files are of different structures, as opposed to simply alternative representations of the same compound.
Alternatively, and less preferred, if the context demands, isomeric mixtures may be described by ambiguous structure representations. In special cases, there might be little or no practical relevance of the exact stereochemistry of a structure. In that case, no special effort should be expended to detail it. For example, if the manuscript refers to “Compound 3c” as having a tetrahydopyranyl protecting group (which has a stereocenter) and no analysis was carried out that determined the stereochemistry of this group, a single CDXML file with no stereochemistry indicated (just the nickname “THP”) can be provided. However, if the manuscript refers to “Compound 3c” as a “3:1 mixture of E/Z isomers”, then two separate CDXML files, one Z and one E must be provided. It is not appropriate to show a single structure with ambiguous notation or to give structures for both isomers in the same file. We need the extraction process to identify both isomers independently, without the need to search files for multiple structures. It is not necessary to describe the extent of the mixture within the structural context. Metadata can be added later to the compound association itself annotating the specifics of the mixture. Future guidance may allow for a more systematic way of describing mixtures, for example, with a mixtures InChI (MInChI) 44 , 45 or using a method designed specifically for IUPAC FAIRSpec Finding Aids.
5 Addition of curated metadata to a FAIRSpec-ready collection
5.1 Internal metadata records
While structure and instrument data representations in a spectroscopic data collection can be significant sources for the automated extraction of descriptive metadata, not all descriptive metadata can be gathered this way. In addition, relational metadata records, such as related sample identifiers, are less easily produced or extracted. In addition, by its very nature, a relational metadata record is not associated with one specific digital item. Thus, its proper place is not “within” any of the digital items it relates to. For example, we have seen how a file structure itself can be the basis of relational metadata. (“This structure is associated with this spectrum because they are in the same directory.”) But, in general, addition of relational metadata will require curation.
Ultimately, key/value metadata associated with an IUPAC FAIRSpec Data Collection (not just a FAIRSpec-ready one) will be expressed primarily in terms of the IUPAC FAIRData (IFD) Model-specified standard. Thus, for example, descriptive metadata associated with an NMR experiment in an IUPAC FAIRSpec Finding Aid might appear as shown in Fig. 7, where the full key name for nmr.expt_solvent, for example, is

Descriptive metadata associated with a Bruker NMR instrument dataset as found in JSON format in an IUPAC FAIRSpec Finding Aid. Several of the values are generated during metadata extraction, including additional solvent identifiers and the spectrometer nominal frequency. Note that units for numerical values are given in the specification of the finding aid, not the finding aid itself. The precision of these numbers does not necessarily represent the experimental precision due to limitations of the JSON format as well as limitations of the originating vendor metadata. For an unambiguous description of data precision, one must inspect the data objects themselves, not just the finding aid.
IFD.property.dataobject.fairspec.nmr.expt_solvent.
It is not expected that a FAIRSpec-ready collection utilizes such standardized keys. And, in fact, it is preferred that metadata such as these, that can be extracted automatically from an instrumental dataset via automation, not be provided explicitly in a FAIRSpec-ready collection at all. For example, the solvent in the originating primary dataset was given as “MeOD”. The extraction software has matched that to an InChI, a SMILES, and a common name, improving searchability of the collection.
It is important to understand that the IUPAC FAIRSpec Finding Aid allows for both standardized and “ad hoc” properties (listed in an IUPAC FAIRSpec Finding Aid as “attributes”). Thus, additional metadata that does not fit easily into the standard can always be added. (Such metadata simply would not have any IFD.property prefix.) To the extent that ad hoc metadata key/value pairs can be mapped to current or future IUPAC FAIRData Standard pairs, that mapping would be done later, during the automated or semi-automated curation process of metadata extraction. Examples of attributes from a third-party software representation of a spectrum are shown in Fig. 8.

Attributes extractable from a third-party vendor that may or may not have equivalences as IFD.property items. Some of these values correlate directly with IFD dataset properties, but many do not. Though not standardized, these additional metadata may be valuable within certain contexts.
We provide here suggestions for adding additional ad hoc metadata based on our implementation tests.
Consistency is important. In all cases, identifiers and keys should be unique and consistently expressed (including spelling and capitalization). Values should use a consistent, if not standardized, vocabulary.
Point-specific metadata files can provide additional descriptive or relational metadata not easily otherwise conveyed. Key: value metadata pairs can be listed in a simple text file accompanying structures or data within a collection. For instrument datasets, this is already the practice of some spectrometer vendors, who have adopted a format resembling the IUPAC JCAMP-DX format, storing metadata specific to that dataset in the form of “private” ##$KEY=VALUE pairs. A generalized format could just be a collection of single lines of the form key=value, (popularized in Java properties files) where the keys should be systematically defined and consistent throughout the collection. For example, If the file metadata.properties (in the form of a Java properties file) containing only
sample_id=RMH-IV-23c
were added to an instrument dataset, then later automated curation could codify that reference as an IFD.property.dataobject.originating_sample_id and use that to create an IUPAC FAIRData Sample object with IFD.property.sample.id “RMH-IV-23c”, thus associating the specified sample with this dataset.
Such a file (perhaps created automatically by an ELN) for a specific structure representation might contain additional metadata such as:
smiles=CC1=CCC(CC1)C(=C)C
inchi=InChI=1S/C10H16/c1-8(2)10-6-4-9(3)5-7-10/h4,10H,1,5–7H2,2–3H3
chebi_id=CHEBI:15384
mf=C10H16
that could be used to complement automated extraction. In addition, such explicitly added metadata can be used to validate accompanying structure representations such as MOL or CDXML files, confirming that the machine reading of a structure has been successful.
Such a file containing
structure_stereochemistry=relative
contained in a structure directory or even in one of the top levels of the collection would be enough to convey the understanding that all chiral structures unless otherwise indicated are to be considered racemic.
A well-organized “primary” spreadsheet can efficiently convey metadata relationships. Metadata items are essentially key:value pairs. A convenient way to represent such pairs is the standard spreadsheet practice where column headers are keys, and each row contains the set of values associated with a given item (generally identified in the first column). Particularly for a whole collection, a primary internal metadata record in the form of a spreadsheet can be efficient. Standard open file formats such as CSV (comma-separated values), 46 TSV (tab-separated values), 47 ODS (OpenDocument spreadsheet), 48 or XLSX (Office Open XML SpreadsheetML file format), 49 are easily extractable via automation for the metadata they contain.
Thus, we might have something like what is shown in Fig. 9, associating publication compound identifiers with lab-local identifiers, database identifiers, and repository PIDs. When (or if) incorporated into an IUPAC FAIRSpec Finding Aid, these properties would be either mapped to standardized IUPAC FAIRSpec metadata keys or included as additional properties.
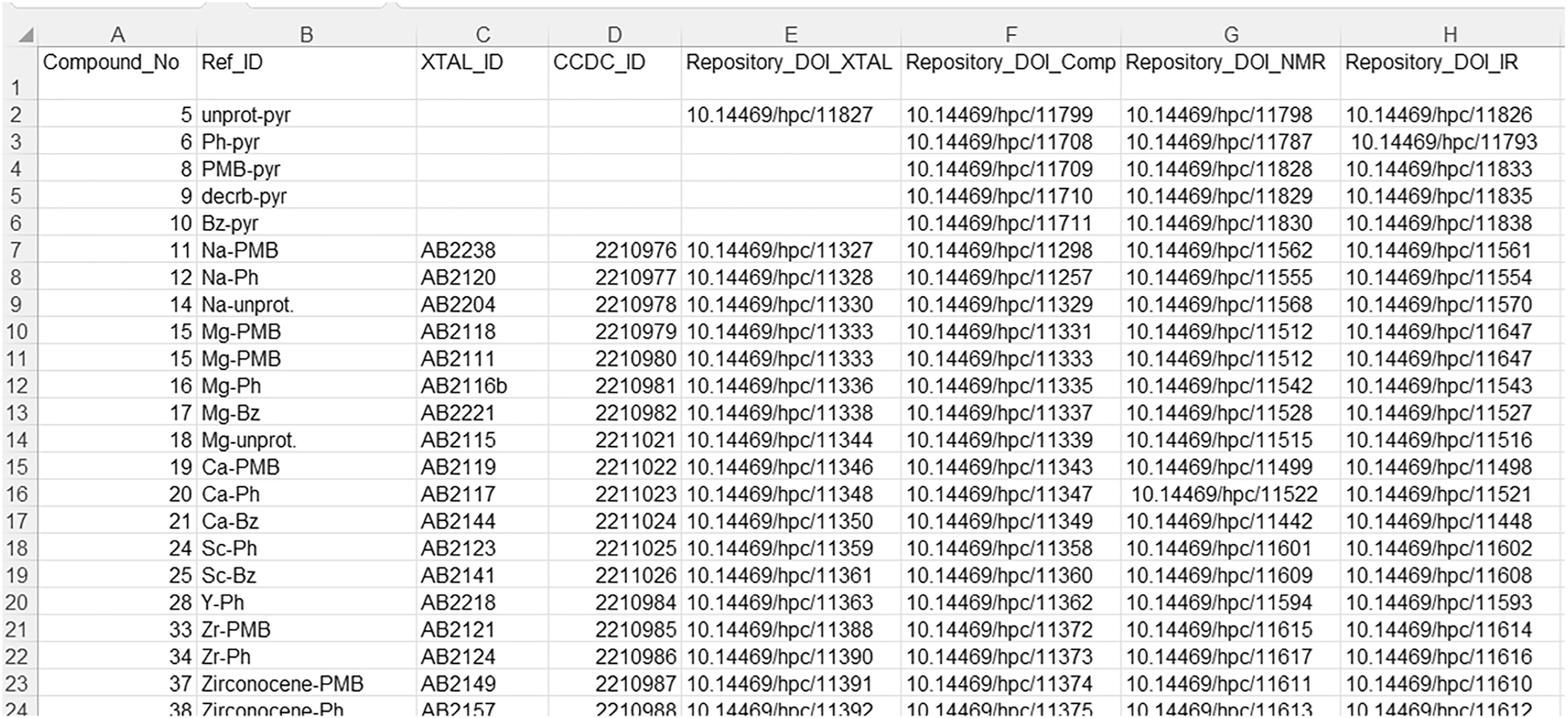
A page in a spreadsheet with metadata that can be extracted using automation for additional metadata attributes associated with compounds in a published IUPAC FAIRSpec Data Collection. The spreadsheet provides both human- and machine-readable content.
5.2 Registered metadata records
Quite possibly, each collection ultimately might be associated with a primary registered metadata record conforming to a declared schema such as the DataCite Schema. 50 This metadata record allows the connection to be made to additional metadata records as appropriate both within the collection and to associated works. These metadata records should be as chemically rich as possible. An example of how a single DOI reference can be used to generate a full IUPAC FAIRSpec Finding Aid for a collection is given on the FAIRSpec GitHub pages. 51
Registering a metadata record for individual instrumental datasets allows for the formation of a unique PID to be associated with individual parts of the dataset, enabling a higher probability of findability via automated search engine “bots”. Such registered records should include the unique path or the database reference to the exact location of the dataset itself - whether located on an institutional, specialist, or generalist repository – thus enabling potentially widespread accessibility to the dataset itself. This also allows for distributed data storage, and it also can include selective privacy settings for both pre- and post-publication.
The criteria for selecting an appropriate repository for registering the dataset metadata record should include some consideration of whether the entry of rich chemical metadata is adequately supported either by the repository human user interface or by a repository application programming interface (API). APIs are essential for use by automated systems such as an ELN. The repository should include processes for automatic generation and then registration of the primary metadata record with an appropriate authority, where the master copy will be kept and indexed to facilitate findability. Any local repository copy of the master metadata record should automatically be kept synchronized with the registered version.
Ideally, the repository would offer an option to produce IUPAC FAIRSpec Finding Aids for its various collections, and these would also be registered with an agency as part of the overall collection. If the repository is associated with a database, it could also produce specialized IUPAC FAIRSpec Finding Aids in response to search queries as a way of standardizing API calls among various repositories, building them only as needed in response to queries.
A more in-depth discussion of metadata registration optimized for spectroscopy will be presented in a future publication that discusses specific implementations that we have worked on during the development of these guidelines.
6 Guidance for managers of laboratory instrument management systems and electronic laboratory notebooks
Facilities implementing laboratory instrument management systems (LIMS) and/or ELNs require specific guidance. When a laboratory user creates a spectroscopic dataset within an electronic tracking environment, they may not have (or want!) access to the raw data files themselves. In addition, in principle, these systems are generally a major part of the workflow all the way from acquisition to publication (and possibly beyond, if a public institutional data repository is also part of the system). In such cases, it may be that an experimentalist might hand-curate a collection by retrieving datasets and structures, creating “personal” file-based collections such as described in Section 4.2. But that is not ideal.
Effectively, a LIMS or ELN presents a unique opportunity, with data-management capabilities potentially already maintaining a FAIRSpec-ready collection. Spectra are keyed to sample IDs; sample provenance is well described. Structures are allowed to be associated with spectra by internal database associations. Metadata intended for local searching of the database is likely already being extracted. All of the necessary components are there. What is needed is a mechanism for generating an IUPAC FAIRSpec Finding Aid automatically from such a system.
Managers and developers of such systems are encouraged to read these guidelines, provide guidance for future IUPAC efforts in this area, and ensure that their systems are “FAIRSpec-ready” – that they maintain sufficient capabilities to export sample, structure, and spectroscopic metadata along with the spectroscopic data itself in a manner that is consistent with these guidelines. In addition, these systems need to allow for appropriate inputs. For example, users need to be trained and encouraged to create conforming structural representations (as described in Section 2) so that when the time comes, a clear and unambiguous association can be made between structure and spectrum.
Software development in this area will be important and potentially quite valuable. A LIMS or ELN that exports an IUPAC FAIRSpec Finding Aid that includes API references to data objects in its database can significantly improve the system’s potential for overall findability, accessibility, interoperability, and reusability, both at the private/local and public/global levels. Thus, an ELN could automatically create local IUPAC FAIRSpec Finding Aids. These could provide real-time snapshots of the stored sample-based data focused on specific compounds or reactions, research group members, or any other association that can be queried. To the extent that these references are later redirected to a public institutional or generalist repository database, publication-ready compound-based products such as full data packages with accompanying IUPAC FAIRSpec Finding Aids could be generated automatically. In fact, provided the references are to a public repository, all that would need to be published alongside a manuscript would be the PID of a landing page for an IUPAC FAIRSpec Finding Aid. The data would never have to be “collected” in the form of a monolithic data file at all.
Finally, LIMS and ELN production of IUPAC FAIRSpec Finding Aids for local use can be the basis for straightforward exchange of data and metadata among otherwise disparate systems, since an IUPAC FAIRSpec Finding Aid can express any additional metadata that is needed, not just “IUPAC-defined” metadata.
7 Summary
We have provided a set of guidelines for the FAIR management of spectroscopic data collections specific to the domain of chemistry. These guidelines cover the generation of machine-readable structure and dataset representations, the organization of what we refer to as the FAIRSpec-ready collection, and suggestions for ways to incorporate additional metadata into the collection. This collection optimally would be able to be curated automatically by software to create an IUPAC FAIRSpec Finding Aid.
We have emphasized that private local FAIRSpec-ready collections can be developed automatically or semi-automatically concurrently with laboratory research, not just at the time of publication. The potential benefits of this “best practice” include the ability on an ongoing basis, with or without use of an ELN or LIMS, structure or substructure searches for related spectroscopic data, the ability to filter a collection for spectra of a certain sort or with a given set of property values, and many other possibilities. In fact, the local benefit of even the most minimal curation (just associating instrument datasets and analysis with specific samples or chemical structures) can be significant.
Researchers, authors, and data managers do not have to wait until the overall IUPAC FAIRSpec Metadata Model is formalized or do they ever have to become experts in its implementation to benefit from these simple guidelines. The key premise here is that a small amount of forward-thinking organization and consistency can go a long way to optimizing the findability, interoperability, accessibility, and reusability of spectroscopic data collections. As a bonus, publishing of IUPAC FAIRSpec Data Collections and their associated IUPAC FAIRSpec Finding Aids allows for longer-term and more diverse exposure for both researchers and their publications and allows for the future reuse of data for purposes in creative ways not imagined by their originator.
-
Research ethics: Not applicable.
-
Informed consent: Not applicable.
-
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
-
Use of Large Language Models, AI and Machine Learning Tools: None declared.
-
Conflict of interest: The authors state no conflict of interest.
-
Research funding: None declared.
-
Data availability: Not applicable.
Appendix: Terminology
In this appendix, we refer to several terms that have multiple meanings in common usage but specific meanings within this context:
Compound association
A metadata object with a unique identifier associating one or more spectra with one or more structures within an IUPAC FAIRSpec Data Collection.
Note 1: Definitions of “compound” abound. The National Cancer Institute defines a compound as “a substance made from two or more different elements that have been chemically joined”. 52 PubChem describes a “Compound record” as pointing to “at least one Substance record”. 53 The IUPAC Gold Book defines a “racemic compound” as a “crystalline racemate in which the two enantiomers [chirally related “molecular entities”] are present in equal amounts in a well-defined arrangement within the lattice of a homogeneous crystalline addition compound”. 54 Common parlance in published works in the area of organic and inorganic chemistry refer to “Compound XXX, which was a mixture of diastereomers” and “pure compounds” (implying “a compound” can be “impure”). A search at an international patent site for “polymer compound” within the “front page” field returns over 1300 results. 55 Notice that none of these definitions define specifically what makes one compound different from another. None answer some of the most basic questions: Can a compound be a mixture of compounds? Is a mixture of polymers a compound? We opt in these guidelines for a practical, inclusive definition of compound specifically in the context of compound associations.
Note 2: The unique identifier may be as simple as a sequential number – 1, 2, 3, …. Alternatively, the originator of the collection might choose to provide a meaningful identifier within the context of the collection. For example, the compound association ID might refer to a compound number in a related publication (“2a”) or a mixture of compounds (“2a+2b”) associated with one or more spectra. Within an association that relates to more than one structure, the structure representations referred to in the association will provide the details of that relationship – whether these are diastereomers, enantiomers, or completely constitutionally different compounds. If a mixture, regardless of the ID, the structure representations will provide the details of that relationship – whether these are diastereomers, enantiomers, or completely constitutionally different compounds.
Dataset
A digital item or a collection of digital items that is derived from laboratory analysis (see Instrumental Dataset, below) or from computation. The two general forms of computed datasets are spectroscopic predictions based on one or more proposed chemical structures (such as can be generated at the nmrdb.org website 56 ) and simulations based on experimental or predicted parameters, such as NMR frequencies, chemical shifts, and coupling constants.
Digital item
A collection of bytes that may be local to a digital collection or may be part of a remotely accessed collection.
Note 1: Digital items are commonly implemented as “files” on a filesystem or named items within a compressed archive (for example, an archive with ZIP, TAR, or TGZ format).
Digital object
A digital item that has associated metadata.
Note 1: In the current context, we make the distinction between the digital items in FAIRSpec-ready data collections that do not have associated IUPAC FAIRSpec metadata, and digital objects in IUPAC FAIRSpec Data Collections, which do.
Instrument dataset
A dataset that comprises the raw or minimally transformed data arising from laboratory analysis.
Example 1: A file directory containing NMR FID data or real- and imaginary-valued transformed data, parameter files, and accompanying metadata.
Example 2: A ZIP archive of such a file directory.
IUPAC FAIRSpec data collection
A collection of digital objects referenced by an IUPAC FAIRSpec Finding Aid and consisting of one or more representations relating to sample, structure, spectral data, or analysis, and conforms with specifications as determined by IUPAC.
Note 1: An IUPAC FAIRSpec Data Collection is generally sample-based or compound-based, depending upon its contexts. Sample-based collections make clear associations between laboratory samples and spectral data on a one-to-many basis. Compound-based collections associate specific chemical structures with spectral data on a many-to-many basis.
Note 2: An IUPAC FAIRSpec Data Collection need not be limited to the types of representation mentioned in Note 1. Any sort of data or metadata may be included. For example, data need not be limited to experimental spectroscopic data. Results of calculations and simulations can be part of the collection. The expectation, however, is that such results are ultimately relevant to spectroscopy.
IUPAC FAIRSpec Finding Aid
A digital item consisting of metadata that summarizes the contents of an IUPAC FAIRSpec Data Collection listing and making associations among representations relating to sample, structure, spectral data, and analyses, and conforms with specifications as determined by IUPAC.
Note 1: In general, a finding aid is a document that assists users in locating items in a digital archive. A widely used standard developed by the US Library of Congress exists for archive-related digital finding aids and is used extensively throughout the archival community. 57 We extend that concept to locating digital items in a spectroscopic data collection.
Note 2: An IUPAC FAIRSpec Finding Aid need not be limited to IUPAC-defined metadata. The format allows for any amount of additional metadata to be included.
Note 3: An IUPAC FAIRSpec Finding Aid may contain representations as well as metadata. The format allows for reasonably short representations such as character strings, images, and individual-spectrum PDF documents to be integrated directly into the finding aid for better findability and reusability.
Persistent identifier
A publicly registered character string providing a long-lasting reference to a digital object and its associated metadata.
Example 1: 10.1515/pac-2021-2009
Representation
A digital object present within a collection and identifiable using an identifier that is unique within the context of the collection.
Note 1: Representations may be individual digital items within a collection referenced by the IUPAC FAIRSpec Finding Aid. Alternatively, they can be character strings (including Base64-encoded 58 byte arrays) that are contained within the finding aid itself.
Example 1: A MOL file may be present within the collection and have the unique file name “3aa/structures/3aa.mol”
Example 2: A PNG (portable network graphics) image may reside as a file or be included within an IUPAC FAIRSpec Finding Aid as a Base64-encoded string so that is more easily accessible or because it has been extracted from a more complex dataset.
Example 3: A SMILES string representation such as “C(O)CCC” need not be in a file of its own; it can be provided in the IUPAC FAIRSpec Finding Aid as the data value of an IFD.representation.structure.SMILES representation.
List of abbreviations
- API
-
Application programming interface
- ASCII
-
American standard code for information interchange
- CDX
-
ChemDraw exchange format
- CDXML
-
ChemDraw XML format
- DOI
-
Digital object identifier
- ELN
-
Electronic laboratory notebook
- ESI
-
Electronic supplementary information
- FAIR
-
Findable, accessible, interoperable, and reusable
- FID
-
Free induction decay
- HELM
-
Hierarchical editing language for macromolecules
- HRMS
-
High-resolution mass spectrometry
- IFD
-
IUPAC FAIRData
- IR
-
Infrared
- JCAMP-DX
-
Joint Committee on Atomic and Molecular Physical Data-Data Exchange format
- JSON
-
JavaScript object notation
- LIMS
-
Laboratory instrument management system
- MDL
-
Molecular Design Limited
- MInChI
-
Mixtures InChI
- mmCIF
-
Macromolecular crystallographic information file
- MOL
-
Molfile
- NMR
-
Nuclear magnetic resonance
- ODS
-
OpenDocument spreadsheet
-
Portable document format
- PID
-
Persistent identifier
- PNG
-
Portable network graphics
- SDF
-
Structure-data file
- SMILES
-
Simplified molecular input line entry system
- URL
-
Uniform resource locator
- UTF-8
-
Unicode transformation format – 8-bit
- UVVIS
-
Ultraviolet–visible spectroscopy
- XLSX
-
Office Open XML SpreadsheetML
- XML
-
Extensible markup language
References
1. Development of a Standard for FAIR Data Management of Spectroscopic Data. Project Details. https://iupac.org/project/2019-031-1-024 (accessed 2025-07-29).Suche in Google Scholar
2. IUPAC/IUPAC-FAIRSpec GitHub project. https://github.com/IUPAC/IUPAC-FAIRSpec (accessed 2025-07-29).Suche in Google Scholar
3. Several Examples of IUPAC FAIRSpec Finding Aids and Their Associated Landing Pages Can Be Found at IUPAC/IUPAC-FAIRSpec GitHub Project Site Web Pages. https://iupac.github.io/IUPAC-FAIRSpec (accessed 2025-07-29).Suche in Google Scholar
4. Hanson, R. M.; Jeannerat, D.; Archibald, M.; Bruno, I. J.; Chalk, S. J.; Davies, A. N.; Lancashire, R. J.; Lang, J.; Rzepa, H. S. IUPAC Specification for the FAIR Management of Spectroscopic Data in Chemistry (IUPAC FAIRSpec) – Guiding Principles. Pure Appl. Chem. 2022, 94 (6), 623–636. https://doi.org/10.1515/pac-2021-2009.Suche in Google Scholar
5. Chapter II: Proposal Preparation Instructions - Proposal & Award Policies & Procedures Guide (PAPPG) (NSF 23-1) | NSF – National Science Foundation. 2023. https://new.nsf.gov/policies/pappg/23-1/ch-2-proposal-preparation (accessed 2025-07-29).Suche in Google Scholar
6. Dissemination and Sharing of Research Results | NSF – National Science Foundation. https://www.nsf.gov/funding/data-management-plan#:∼:text=funded%20investigators%20are%20expected%20to%20share (accessed 2025-07-29).Suche in Google Scholar
7. CHE Data Management and Sharing Plan Guidance. https://www.nsf.gov/mps/che/data-management-sharing-plans (accessed 2025-07-29).Suche in Google Scholar
8. Data sharing: Guidance for Best Practice and Reproducibility of Experimental Data, Royal Society of Chemistry https://www.rsc.org/publishing/publish-with-us/publish-a-journal-article/data-sharing (accessed 2025-07-29).Suche in Google Scholar
9. Research Data Sharing, Springer Nature https://support.springernature.com/en/support/solutions/folders/6000238326 (accessed 2025-07-29).Suche in Google Scholar
10. ACS Research Data Guidelines https://researcher-resources.acs.org/publish/data_guidelines#organic_chem_data (accessed 2025-07-29).Suche in Google Scholar
11. US Government Code of Federal Regulations 21 CFR 11.10 Electronic Records – Controls for Closed Systems https://www.ecfr.gov/current/title-21/chapter-I/subchapter-A/part-11/subpart-B/section-11.10 (accessed 2025-07-29).Suche in Google Scholar
12. Patel, K. T.; Chotai, N. P. Documentation and Records: Harmonized GMP Requirements. J. Young Pharm. 2011, 3 (2), 138–150. Available as a National Library of Medicine online article as https://pmc.ncbi.nlm.nih.gov/articles/PMC3122044 (accessed 07 29, 2025).10.4103/0975-1483.80303Suche in Google Scholar PubMed PubMed Central
13. Wilkinson, M. D.; Dumontier, M.; Aalbersberg, Ij. J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L. B.; Bourne, P. E.; Bouwman, J.; Brookes, A. J.; Clark, T.; Crosas, M.; Dillo, I.; Dumon, O.; Edmunds, S.; Evelo, C. T.; Finkers, R.; Gonzalez-Beltran, A.; Gray, A. J. G.; Groth, P.; Goble, C.; Grethe, J. S.; Heringa, J.; ’t Hoen, P. A. C.; Hooft, R.; Kuhn, T.; Kok, R.; Kok, J.; Lusher, S. J.; Martone, M. E.; Mons, A.; Packer, A. L.; Persson, B.; Rocca-Serra, P.; Roos, M.; van Schaik, R.; Sansone, S.-A.; Schultes, E.; Sengstag, T.; Slater, T.; Strawn, G.; Swertz, M. A.; Thompson, M.; van der Lei, J.; van Mulligen, E.; Velterop, J.; Waagmeester, A.; Wittenburg, P.; Wolstencroft, K.; Zhao, J.; Mons, B. The FAIR Guiding Principles for Scientific Data Management and Stewardship. Sci. Data 2016, 3 (1), 160018. https://doi.org/10.1038/sdata.2016.18.Suche in Google Scholar PubMed PubMed Central
14. The FAIR2 Specification https://www.fair2.ai/specification (accessed 2025-07-29).Suche in Google Scholar
15. DOI Foundation, https://www.doi.org (accessed 2025-07-29).Suche in Google Scholar
16. Tremouilhac, P.; Nguyen, A.; Huang, Y.-C.; Kotov, S.; Lütjohann, D. S.; Hübsch, F.; Jung, N.; Bräse, S. Chemotion ELN: an Open Source Electronic Lab Notebook for Chemists in Academia. J. Cheminf. 2017, 9 (1), 54. https://doi.org/10.1186/s13321-017-0240-0.Suche in Google Scholar PubMed PubMed Central
17. DataCite: Connecting Research, Advancing Knowledge. https://www.datacite.org (accessed 2025-07-29).Suche in Google Scholar
18. Dalby, A.; Nourse, J. G.; Hounshell, W. D.; Gushurst, A. K. I.; Grier, D. L.; Leland, B. A.; Laufer, J. Description of Several Chemical Structure File Formats Used by Computer Programs Developed at Molecular Design Limited. J. Chem. Inf. Comput. Sci. 1992, 32 (3), 244–255. https://doi.org/10.1021/ci00007a012.Suche in Google Scholar
19. CTfile Formats. https://www.daylight.com/meetings/mug05/Kappler/ctfile.pdf (accessed 2025-07-29).Suche in Google Scholar
20. CDX Format Specification. https://iupac.github.io/IUPAC-FAIRSpec/cdx_sdk (accessed 2025-07-29).Suche in Google Scholar
21. Weininger, D. SMILES, A Chemical Language and Information System. 1. Introduction to Methodology and Encoding Rules. J. Chem. Inf. Model. 1988, 28 (1), 31–36. https://doi.org/10.1021/ci00057a005.Suche in Google Scholar
22. Daylight Theory: SMILES. https://www.daylight.com/dayhtml/doc/theory/theory.smiles.html (accessed 2025-07-29).Suche in Google Scholar
23. OpenSMILES Specification. http://opensmiles.org/opensmiles.html (accessed 2025-07-29).Suche in Google Scholar
24. Heller, S.; McNaught, A.; Stein, S.; Tchekhovskoi, D.; Pletnev, I. InChI - the Worldwide Chemical Structure Identifier Standard. J. Cheminf. 2013, 5 (1), 7. https://doi.org/10.1186/1758-2946-5-7.Suche in Google Scholar PubMed PubMed Central
25. IUPAC SMILES+ Specification. IUPAC | International Union of Pure and Applied Chemistry. https://iupac.org/project/2019-002-2-024 (accessed 2025-07-29).Suche in Google Scholar
26. InChI Requirements for Representation of Organometallic and Coordination Compound Structures. IUPAC | International Union of Pure and Applied Chemistry. https://iupac.org/project/2009-040-2-800 (accessed 2025-07-29).Suche in Google Scholar
27. Enhanced recognition and Encoding of Stereoconfiguration by InChI Tools. IUPAC | International Union of Pure and Applied Chemistry. https://iupac.org/project/2019-017-2-800 (accessed 2025-07-29).Suche in Google Scholar
28. Brecher, J. Graphical Representation Standards for Chemical Structure Diagrams (IUPAC Recommendations 2008). Pure Appl. Chem. 2008, 80 (2), 277–410. https://doi.org/10.1351/pac200880020277.Suche in Google Scholar
29. Brecher, J. Graphical Representation of Stereochemical Configuration (IUPAC Recommendations 2006). Pure Appl. Chem. 2006, 78 (10), 1897–1970. https://doi.org/10.1351/pac200678101897.Suche in Google Scholar
30. Committee on Publications and Cheminformatics Data Standards. https://iupac.org/body/024 (accessed 2025-07-29).Suche in Google Scholar
31. Apodaca, R. L. Stereochemistry and the V2000 Molfile Format, 2021. http://depth-first.com/articles/2021/12/29/stereochemistry-and-the-v2000-molfile-format (accessed 2025-07-29).10.59350/qvrve-21506Suche in Google Scholar
32. Atomic Coordinate Entry Format Version 3.3. https://www.wwpdb.org/documentation/file-format-content/format33/v3.3.html (accessed 2025-07-29).Suche in Google Scholar
33. Bourne, P. E.; Berman, H. M.; McMahon, B.; Watenpaugh, K. D.; Westbrook, J. D.; Fitzgerald, P. M. D. Macromolecular Crystallographic Information File, In Methods in Enzymology; Macromolecular Crystallography Part B; Academic Press, Vol. 277, 1997; pp. 571–590.10.1016/S0076-6879(97)77032-0Suche in Google Scholar PubMed
34. Molstar/BinaryCIF, 2024. https://github.com/molstar/BinaryCIF (accessed 2025-07-29).Suche in Google Scholar
35. Zhang, T.; Li, H.; Xi, H.; Stanton, R. V.; Rotstein, S. H. HELM: A Hierarchical Notation Language for Complex Biomolecule Structure Representation. J. Chem. Inf. Model. 2012, 52 (10), 2796–2806. https://doi.org/10.1021/ci3001925.Suche in Google Scholar PubMed
36. IUPAC Subcommittee on HELM. https://iupac.org/body/803 (accessed 2025-07-29).Suche in Google Scholar
37. FAIR Principles. GO FAIR. https://www.go-fair.org/fair-principles (accessed 2025-07-29).Suche in Google Scholar
38. Grasselli, J. G. JCAMP-DX, a Standard Format for Exchange of Infrared Spectra in Computer Readable Form (Recommendations 1991). Pure Appl. Chem. 1991, 63 (12), 1781–92. https://doi.org/10.1351/pac199163121781.Suche in Google Scholar
39. Ulrich, E. L.; Baskaran, K.; Dashti, H.; Ioannidis, Y. E.; Livny, M.; Romero, P. R.; Maziuk, D.; Wedell, J. R.; Yao, H.; Eghbalnia, H. R.; Hoch, J. C.; Markley, J. L. NMR-STAR: Comprehensive Ontology for Representing, Archiving and Exchanging Data from Nuclear Magnetic Resonance Spectroscopic Experiments. J. Biomol. NMR 2019, 73 (1), 5–9. https://doi.org/10.1007/s10858-018-0220-3.Suche in Google Scholar PubMed PubMed Central
40. nmrML - Home https://nmrml.org (accessed 2025-07-29).Suche in Google Scholar
41. IUPAC_FAIRSpec_Specification_draft.pdf in https://github.com/IUPAC/IUPAC-FAIRSpec/tree/main/documents/specifications (accessed 2025-07-29).Suche in Google Scholar
42. IUPAC FAIRSpec GitHub Example Icl-13086, https://iupac.github.io/IUPAC-FAIRSpec#icl-10386 (accessed 2025-07-29).Suche in Google Scholar
43. Mies, T.; White, A. J. P.; Rzepa, H. S.; Barluzzi, L.; Devgan, M.; Layfield, R. A.; Barrett, A. G. M. Syntheses and Characterization of Diverse Main Group, Transition, Lanthanide and Actinide Metal Complexes of Ethyl-3-Oxo-2,3-Dihydro-1h-Pyrazole-4-Carboxylate and Related Bidentate Ligands. Inorg. Chem. 2023, 62, 13253–76. https://doi.org/10.1021/acs.inorgchem.3c01506.Suche in Google Scholar PubMed PubMed Central
44. InChI Extension for Mixture Composition. IUPAC | International Union of Pure and Applied Chemistry. https://iupac.org/project/2015-025-4-800 (accessed 2025-07-29).Suche in Google Scholar
45. Clark, A. M.; McEwen, L. R.; Gedeck, P.; Bunin, B. A. Capturing Mixture Composition: an Open Machine-Readable Format for Representing Mixed Substances. J. Cheminf. 2019, 11 (1), 33. https://doi.org/10.1186/s13321-019-0357-4.Suche in Google Scholar PubMed PubMed Central
46. Common Format and MIME Type for Comma-Separated Values (CSV) Files. https://www.ietf.org/rfc/rfc4180.txt (accessed 2025-07-29).Suche in Google Scholar
47. Definition of Tab-Separated-Values (tsv). https://www.iana.org/assignments/media-types/text/tab-separated-values (accessed 2025-07-29).Suche in Google Scholar
48. OpenDocument Spreadsheet (ODS). https://esco.ec.europa.eu/en/about-esco/escopedia/escopedia/opendocument-spreadsheet-ods (accessed 2025-07-29).Suche in Google Scholar
49. Structure of a SpreadsheetML Document. 2023. https://learn.microsoft.com/en-us/office/open-xml/spreadsheet/structure-of-a-spreadsheetml-document (accessed 2025-07-29).Suche in Google Scholar
50. DataCite Metadata Working Group. DataCite Metadata Schema Documentation for the Publication and Citation of Research Data and Other Research Outputs v4.5. 2024. https://doi.org/10.14454/G8E5-6293. An example of a DataCite metadata record can be found at https://data.datacite.org/application/vnd.datacite.datacite+json/10.14469/hpc/10386 (accessed 2025-07-29).Suche in Google Scholar
51. Demo 2025.5 examples2/icl-10386-crawl-only. https://iupac.github.io/IUPAC-FAIRSpec/#10386co (accessed 2025-07-29).Suche in Google Scholar
52. Compound. https://www.cancer.gov/publications/dictionaries/cancer-terms/def/compound (accessed 2025-07-29).Suche in Google Scholar
53. PubChem Compounds https://pubchem.ncbi.nlm.nih.gov/docs/compounds (accessed 2025-07-29).Suche in Google Scholar
54. IUPAC Gold Book Racemic Compound. https://goldbook.iupac.org/terms/view/R05027 (accessed 2025-07-29).Suche in Google Scholar
55. WIPO Patentscope https://patentscope.wipo.int (accessed 2025-07-29).Suche in Google Scholar
56. nmrdb.org Tools for NMR Spectroscopists https://www.nmrdb.org (accessed 2025-07-29).Suche in Google Scholar
57. EAD: Encoded Archival Description https://www.loc.gov/ead (accessed 2025-07-29).Suche in Google Scholar
58. RFC 47648: The Base16, Base32, and Base64 Data Encodings https://datatracker.ietf.org/doc/html/rfc4648 (accessed 2025-07-29).Suche in Google Scholar
© 2025 IUPAC & De Gruyter
This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Artikel in diesem Heft
- Frontmatter
- IUPAC Recommendations
- Experimental methods and data evaluation procedures for the determination of radical copolymerization reactivity ratios from composition data (IUPAC Recommendations 2025)
- IUPAC Technical Reports
- Kinetic parameters for thermal decomposition of commercially available dialkyldiazenes (IUPAC Technical Report)
- FAIRSpec-ready spectroscopic data collections – advice for researchers, authors, and data managers (IUPAC Technical Report)
- Review Articles
- Are the Lennard-Jones potential parameters endowed with transferability? Lessons learnt from noble gases
- Quantum mechanics and human dynamics
- Quantum chemistry and large systems – a personal perspective
- The organic chemist and the quantum through the prism of R. B. Woodward
- Relativistic quantum theory for atomic and molecular response properties
- A chemical perspective of the 100 years of quantum mechanics
- Methylene: a turning point in the history of quantum chemistry and an enduring paradigm
- Quantum chemistry – from the first steps to linear-scaling electronic structure methods
- Nonadiabatic molecular dynamics on quantum computers: challenges and opportunities
- Research Articles
- Alzheimer’s disease – because β-amyloid cannot distinguish neurons from bacteria: an in silico simulation study
- Molecular electrostatic potential as a guide to intermolecular interactions: challenge of nucleophilic interaction sites
- Photophysical properties of functionalized terphenyls and implications to photoredox catalysis
- Combining molecular fragmentation and machine learning for accurate prediction of adiabatic ionization potentials
- Thermodynamic and kinetic insights into B10H14 and B10H14 2−
- Quantum origin of atoms and molecules – role of electron dynamics and energy degeneracy in atomic reactivity and chemical bonding
- Clifford Gaussians as Atomic Orbitals for periodic systems: one and two electrons in a Clifford Torus
- First-principles modeling of structural and RedOx processes in high-voltage Mn-based cathodes for sodium-ion batteries
- Erratum
- Erratum to: Furanyl-Chalcones as antimalarial agent: synthesis, in vitro study, DFT, and docking analysis of PfDHFR inhibition
Artikel in diesem Heft
- Frontmatter
- IUPAC Recommendations
- Experimental methods and data evaluation procedures for the determination of radical copolymerization reactivity ratios from composition data (IUPAC Recommendations 2025)
- IUPAC Technical Reports
- Kinetic parameters for thermal decomposition of commercially available dialkyldiazenes (IUPAC Technical Report)
- FAIRSpec-ready spectroscopic data collections – advice for researchers, authors, and data managers (IUPAC Technical Report)
- Review Articles
- Are the Lennard-Jones potential parameters endowed with transferability? Lessons learnt from noble gases
- Quantum mechanics and human dynamics
- Quantum chemistry and large systems – a personal perspective
- The organic chemist and the quantum through the prism of R. B. Woodward
- Relativistic quantum theory for atomic and molecular response properties
- A chemical perspective of the 100 years of quantum mechanics
- Methylene: a turning point in the history of quantum chemistry and an enduring paradigm
- Quantum chemistry – from the first steps to linear-scaling electronic structure methods
- Nonadiabatic molecular dynamics on quantum computers: challenges and opportunities
- Research Articles
- Alzheimer’s disease – because β-amyloid cannot distinguish neurons from bacteria: an in silico simulation study
- Molecular electrostatic potential as a guide to intermolecular interactions: challenge of nucleophilic interaction sites
- Photophysical properties of functionalized terphenyls and implications to photoredox catalysis
- Combining molecular fragmentation and machine learning for accurate prediction of adiabatic ionization potentials
- Thermodynamic and kinetic insights into B10H14 and B10H14 2−
- Quantum origin of atoms and molecules – role of electron dynamics and energy degeneracy in atomic reactivity and chemical bonding
- Clifford Gaussians as Atomic Orbitals for periodic systems: one and two electrons in a Clifford Torus
- First-principles modeling of structural and RedOx processes in high-voltage Mn-based cathodes for sodium-ion batteries
- Erratum
- Erratum to: Furanyl-Chalcones as antimalarial agent: synthesis, in vitro study, DFT, and docking analysis of PfDHFR inhibition

