Abstract
Triple-negative breast cancer (TNBC) is a highly metastatic subtype of breast cancer. Due to the absence of obvious therapeutic targets, microRNAs (miRNAs) provide possible hope to treat TNBC. Withaferin A (WA), a steroidal lactone, possesses potential anticancer activity with lesser side effects. The present study identifies hub genes (CDKN3, TRAF6, CCND1, JAK1, MET, AXIN2, JAG1, VEGFA, BRCA1, E2F3, WNT1, CDK6, KRAS, MYB, MYCN, TGFβR2, NOTCH1, SIRT1, MYCN, NOTCH2, WNT3A) from the list of predicted targets of the differentially expressed miRNAs (DEMs) in WA-treated MDA-MB-231 cells using in silico protein–protein interaction network analysis. CCND1, CDK6, and TRAF6 hub genes were predicted as targets of miR-34a-5p and miR-146a-5p, respectively. The study found the lower expression of miR-34a-5p and miR-146a-5p in MDA-MB-231 cells, and further, it was observed that WA treatment effectively restored the lost expression of miR-34a-5p and miR-146a-5p in MDA-MB-231 cells. An anti-correlation expression pattern was found among the miR-34a-5p and miR-146a-5p and the respective target hub genes in WA-treated TNBC cells. In conclusion, WA might exert anti-cancer effect in TNBC cells by inducing miR-34a-5p and miR-146a-5p expressions and decreasing CCND1, CDK6, and TARF6 target hub genes in TNBC cells.
Introduction
Triple-negative breast cancer (TNBC) has negative expression of estrogen (ER), progesterone (PR), and human epidermal growth factor receptor 2 (HER2) and comes under the subgroup of breast cancer (BCa) [1]. It appears in younger women and very frequently develops distant metastasis in patients. It accounts for 15–20% of all BCa cases, worldwide. TNBC patients display greater rates of reoccurrence compared to other subgroups of BCa. Mostly, BCa patients are treated with hormonal therapy, but in the case of TNBC subgroup of BCa, hormonal therapy is not beneficial due to the absence of hormonal therapy-targeted hormone receptors such as ER, PR, and HER2. Therefore, chemotherapy remains the primary care of TNBC patients; however, TNBC patients develop drug resistance against chemotherapy; thus, patients show lesser 5-year survival chances compared to the other subgroup of BCa. Food and Drug Administration and European Medicines Agency still have not optimized and approved any liable therapy for TNBC [2].
microRNAs (miRNAs) are non-coding, short segments of nucleotides (18–24 nt) found in cell cytoplasm and their biogenesis initiates in the nucleus, where they are transcribed into pri-miRNA which cleaves into pre-miRNA. The pre-miRNA translocates into the cytoplasm, where it converts into a miRNA duplex. The miRNA duplex gives a mature guide strand miRNA with proper 5′ and 3′ untranslated regions. The miRNAs [3] come under the category of non-coding RNA and they usually perform their biological functions by regulating the gene expression through the degradation of mRNA and also by controlling the transcription and translation via canonical and non-canonical mechanisms [4]. A single miRNA can regulate the expression of one or more than one gene at the same time inside the cell [5,6]. miRNAs regulate the expression of gene(s) involved in tumor aggressiveness and metastasis besides other hallmarks of the cancer [7]. Accumulated evidences indicate that altered expressions of miRNAs are actively involved in cancer tumorigenesis, and therefore they have been considered attractive anti-cancer therapeutic agents [2,7]. Cumulative studies suggest the clinical significance of miRNA as biomarkers for diagnosis, pathogenesis, and prognosis of cancer as well as modulators of drug resistance in cancer [8,9]
Besides this, various studies reported that expression modulation of dysregulated miRNAs by the treatment of natural compounds has anticancer effect. Moreover, natural compounds are choice of drugs nowadays due to their minor toxicity, side effects, and cost-effectiveness although chemotherapy causes severe side effects to other parts of the body [10,11,12,13,14]. But natural compounds or products do not cause any severe side effects toward the other organs of the body. Withaferin A (WA) is a 28-carbon-containing anolide compound that is extracted from Withania somnifera (Solanaceae). WA is a predominant pharmacological active compound of W. somnifera. It has been proved that WA has anticancer potential (in vitro and in vivo) against different cancers by targeting important molecular players playing crucial role in cellular proliferation, differentiation, angiogenesis, metastasis, and drug-resistance in cancer cells [15]. Recently, it has been reported that WA has potential to silence hormone receptor (HER2/PR/ESR)-mediated downstream gene expression events to suppress highly invasive characteristics of TNBC [16,17]. The author indicated that WA might be considered a striking naturally occurring compound for TNBC treatment. The same author in 2014 reported that WA can induce cancer cell death by arresting the cell cycle in metastatic cells of TNBC. Recently, our research group first time explored the miRNA differential expression modulation potential of WA in TNBC cells and deduced the anticancer mechanism of WA at the miRNA level in TNBC cells [7].
It has been extensively reported in the literature that the miRNA-mediated hub gene expression modulation shows better anticancer effect compared to non-hub genes due to its ability to interact with a number of target genes and regulate their functions [18,19,20]. Therefore, the present study aimed to identify the hub genes within predicted target genes of differentially expressed miRNAs (DEMs) in WA-treated TNBC cells previously reported by our research group [7]. Thus, in the present study, first, the miRNA–mRNA hub gene regulatory network was formed and, then, an anti-correlation between miRNA and mRNA-hub genes was predicted and validated using molecular techniques in TNBC cells.
Materials and methods
Cell culture and WA treatment
TNBC, MDA-MB-231 cells were purchased from NCCS Pune, India. Cells were cultivated in cell culture-specific incomplete media and the media was complemented with 10% fetal bovine serum and a combination of two different antibiotics (streptomycin and penicillin). Cells were grown at 37°C in a 5% carbon dioxide environment in an incubator. Cultivated TNBC (MDA-MB-231) cells were treated with WA with ≈ 2 µM concentration and 0.1% DMSO and kept for 24 h in triplicate. WA-treated and DMSO-treated TNBC and MDA-M-231 cell samples were considered experimental and control groups, respectively [21].
Hub gene identification
We performed next-generation sequencing (NGS) of small RNA in WA-treated and DMSO-treated MDA-MB-231 cell samples in our previous study [7]. Identified (p < 0.05) DEMs from NGS were subjected to their target gene prediction using an online miRSystem database. The predicted target genes that followed hit ≥3 and experimentally validated (mentioned in miRsystem) were sorted and used for hub gene identification [22].
To determine the hub genes, predicted target genes were submitted to the STRING database to create the functional protein–protein (PPI) network. The full PPI network of sorted target genes was constructed by applying an interaction score (medium confidence >0.4). A full PPI network was used to extract the candidate hub genes. The PPI network was analyzed by Cytoscape software v 3.38.1 plugin with the cytoHubba app [2]. Furthermore, the node degree ranked scoring method “Maximal Clique Centrality” was applied to recognize the more essential hub genes in PPI network using the cytoHubba plugin [23]. The top 20 hub genes were sorted by applying nodes degree score ≥10.
The miRNA–mRNA regulatory network construction
Identified hub genes (node degree ≥10) from PPI network analysis were undertaken for miRNA–mRNA regulatory network construction. For this, a list of hub genes and their regulatory DEMs were submitted into the Cytoscape software v 3.38.1 and a regulatory network was constructed.
Total miRNA extraction and cDNA synthesis
The total miRNA was withdrawn from WA-treated (experimental group) and vehicle-treated (control group) in triplicate using mirVana™ miRNA isolation kit (Catalog: 15604) as per manufacturer’s protocols. The miRNA’s quantity and quality were analyzed with the help of NanoDrop. The miRNA content (500 ng) was polyadenylated and back converted into complementary DNA at 37°C Temp with miRNA cDNA synthesis-specific kit as per standard protocol in the PCR machine [7].
Total RNA extraction and cDNA synthesis
Total RNA was withdrawn from WA and vehicle-treated samples using Triozol reagent (Catalog: 15596018). A NanoDrop instrument was utilized to quantify and quality of the RNA content in the experimental and control samples. A total of 1,000 ng of RNA content was biochemically converted into complementary DNA at 66°C for 20 min with RNA-specific cDNA synthesis kit (Catalog: 1708891) in the PCR machine as per standard protocol [7].
Quantitative RT-PCR analysis of miRNA–mRNA regulatory network
The SYBR green-based qRT-PCR expression analysis of identified hub genes (CCND1, NOTCH2, KRAS, VEGFA, CDK6, WNT3A, WNT1, JAG1, JAK1, and TRAF6) and their respective regulatory significant DEMs were used to confirm the anti-correlation between miRNA and mRNA hub genes in WA-treated MDA-MB-231 cells. The qRT-PCR was performed in triplicate using miRNAs and mRNA-specific 5′ forward and 3′ backward primers (Table 1). The two separate qRT-PCR setups were performed for miRNA and mRNA. The reaction mixture volume used in the qRT-PCR experiment for test miRNA and mRNA was 12.5 µL and 10 µL respectively. The detailed methodology for qRT-PCR expression of miRNA and gene was adopted from our previously published research paper [7]. The 2−∆∆CT method was used to estimate the fold change expression of miRNA and mRNA gene [7].
Primer nucleotide sequences of miRNA and target hub genes
| MicroRNAs | Forward-specific primers | Reverse-specific primers |
|---|---|---|
| hsa-miR-34a-5p | GGT AGT GTC TTA GCT GGT TGT | *mRQ3′ |
| hsa-miR-146a-5p | ACA TTC AAC CTG TCG GTG AGT | mRQ3′ |
| Genes | ||
| CCND1 | CCTGGTGAACAAGCTCAAGT | GTGTTTGCGGATGATCTGTTTG |
| NOTCH1 | CTGGCGGTGCACACTATT | TCCACAGGCGAGGAGTAG |
| KRAS | GCCTTGACGATACAGCTAATTC | GTATCAAAGAATGGTCCTGCAC |
| VEGFA | GAGCTTCCTACAGCACAACA | CCAGGACTTATACCGGGATTTC |
| CDK6 | GAACTGGTTTCGCTGTCATTC | GGGCAGGTTCCCTTCATTAT |
| WNT3A | TTGCAGTGACACGCTCAT | GACACCATCCCACCAAACT |
| WNT1 | GTCCTCCTAAGTCCCTTCCTAT | GTAACCTCCTGCTTCAGCTAC |
| JAG1 | ACTGCTCACACCTGAAAGAC | CCACAGACGTTGGAGGAAATA |
| JAK1 | GGATTACAAGGATGACGAAGGA | CGAAGAAGGCCAGGGAAATA |
| TRAF6 | CTCCTGTAGCGCTGTAACAAA | TCCCTGGATCTCCTCCATAAA |
*mRQ3′: 3′ universal primers provided within miRNA first strand cDNA synthesis kit.
Western blotting analysis
Immunoblot technique was utilized to assess the level of cyclin D1 in WA-treated and vehicle-treated MDA-MB-231 cells. For this, the cells were cultivated and treated with WA at ≈2 μM concentration and incubated for 24 h. Total protein content was withdrawn from samples and estimated using the Bradford method. The wells of 10% SDS-PAGE were packed with each 30 µg protein content and resolved successfully. Resolved protein contents were electrophoretically transferred to PVDF membrane using the western blotting protocol. Afterwards, PVDF membrane was incubated with cyclin D1 monoclonal antibody (Catalog: MA514512) and β-actin monoclonal antibody (Catalog: MA516410). Western blotting experiment was performed as per the given methodology in our previously published research paper [7]. Chemo doc imaging instrument (Bio-Rad) was used for image capture. The protein immunoblots were processed through image lab software.
Survival analysis of miRNAs
To accomplish the overall survival (OS) analysis in the TNBC patients, we selected the TNBC molecular subtype samples in the TCGA database for the survival analysis of test miRNAs. Expression levels and OS rate relationship of the test miRNAs were determined using the Kaplan–Meier method [24].
Statistical analysis
All the obtained data are statistically tested and significant at a probability level of p value < 0.05. GraphPad prism v.5.0 was used for all the statistical analysis. Each experiment independently was performed in the triplicate.
Results
Identified hub gene among the DEMs target genes
Database-validated predicted target genes of NGS-identified WA treatment (IC50 2 µM) induced DEMs compared to vehicle (0.1% DMSO)-treated cells were used to create the gene–gene interaction network to determine the potential hub genes [7]. The physical interactive network showed a total of 215 edges and 58 nodes with PPI enrichment p-value <1.0 × 10−16 (Figure 1). Generally, the gene having more edges may actively involve in the regulation of various biological functions. Thus, keeping this fact in mind, the “degree centrality” parameter was used to calculate the edge count of each single gene in a PPI network using cytoHubba tool in the Cytoscape software. The top 20 hub genes in the network were sorted according to the number of degrees of centrality and considered hub genes (Figure 1).
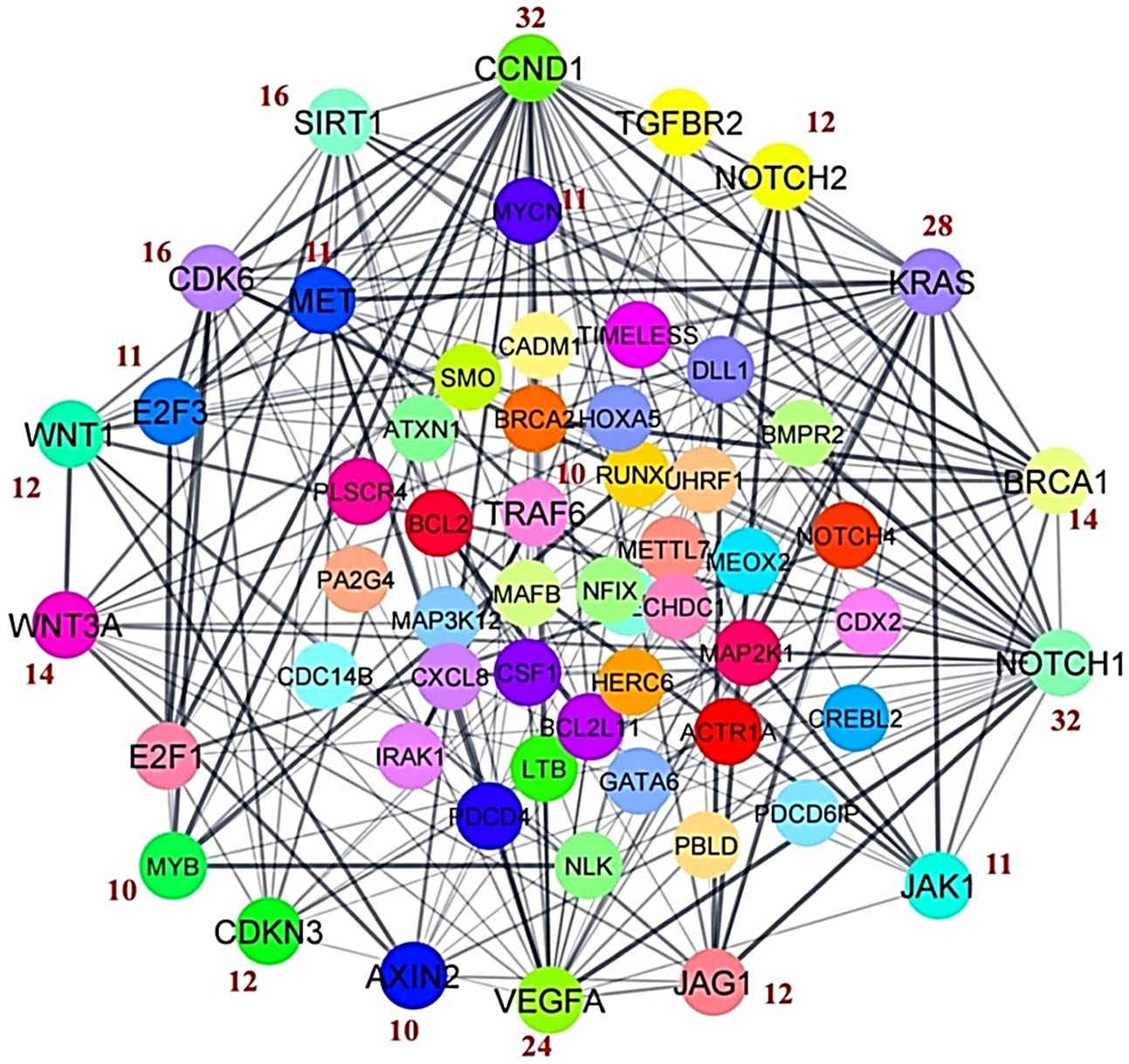
A physical interactive network of predicted target genes of DEM. The top 20 hub genes were selected on the basis of degree centrality. The numbers mentioned in red color indicate the number of degrees of respective hub genes.
The regulatory miRNAs sorted for their associated hub genes
The significantly identified DEMs in WA-treated samples from the previous study and the identified hub genes obtained from network analysis were intersected to construct the miRNA–mRNA regulatory network. The results revealed that four up-expressed miRNAs (miR-34a-5p, miR-181c-5p, miR-15a-5p, and miR146a-5p) and one down-expressed (miR-20a-5p) miRNA are involved in the regulation of top 20 hub genes. The regulatory miRNAs and their targeting hub genes are displayed in Figure 2.
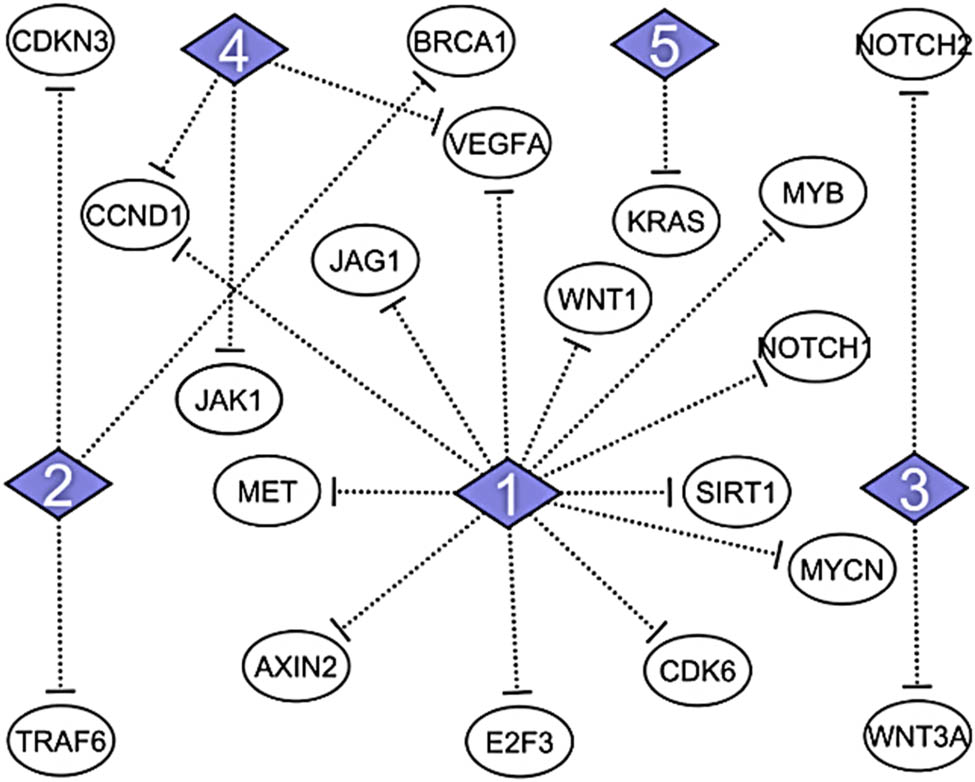
Regulatory network of DEMs and their predicted target genes. The top 20 hub genes and their regulatory miRNAs. 1 = hsa-miR-34a-5p; 2 = hsa-miR-146a-5p; 3 = hsa-miR-15a-5p; 4 = hsa-miR-20a-5p; 5 = hsa-miR-181c-5p.
qRT-PCR validation of the identified regulatory miRNAs and mRNA hub genes
The qRT-PCR expression analysis of the significant regulatory DEMs (miR-34a-5p, miR-181c-5p, miR-15a-5p, miR146a-5p, and miR-20a-5p) and their potential hub genes (CCND1, NOTCH2, KRAS, VEGFA, CDK6, WNT3A, WNT1, JAG1, JAK1, and TRAF6) was performed to check the inverse relation between them in WA-treated samples compared to vehicle-treated cells samples. The primer sequences of miRNA and mRNA hub genes are given in Table 1. Results found that the expression levels of miR-34a-5p and miR-146a-5p were higher in WA-treated TNBC cells when we compared the same with vehicle-treated TNBC cells (Figure 3). Conversely, the expression levels of CCND1, CDK6, and TRAF6 were lower in the WA-treated TNBC cell samples in regards to vehicle-treated TNBC cell samples while the expression levels of NOTCH2, KRAS, VEGFA, WNT3A, WNT1, JAG1, and JAK1 were higher in WA-treated cells compared to vehicle-treated cells. Therefore, we found the anti-correlation relationship between miR-34a-5p and CCND1; miR-34a-5p and CDK6; and miR-146a-5p and TRAF6 (Figure 4).
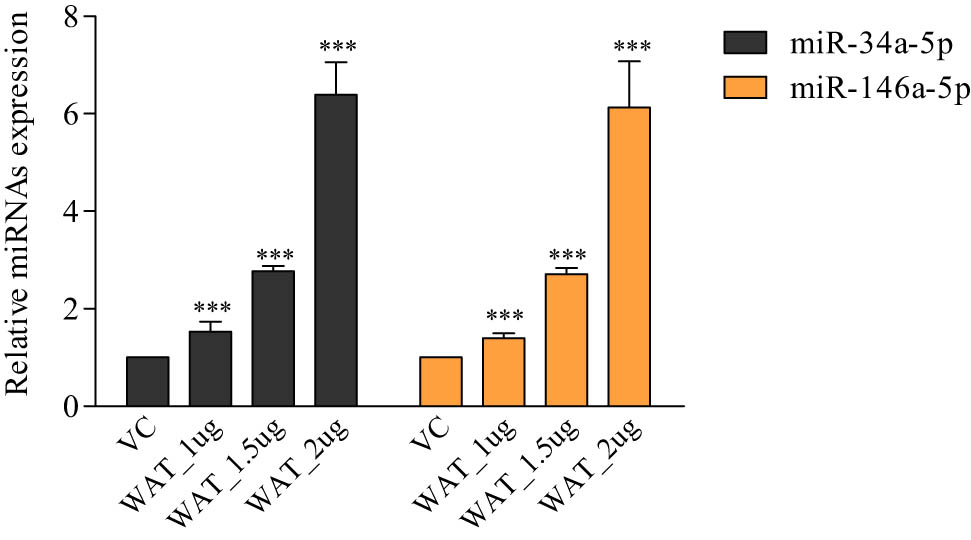
Quantitative RT-PCR analysis of lead miRNAs in WA-treated TNBC cells. The miR-34a-5p and miR-146a-5p expression profiles in WA-exposed MDA-MB-231 cells. Results were compared with vehicle (0.1% DMSO)-treated test cells. VC: vehicle (0.1% DMSO)-treated samples. WAT_1µg: WA-treated samples at 1 µg concentration. WAT_1.5 µg: WA-treated samples at 1.5 µg concentration. WAT_2 µg: WA-treated samples at 2 µg concentration. The qRT-PCR was performed in triplicate (n = 3).
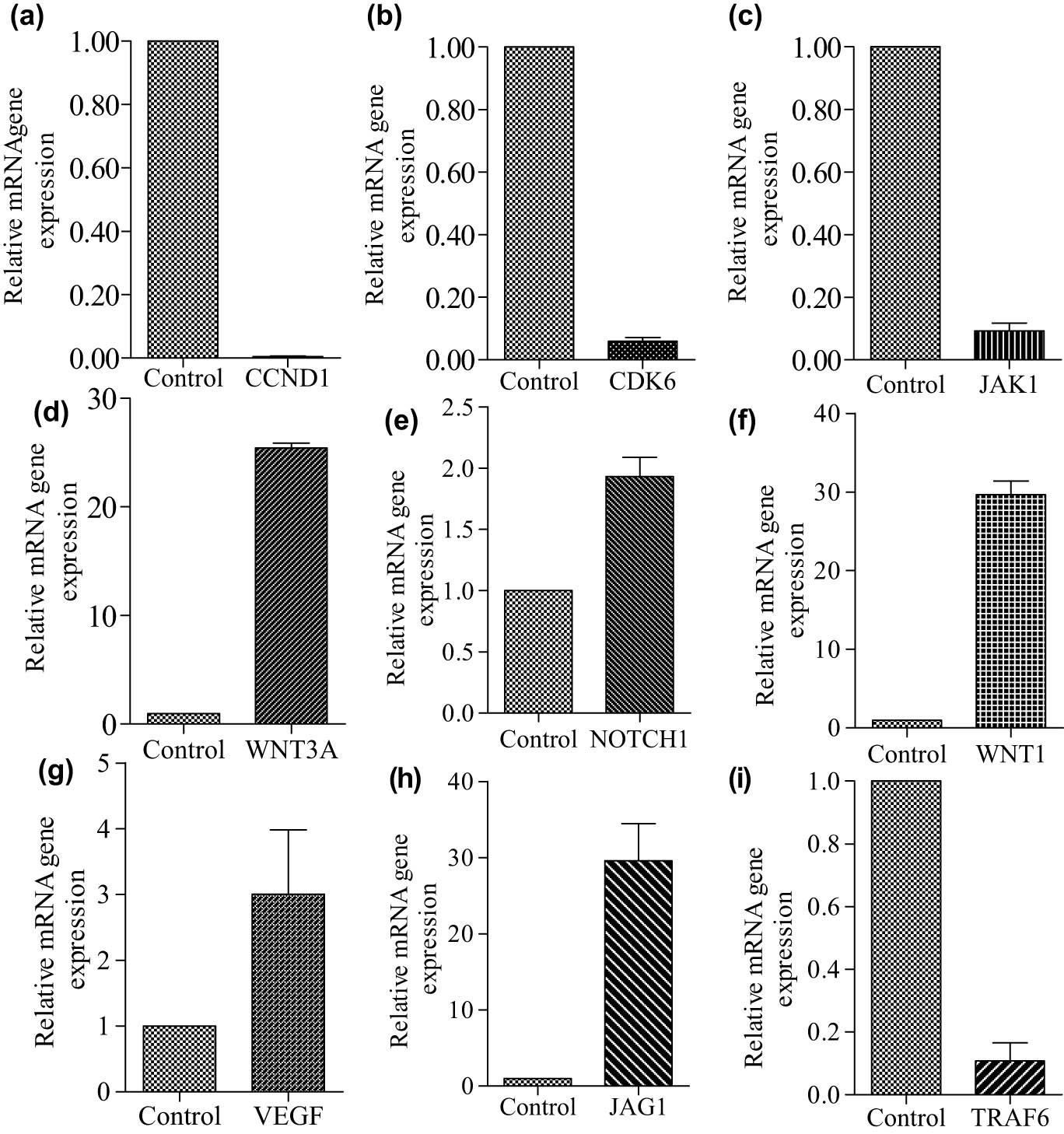
Quantitative RT-PCR analysis of potential candidate hub genes in WA treated in comparison to vehicle (0.1% DMSO)-treated (control) TNBC cells. The expression levels of CCND1 (a), CDK6 (b), JAK1 (c), WNT3A (d), NOTCH1 (e), WNT1 (f), VEGFA (g), JAG1 (h), and TRAF6 (i). The qRT-PCR was performed in triplicate (n = 3).
Anti-correlation between the identified miRNA and hub genes mRNA
To examine whether the overexpression of miR-34a-5p and miR-146a-5p suppresses the CCND1, CDK6, and TRAF6 expression or not, respectively, the qRT-PCR and western blot techniques were utilized. In the qRT-PCR investigation, it was noticed that the up-regulation of miR-34a-5p was inversely associated with the expression of CCND1 (cyclin D1) and CDK6 hub genes in WA-treated TNBC MDA-MB-231 cells compared to control samples (Figure 5a). Similarly, we observed that higher expression of miR-146a-5p was negatively associated with the expression of TRAF6 hub gene in the same set of samples (Figure 5b). Further to check the expression of CCND1 at the protein level, we performed the western blotting analysis; we observed a low density of protein band of cyclin D1 in WA-treated cell samples compared to vehicle-treated cell samples (Figure 5c and d).
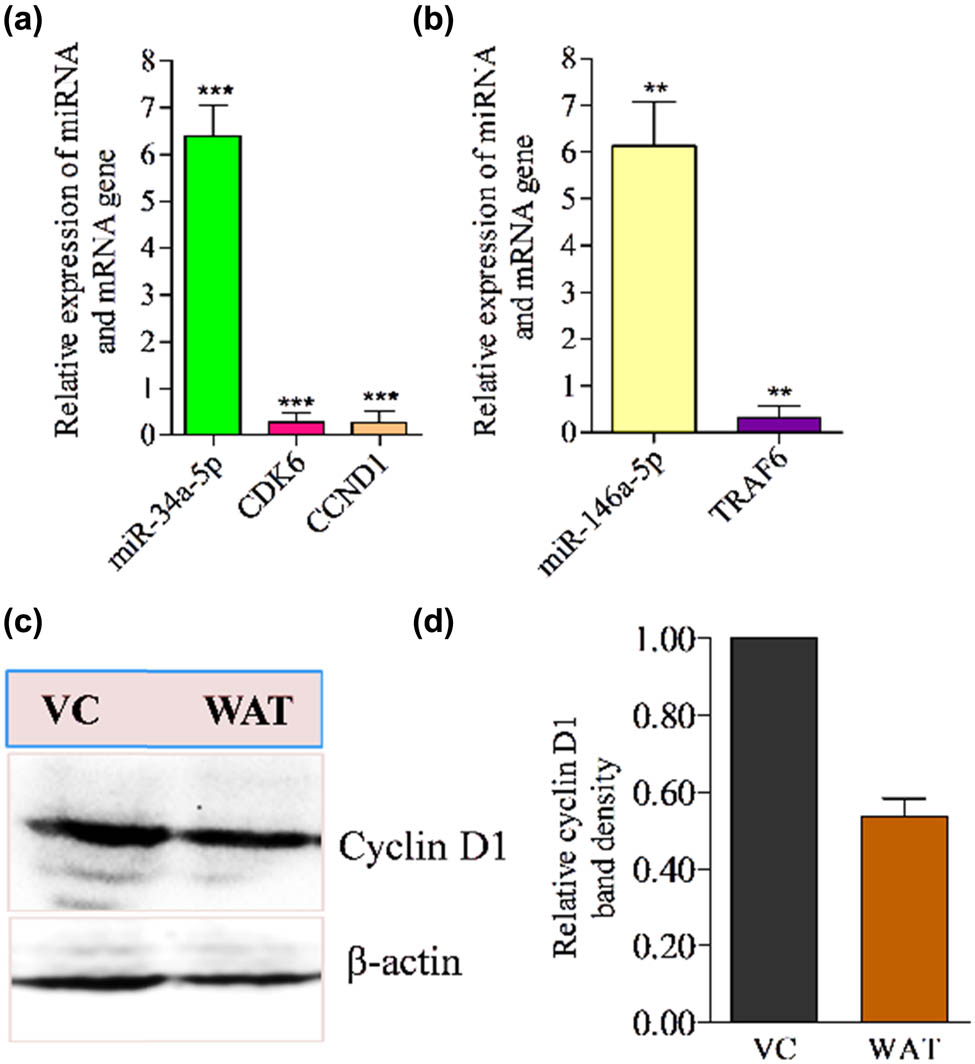
Quantitative RT-PCR analysis of lead miRNAs and western blotting analysis of cyclin D1 in WA-treated TNBC cells. (a) Anti-correlation between miR-34a-5p and their target gene (CDK6 and CCND1). (b) Anti-correlation between miR-146a-5p and its target gene (TRAF6) in WA-treated MDA-MB-231 cells. (c) Western blot results of cyclin D1 in WA-treated MDA-MB-231 cell samples. (d) Densitometry of cyclin D1 protein band. Results were compared with vehicle (0.1% DMSO) treated test cells. The qRT-PCR and western blot experiments independently were performed in triplicate (n = 3).
Survival analysis of the lead-identified DEMs
To get an idea about the translational potential of the present study, we tried to determine the association of the lead miRNAs (miR-34a-5p and miR146a-5p) with OS in TNBC patients. The OS analysis of each lead miRNA was performed in the TNBC subgroup of BCa samples using publically available data in the TCGA database through the Kaplan–Meier survival method. The analysis revealed that high expression of hsa-miR-34a-5p and hsa-miR-146a-5p was related to overall higher survival in TNBC patients. The OS rate, HR ratio, and significant logrank p values of each analysis are depicted in Figure 6. Results showed significant association of the miRNAs with the OS of TNBC patients.
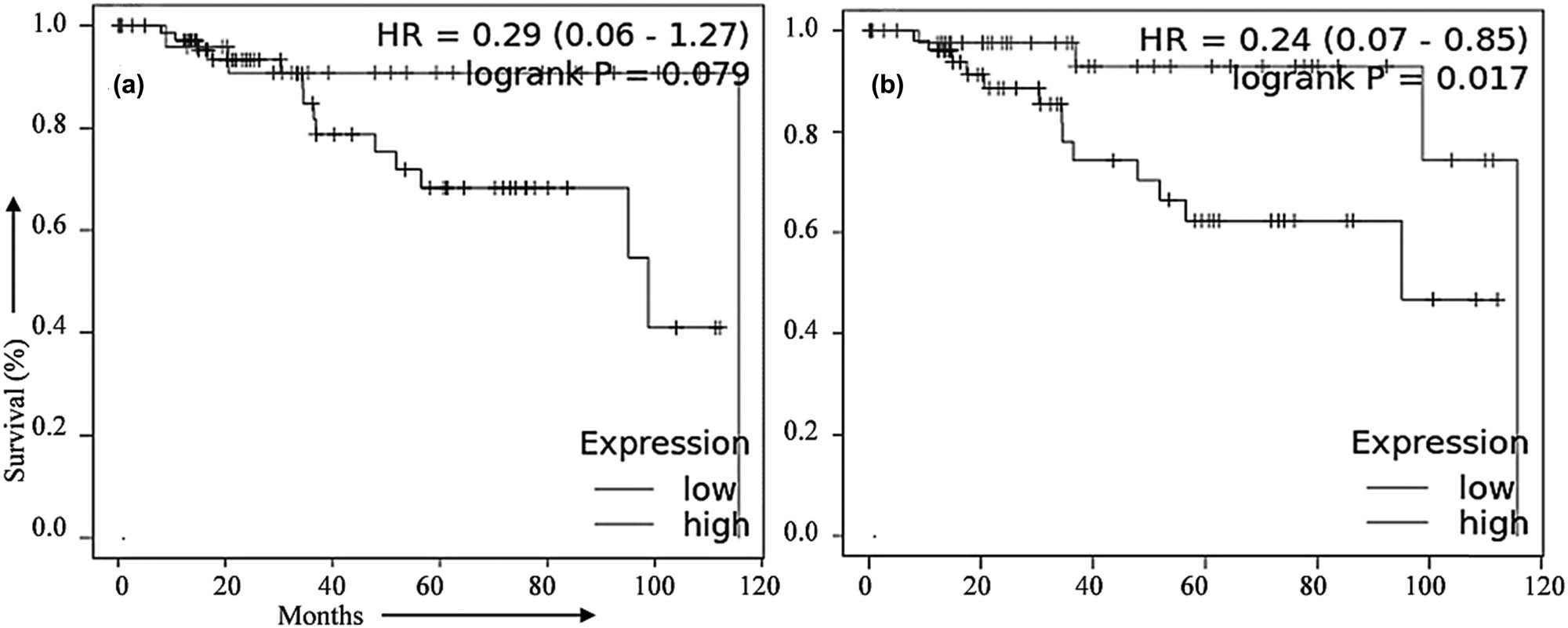
Association of validated miRNAs with the overall survival (OS) in TNBC patients. Expression pattern of (a) miR-146a-5p and (b) miR-34a-5p associated with the OS rate in TNBC patients. Data available in the TCGA database were used for the analysis and the Kaplan–Meier method was applied.
Discussion
Treatment management of TNBC is very difficult because most TNBC patients show a high degree of invasiveness, metastasis, distant metastasis, and reoccurrence of the disease [21]. Besides this, the severe side effects of chemotherapy such as toxicity are also closely linked with the poor treatment management of this disease. Therefore, nowadays naturally occurring anticancer compounds are excellent because of their minor side effects and lesser cost. Moreover, it has been investigated that natural anticancer compounds can regulate the miRNA expression in cancers [6,7,22,3]. In this present piece of work, the potential lead hub genes (CCND1, CDK6, and TRAF6) were identified from the interactive network analysis of PPI of predicted target genes of those significant DEMS who were identified through NGS in WA-treated in vitro metastatic cell line model of TNBC disease and that DEMs have been reported in our previous study [7]. The miRNA–mRNA network analysis showed that a single regulatory miRNA has interactions with more numbers of hub genes (Figure 2). Therefore, expression regulation of hub genes via miRNAs has the greatest anticancer effect by controlling the various cellular processes in the cancer cells [18,19,20]. Interestingly, in our miRNA–mRNA (hub) gene interaction network analysis, we found that miR-34a and miR-146a individually have more regulatory physical interactions with more numbers of hub genes (Figure 2). However, overexpression of miR-34a and miR-146a induces apoptosis, seizes the cell cycle, and suppresses the EMT in TNBC [24–27]. Interestingly, we observed the greater expression of these miRNAs in WA-treated TNBC metastatic cells compared to vehicle-treated cells (Figure 3). Several other studies have reported that overexpression of these miRNAs (miR-34a-5p and miR-146a-5p) expands the survival of TNBC patients [28–31]. The present study also found that the higher expression of miR-34a-5p and miR-146a-5p was significantly linked with the higher survival in TNBC patients (Figure 6). The survival results of the present study are in line with the results from these previous studies which indicate that overexpression of miR-34a-5p and miR-146a-5p via WA treatment could be advantageous in the better survival of TNBC patients. It has been reported that TRAF6 is a potential target of miR-146a-5p and several studies reported that overexpression of miR-146a-5p induces apoptosis and halts the EMT process in cancers [32,33,34]. We also found TRAF6 as a target of miR-146a-5p. The qRT-PCR expression analysis revealed that up-expression of miR-146a-5p in response to WA treatment significantly decreased TRAF6 levels in MDA-MB-231 TNBC cells (Figure 5c). Several other studies reported that higher expression of TRAF6 is actively involved in tumor initiation, progression, invasion, and metastasis in various cancers [35,36,37]. Present study results revealed that establishing the anti-correlation among the expression of TRAF6 and miR-146a-5p through WA treatment may be a better therapeutic strategy for TNBC patients. Further, in the present study, CCND1 and CDK6 were also found the downstream targets of another miRNA that is miR-34a-5p. Studies reported that overexpression of miR-34a-5p effectively arrests the cell cycle progression at the G1 phase by suppressing the expression of CCND1 and CDK6 in cancer cells [38,39]. In the present study, we also found that overexpression of miR-34a-5p remarkably inhibits the higher expression of CCND1 and CDK6 in MDA-MB-231 cells (Figure 5a, c, and d). The anti-correlation among the miR-34a-5p and CCND1 (Figure 5b and c) expression indicates that WA treatment may induce cell cycle arrest in TNBC cells. The study indicates that the inverse correlation between miR-34a-5p and CCND1, CDK6 could serve as an important strategy for anti-cancer therapy in TNBC disease.
Conclusion
WA treatment remarkably increased the expression of tumor suppressive miRNAs (miR-146a-5p and miR-34a-5p) in metastatic MDA-MB-231 cells compared to vehicle-treated MDA-MB-231 cells. The altered expression of miR-34a-5p and miR-146a-5p was negatively correlated with the expression of TRAF6, CCND1, and CDK6 hub genes in WA-treated samples. Therefore, down-expression of TRAF6, CCND1, and CDK6 genes by up-expression of miR-146a-5p and miR-34a-5p in response to WA treatment in MDA-MB-231 cells might be considered novel target-based strategy for the treatment of TNBC patients.
Acknowledgement
SK acknowledges the Department of Science and Technology, India, for providing financial support in the form DST-SERB Grant [EEQ/2016/000350]. SK also acknowledges DST India for providing a Departmental grant to the Department of Biochemistry, Central University of Punjab, Punjab, in the form of DST-FIST grant.
-
Funding information: This research was funded by a grant received from DST-SERB Grant [EEQ/2016/000350], Department of Science and Technology, India.
-
Author contributions: Conceptualization: S.K.; methodology: S.K.; M.S., and S.C.; validation: S.K., M.S., and S.C.; formal analysis: S.K.; M.S., and S.C.; investigation: S.K., M.S., and S.C.; writing – original draft: M.S.; writing – review and editing: S.K.; visualization: M.S., and S.C.; supervision: S.K.; project administration: S.K.; funding acquisition: S.K.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The study-related NGS data are available at NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE183395.
References
[1] Yin L, Duan JJ, Bian XW, Yu SC. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020;22:61.10.1186/s13058-020-01296-5Search in Google Scholar PubMed PubMed Central
[2] Shuaib M, Prajapati KS, Singh AK, Kushwaha PP, Waseem M, Kumar S. Identification of miRNAs and related hub genes associated with the triple negative breast cancer using integrated bioinformatics analysis and in vitro approach. J Biomol Struct Dyn. 2021;40:1–15.10.1080/07391102.2021.1961869Search in Google Scholar PubMed
[3] Pordel S, Khorrami M, Saadatpour F, Rezaee D, Cho WC, Jahani S, et al. The role of microRNA-185 in the pathogenesis of human diseases: A focus on cancer. Pathol Res Pract. 2023;249:154729.10.1016/j.prp.2023.154729Search in Google Scholar PubMed
[4] Najafi S. Circular RNAs as emerging players in cervical cancer tumorigenesis; A review to roles and biomarker potentials. Int J Biol Macromol. 2022;206:939–53.10.1016/j.ijbiomac.2022.03.103Search in Google Scholar PubMed
[5] Waseem M, Ahmad MK, Srivatava VK, Rastogi N, Serajuddin M, Kumar S, et al. Evaluation of miR-711 as novel biomarker in prostate cancer progression. Asian Pac J Cancer Prev. 2017;18:2185–91.Search in Google Scholar
[6] Prajapati KS, Shuaib M, Kushwaha PP, Singh AK, Kumar S. Identification of cancer stemness related miRNA(s) using integrated bioinformatics analysis and in vitro validation. 3 Biotech. 2021;11:446.10.1007/s13205-021-02994-3Search in Google Scholar PubMed PubMed Central
[7] Shuaib M, Prajapati KS, Gupta S, Kumar S. Natural steroidal lactone induces G1/S phase cell cycle arrest and intrinsic apoptotic pathway by up-regulating tumor suppressive miRNA in triple-negative breast cancer cells. Metabolites. 2022;13:29.10.3390/metabo13010029Search in Google Scholar PubMed PubMed Central
[8] Fattahi M, Shahrabi S, Saadatpour F, Rezaee D, Beyglu Z, Delavari S, et al. microRNA-382 as a tumor suppressor? Roles in tumorigenesis and clinical significance. Int J Biol Macromol. 2023;250:125863.10.1016/j.ijbiomac.2023.125863Search in Google Scholar PubMed
[9] Fattahi M, Rezaee D, Fakhari F, Najafi S, Aghaei-Zarch SM, Beyranvand P, et al. microRNA-184 in the landscape of human malignancies: a review to roles and clinical significance. Cell Death Discov. 2023;9:423.10.1038/s41420-023-01718-1Search in Google Scholar PubMed PubMed Central
[10] Kumar S, Chashoo G, Saxena AK, Pandey AK. Parthenium hysterophorus: a probable source of anticancer, antioxidant and anti-HIV agents. BioMed Res Int. 2013;2013:810734.10.1155/2013/810734Search in Google Scholar PubMed PubMed Central
[11] Kumar S, Pandey S, Pandey AK. In vitro antibacterial, antioxidant, and cytotoxic activities of Parthenium hysterophorus and characterization of extracts by LC-MS analysis. BioMed Res Int. 2014;2014:495154.10.1155/2014/495154Search in Google Scholar PubMed PubMed Central
[12] Kushwaha PP, Singh AK, Prajapati KS, Shuaib M, Fayez S, Bringmann G, et al. Induction of apoptosis in breast cancer cells by naphthylisoquinoline alkaloids. Toxicol Appl Pharmacol. 2020;409:115297.10.1016/j.taap.2020.115297Search in Google Scholar PubMed
[13] Kushwaha PP, Maurya SK, Singh A, Prajapat KS, Singh AK, Shuaib M, et al. Bulbine frutescens phytochemicals as novel ABC-transporter inhibitor: A molecular docking and molecular dynamics simulation study. J Cancer Metastasis Treat. 2021;7:2.10.20517/2394-4722.2020.92Search in Google Scholar
[14] Kumar S, Gupta S. Dietary phytochemicals and their role in cancer chemoprevention. J Cancer Metastasis Treat. 2021;7:51.10.20517/2394-4722.2021.125Search in Google Scholar PubMed PubMed Central
[15] Hahm ER, Kim SH, Singh KB, Singh K, Singh SV. A comprehensive review and perspective on anticancer mechanisms of withaferin A in breast cancer. Cancer Prev Res. 2020;13:721–34.10.1158/1940-6207.CAPR-20-0259Search in Google Scholar PubMed PubMed Central
[16] Szarc Vel Szic K, Declerck K, Crans R, Diddens J, Scherf DB, Gerhäuser C, et al. Epigenetic silencing of triple negative breast cancer hallmarks by Withaferin A. Oncotarget. 2017;8:40434–53.10.18632/oncotarget.17107Search in Google Scholar PubMed PubMed Central
[17] Szarc vel Szic K, Op de Beeck K, Ratman D, Wouters A, Beck IM, Declerck K, et al. Pharmacological levels of Withaferin A (Withania somnifera) trigger clinically relevant anticancer effects specific to triple negative breast cancer cells. PLoS One. 2014;9:e87850.10.1371/journal.pone.0087850Search in Google Scholar PubMed PubMed Central
[18] Tian Z, He W, Tang J, Liao X, Yang Q, Wu Y, et al. Identification of important modules and biomarkers in breast cancer based on WGCNA. Onco Targets Ther. 2020;13:6805–17.10.2147/OTT.S258439Search in Google Scholar PubMed PubMed Central
[19] Wang J, Yu H, Yili A, Gao Y, Hao L, Aisa HA, et al. Identification of hub genes and potential molecular mechanisms of chickpea isoflavones on MCF-7 breast cancer cells by integrated bioinformatics analysis. Ann Transl Med. 2020;8:86.10.21037/atm.2019.12.141Search in Google Scholar PubMed PubMed Central
[20] Yan LR, Wang A, Lv Z, Yuan Y, Xu Q. Mitochondria-related core genes and TF-miRNA-hub mrDEGs network in breast cancer. Biosci Rep. 2021;41:BSR20203481.10.1042/BSR20203481Search in Google Scholar PubMed PubMed Central
[21] Kushwaha PP, Vardhan PS, Kapewangolo P, Shuaib M, Prajapati SK, Singh AK, et al. Bulbine frutescens phytochemical inhibits notch signaling pathway and induces apoptosis in triple negative and luminal breast cancer cells. Life Sci. 2019;234:116783.10.1016/j.lfs.2019.116783Search in Google Scholar PubMed
[22] Lu TP, Lee CY, Tsai MH, Chiu YC, Hsiao CK, Lai LC, et al. miRSystem: an integrated system for characterizing enriched functions and pathways of microRNA targets. PLoS One. 2012;7:e42390.10.1371/journal.pone.0042390Search in Google Scholar PubMed PubMed Central
[23] Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.10.1101/gr.1239303Search in Google Scholar PubMed PubMed Central
[24] Győrffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J. 2021;19:4101–9.10.1016/j.csbj.2021.07.014Search in Google Scholar PubMed PubMed Central
[25] Hanieh H. Aryl hydrocarbon receptor-microRNA-212/132 axis in human breast cancer suppresses metastasis by targeting SOX4. Mol Cancer. 2015;14:172.10.1186/s12943-015-0443-9Search in Google Scholar PubMed PubMed Central
[26] Venkatadri R, Muni T, Iyer AK, Yakisich JS, Azad N. Role of apoptosis-related miRNAs in resveratrol-induced breast cancer cell death. Cell Death Discov. 2016;7:e2104.10.1038/cddis.2016.6Search in Google Scholar PubMed PubMed Central
[27] Si C, Yu Q, Yao Y. Effect of miR-146a-5p on proliferation and metastasis of triple-negative breast cancer via regulation of SOX5. Exp Ther Med. 2018;15:4515–21.10.3892/etm.2018.5945Search in Google Scholar PubMed PubMed Central
[28] Engkvist ME, Stratford EW, Lorenz S, Meza-Zepeda LA, Myklebost O, Munthe E. Analysis of the miR-34 family functions in breast cancer reveals annotation error of miR-34b. Sci Rep. 2017;7:9655.10.1038/s41598-017-10189-1Search in Google Scholar PubMed PubMed Central
[29] Haghi M, Taha MF, Javeri A. Suppressive effect of exogenous miR-16 and miR-34a on tumorigenesis of breast cancer cells. J Cell Biochem. 2019;120:13342–53.10.1002/jcb.28608Search in Google Scholar PubMed
[30] Maroni P, Puglisi R, Mattia G, Carè A, Matteucci E, Bendinelli P, et al. In bone metastasis miR-34a-5p absence inversely correlates with Met expression, while Met oncogene is unaffected by miR-34a-5p in non-metastatic and metastatic breast carcinomas. Carcinogenesis. 2017;38:492–503.10.1093/carcin/bgx027Search in Google Scholar PubMed
[31] Mackiewicz M, Huppi K, Pitt JJ, Dorsey TH, Ambs S, Caplen NJ. Identification of the receptor tyrosine kinase AXL in breast cancer as a target for the human miR-34a microRNA. Breast Cancer Res Treat. 2011;130:663–79.10.1007/s10549-011-1690-0Search in Google Scholar PubMed PubMed Central
[32] Han R, Zhao J, Lu L. MicroRNA‑34a expression affects breast cancer invasion in vitro and patient survival via downregulation of E2F1 and E2F3 expression. Oncol Rep. 2020;43:2062–72.10.3892/or.2020.7549Search in Google Scholar PubMed
[33] Mansoori B, Silvestris N, Mohammadi A, Khaze V, Baghbani E, Mokhtarzadeh A, et al. miR-34a and miR-200c have an additive tumor-suppressive effect on breast cancer cells and patient prognosis. Genes (Basel). 2021;12:267.10.3390/genes12020267Search in Google Scholar PubMed PubMed Central
[34] Zavala V, Pérez-Moreno E, Tapia T, Camus M, Carvallo P. miR-146a and miR-638 in BRCA1-deficient triple negative breast cancer tumors, as potential biomarkers for improved overall survival. Cancer Biomark. 2016;6(1):99–107.10.3233/CBM-150545Search in Google Scholar PubMed
[35] Panoutsopoulou K, Liu Y, Avgeris M, Dreyer T, Dorn J, Magdolen V, et al. Repression of miR-146a in predicting poor treatment outcome in triple-negative breast cancer. Clin Biochem. 2023;114:43–51.10.1016/j.clinbiochem.2022.12.004Search in Google Scholar PubMed
[36] Meng Q, Liang C, Hua J, Zhang B, Liu J, Zhang Y, et al. A miR-146a-5p/TRAF6/NF-kB p65 axis regulates pancreatic cancer chemoresistance: functional validation and clinical significance. Theranostics. 2020;10:3967–79.10.7150/thno.40566Search in Google Scholar PubMed PubMed Central
[37] Xu C, Wang P, Guo H, Shao C, Liao B, Gong S, et al. MiR-146a-5p deficiency in extracellular vesicles of glioma-associated macrophages promotes epithelial-mesenchymal transition through the NF-κB signaling pathway. Cell Death Discov. 2023;9:206.10.1038/s41420-023-01492-0Search in Google Scholar PubMed PubMed Central
[38] Yang X, Liu R. Long non-coding RNA HCG18 promotes gastric cancer progression by regulating miRNA-146a-5p/tumor necrosis factor receptor-associated factor 6 axis. Bioengineered. 2022;13:6781–93.10.1080/21655979.2022.2034565Search in Google Scholar PubMed PubMed Central
[39] Kim MJ, Min Y, Jeong SK, Son J, Kim JY, Lee JS, et al. USP15 negatively regulates lung cancer progression through the TRAF6-BECN1 signaling axis for autophagy induction. Cell Death Dis. 2022;13:348.10.1038/s41419-022-04808-7Search in Google Scholar PubMed PubMed Central
© 2024 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Research Articles
- Antitumor activity of 5-hydroxy-3′,4′,6,7-tetramethoxyflavone in glioblastoma cell lines and its antagonism with radiotherapy
- Digital methylation-specific PCR: New applications for liquid biopsy
- Synergistic effects of essential oils and phenolic extracts on antimicrobial activities using blends of Artemisia campestris, Artemisia herba alba, and Citrus aurantium
- β-Amyloid peptide modulates peripheral immune responses and neuroinflammation in rats
- A novel approach for protein secondary structure prediction using encoder–decoder with attention mechanism model
- Diurnal and circadian regulation of opsin-like transcripts in the eyeless cnidarian Hydra
- Withaferin A alters the expression of microRNAs 146a-5p and 34a-5p and associated hub genes in MDA-MB-231 cells
- Toxicity of bisphenol A and p-nitrophenol on tomato plants: Morpho-physiological, ionomic profile, and antioxidants/defense-related gene expression studies
- Review Articles
- Polycystic ovary syndrome and its management: In view of oxidative stress
- Senescent adipocytes and type 2 diabetes – current knowledge and perspective concepts
- Seeing beyond the blot: A critical look at assumptions and raw data interpretation in Western blotting
- Biochemical dynamics during postharvest: Highlighting the interplay of stress during storage and maturation of fresh produce
- A comprehensive review of the interaction between COVID-19 spike proteins with mammalian small and major heat shock proteins
- Exploring cardiovascular implications in systemic lupus erythematosus: A holistic analysis of complications, diagnostic criteria, and therapeutic modalities, encompassing pharmacological and adjuvant approaches
Articles in the same Issue
- Research Articles
- Antitumor activity of 5-hydroxy-3′,4′,6,7-tetramethoxyflavone in glioblastoma cell lines and its antagonism with radiotherapy
- Digital methylation-specific PCR: New applications for liquid biopsy
- Synergistic effects of essential oils and phenolic extracts on antimicrobial activities using blends of Artemisia campestris, Artemisia herba alba, and Citrus aurantium
- β-Amyloid peptide modulates peripheral immune responses and neuroinflammation in rats
- A novel approach for protein secondary structure prediction using encoder–decoder with attention mechanism model
- Diurnal and circadian regulation of opsin-like transcripts in the eyeless cnidarian Hydra
- Withaferin A alters the expression of microRNAs 146a-5p and 34a-5p and associated hub genes in MDA-MB-231 cells
- Toxicity of bisphenol A and p-nitrophenol on tomato plants: Morpho-physiological, ionomic profile, and antioxidants/defense-related gene expression studies
- Review Articles
- Polycystic ovary syndrome and its management: In view of oxidative stress
- Senescent adipocytes and type 2 diabetes – current knowledge and perspective concepts
- Seeing beyond the blot: A critical look at assumptions and raw data interpretation in Western blotting
- Biochemical dynamics during postharvest: Highlighting the interplay of stress during storage and maturation of fresh produce
- A comprehensive review of the interaction between COVID-19 spike proteins with mammalian small and major heat shock proteins
- Exploring cardiovascular implications in systemic lupus erythematosus: A holistic analysis of complications, diagnostic criteria, and therapeutic modalities, encompassing pharmacological and adjuvant approaches

