Abstract
Coronavirus disease 2019 (COVID-19) is a novel disease that had devastating effects on human lives and the country’s economies worldwide. This disease shows similar parasitic traits, requiring the host’s biomolecules for its survival and propagation. Spike glycoproteins severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2 spike protein) located on the surface of the COVID-19 virus serve as a potential hotspot for antiviral drug development based on their structure. COVID-19 virus calls into action the chaperonin system that assists the attacker, hence favoring infection. To investigate the interaction that occurs between SARS-CoV-2 spike protein and human molecular chaperons (HSPA8 and sHSP27), a series of steps were carried out which included sequence attainment and analysis, followed by multiple sequence alignment, homology modeling, and protein–protein docking which we performed using Cluspro to predict the interactions between SARS-CoV-2 spike protein and human molecular chaperones of interest. Our findings depicted that SARS-CoV-2 spike protein consists of three distinct chains, chains A, B, and C, which interact forming hydrogen bonds, hydrophobic interactions, and electrostatic interactions with both human HSPA8 and HSP27 with −828.3 and −827.9 kcal/mol as binding energies for human HSPA8 and −1166.7 and −1165.9 kcal/mol for HSP27.
Introduction
Viruses show parasitic traits or tendencies in which their infection into the host benefits the virus by increasing its pathogenesis [1]. The coronavirus disease 2019, also known as COVID-19, shows similar parasitic traits in which it requires the host’s biomolecules for its survival and propagation, and molecular chaperons are among the biomolecules targeted by the virus. This review aims to review the role played by molecular chaperons upon the invasion by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus to the human host, as these molecules are wildly associated with viral development [1].
COVID-19, a highly pathogenic novel disease caused by coronavirus belonging to the Coronaviridae family, was first discovered in China, Wuhan city, in December 2019 [2]. SARS-CoV-2, like any human coronaviruses (hCoVs), is a single-stranded, positive sense RNA-enveloped virus with a genome size of approximately 29.9 kb [3].
The single-stranded RNA of the viral genome consists of 14 open reading frames (ORFs) that encode for 16 nonstructural proteins (nsp1–16), 9 accessory proteins (ORF), and 4 structural proteins, which are nucleocapsid (N), membrane (M), envelope (E), and spike (S) protein also known as surface glycoproteins (Figure 1) [4]. The nucleocapsid above forms a capsid that plays a role in delivering the genome of the virus into the interior of the host cells and in protecting the viral genome from extensive environmental conditions.
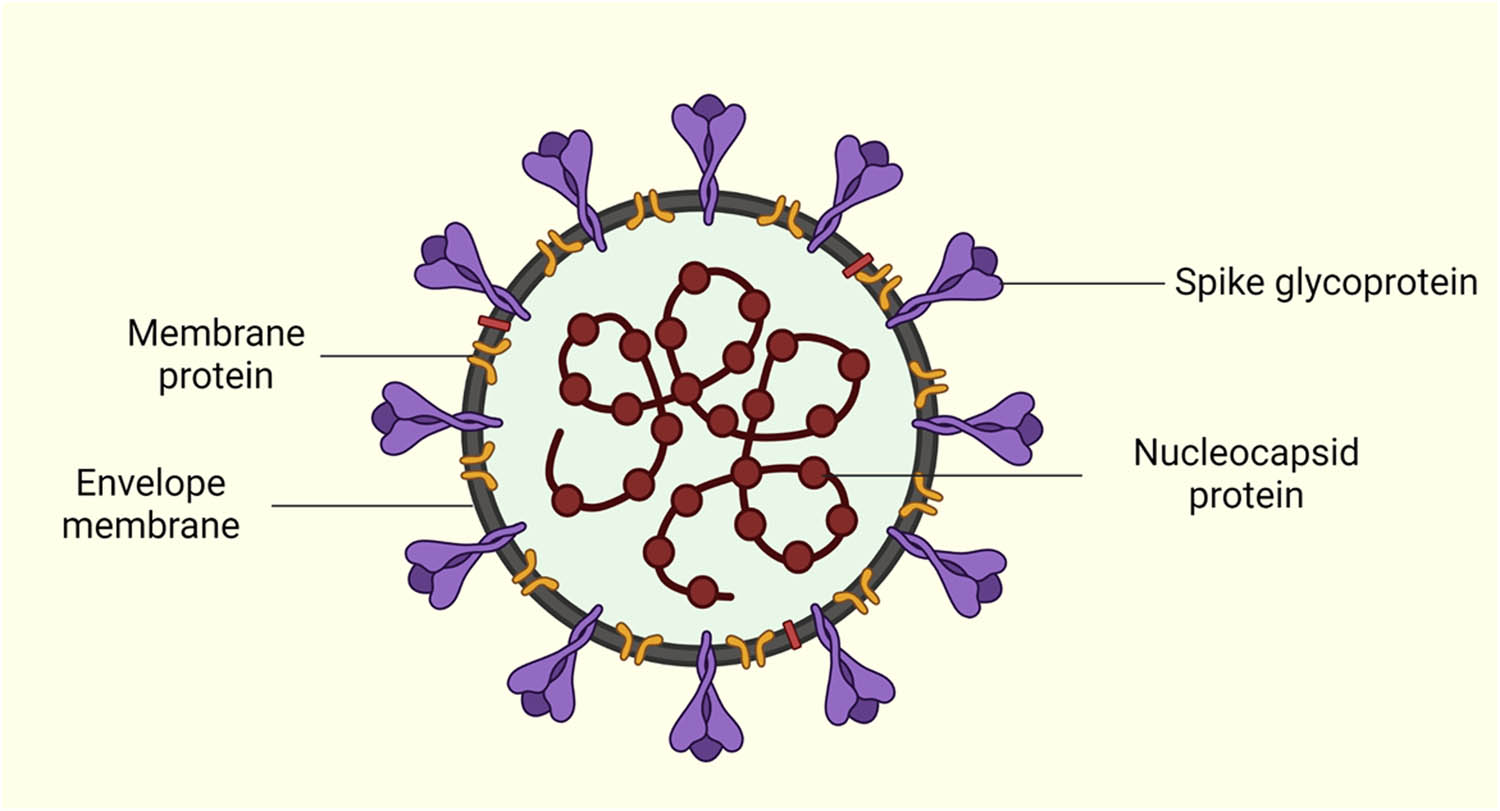
SARS-CoV-2 virus depicting the location of the nucleocapsid (N), membrane (M), envelope (E), and spike (S) protein. “Created by Biorender.com”.
Spike glycoproteins
Spike glycoproteins/spike proteins (SARS-CoV-2 spike proteins), as one of the SARS-CoV-2 proteins, potentially serve as hotspots for antiviral drug development based on their structure. SARS-CoV-2 spike proteins are transmembrane glycoproteins that exist as trimeric proteins that protrude from the virus’s surface. The SARS-CoV-2 spike proteins mediate SARS-CoV-2 entry into the host cells and subsequent pathogenesis; hence, it is a potential antiviral target [3]. SARS-CoV-2 spike proteins consist of two functional subunits, S1 and S2 (Figure 2). The S1 subunit contains a receptor-binding domain (RBD) and an N-terminal domain. The pivotal role of the S1 subunit in COVID-19 viral entry and subsequent pathogenesis is to identify and bind to the host cell receptors (viral attachment) [3,5]. The S2 subunits comprise fusion peptides, including heptad repeat 1 (HR1) and heptad repeat 2 (HR2) hence the trimeric protein. The role of the S2 subunit is to facilitate the in-between fusion of the host cell membrane and the viral-enveloped membrane [5].
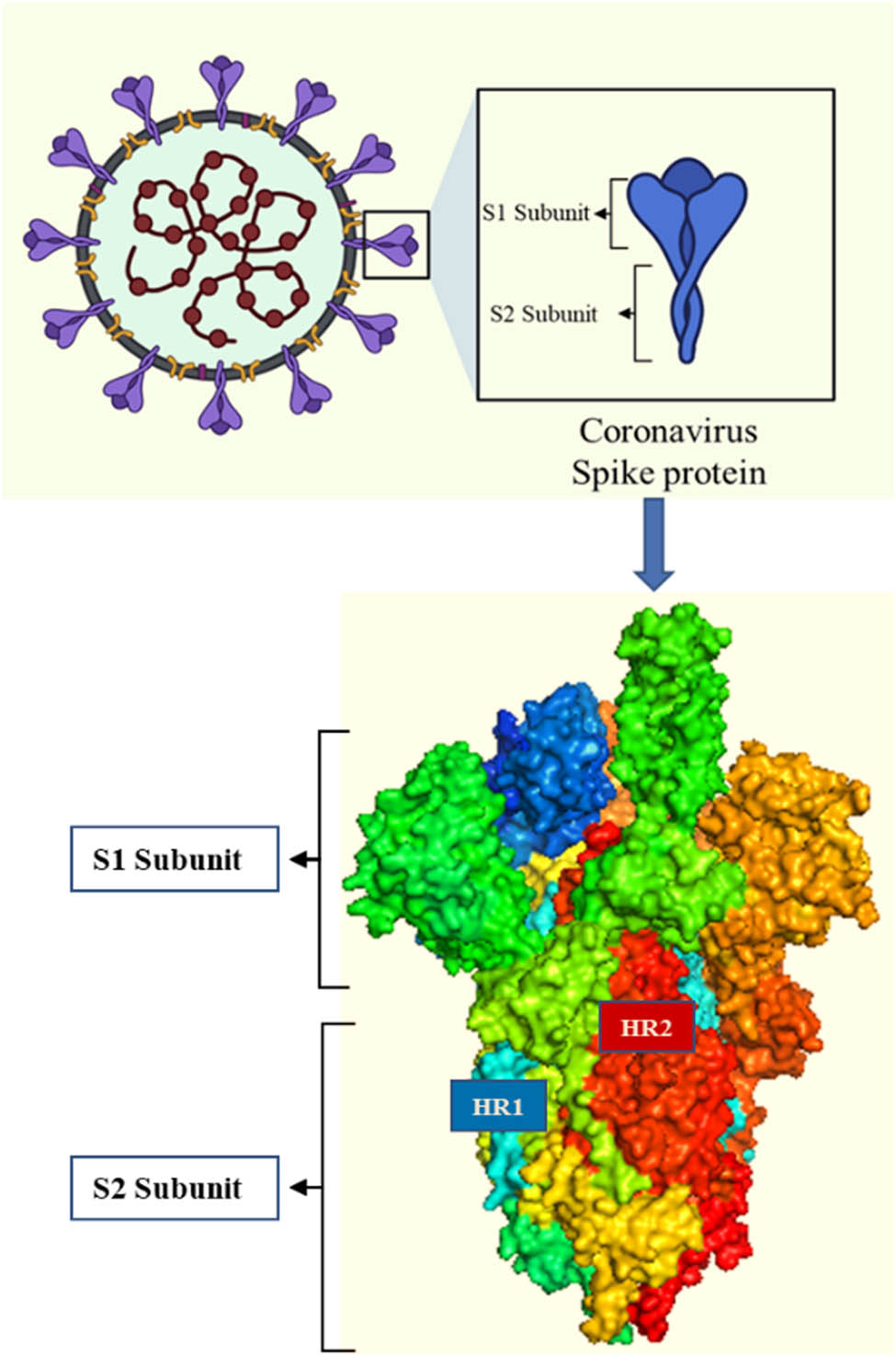
Trimeric structure of the SARS-CoV-2 spike protein (created by Biorender.com).
Spike proteins are essential for viral entry into the host cell. According to a study by Hu et al. [6], coronavirus exhibits spike proteins (SARS-CoV-2 spike protein) that cover the virus’s surface. The spike proteins mediate viral entrance by attaching to angiotensin-converting enzyme 2 (ACE2), which acts as the host’s cell receptor [6]. Spike proteins exist in a metastable prefusion conformation. When SARS-CoV-2 interacts with a host cell, external structural rearrangement of the spike proteins happens, which permits the virus to combine with the host’s cell membrane [7].
According to recent studies, SARS-CoV-2 utilizes trimetric spike proteins to attach to the host’s ACE2 (receptor) and combine with the cell membrane to gain cell entry [8]. The fusion of spike proteins and the host cell membrane process and gaining cell entrance is a multi-step process involving three different and separate spike protein cleavage events that prime the SARS-CoV-2 for interaction with ACE2, membrane fusion, and cell entry [8]. Huang et al. [7] conducted a study on SARS-CoV-2 spike protein proteins’ structural and functional capabilities in developing potential antiviral drugs for COVID-19. The research study highlighted the current research advances in the arrangement, utility, and progress of antivirus drugs aiming at the coronavirus spike proteins [7].
No paucity of research investigates the establishment of binding sites of the SARS-CoV-2 spike protein that mediate the viral entry with interactions with human HSPA8 and HSP27 or heat shock proteins (HSPs) for that matter toward the synthesis of COVID-19 treatment that will target those binding sites and the spike proteins and prevent viral entry in the first place. With SARS-CoV-2 virus still posing imminent threats due to lack of treatment, drug development needs more attention, specifically working on drugs that treat the virus [9]. Therefore, it seemed potent to carry out a research study that primarily investigates the establishment of the binding sites of the spike proteins from SARS-CoV-2 through the interaction with human HSPA8 and HSP27 through sequence analysis and alignment, homology modeling, and protein–protein docking toward the synthesis of COVID-19 treatment.
COVID-19 virus, like other viruses and infectious agents, calls into action the chaperonin system, which under normal circumstances defends the host organism being infected. Still, however, in the event of infection, they may assist the attacker. This scenario is called chaperonopathy by mistake, in which the infectious agent (COVID-19 virus) commands the chaperoning systems of the host organism, redirecting the usual activities to favor infection [10].
Different viruses exploit different strategies for viral attachment and endocytosis, viral penetration, and uncoating, viral assembly and budding, and lastly, viral transcription and replication within the host [3]. Despite all the differences, there is one emerging common principle that is viruses co-opt the host’s molecular chaperons to proselytize distinct entry steps. In the case of the SARS-CoV-2 virus, the role of chaperones and the mechanism by which the virus hijacks the host’s molecular chaperones are still poorly known [1]. From recent studies, SAR-CoV and SARS-CoV-2 have been found to enter the host cells using the same receptor which is ACE2 via the RBD located on spike proteins; hence, it is presumed that the two viruses might share a similar mechanism in hijacking the host’s chaperoning system [1,3].
The binding of the spike proteins on the receptor initiates endocytosis of the viral particles, targeting the endosome and resulting in the subsequent release of the nucleocapsid into the cytosol [11]. Within the cytosol, the endoplasmic reticulum (ER) then assumes a vital role during the replication cycle of the virus, with the virus expropriating the activities of the ER-associated chaperones that are generated via viral-ER-induced stress [1,11]. In fact, there have been speculations that ER molecular chaperones facilitate hCoVs’ entry into host cells. For example, both bCoV-HKU9 beta coronavirus and MERS-CoV’s spike proteins are known to engross the chaperone BiP, which normally mediates protein degradation and folding pathways in the ER, but in this case, it assists in cellular uptake of the viruses [1,11]. The viral RNA is translated and transcribed via the interaction of ER molecular chaperones (for example, calnexin) ensuring correct and proper folding and then the nucleocapsid proteins and the genomic RNA combine forming new viral particles [1].
From recent studies, it is presumed that the viral load is impacted by the level of expression of chaperones. Due to viral replication, there is an increase in the accumulation of unfolded, nascent viral polypeptide in the ER exceeding its folding capacity which is the result of the ER stress [1,11]. This ER stress activates cell-signaling pathways that regulate gene expression at both translational and transcriptional levels, which raises the level of chaperones. If gene-encoding chaperones are activated and the level of chaperone expression in ER increases, the maturation, folding, and degradation of proteins resumes and this includes the viral ones [1]. This in turn results in the restoration of homeostasis, thereby assuring viral survival.
Molecular chaperones – HSPs
HSPs also known as stress proteins were discovered in a heat shock response by Ritossa in 1962. These stress proteins were named HSPs based on their increased synthesis after heat shock in houseflies [12]. HSPs are ubiquitous that is, they exist in all organisms, from prokaryotes to eukaryotes, and they come in distinct forms categorized into different families [13]. As multimolecular complexes, HSPs are expressed constitutively, 5–10% of the existing total protein under normal growth conditions [12,14]. There is compelling evidence that HSPs play essential cellular physiological roles in situations involving systemic and cellular stress and also under normal circumstances [13]. These roles include protein folding, refolding of misfolded proteins, degradation or repair of proteins, and intracellular transportation of proteins in the cytosol, mitochondria, and the ER [12].
Eukaryote HSP genes are divided into several families or categories and named based on their molecular mass in Daltons (Da) (for example, small heat shock proteins [sHSPs], HSP40, HSP60, HSP70, HSP90, and HSP100), function, and sequence homology [15]. Several of these family members consist of counterparts that may be referred to as heat shock cognates expressed under non-stress normal conditions, for example, heat shock cognate 70 also known as heat shock 70 kDa protein 8 (HSPA8) [15] (Table 1).
Cellular locations and functions of HSPs in mammals [15]
| Protein | Molecular weight (kDa) | Cell localization | Function |
|---|---|---|---|
| sHSPs | 10–40 | Nucleus, cytoplasm | Antiapoptotic (apoptosis inhibitors), protection against stress. Cytoskeleton/microfilament stabilization |
| HSP60 | 58–65 | Cytoplasm, mitochondria | Protein refolding, prevention of protein aggregation of denatured proteins, chaperonin, and required in growth at elevated temperatures. |
| HSP70 | 66–78 | ER, nucleus, mitochondria, cytoplasm | Required for assembling proteins, transportation of proteins into the ER, protein secretion, protein folding and refolding, cytoprotection, and downregulation of HSF1. |
| HSP90 | 82–90 | Nucleus, cytoplasm | Crucial in the translocation of proteins, viability, and essential in steroid hormone receptor regulation. |
| HSP100 | 97–120 | Nucleolus, cytoplasm, chloroplast, nucleus | Folding of proteins, thermotolerance, |
Generally, HSP expression can be inducive or constitutive based on cellular conditions. Under stress conditions, heat shock factors (HSFs) generated as a portion of a heat shock response upregulate inducible HSPs that maintain cell homeostasis and generate cellular survival functions [16]. Under physiological conditions, HSPs function as molecular chaperones assisting in proper folding of newly synthesized proteins and employing housekeeping functionality [16].
Small HSPs
sHSPs are a unique family of molecular chaperones that are devoid of the ATPase domain. These are classified as tiny proteins, ranging in size from 10 to 40 kDa, with a-crystallin domain at their core and varying N- and C-terminal domains on each side. The crystalline domain, which is present in sHSPs, has subunit molecular weights of 10–40 kDa. Both HSP27, alpha-crystalline and beta-crystallin, are sHSP family members that form sizable oligomeric species. The focus of this review is mostly on mammalian sHSPs. According to sequence homology, the human genome contains ten sHSPs (designated HSPB1 to HSPB10). For ease of identification, HSPB1 was referred to as HSP27, HSPB4 as alpha-crystallin, HSPB5 as beta-crystallin, HSPB6 as HSP20, and HSPB8 which is their previous nomenclature [17]. The sHSPs have been divided into Classes I or II [18]. Class I sHSPs include HSP27, beta-crystallin, HSP20, and HSP22. These are extensively distributed, present in a variety of tissues (such as eye lenses, muscles, and heart), and predominately heat-inducible (HSP22 heat-inducibility is cell type dependent, for example, in the heart the functionality of HSP22 is dependent on BCL-2-associated athanogene 3 [BAG3]) [19] and crucial for cell survival in high-stress situations [20]. The Class II sHSPs consist of a tissue-specific pattern of expression (for example, alpha-crystallin is expressed in genomic stability, chaperone activity, and refractive index of the eye lens; the chaperons HSPB3, HSPB7, and HSPB2 are expressed in the maintenance of myofibrillar integrity) [17,19]. Members of the class II sHSPs include beta-crystallin, HSPB2, HSPB7, HSPB3, HSPB9, and HSPB10. It is thought that Class II sHSPs have a significant impact on development, differentiation, and specialized tissue-specific functions [18].
In addition to their molecular chaperone-like function in the prevention of peptide and protein aggregation, sHSPs like B-crystallin and HSP27 are involved in a variety of cellular processes including cytoskeletal integrity maintenance, protein folding, degradation of proteins, stress tolerance, cell death, differentiation, cell cycle, signal transduction, and growth [21]. Proteins belonging to the HSP family have interactions with multiple substrates and display cardio and neuroprotection, pro-angiogenic property, powerful anti-apoptotic activity, and anti-inflammatory function. Uncertainty surrounds the underlying molecular mechanism driving the promiscuous interactions and pleiotropic activities of sHSPs. These functions of sHSPs have significant effects on both general health and illness situations. Therapeutic targets have been suggested to include HSP27 and alpha- beta-crystallins [22].
Functions of sHSP27
Several factors, including heat shock, ischemia, and hemodynamics, can cause physiological stress, which alters a cell’s structure and metabolic functions. The capacity of HSP27 to speed up protein synthesis and RNA synthesis after heat shock exposure may give the cell a survival edge [23]. Stress proteins’ ability to act as molecular chaperones is regarded to be cytoprotective in nature. Particularly, it appears that HSPS90, HSPS70, HSPS47, HSPS32, and HSPS27 play a significant role after cardiac ischemia, ischemic preconditioning, vascular wall injury, and cardiac hypertrophy [23].
Bacteria and eukaryotic cells have HSPs while viruses do not have HSPs and they rely on the HSP from the host cells for the viral folding of proteins. HSP27 was discovered to be an infection factor for many viruses including Zika Virus and SARS-Cov-2 infection where the researchers induced and suppressed HSP27 expression and resulted in increasing and decreasing the production of the Zika virus particles [24]. They discovered that the HSP27 molecular chaperone plays an important role in cellular processes like protein folding, translation and degradation, and replications of viruses. Since SARS-CoV-2 cannot manufacture proteins on its own when it infects a human host, it takes advantage of the host to produce proteins like spike proteins [24].
HSP27 promotes chaperonin activity, assisting in the refolding of partially denatured proteins into active conformations [25,26,27]. HSP27 interferes with F-actin regulation, prevents F-actin polymerization [28,29], and acts by obstructing the cell death pathway to protect the cell against apoptosis. HSP27 is required for the presentation of oxidized proteins to the proteasome breakdown mechanism [30].
Coronavirus and the selected HSP27
HSPs can be produced when pathogenic bacteria enter the host cell. According to a recent study, the chaperone HSP27 stabilizes the expression of melanoma differentiation-associated gene 5 (MDA5) to prevent viral replication, positively regulating the RLR/MDA5 signaling pathway that is activated by the encephalomyocarditis virus. Large oligomeric species are formed by several sHSPs’ family members, including HSP27, alpha-crystallin, and beta-crystallin. It has been suggested that sHSPs participate in viral replication. According to research, HSP27 was reported to be rapidly elevated in coronaviruses and other viruses, indicating its function in early replication. Little has been recognized about HSP27’s role in the SARS-CoV-2 infectious cycle, but it has been suggested as a possible drug target for SARS-CoV-2 due to its participation in the stabilization of MDA5 in the prevention of viral replication [31]. HSP27 is speculated to interact with MDA5 and it has been noted that an over-expression of HSP27 significantly increases the expression of MDA5. Hence, it was postulated that HSP27 specifically stabilizes MDA5 [31]. It is crucial to consider how viruses, in general, hijack chemicals found in the human host to create potent and long-lasting treatments for COVID-19.
HSP family 70
HSP 70 (HSP70) family is the most relevant HSP of all the HSP families comprising stress-inducible HSP70 proteins (for example, 72-kDa protein also known as HSP72), nuclear and cytosolic constitutive HSP70 proteins (for example, HSC70/HSP73/HSPA8), mitochondrial mtHSP70 (for example, Grp75), and the endoplasmic BiP (for example, GRP78) [16]. HSP70s are found in all surviving domains of life. Structurally and at a molecular level, HSP70s contain three functional domains, a nucleotide-binding domain (NBD) connected to a substrate-binding domain (SBD) by a flexible and long hydrophobic linker, and a C-terminal peptide-binding domain [32,33]. Additionally, cytosolic eukaryotic HSP70s consist of a particular Glu-Glu-Val-Asp motif at the C-terminal extremes that is vital for interaction with the co-chaperones for HSP that regulates the HSP70 ATPase activity and its capabilities of binding to a substrate [34,35].
HSP70 molecular chaperones are involved in a variety of processes that include maintaining cell homeostasis, folding newly synthesized proteins, subcellular transport of proteins, formation and dissociation of complexes, stabilization of protein substrates against denaturation, or aggregation under adverse conditions, such as viral infection, heat, or acid. They rarely perform these processes on their own that is they require assistance from co-chaperones that speed up ATP hydrolysis and stabilize HSP70-substrate interactions. Some co-chaperones include J-domain containing HSP40, HSP90, eukaryotic HSP110, and nucleotide exchange factors [36]. HSP70s have been a hot spot for extensive study due to their different functions according to their location, for example, extracellular HSP70s carry out immunomodulatory tasks that activate tolerance responses and immunological responses, whereas intracellular HSP70s carry out cytoprotective functions as molecular chaperon proteins [16].
Apart from the diverse cellular functions performed by HSP70s, they are hijacked by viruses and included in their life cycle mediating viral attachment and endocytosis, viral penetration and uncoating, viral assembly and budding, and lastly viral transcription and replication within the host. Because of these extensive roles of HSP70, it has been the antiviral target to inhibit virus infection [3].
Coronavirus and selected human HSP70
Amongst members of the HSP70 family, heat shock 70 kDa protein 8 (HSPA8), also known as heat shock cognate (Hsc70), is reportedly involved in viral life cycle regulation. HSPA8 as a constitutive HSP70 protein plays an essential role in targeting proteins to lysosome machinery for degradation, protein translocation, and protein refolding [35]. Apart from the previously mentioned diverse cellular functions of HSPA8, it, however, mediates viral attachment and endocytosis, viral penetration and uncoating, viral assembly and budding, and lastly, viral transcription and replication in the virus’s life cycle within the host. Because of these extensive roles of HSPA8, it has been the antiviral target to inhibit virus infection [3].
Methods and materials
To investigate the interaction that occurs between SARS-CoV-2 spike protein (accession number YP_009724390.1) and the selected HSPs, human HSP27 (accession number P04792) and human HSPA8 (accession number AAH19816.1), a series of steps were carried out which included sequence attainment and analysis performed using NCBI [36] and UniProt [4]. Sequences of the SARS-CoV-2 spike protein, human HSP27, and human HSPA8 with the previously mentioned accession numbers attained and analyzed. This was followed by multiple sequence alignment of sequences of spike proteins of known hCoVs using Bioedit software [37]. Multiple sequence alignment of sequences of selected human HSP27 and human HSPA8 was performed against those sequences that are closely related to human HSP27 in case of HSP27 and those that are closely related to human HSPA8 for HSPA8 obtained from BLAST under NCBI [36]. Homology modeling was performed using Phyre2 [38], to attain 3D structures of the spike proteins, human HSP27 and human HSPA8, which were visualized using Chimera, PyMOL [39], and Discovery studio [40]. Protein–protein docking was performed using Cluspro to predict the interactions between SARS-CoV-2 spike protein and human HSPA8, and SARS-CoV-2 spike protein and human HSP27. Both human HSP27 and human HSPA8 were docked against each individual chain of the SARS-CoV-2 spike protein and the interactions were analyzed using BIOVIA Discovery studio [40].
Results and discussion
Interactions of human HSP27 with SARS-CoV-2 spike protein
The multiple sequence alignment result of the human HSP27 from different mammals Homo sapiens, Mus musculus, Sus scrofa, Bos taurus, Macaca mulatta, Equus caballus, Gorrilla gorilla, Felis catus, Canis lupus, Poeciliopsis lucida, Cricetulus longicaudatus, and Rattus norvegicus that indicate that the amino acid residues of HSP27 are highly conserved (in black) which indicates that they have the same biochemical functions and similar structures (Figures 3 and 4) [37].
![Figure 3
Sequence alignment of mammalian HSP27 from 12 different mammalian species (amino acid residues 1–215); Homo sapiens (human), Mus musculus (Mouse), Sus scrofa (pig), Bos taurus (cow), Macaca mulatta (Rhesus macaque), Equus caballus (Horse), Gorrilla gorilla (Gorilla), Felis catus (Cat), Canis lupus (Dog), Poeciliopsis lucida (Desert topminnow), Cricetulus longicaudatus (Chinese Hamster), and Rattus norvegicus (laboratory rat) to show 161 the similarities between the protein sequences [37].](/document/doi/10.1515/bmc-2022-0027/asset/graphic/j_bmc-2022-0027_fig_003.jpg)
Sequence alignment of mammalian HSP27 from 12 different mammalian species (amino acid residues 1–215); Homo sapiens (human), Mus musculus (Mouse), Sus scrofa (pig), Bos taurus (cow), Macaca mulatta (Rhesus macaque), Equus caballus (Horse), Gorrilla gorilla (Gorilla), Felis catus (Cat), Canis lupus (Dog), Poeciliopsis lucida (Desert topminnow), Cricetulus longicaudatus (Chinese Hamster), and Rattus norvegicus (laboratory rat) to show 161 the similarities between the protein sequences [37].
Table S1 shows the types of intermolecular bonds involved in the three chains of SARS-CoV-2 spike protein with the HSP27 and the binding energy (kcal/mol) involved in the interaction of HSP27 with each chain of SARS-CoV-2 spike protein in which the interactions were the same and results were obtained after docking the two proteins and visualized by Discovery Studio [40].
Amino acids of SARS-CoV-2 spike protein with multiple interactions were identified interacting with amino acids of HSP27 and visualized the segments of amino acids residue of SARS-CoV-2 spike protein with amino acids interacting with HSP27 showing the type of intermolecular bonds involved shown in Table S2 [40].
The 2D diagrams, as shown above in Figure 5a, show interactions between amino acids from the SARS-CoV-2 spike protein forming intermolecular bonds with amino acids from human HSP27. Part of the amino acids from SARS-CoV-2 spike protein forming bonds with HSP27 was selected, consisting of amino acids from positions 710–714 (Arg20, Tyr23, Trp22) that formed conventional hydrogen bonds, carbon hydrogen bond, and electrostatic interaction with the amino acids from the human HSP27. Amino acid Ser711 formed carbon–hydrogen bonds with Trp22, amino acid Ile712 formed hydrophobic (Alkyl) interactions with Arg20, and amino acid Ala713 formed conventional hydrogen bonds with Trp22, Typ23, and Arg20 from the human HSP27. This contributes to the formation of a stable complex between the two proteins [40].
![Figure 5
(a) The intermolecular bonds involved around amino acids 710, 711, 712, 713, and 714 of SARS-CoV-2 spike protein interacting with amino acids (Arg20, Tyr23, Trp22) humans HSP27. (b) The intermolecular bonds involved around amino acids 1,037, 1,038, 1,039, 1,040, and 1,041 of SARS-CoV-2 spike protein interacting with amino acids (Pro39, Gln44, Glu41, Trp45, Leu38, Gly48) human HSP27. (c) The intermolecular bonds involved around amino acids 703, 704, 705, 706, and 707 of SARS-CoV-2 spike protein interacting with amino acids (Asp21, Trp22, Tyr23) human HSP27 [40].](/document/doi/10.1515/bmc-2022-0027/asset/graphic/j_bmc-2022-0027_fig_005.jpg)
(a) The intermolecular bonds involved around amino acids 710, 711, 712, 713, and 714 of SARS-CoV-2 spike protein interacting with amino acids (Arg20, Tyr23, Trp22) humans HSP27. (b) The intermolecular bonds involved around amino acids 1,037, 1,038, 1,039, 1,040, and 1,041 of SARS-CoV-2 spike protein interacting with amino acids (Pro39, Gln44, Glu41, Trp45, Leu38, Gly48) human HSP27. (c) The intermolecular bonds involved around amino acids 703, 704, 705, 706, and 707 of SARS-CoV-2 spike protein interacting with amino acids (Asp21, Trp22, Tyr23) human HSP27 [40].
The interactions are also shown in Figure 5b using amino acids from the SARS-CoV-2 spike protein from position 1,037 to1,040 (Pro39, Glu41, Leu38, and Gly48) interacting with amino acids from human HSP27 where amino acid Lys1038 on the spike protein formed conventional hydrogen bonds with Gln44, carbon–hydrogen bond with Gly48, and electrostatic (attractive charge) interactions with Trp45, and Arg1039 formed electrostatic (attractive charge) interactions with Glu41 and formed conventional hydrogen bonds with Pro39. VAL1040 formed hydrophobic (Alkyl) interactions with Leu38, and lastly, Phe1042 formed conventional hydrogen bonds with Pro39 from human HSP27 [40].
Amino acids from position 703–707 (Asp21, Trp22, Tyr23) from spike protein were used to show other interactions between the two proteins. The amino acid residues Ser704 and Ala706 formed conventional hydrogen bonds with Tyr23 from human HSP27, Val705 formed hydrophobic (pi-sigma) interactions with Trp22 and Tyr3 of HSP27, and Tyr707 formed conventional hydrogen bonds with Asp21 [40].
Interactions between human HSPA8 and SARS-CoV-2 spike protein
Sequence analysis of human HSPA8 (with accession number AAH19816.1) indicated that the molecular chaperon is located on chromosome 11 and has 646 amino acid residues (Table S3). A heat shock 70 kDa protein domain was depicted at positions 1–613 and the chaperon has an N-terminal NBD at positions 2–386, and an SBD at positions 349–509 (Table S3) [41] that makes part of a class of proteins associated with membrane protein complexes for signal transduction or transport.
From previous preliminary studies, HSPA8 has been known to be localized in the cytoplasm and nucleus and shuttles between the cytoplasm and nucleus to perform various functions [3]. However, the results from sequence analysis of the HSPA8 postulated that the human HSPA8 is localized in the melanosomes and the cell surface membrane (Table S3) where it acts as a membrane-anchored protein expressed in the cytoplasmic membranes of distinctive cell linings that include smooth muscle cells, respiratory tract cells, and the epithelia of the small intestine [1]. The location of the human HSPA8 on the cell surface membrane is convenient for the molecular chaperon to act as a receptor that binds with the SARS-CoV-2 spike protein, thereby mediating viral entry into the host cell [42].
Sequence analysis of the SARS-CoV-2 spike protein (accession number YP_009724390.1) (Table S4) indicated that the surface glycoprotein consists of 1273 amino acids [43,8] with the RBD at positions 319–541, an N-terminal domain at positions 14–303, and a C-terminal domain at positions 334–527 [32,33,34,35,36,37,38,39,40,41,42,43] (Figure 6). The previously mentioned domains make part of the S1 subunit of the spike protein [7] that major functions in viral attachment during the initial stages of viral infection.
![Figure 6
3D structures of the human HSPA8 and SARS-CoV-2 spike protein. (a) 3D structure of human HSPA8 obtained from Phyre 2 as visualized by PyMOL indicating the N-terminal NBD and SBD. (b) Ramachandran plot for human HSPA8. (c) 3D structure of the SARS-CoV-2 spike protein obtained from Phyre 2 as visualized by PyMOL indicating RBD, C-terminal domain, N-terminal domain, fusion peptides (FP1 and FP2), and HR units (HR1 and HR2). (d) Ramachandran plot for SARS-CoV-2 spike protein obtained from PDBsum [44,39].](/document/doi/10.1515/bmc-2022-0027/asset/graphic/j_bmc-2022-0027_fig_006.jpg)
3D structures of the human HSPA8 and SARS-CoV-2 spike protein. (a) 3D structure of human HSPA8 obtained from Phyre 2 as visualized by PyMOL indicating the N-terminal NBD and SBD. (b) Ramachandran plot for human HSPA8. (c) 3D structure of the SARS-CoV-2 spike protein obtained from Phyre 2 as visualized by PyMOL indicating RBD, C-terminal domain, N-terminal domain, fusion peptides (FP1 and FP2), and HR units (HR1 and HR2). (d) Ramachandran plot for SARS-CoV-2 spike protein obtained from PDBsum [44,39].
Other essential segments or regions within the SARS-CoV-2 spike protein predicted from sequence analysis included the integrin-binding motif that is essential for cell-to-cell adhesion [45], a receptor-binding motif that binds to human host receptors, [46], fusion peptides, and HR units. These regions make part of the S2 membrane-fusion subunit and assist in viral fusion to the host cell membrane allowing viral genomes to penetrate the host cells. The presence of the S1 and S2 subunits substantiates that the spike protein is essential for viral attachment and fusion, the initial stages of viral infection since it has the necessary commodities required to carry out the function [47].
In the study, Bioedit software was utilized to perform multiple sequence alignment of the SARS-CoV-2 spike proteins with spike proteins from other known hCoVs and multiple sequence alignment of human HSPA8 with other closely related mammalian species attained from BLAST. The purpose of multiple sequence alignment was to align all sequences in the query set to determine and identify conserved domains across a group of evolutionarily related sequences [48].
Sequences of HSPA8 were attained from nine mammalian species that are closely related to the HSPA8 from H. sapiens using BLAST and the max score of the results. Figure 7 shows a section of multiple sequence alignment of the 10 sequences of mammalian HSPA8 (including one from H. sapiens), from amino residues 403–545 as they are the amino acid residues involved in the interaction with the amino acid residues from SARS-CoV-2 spike protein. The multiple sequence alignment (Figure 7) indicates that the amino acid residues of the functional domain in the human HSPA8, SBD at positions 349–509 (Table S3), are highly conserved throughout all the 10 different mammalian species of the same HSP from the selected amino acid residue region (from position 403 to 545) indicating that the SBD is more likely to be essential for the function of the HSPA8 [49].
![Figure 7
Sequence alignment of HSPA8 from 10 different mammalian species (amino residues 403–545); Homo sapiens, Fukomys damarensis, Manis javanica, Marmota monax, Equus caballus, Mus musculus, Bos taurus, and Microtus ochrogaster [37].](/document/doi/10.1515/bmc-2022-0027/asset/graphic/j_bmc-2022-0027_fig_007.jpg)
Sequence alignment of HSPA8 from 10 different mammalian species (amino residues 403–545); Homo sapiens, Fukomys damarensis, Manis javanica, Marmota monax, Equus caballus, Mus musculus, Bos taurus, and Microtus ochrogaster [37].
Multiple sequence alignment for the spike proteins (Figure 8) was performed from sequences of the SARS-CoV-2 spike proteins and the spike proteins from known hCoVs [50] using Bioedit. Figure 8 indicates the sequence alignment of the spike proteins from the coronaviruses from amino acid residues 1–245. The residues highlighted in red are the residues involved in the protein–protein interaction between the spike protein and human HSPA8 in Figure 5. For most of the sequence alignment (Figure 8), the amino acids are non-conserved with other regions being conserved and semi-conserved which are essential in viral entry and interaction with the host cell receptor.
![Figure 8
Sequence alignment of spike proteins from the six known hCoV, middle east respiratory syndrome-related coronavirus spike protein, human coronavirus HKU1 spike protein, human coronavirus 229E spike protein, human coronavirus NL63 spike protein, and human coronavirus OC43 spike protein. Amino residues in red are the residues involved in intermolecular interaction with human HSPA8 (amino acid residues 27–245) [37].](/document/doi/10.1515/bmc-2022-0027/asset/graphic/j_bmc-2022-0027_fig_008.jpg)
Sequence alignment of spike proteins from the six known hCoV, middle east respiratory syndrome-related coronavirus spike protein, human coronavirus HKU1 spike protein, human coronavirus 229E spike protein, human coronavirus NL63 spike protein, and human coronavirus OC43 spike protein. Amino residues in red are the residues involved in intermolecular interaction with human HSPA8 (amino acid residues 27–245) [37].
SARS-CoV-2 spike protein consists of three distinct chains, chains A, B, and C. These chains were docked separately with the human HSPA8 to predict whether the human HSPA8 interacts with each individual chain [51]. The amino acid residues from human HSPA8 (from positions 403 to 545) interacted with each individual chain of the spike protein forming hydrogen bonds, hydrophobic interactions, and electrostatic interactions. The binding energy from the interaction between human HSPA8 and chain A for the SARS-CoV-2 spike was −828.3 kcal/mol, and that between human HSPA8 and chain B, chain C for the SARS-CoV-2 spike was −827.9 kcal/mol (Table S5).
The negative binding energy value indicated that the interaction between the human HSPA8 and the SARS-CoV-2 spike protein occurs spontaneously without consuming energy. The stability of a protein complex is related to its binding energy that is the higher the binding energy the more stable the protein complex. Because the binding energy values of the docked molecules were high, it can be postulated and concluded that the complex formed from the interaction between the HSPA8 and SARS-CoV-2 spike protein is stable [52].
To better comprehend and visualize the chemical interactions between SARS-CoV-2 spike proteins and human HSPA8, 2D diagrams were generated for each individual chain of the spike protein (Figure 9). The 2-dimensional structure of Figure 9a was constructed using amino acids from the SARS-CoV-2 spike protein with the most interactions with amino acids from human HSPA8 [33]. A small chain of amino acids from SARS-CoV-2 spike protein chain A was selected, which consisted of amino acids from positions 136–140 (Cys136, Asn137, Asp138, Pro139, and Phe140) that formed conventional hydrogen bonds and electrostatic interaction with the amino acids from the human HSPA8. Amino acid Asn137 formed conventional hydrogen bonds with Arg469, and amino acid Asp138 formed electrostatic interactions with Arg469 and conventional hydrogen bonds with Arg469 and Asp433 from the human HSPA8. This indicates that the amino acids in the spike protein form extensive bonds with those from the human HSPA8, thereby contributing to a stable complex from the interactions [33].
![Figure 9
2D diagrams indicating the chemical interactions between SARS-CoV-2 spike proteins and human HSPA8. (a) Intermolecular interactions between selected amino acid residues of the SARS-CoV-2 spike proteins in chain A (Cys136, Asn137, Asp138, Pro139, and Phe140) and human HSPA8. (b) Intermolecular interactions between selected amino acid residues of the SARS-CoV-2 spike proteins in chain B (Phe65, His66, Ala67 Ile68, and His69) and human HSPA8. (c) Intermolecular interactions between selected amino acid residues of the SARS-CoV-2 spike proteins in chain C (Leu212, Val213, Arg214, Asp215, and Leu216) and human HSPA8 [33].](/document/doi/10.1515/bmc-2022-0027/asset/graphic/j_bmc-2022-0027_fig_009.jpg)
2D diagrams indicating the chemical interactions between SARS-CoV-2 spike proteins and human HSPA8. (a) Intermolecular interactions between selected amino acid residues of the SARS-CoV-2 spike proteins in chain A (Cys136, Asn137, Asp138, Pro139, and Phe140) and human HSPA8. (b) Intermolecular interactions between selected amino acid residues of the SARS-CoV-2 spike proteins in chain B (Phe65, His66, Ala67 Ile68, and His69) and human HSPA8. (c) Intermolecular interactions between selected amino acid residues of the SARS-CoV-2 spike proteins in chain C (Leu212, Val213, Arg214, Asp215, and Leu216) and human HSPA8 [33].
The 2-dimensional structure of Figure 9b was constructed using amino acids from the SARS-CoV-2 spike protein from positions 65–69 (Phe65, His66, Ala67 Ile68, and His69) with the most interactions with amino acids from human HSPA8 [33]. Multiple extensive bonds were depicted from the analysis showing multiple conventional hydrogen bonds and hydrophobic interactions. Amino acid HIS69 on the spike protein formed multiple conventional hydrogen bonds with Glu404, Gln441, and Tyr545 from human HSPA8 also forming hydrophobic (pi-alkyl and alkyl) interactions with amino acids Ile403 and Ile440. Other amino acid residues interacted with the human HSPA8 that is His66 from chain B interacted with Gly407 using hydrogen bonds, and Ala67 from chain B interacted with Leu542 and Tyr545 via hydrophobic interactions [33]. These interactions are highly extensive therefore more energy will be required to break these bonds making the protein complex formed from the interaction more stable. The interaction between human HSPA8 and chain B of the SARS-CoV-2 spike protein yields the most stable complex due to these multiple extensive bonds.
Chains A and C showed roughly similar interactions; hence, the complex formed from the interactions of the two chains and the human HSPA8 may be deemed roughly the same in terms of stability. From Figure 9c, amino acids from positions 212–216 (Leu212, Val213, Arg214, Asp215, and Leu216) from chain C from spike proteins were used to construct the 2D diagram showing the interactions between the two proteins of interest forming conventional hydrogen bonds, hydrophobic interactions, and electrostatic interactions[33]. The amino acid residues Val213 and Arg214 formed conventional hydrogen bonds with Lys451 from human HSPA8, Val213 interacts with Phe547 via hydrophobic interactions, Arg214 interacts with Asp452 via electrostatic interactions, and Asp215 interacts with Lys451 via hydrophobic interactions.
Conclusion
Similar to other viruses, COVID-19 targets the host’s chaperone system viral protein folding, viral assembly, and other essential steps in the virus’s life cycle with the host’s chaperones being the means necessary for viral infection, survival, and its spreading [1]. With the novelty of the virus and little knowledge on how COVID-19 targets the chaperon system, it is therefore pertinent to carry out a research that investigates the interactions between HSPs (HSP27 and HSPA8) with SARS-CoV-2 spike proteins resulting in the establishment of binding sites toward the synthesis of long-lasting treatment.
From the research findings, the aim to investigate the interaction between SARS-CoV-2 spike protein, and molecular chaperons, human HSP27, and HSPA8 was achieved successfully. The multiple sequence alignment indicated that the amino residues of the functional domains in the human HSPA8 and human HSP27 (that are conserved throughout all species) and those in SARS-CoV-2 spike proteins might perform similar functions in mammalian species and other known hCoVs [37]. Protein–protein docking produced highly detailed interactions between the molecular chaperones (human HSPA8 and human HSP27) and the SARS-CoV-2 spike protein chains. Further analysis of the interactions indicated that the binding energies were −828.3 and −827.9 kcal/mol for HSPA8 and −1166.7 and −1165.9 kcal/mol for HSP27. In conjunction with the multiple extensive bonds formed, it can be concluded that the protein complex formed from the interaction is stable; hence, a conclusion can be drawn that SARS-CoV-2 spike proteins target human HSPA8 and human HSP27 for its entry into host cells and its survival within the host.
This study provided proof of promising molecular targets (spike proteins, human HSP27, and human HSPA8) with high potential activity for further in vivo and in vitro experiments toward the synthesis of COVID-19 treatment. The identification of the interactions between SARS-CoV-2 spike proteins and human HSP27 and those between SARS-CoV-2 spike proteins and human HSPA8 provided the target regions that are required in finding appropriate and effective inhibitors for the interactions, toward drug development.
Acknowledgements
The authors would like to acknowledge the support and assistance from SA-MRC-Self-Initiated Research Grants.
-
Funding information: This work was funded by the South African Medical Research Council-Self Initiated Research Grants.
-
Author contributions: Liberty T. Navhaya, as the first author put together the concept that I as supervisor came with Dzveta Mutsawashe Blessing contributed almost equally with the first author. Mthembu Yamnkela contributed to the writing of some sections of the paper. Sesethu Godlo contributed in writing some sections of the manuscript and Xolani Henry Makhoba, I came up with the concept of the manuscript as a supervisor and assisted in structuring the manuscript.
-
Conflict of interest: The authors state no conflict of interest.
-
Data availability statement: Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References
[1] Paladino L, Vitale AM, Caruso Bavisotto C, Conway de Macario E, Cappello F, Macario AJ, et al. The role of molecular chaperones in virus infection and implications for understanding and treating COVID-19. J Clin Med. 2020 Oct;9(11):3518.10.3390/jcm9113518Search in Google Scholar PubMed PubMed Central
[2] Ciotti M, Ciccozzi M, Terrinoni A, Jiang WC, Wang CB, Bernardini S. The COVID-19 pandemic. Crit Rev Clin Lab Sci. 2020 Aug;57(6):365–88.10.1080/10408363.2020.1783198Search in Google Scholar PubMed
[3] Wang Z, Li Y, Yang X, Zhao J, Cheng Y, Wang J. Mechanism and complex roles of HSC70 in viral infections. Front Microbiol. 2020 Jul;11:1577.10.3389/fmicb.2020.01577Search in Google Scholar PubMed PubMed Central
[4] Ramasamy S, Subbian S. Critical determinants of cytokine storm and type I interferon response in COVID-19 pathogenesis. Clin Microbiol Rev. 2021 Jun;34(3):e00299-20.10.1128/CMR.00299-20Search in Google Scholar PubMed PubMed Central
[5] Velusamy P, Kiruba K, Su CH, Arun V, Anbu P, Gopinath SC, et al. SARS-CoV-2 spike protein: Site-specific breakpoints for the development of COVID-19 vaccines. J King Saud Univ-Sci. 2021 Dec;33(8):101648.10.1016/j.jksus.2021.101648Search in Google Scholar PubMed PubMed Central
[6] Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021 Mar;19(3):141–54.10.1038/s41579-020-00459-7Search in Google Scholar PubMed PubMed Central
[7] Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020 Sep;41(9):1141–9.10.1038/s41401-020-0485-4Search in Google Scholar PubMed PubMed Central
[8] Xia X. Domains and functions of spike protein in Sars-Cov-2 in the context of vaccine design. Viruses. 2021 Jan;13(1):109.10.3390/v13010109Search in Google Scholar PubMed PubMed Central
[9] Rabaan AA, Al-Ahmed SH, Sah R, Tiwari R, Yatoo M, Patel SK, et al. SARS-CoV-2/COVID-19 and advances in developing potential therapeutics and vaccines to counter this emerging pandemic. Ann Clin Microbiol Antimicrob. 2020 Dec;19(1):1–37.10.1186/s12941-020-00384-wSearch in Google Scholar PubMed PubMed Central
[10] Macario AJ, de Macario EC. Chaperone proteins and chaperonopathies. In: Stress: physiology, biochemistry, and pathology. Academic Press; 2019 Jan. p. 135–52.10.1016/B978-0-12-813146-6.00012-6Search in Google Scholar
[11] Speckhart K, Williams JM, Tsai B. How DNA and RNA viruses exploit host chaperones to promote infection. Viruses. 2021 May;13(6):958.10.3390/v13060958Search in Google Scholar PubMed PubMed Central
[12] Dubey A, Prajapati KS, Swamy M, Pachauri V. Heat shock proteins: a therapeutic target worth to consider. Vet World. 2015 Jan;8(1):46.10.14202/vetworld.2015.46-51Search in Google Scholar PubMed PubMed Central
[13] Kregel KC. Invited review: heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol. 2002 May;92(5):2177–86.10.1152/japplphysiol.01267.2001Search in Google Scholar PubMed
[14] Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988 Dec;22(1):631–77.10.1146/annurev.ge.22.120188.003215Search in Google Scholar PubMed
[15] Liang P, MacRae TH. Molecular chaperones and the cytoskeleton. J Cell Sci. 1997 Jul;110(13):1431–40.10.1242/jcs.110.13.1431Search in Google Scholar PubMed
[16] Mansilla MJ, Montalban X, Espejo C. Heat shock protein 70: roles in multiple sclerosis. Mol Med. 2012 Jun;18(6):1018–28.10.2119/molmed.2012.00119Search in Google Scholar PubMed PubMed Central
[17] Jahn M, Rehn A, Pelz B, Hellenkamp B, Richter K, Rief M, et al. The charged linker of the molecular chaperone HSP90 modulates domain contacts and biological function. Proc Natl Acad Sci. 2014 Dec;111(50):17881–6.10.1073/pnas.1414073111Search in Google Scholar PubMed PubMed Central
[18] Wendt R, Lingitz MT, Laggner M, Mildner M, Traxler D, Graf A, et al. Clinical relevance of elevated soluble ST2, HSP27 and 20S proteasome at hospital admission in patients with COVID-19. Biology. 2021 Nov;10(11):1186.10.3390/biology10111186Search in Google Scholar PubMed PubMed Central
[19] Sun X, Siri S, Hurst A, Qiu H. Heat Shock Protein 22 in physiological and pathological hearts: small molecule, large potentials. Cells. 2021 Dec;11(1):114.10.3390/cells11010114Search in Google Scholar PubMed PubMed Central
[20] Salzberger B, Glück T, Ehrenstein B. Successful containment of COVID-19: the WHO-Report on the COVID-19 outbreak in China. Infection. 2020 Apr;48(2):151–3.10.1007/s15010-020-01409-4Search in Google Scholar PubMed PubMed Central
[21] Zhang Z, Jing J, Ye Y, Chen Z, Jing Y, Li S, et al. Characterization of the dual functional effects of heat shock proteins (HSPs) in cancer hallmarks to aid development of HSP inhibitors. Genome Med. 2020 Dec;12(1):1–6.10.1186/s13073-020-00795-6Search in Google Scholar
[22] Pedersen SF, Ho YC. SARS-CoV-2: a storm is raging. J Clin Invest. 2020 May;130(5):2202–5.10.1172/JCI137647Search in Google Scholar
[23] Carper SW, Rocheleau TA, Cimino D, Storm FK. Heat shock protein 27 stimulates RNA and protein synthesis recovery following a heat shock. J Cell Biochem. 1997 Aug;66(2):153–64.10.1002/(SICI)1097-4644(19970801)66:2<153::AID-JCB3>3.3.CO;2-NSearch in Google Scholar
[24] Pujhari S, Macias VM, Nissly RH, Nomura M, Kuchipudi SV, Rasgon JL. Heat shock protein 70 (HSP70) is involved in the Zika virus cellular infection process. BioRxiv. 2017;8(1):8–16.10.1080/22221751.2018.1557988Search in Google Scholar
[25] Ciocca DR, Oesterreich S, Chamness GC, MCGuire WL, Fuqua SA. Biological and clinical implications of heat shock protein 27000 (HSP27): a review. JNCI: J Natl Cancer Inst. 1993 Oct;85(19):1558–70.10.1093/jnci/85.19.1558Search in Google Scholar
[26] Jakob U, Gaestel M, Engel K, Buchner J. Small heat shock proteins are molecular chaperones. J Biol Chem. 1993 Jan;268(3):1517–20.10.1016/S0021-9258(18)53882-5Search in Google Scholar
[27] Rogalla T, Ehrnsperger M, Preville X, Kotlyarov A, Lutsch G, Ducasse C, et al. Regulation of HSP27 oligomerization, chaperone function, and protective activity against oxidative stress/tumor necrosis factor α by phosphorylation. J Biol Chem. 1999 Jul;274(27):18947–56.10.1074/jbc.274.27.18947Search in Google Scholar
[28] Guay J, Lambert H, Gingras-Breton G, Lavoie JN, Huot J, Landry J. Regulation of actin filament dynamics by p38 map kinase-mediated phosphorylation of heat shock protein 27. J Cell Sci. 1997 Feb;110(3):357–68.10.1242/jcs.110.3.357Search in Google Scholar
[29] Lavoie JN, Hickey E, Weber LA, Landry J. Modulation of actin microfilament dynamics and fluid phase pinocytosis by phosphorylation of heat shock protein 27. J Biol Chem. 1993 Nov;268(32):24210–4.10.1016/S0021-9258(20)80512-2Search in Google Scholar
[30] Arrigo A-P. HSP27: novel regulator of intracellular redox State. IUBMB Life. 2001 Dec;52(6):303–7.10.1080/152165401317291156Search in Google Scholar
[31] Wan Q, Song D, Li H, He ML. Stress proteins: the biological functions in virus infectionpresent and challenges for target-based antiviral drug development. Signal Transduct Target Ther. 2020 Jul;5(1):1–40.10.1038/s41392-020-00233-4Search in Google Scholar PubMed PubMed Central
[32] Jeng W, Lee S, Sung N, Lee J, Tsai FT. Molecular chaperones: guardians of the proteome in normal and disease states. F1000Research. 2015;4:1448.10.12688/f1000research.7214.1Search in Google Scholar PubMed PubMed Central
[33] Tukaj S, Sitko K. Heat shock protein 90 (HSP90) and HSP70 as potential therapeutic targets in autoimmune skin diseases. Biomolecules. 2022 Aug;12(8):1153.10.3390/biom12081153Search in Google Scholar PubMed PubMed Central
[34] Makhoba XH, Makumire S. The capture of host cell’s resources: The role of heat shock proteins and polyamines in SARS-COV-2 (COVID-19) pathway to viral infection. Biomol Concepts. 2022 Jan;13(1):220–9.10.1515/bmc-2022-0008Search in Google Scholar PubMed
[35] Leak RK. Heat shock proteins in neurodegenerative disorders and aging. J Cell Commun Signal. 2014 Dec;8(4):293–310.10.1007/s12079-014-0243-9Search in Google Scholar PubMed PubMed Central
[36] Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, et al. The NCBI biosystems database. Nucleic Acids Res. 2010 Jan;38(suppl_1):D492–6.10.1093/nar/gkp858Search in Google Scholar PubMed PubMed Central
[37] Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic acids symposium series. Vol. 41. No. 41, London: Information Retrieval Ltd; 1999 Jan. p. 95–8. c1979-c2000.Search in Google Scholar
[38] Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction, and analysis. Nat Protoc. 2015 Jun;10(6):845–58.10.1038/nprot.2015.053Search in Google Scholar PubMed PubMed Central
[39] Schrödinger L, DeLano WJ. PyMOL. The PyMOL molecular graphics system, version. 2. New York, NY, USA: Schrödinger, LLC; 2020.Search in Google Scholar
[40] BIOvIA DS. Discovery studio modeling environment 4. San Diego: Dassault Syst. Release; 2015.Search in Google Scholar
[41] Bonam SR, Ruff M, Muller S. HSPA8/HSC70 in immune disorders: a molecular rheostat that adjusts chaperone-mediated autophagy substrates. Cells. 2019 Aug;8(8):849.10.3390/cells8080849Search in Google Scholar PubMed PubMed Central
[42] Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 2022 Jan;23(1):3–20.10.1038/s41580-021-00418-xSearch in Google Scholar PubMed PubMed Central
[43] Cueno ME, Imai K. Structural comparison of the SARS CoV 2 spike protein relative to other human-infecting coronaviruses. Front Med. 2021 Jan;7:594439.10.3389/fmed.2020.594439Search in Google Scholar PubMed PubMed Central
[44] Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993 Apr;26(2):283–91.10.1107/S0021889892009944Search in Google Scholar
[45] Beaudoin CA, Hamaia SW, Huang CL, Blundell TL, Jackson AP. Can the SARS-CoV-2 spike protein bind integrins independent of the RGD sequence? Front Cell Infect Microbiol. 2021;11:765300.10.3389/fcimb.2021.765300Search in Google Scholar PubMed PubMed Central
[46] Petrenko VA, Gillespie JW, De Plano LM, Shokhen MA. Phage-displayed mimotopes of SARS-CoV-2 spike protein targeted to authentic and alternative cellular receptors. Viruses. 2022 Feb;14(2):384.10.3390/v14020384Search in Google Scholar PubMed PubMed Central
[47] Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci. 2020 May;117(21):11727–34.10.1073/pnas.2003138117Search in Google Scholar PubMed PubMed Central
[48] Thompson JD, Linard B, Lecompte O, Poch O. A comprehensive benchmark study of multiple sequence alignment methods: current challenges and future perspectives. PLoS One. 2011 Mar;6(3):e18093.10.1371/journal.pone.0018093Search in Google Scholar PubMed PubMed Central
[49] Objectives. (n.d.). http://www.ncbi.nlm.nih.gov/Class/Structure/aa/aa_explorer.cgi.Search in Google Scholar
[50] Forni D, Cagliani R, Clerici M, Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017 Jan;25(1):35–48.10.1016/j.tim.2016.09.001Search in Google Scholar PubMed PubMed Central
[51] Ziegler CG. Deconstructing cell intrinsic immunity and host-pathogen interactions using single-cell genomics. Doctoral Dissertation. Harvard University Graduate School of Arts and Sciences; 2021.Search in Google Scholar
[52] Piplani S, Singh PK, Winkler DA, Petrovsky N In silico comparison of SARS-CoV-2 spike protein-ACE2 binding affinities across species and implications for virus origin. Sci Rep. 2021 Jun;11(1):1–3.10.1038/s41598-021-92388-5Search in Google Scholar PubMed PubMed Central
© 2024 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Research Articles
- Antitumor activity of 5-hydroxy-3′,4′,6,7-tetramethoxyflavone in glioblastoma cell lines and its antagonism with radiotherapy
- Digital methylation-specific PCR: New applications for liquid biopsy
- Synergistic effects of essential oils and phenolic extracts on antimicrobial activities using blends of Artemisia campestris, Artemisia herba alba, and Citrus aurantium
- β-Amyloid peptide modulates peripheral immune responses and neuroinflammation in rats
- A novel approach for protein secondary structure prediction using encoder–decoder with attention mechanism model
- Diurnal and circadian regulation of opsin-like transcripts in the eyeless cnidarian Hydra
- Withaferin A alters the expression of microRNAs 146a-5p and 34a-5p and associated hub genes in MDA-MB-231 cells
- Toxicity of bisphenol A and p-nitrophenol on tomato plants: Morpho-physiological, ionomic profile, and antioxidants/defense-related gene expression studies
- Review Articles
- Polycystic ovary syndrome and its management: In view of oxidative stress
- Senescent adipocytes and type 2 diabetes – current knowledge and perspective concepts
- Seeing beyond the blot: A critical look at assumptions and raw data interpretation in Western blotting
- Biochemical dynamics during postharvest: Highlighting the interplay of stress during storage and maturation of fresh produce
- A comprehensive review of the interaction between COVID-19 spike proteins with mammalian small and major heat shock proteins
- Exploring cardiovascular implications in systemic lupus erythematosus: A holistic analysis of complications, diagnostic criteria, and therapeutic modalities, encompassing pharmacological and adjuvant approaches
Articles in the same Issue
- Research Articles
- Antitumor activity of 5-hydroxy-3′,4′,6,7-tetramethoxyflavone in glioblastoma cell lines and its antagonism with radiotherapy
- Digital methylation-specific PCR: New applications for liquid biopsy
- Synergistic effects of essential oils and phenolic extracts on antimicrobial activities using blends of Artemisia campestris, Artemisia herba alba, and Citrus aurantium
- β-Amyloid peptide modulates peripheral immune responses and neuroinflammation in rats
- A novel approach for protein secondary structure prediction using encoder–decoder with attention mechanism model
- Diurnal and circadian regulation of opsin-like transcripts in the eyeless cnidarian Hydra
- Withaferin A alters the expression of microRNAs 146a-5p and 34a-5p and associated hub genes in MDA-MB-231 cells
- Toxicity of bisphenol A and p-nitrophenol on tomato plants: Morpho-physiological, ionomic profile, and antioxidants/defense-related gene expression studies
- Review Articles
- Polycystic ovary syndrome and its management: In view of oxidative stress
- Senescent adipocytes and type 2 diabetes – current knowledge and perspective concepts
- Seeing beyond the blot: A critical look at assumptions and raw data interpretation in Western blotting
- Biochemical dynamics during postharvest: Highlighting the interplay of stress during storage and maturation of fresh produce
- A comprehensive review of the interaction between COVID-19 spike proteins with mammalian small and major heat shock proteins
- Exploring cardiovascular implications in systemic lupus erythematosus: A holistic analysis of complications, diagnostic criteria, and therapeutic modalities, encompassing pharmacological and adjuvant approaches

![Figure 4
The image shows a 3D model showing the interaction between SARS-CoV-2 spike proteins and the human HSP27 [40,39].](/document/doi/10.1515/bmc-2022-0027/asset/graphic/j_bmc-2022-0027_fig_004.jpg)
