Abstract
Opsins play a key role in the ability to sense light both in image-forming vision and in non-visual photoreception (NVP). These modalities, in most animal phyla, share the photoreceptor protein: an opsin-based protein binding a light-sensitive chromophore by a lysine (Lys) residue. So far, visual and non-visual opsins have been discovered throughout the Metazoa phyla, including the photoresponsive Hydra, an eyeless cnidarian considered the evolutionary sister species to bilaterians. To verify whether light influences and modulates opsin gene expression in Hydra, we utilized four expression sequence tags, similar to two classic opsins (SW rhodopsin and SW blue-sensitive opsin) and two non-visual opsins (melanopsin and peropsin), in investigating the expression patterns during both diurnal and circadian time, by means of a quantitative RT-PCR. The expression levels of all four genes fluctuated along the light hours of diurnal cycle with respect to the darkness one and, in constant dark condition of the circadian cycle, they increased. The monophasic behavior in the L12:D12 cycle turned into a triphasic expression profile during the continuous darkness condition. Consequently, while the diurnal opsin-like expression revealed a close dependence on light hours, the highest transcript levels were found in darkness, leading us to novel hypothesis that in Hydra, an “internal” biological rhythm autonomously supplies the opsins expression during the circadian time. In conclusion, in Hydra, both diurnal and circadian rhythms apparently regulate the expression of the so-called visual and non-visual opsins, as already demonstrated in higher invertebrate and vertebrate species. Our data confirm that Hydra is a suitable model for studying ancestral precursor of both visual and NVP, providing useful hints on the evolution of visual and photosensory systems.
Introduction
Opsins play a key role in photoreception, which is the first key step in seeing in Metazoa [1,2]. Opsins are integral membrane proteins of photoreceptor cells that absorb photon energy and ultimately convert it into an electrical signal toward the central nervous system. Photoreception takes place in photoreceptor cells (“ciliary” and “rhabdomeric”, respectively, in vertebrates and invertebrates) capable to sense directly ambient light [3,4]. Photoreception is phylogenetically one of the oldest sensory modalities thanks to the amazing ubiquity in all animal phyla of morphological, functional, and molecular systems (from simple invertebrate light-sensitive cells to more complex vertebrate eyes) that respond to environmental luminous stimuli [5,6]. Although framed in different structure-function relationships, in vertebrate and invertebrate visual cells, photoreception starts with the photoisomerization of the retinal chromophore of the photopigment, usually an opsin-based pigment. This process triggers the binding of the opsin with a G-protein that leads to an enzymatic visual cascade, culminating in a final messenger, which gates light-sensitive ion channels to modulate and shape the electric signal toward the nervous system [7,8].
In addition to conventional eyed structures, vertebrates and invertebrates have supplementary non-visual photoreception (NVP) systems for non-image-forming vision or circadian vision [9,10,11]. Photic information mediated by NVP integrates visual activity and is involved in temporal (time-of-day) and behavioral physiology of the animal (e.g., photoperiodism, timing, and photoentrainment of circadian rhythms) [12,13,14].
NVP cells, formerly named extraretinal or extraocular photoreceptors, are currently termed non-visual (non-image-forming) photosensitive cells in invertebrates, and non-rod non-cone photoreceptors in vertebrates (after the discovery of photosensitive retinal ganglion cells) [12]. NVP cells are mainly located within nervous system districts and share with retinal photoreceptors common evolutionary origin and light-sensing modalities, above all the same superfamily of opsin-based photoreceptor proteins. Therefore, NVP opsins have been identified in cells beyond the retinal photoreceptors in several vertebrates and in extraretinal tissues of some invertebrates [12,15].
Focusing the attention on both opsin-based pigments and their structure/function relationship, the search for novel opsins supplying image- and nonimage-forming photoreception is a well-established field in photosensory biology and in vision research. It provides new insights into the evolution of the visual systems and of the visual ecology.
Accordingly, for more than 30 years, we were using the eyeless freshwater Hydra (Cnidaria, Hydrozoa), considered the evolutionary sister to bilaterians [16], as animal model to identify the presence and the co-existence of different opsins, classified in phylogenetic higher invertebrate and vertebrate species as serving visual and non-visual functions, and to study their expression and functional role [1,17,18,19,20].
Hydra, the first metazoan in which a primitive nervous system in the form of a diffuse nerve net can be found, has no eyes or eye-like structures; nevertheless, it is photosensitive, responding to light by slow contraction and elongation movements. The photic modulation of its periodic behavior has been demonstrated [21,22,23,24] as well as the identification and the localization of opsin-based pigments responsible for its photosensitivity [16,19,25,26]. However, many questions remain unanswered due to the large number of opsin genes identified, with respect to those certainly expressed [17], and their molecular evolution [18].
Furthermore, during evolution, regular light/dark changes have had a profound impact on visual systems including the evolution of regular rhythms in biological processes ranging from behavioral to biochemical. Although a key evolutionary trait is the adaptation to day/night alternation and its impact on integrated physiology, the photic regulation of opsin gene expression remains rather understudied.
So far, evidence for diurnal and circadian regulation of opsin expression patterns has been reported in very few species of invertebrates [27,28,29] and vertebrates [30,31,32,33]. We directed our efforts on the possible photic regulatory effect of the diurnal and circadian regimes on the expression of selected opsin-like expressed sequence tags (ESTs) in Hydra, similar to visual and non-visual opsins, demonstrating that both cycles are able to tune and/or to regulate the expression of the transcripts. These results suggest that opsin rhythms are probably endogenous and that putative circadian oscillators could drive the opsin transduction. Moreover, the different opsin mRNA levels observed in the circadian regime with respect to those obtained in the diurnal one suggest that an internal mechanism could modulate those processes.
Our data in Hydra show that a strong regulation by light on the opsin gene expression of selected opsin-like photopigments occurs and that the co-existence of classic and unconventional opsins indicates this cnidarian as a putative common precursor of both visual and non-visual photosensitive modalities, confirming the suitability of this animal model for the study of the evolution of the visual function.
Methods
Animals
Hydra vulgaris specimens were asexually cultured in Hydra medium (HM) (1 mM CaCl2 and 0.1 mM NaHCO3, pH 7) according to protocols previously reported [15,22,25]. Animals were kept in a thermostatic cabinet at 17 ± 1°C under controlled lighting conditions by a 12:12 light–dark (LD) cycle where lights went on at 8:00 am (ZT0) and turned off at 8:00 pm (ZT12). Circadian regime was applied keeping the animals in total darkness for 24-h cycle (dark–dark). Cultures were fed with Artemia nauplii daily and washed 6 h after feeding. Animals used for the experimental procedure were starved for 3 days before use to avoid any contamination by Artemia’s photopigments, carotenoids, and other photoreceptor proteins.
Hydra target sequence collection
Hydra ESTs of opsin-like photopigments, similar to visual and non-visual opsins, were collected from dbEST database (https://www.ncbi.nlm.nih.gov/genbank/dbest/). The sequences, showing strong similarity to photopigment cDNA sequences of other vertebrate and invertebrate organisms, were aligned at NCBI Gene (http://www.ncbi.nlm.nih.gov/gene/) and common primers were designed to obtain optimized primers for quantitative PCR analysis. In particular, the following cDNA sequences were used: DT617488 acc. (similar to putative photopigment melanopsin); CN554795 acc. (similar to rhodopsin); CN775258 acc. (similar to blue-sensitive opsin); CB073527.1 acc. (similar to visual pigment-like receptor peropsin-like); and CX055471 acc. (Beta-actin), as reference. Details of the expressed sequences and their related forward and reverse primers are given in Tables 1 and 2.
RT-PCR primers characteristics
| Locus definition | GeneBank accession | Forward primer | Reverse primer | PCR product size (bp) |
|---|---|---|---|---|
| Melanopsin-like | DT617488 | 5′AATTCGTTTGTCATTATTACTATATTG3′ | 5′AGAGTCCATATTGATGTTGTTAAG 3′ | 221 |
| Peropsin-like | CB073527 | 5′ACCATACTAGCAAAGGGAAACAC3′ | 5′GCAATAGCAGTTAAGGCAAGG 3′ | 200 |
| Blue-sensitive opsin-like | CN775258 | 5′CGTTTGAGCAAGCACCTGATTC3′ | 5′CCGTAGCCGTTTCTTTAGTCTTATATTTAG3′ | 114 |
| Rhodopsin-like | CN554795 | 5′CTTATTGTGCTTTGTTTGCTAAATCATC3′ | 5′ACTGTTGGGTTATTGAAGAGTTTCC3′ | 124 |
| Beta actin | CX055471 | 5′CTCCGTGTTGCTCCAGAAG3′ | 5′AGACACCATCTCTTGAATAAAG3′ | 199 |
Gene expressed sequence tags (ESTs) nomenclature, GenBank accession number, forward and reverse primer sequences, and predicted size of the amplified product.
cDNA FASTA sequences and the relative primers sequences
| EST definition | GeneBank accession no. | FASTA sequence |
|---|---|---|
| ACAH-aaa95g08.g1 Hydra_EST_UCI-10 Hydra vulgaris | DT617488 | ACCCACGCGTCCGGCGAATGATGCTTTCTAAAGATATTACTAAAATTACATCCGTACATTATTCCATATGTATATTCCTTGGTATAATTATTAATTCGTTTGTCATTATTACTATATTGAAAAATAAAAAATTGCAAACTACAACAAACTTCTTCATTTTAAATTTGGCAATAGCTGATCTTTTATTTACTGTGTTTGGCATATCGGCAATTTTTTATAAAGGCTTTGCAAAAACATTGTCGAAAGAAAACTCTATTTGTGTGGTTATAGGTTTTTTTACATTACTTTTCTTAACAACATCAATATGGACTCTTGTTATGATTAGCATCAACAGATACTTAAATGTTGCAAAAGCCAACATTATAAAAACACTTTACACAAGAAAAAAAACTGTTTTAATTATTGCAGGCGTTTGGATATTTTCTGTATTTATTTCAGCTCCACCTTTGATTGGTTGGAGCGAATTTAAATCAACTTCCAGTTTTTGTACCATTAATGGCAAAAAAAATATATCGTACACAGTTTTTCTTGGTTTATTAGTTTATATAATACCAATGGTTTTTTTGGCAAGTTTGTACATGCGCATTTTTTTTTTTGTTAAACAAAGTCGAAAAAGAGA AACTGAGAAGAAGTA TAAACTC |
| CB073527.1 taa17c02.y1 Hydra EST - II Hydra vulgaris | CB073527 | GGACAGACATTTGTGTGATCAATGGCATTTGTTTTTATTATCGTATTTTTATCGTTCTTATGTGGATTTTCTGTGATATTAAACGTCACTGTAGTCTTAACCATACTAGCAAAGGGAAACACTAAAAACACTCGAGATGTAATTTTGATGAGTCTTGCAATATGTGATGGCGTACAGTGCACTATAGGATATCCGGTCGAACTGTTTGGTTATGCTAATTACAAAAATCCATCACTATCCGAAAAGTTTTGCAAACCGAGTGGTTTCATTGTTATGTACCTTGCCTTAACTGCTATTGCACATTTGGTTTGTTTATGTATATATCGCTATTTAACTATTGTATATCCACTAAAACTACAGATATTTCTCACAAAATCGAATTGGAGTGCATGTGGTTGTATTGCCTTTTGCTGGATCTATGGTTTGTTTTGGTCATTAAGTCCTCTACTCGGTTGGAATGAAATAGTGCGAGAAAACAAGGATACCTACAAATGCTCAATTAAT |
| CN775258.1 tae71c10.y1 Hydra EST Darmstadt I Hydra vulgaris | CN775258 | TTGAAAAGTAATATAAACTCTATTGAATATTCTGAGCAATCGGAAAATGTATCGACGTTTGAGCAAGCACCTGATTCATCAGTTCCACAACAAGCACGAAACGAAACACTAAACGACAAAAAAGTTACAGTTTACTTTTCTAAATATAAGACTAAAGAAACGGCTACGGTAGACAGTAAATCCCCTTTAATTCAAAATAACTTTCTTGAAGCAGAAAAAAAGAAAGAAAAGCGTCAAATGGTCGCTATTGCAATACATTTGAAGCAAGTTCGAGTTACAAAAATGCTTATGATTTTAGTGTTAAGCTTCTTTTTTTGTTGGACACCATTTTTTGTTGGAGCATTGTACCACGTTTATCACAAAAAAATTAACGGTTTTCAAGTGACAACTTTTGGAATAATGTGCGCTTGTTTAAATTGCATTCTAAATCCTTTTATTTATTCTATTATGAATCGTAGTTTTCGAAAATGCGTGGTAAAAATGTGGAAAAACCTAATTTCATCTTANTTATTATTCAGA |
| CN554795.1 tae29c05.y1 Hydra EST Darmstadt I Hydra vulgaris | CN554795 | CAGGATACCTACAGATGCTCAATTAATTTATATCCGGATAATGAGATAAAAAGTAGTTATTTATACGCTCTGGCAATATTTTGTTACCTTATTCCTCTAATAATAATTATTTACTGTAGTTTAAAAGTCCGCTCAGAACTTCGCAACATGTTAAAAATGTGCAAACAAATTTCTGGTGTTGAAGCAAATATTACAAAAGTTACATATCGAATAGAAAAACAAGATTTTATATCTGTAAGTTTTATAATAGCATCATTTTTTACTGTTTGGACTCCATATGCCGTATGTGTTTTTTATTTGACAATTGGAAAAAAACTACCTCCTAGTTTTTTAACTTATTGTGCTTTGTTTGCTAAATCATCAACAATTCTGAACCCTATCATTTATTGTCTCATGTATAAAAAATTTCGCCAAACATTACAAAGTAAATTTGGGAAACTCTTCAATAACCCAACAGTTACACCAGCTGTTTAAAAAAAAGTTCTGATGTTTGGATACCTCAAGATTGAAAGCAAGTCGCTGCTTTGTAC |
| CX055471.1 tai90d12.y2 Hydra EST UCI 5 ALP Hydra vulgaris | CX055471 | CAAGAGCTGTTTTCCCATCTATTGTTGGACGTCCTCGTCATCAAGGAGTCATGGTTGGTATCTTACAGAAGGATTCCTATGTCGGTGACGAAGCTCAGAGCAAACGTGGTATCTTAACTTTGAAATACCCAATTGAACACGGAATTGTAACTAACTGGGATGATATGGAAAAAATTTGGCATCACACTTTTTACAATGAGCTCCGTGTTGCTCCAGAAGAACACCCTGTCCTTCTTACTGAAGCTCCCCTGAATCCCAAAGCAAATCATGAAAAAATGACCCAAATCATGTTTGAGACATTTAACTCTCCTGCAATGTATGTTGTTATTTACGCCGTCTTATCCTTGTATGCTTCTGGTCTTACCACTGTTATTGTACTTTATTCAAGAGATGGTGTCTCTTACTCAGTACCAATC |
The italic typed letters show the nucleotide sequences for forward primers while bold typed letters show those for reverse primers.
Animal sample collection
The animal collection procedure was done in light conditions identical to the correspondent diurnal phase (housing conditions). The procedure in darkness was done by using an infrared viewer or red-light illumination for photographic development. One hundred animals were collected from a homogeneous population at each time and for each experimental light condition. The whole experimental procedure was performed twice for each light regime. Animal samples were harvested and homogenized in TRIzol Reagent (Invitrogen) for 1 day every 3 h, starting from 9:00 am, in two different light condition regimes: (1) a Zeitgeber cycle or diurnal cycle (represented by the alternation of 12 h light and 12 h darkness conditions) and (2) a circadian cycle (represented by total darkness along 24 h). Considering that T0 and T12 were fixed at 8:00 am and 8:00 pm, the ZT/CT (Zeitgeber Time/Circadian Time), numbered with 1, 4, 7, 10, 13, 16, 19, and 22, corresponded, respectively, to the harvesting made at 9:00 am, 12:00 am, 3:00 pm, 6:00 pm, 9:00 pm, 12:00 pm, 3:00 am, and 6:00 am.
RNA extraction and quantitative RT-PCR
TRIzol samples were stored at −80°C before RNA extraction. Total RNA was purified, quantified, characterized, and retrotranscribed as previously reported [34]. For all samples tested, the RNA integrity number (Bionalyzer 2100, Agilent) was greater than eight relating to a 0–10 scale. Quantitative real-time PCR (RT-PCR) was performed by an iCycler-iQ5® (Bio-Rad, Milan, Italy) in a 20 μL reaction mixture containing 10–50 ng of cDNA optimized primers for SYBR-green analysis. Assays were performed in quadruplicate (maximum ΔCt of replicate samples <0.5), and a standard curve from consecutive fivefold dilutions (100–0.16 ng) of a cDNA pool representative of all samples was included for PCR efficiency determination. Optimized primers for SYBR Green analysis and optimum annealing temperatures were designed by the Allele-Id software version 7.0 (Biosoft International, Palo Alto, CA, USA) and were synthesized (HPLC purification grade) by MWG-Biotech (Ebersberg, Germany). For each target, all sequences at NCBI Blast https://blast.ncbi.nlm.nih.gov/Blast.cgi were aligned and common primers were designed (see Tables 1 and 2). Relative expression calculation, correct for PCR efficiency and normalized with respect to reference genes β-actin, was performed by the iQ5 software. Results are expressed as fold expression, compared with control (=1) [35]. Statistical significance was evaluated by the Origin 2022 software (OriginLab Co.) performing a one-way analysis of variance (ANOVA), followed by Bonferroni’s post hoc test. For the sake of clarity of figures, only the more representative differences were reported in the graphics with their significances always represented with one asterisk for both p < 0.05 and p < 0.001; for simplicity, the relative p-value was reported in the caption only if the significativity was discussed in the text, alternatively, only the fewer comparisons that resulted in not significative were indicated [36]. All the measurements were obtained from two independent experiments and values are expressed as the mean ± standard deviation.
Results
The gene sequences studied belonged to opsin-like photopigments similar to opsins serving visual and non-visual tasks in vertebrates and invertebrates, respectively, rhodopsin and blue sensitive opsins, and melanopsin and peropsin (see “Methods” section). We used partial mRNA sequences (ESTs) of these genes to investigate the expression patterns during both the Zeitgeber (ZT) and the circadian (CT) time by means of a quantitative RT-PCR.
In Figure 1, the relative expression of all opsin-like sequences during the ZT and CT cycles is represented. It is evident (Figure 1a) that the expression levels fluctuated along the light hours of diurnal cycle with respect to the darkness ones. In fact, they all appeared synchronized in peaking around ZT7 (3:00 pm), with DT617488 (similar to putative photopigment melanopsin) and CN554795 (similar to rhodopsin) presenting the highest levels. However, while all of them showed a monophasic cyclical alternation, a second steep rise was observed for CN554795 during the dark hours at ZT19 (Figure 1a; ANOVA one way, Bonferroni post hoc test; for statistical significance see figure caption). In addition, in the circadian cycle, the relative expression increased about twofold for all opsins-like sequences and about fourfold for CN554795 (Figures 1b and c; ANOVA one way, Bonferroni post hoc test; for statistical significance see figure caption).
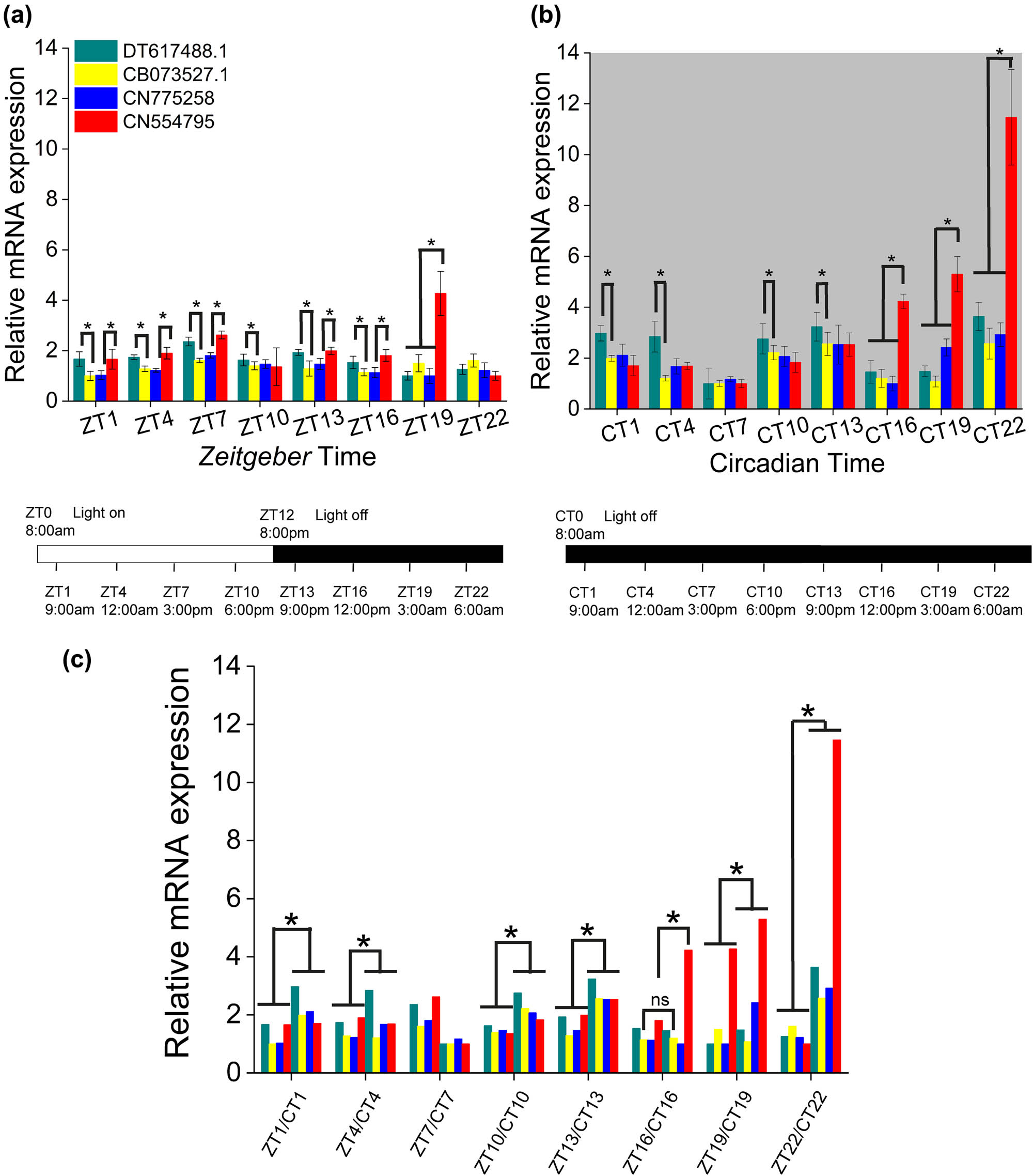
Relative expression level of EST sequences of opsin-like genes. (a) The relative expression level DT617488, CN554795, CN7752589, and CB073527.1 during a Zeitgeber cycle (ZT1 = 9:00 am, ZT4 = 12:00 am, ZT7 = 3:00 pm, ZT10 = 6:00 pm, ZT13 = 9:00 pm, ZT16 = 12:00 pm, ZT19 = 3:00 am, ZT22 = 6:00 am). Significativity was evaluated between the various opsins within each ZT group by means of ANOVA one way, Bonferroni post hoc. All differences were significant (p < 0.05; p < 0.001) except for.ZT1: CN77 vs CB, CN55 vs DT; ZT4: CN77 vs CB; ZT10: CN77 vs CB; ZT13: CN55 vs DT; ZT16: CN77 vs CB; ZT19: CN77 vs DT; and ZT22: CN77 vs DT. (b) The relative expression levels during a circadian cycle (CT1 = 9:00 am, CT4 = 12:00 am, CT7 = 3:00 pm, CT10 = 6:00 pm, CT13 = 9:00 pm, CT16 = 12:00 pm, CT19 = 3:00 am, and CT22 = 6:00 am). Significativity was evaluated between the various opsins-like within each CT group. Differences were significative and DT took the maximum values in CT1-4 and CT10-22 (for DT versus CB: CT1 p = 2.6 × 10−6, CT4 p = 1.5 × 10−12, CT10 p = 1.5 × 10−8, CT13 p = 2.2 × 10−9, CT16 p = 1.68 × 10−5, CT19 p = 4.3 × 10−8, and CT22 p = 2.5 × 10−9) and CN55 in CT16-22 (for CN55 vs DT: CT16 p = 1.13 × 10−16; CT19 p = 4.8 × 10−19, and CT22 p = 1.2 × 10−21). (c) Significative differences between ZT and CT cycle at each time. Differences were significative when comparing the two experimental conditions (for DT: ZT1 vs CT1 p = 6.3 × 10−7, ZT4 vs CT4 p = 4.3 × 10−8, ZT10 vs CT10 p = 4.1 × 10−9, ZT13 vs CT13 p = 1.8 × 10−7, ZT16 vs CT16 p = 0.06, ZT19 vs CT19 p = 3.5 × 10−4, and ZT22 vs CT22 p = 5.3 × 10−11) with CN55 presenting the maximum difference values (CN55 vs DT: ZT16 vs CT16 p = 3.7 × 10−10, ZT19 vs CT19 p = 1.25 × 10−7, and ZT22 vs CT22 p = 1.2 × 10−12). The bars at the base of the plots show respectively the L12:D12 and D12:D12 experimental conditions; relative quantification was carried out by normalization of the values to those of the housekeeping gene β-actin. The real-time PCRs were made in four independent replicate assays. Error bars show the standard error of the mean.
Figure 2 shows the fluctuations of the expression of the selected opsin-like proteins along ZT and CT cycles. The expression of each opsin-like sequence was graphed separately from each other by bar plots (Figure 2a and b) so that they could be easily compared. It clearly emerges that the monophasic behavior of DT617488, CN775258, and CB073527.1 observed during the L12:D12 cycle (Figure 2a; ANOVA one way, Bonferroni post hoc; for statistical significance see figure caption) turned in a three-phasic expression profile during the continuous darkness condition with peaks at the beginning of CT1 (9:00 am), CT10 (6:00 pm), and CT22 (6:00 am), (Figure 2b; ANOVA one way, Bonferroni post hoc test; for statistical significance see figure caption). The increase of CN554795 followed an exponential trend (Figure 2b and c) resulting well modeled by means of a one-phase exponential growth function (Figure 3):
where Y 0 represents the initial expression level, e is the natural logarithm base, A is the expression amount, and K is the growth rate factor.
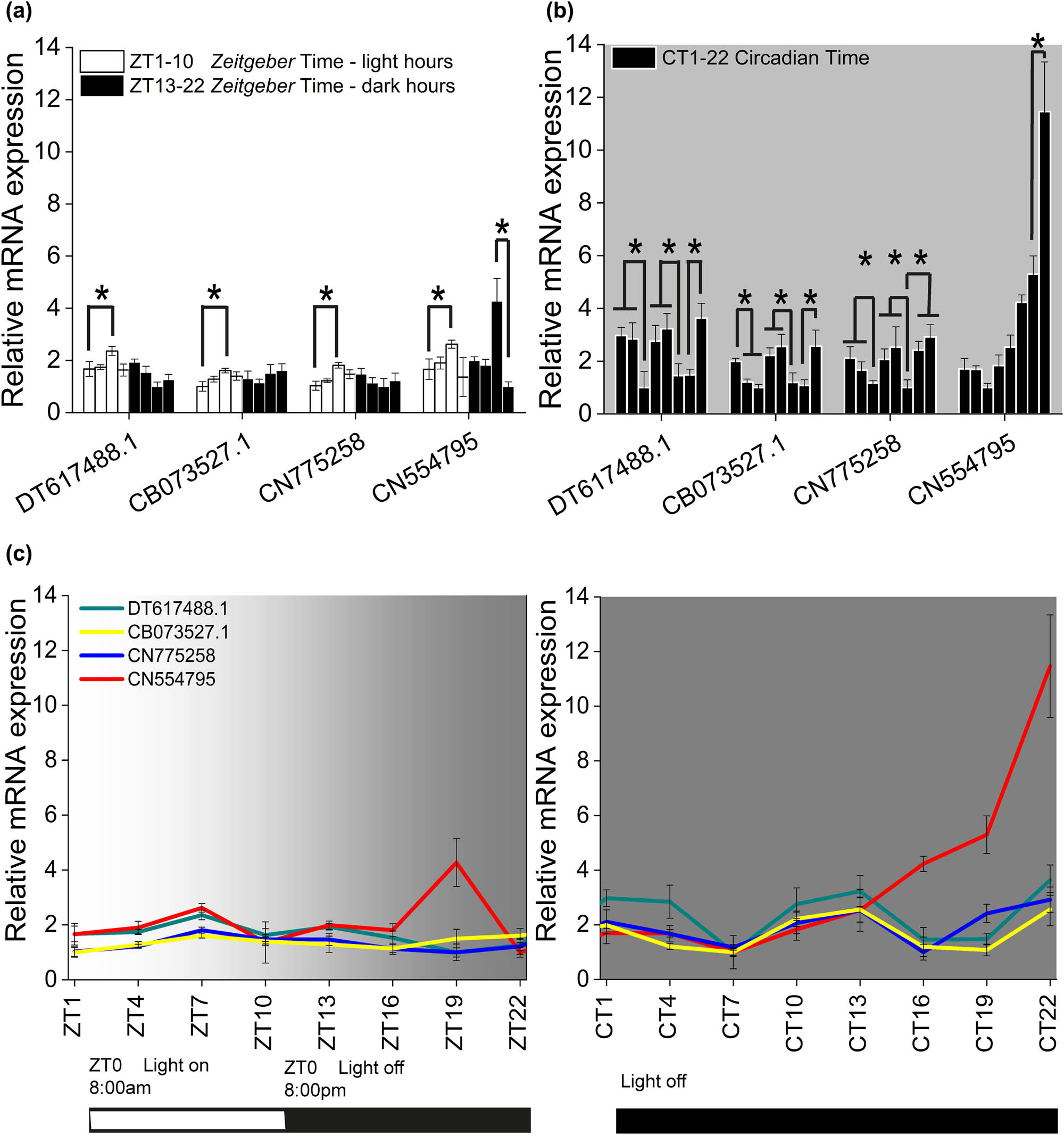
Opsin-like expression fluctuations along ZT and CT. The different trend of opsin-like expression respectively in (a) L12:D12 diurnal cycle, where the maximum difference values were reached at ZT7 (ZT7 vs ZT1: DT p = 1.6 × 10−4, CB p = 3.4 × 10−3, CN77 p = 9.8 × 10−6, CN55 p = 7.4 × 10−7 and ZT19 VS ZT22: CN55 p = 1.5 × 10−9) and in (b) D12:D12 circadian cycle, where DT, CB, CN77 peaked three times during the circadian time and CN55 exponentially rose (DT: CT1 vs CT7 p = 5.3 × 10−9, CT10 vs CT16 p = 1.1 × 10−9, CT22 vs CT19 p = 2.4 × 10−11; CB: CT1 vs CT7 p = 1.4 × 10−6, CT10 vs CT16 p = 4.1 × 10−8, CT22 vs CT19 p = 1.2 × 10−7; CN77: CT1 vs CT7 p = 1.1 × 10−6, CT10 vs CT16 p = 3.2 × 10−6, CT22 vs CT16 p = 1.2 × 10−7; and CN55: CT22 vs CT19 p = 1.9 × 10−12). (c) ZT versus CT expression fluctuations for all opsin-like. The bars at the base of the plots show respectively the L12:D12 and D12:D12 experimental conditions. Error bars show standard error of the mean.
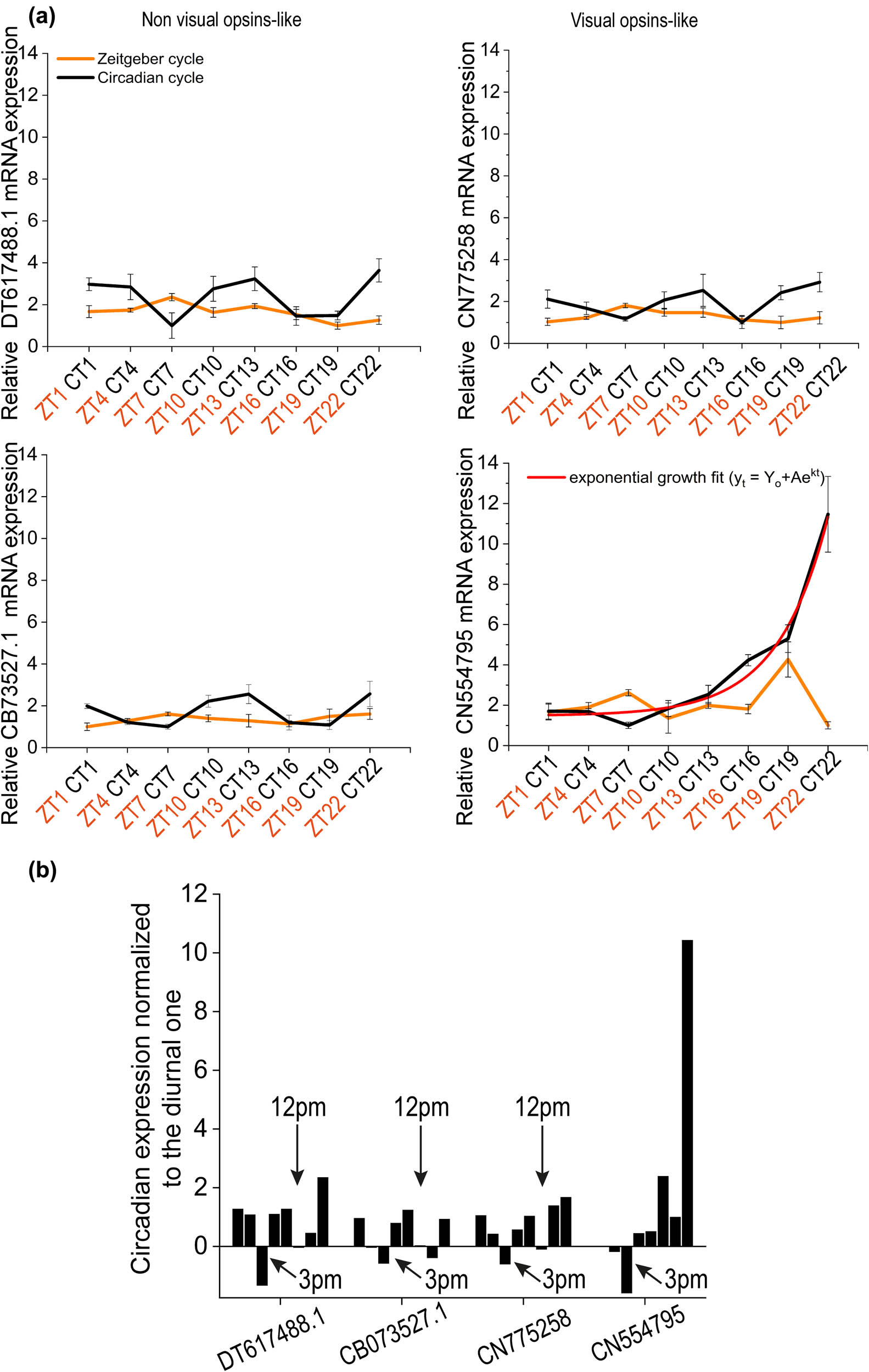
Comparison of ZT and CT for each opsin-like. (a) Relative mRNA expression level for Melanopsin-like (DT617488), SW Rhodopsin-like (CN554795), SW Blue sensitive opsin-like (CN7752589), and Peropsin-like (CB073527.1) in which the continuous darkness conditions was compared respect to the diurnal one. Error bars show standard error of the mean. (b) Circadian expression levels normalized to the corresponding diurnal ones for each opsin-like.
Figure 3 shows the comparison between ZT with CT for each opsin-like proteins employed. The expression of transcripts for each opsin-like was always higher during continuous darkness conditions with respect to those in the light/dark cycle (Figure 3a) except at 3:00 pm where the trend was inverted. Moreover, at 12:00 pm, DT617488, CN775258, and CB073527.1 presented approximately the same expression level both in diurnal and circadian time, which could indicate a constitutive level of the opsin expression. These considerations are better represented in Figure 3b, where each CT expression value was normalized to the corresponding ZT value.
Discussion
Nowadays, it is well established that opsin gene expression is strictly correlated with the circadian regulation of the photic input in animals. Therefore, evidence for diurnal and circadian regulation of opsin expression patterns has been reported in photoreceptors of invertebrates, Limulus [27,28], Apis [29], and (lower) vertebrates [30,31,32,33].
In recent years, light-regulated gene expression of photosensitive proteins has gained topicality due to the potential implications with development, disease onset progression, and consequent timing of therapy [37]. For example, strong correlations have been found in mice between mutated opsins with photoreceptor cell death [38], in Drosophila between daily blue light exposure with the onset of brain neurodegeneration [39], and in the honeybee Apis between temporal expression of all opsins with the maturation of the retina during pupal development [40]. Furthermore, very recently, in the seahorse Hippocampus, it has been shown that specific wavelengths affect opsin mRNA levels whose changes were life-state dependent, allowing interference of opsin ontogeny and of the animal development stages [41].
Thus, consistent with the above novel research frame, our study reports the first evidence of a diurnal and circadian regulation of selected opsin transcripts in the eyeless cnidarian Hydra, which has the first elementary nervous system in the form of a diffused net [42]. The main result that emerged from this study is that the expression level of the examined opsins showed a similar trend in the same experimental condition (ZT or CT) but not when ZT fluctuations were compared versus CT fluctuations. In fact, the opsin-like expression during the diurnal time (L12:D12) revealed a close dependence on the light hours with an increment for all opsins at 3:00 pm (ZT7) and only for SW opsin-like with a further rise at 3:00 am (ZT19).
Moreover, the condition of continuous darkness (D12:D12) unveiled that, with no external light stimuli, the expression was higher and forced to cycle three times. Therefore, the irradiance (i.e., the ambient light intensity) during the Zeitgeber time (ZT), acting as the main temporal indicator of the daytime, would be able to mute the circadian mechanism to better optimize the opsins expression to the variable demands of night and day. At the same time, other mechanisms, to date still unidentified, would intervene in the endogenous expression.
These data are consistent with similar findings reported in other invertebrates: e.g., the horseshoe crab Limulus [43], the crab Gelasimus [44], the aphid Acyrthosiphon [45], and the moth Helicoverpa [46]. In addition, both the light regulation of opsin expression and the diurnal rhythm have strong implications for the behavioral output [47,48,49]. Opposite regulation exerted by the light of opsin and clock gene expression, respectively, up-regulated for an opsin OPN4 gene and down-regulated for CLOCK and CRY clock light-sensitive genes, has been very recently reported in eyespot structures of the bivalve mollusk Mytilus [50].
Accordingly, the LD fluctuations observed in our study completely mirror the day/night oscillatory slope of the behavioral spectra in Hydra representing the diurnal variation of a biophysical parameter of the bioelectrical activity (i.e., the contraction bursts and rhythmic pulses) underlying the rhythmic body contraction and elongation which determine the simple and restricted Hydra behaviors such as osmoregulation, locomotion, and feeding and are more elevated during the day respect to the night [51,52,53].
Moreover, the condition of continuous darkness in the circadian cycle unveiled that an internal mechanism autonomously expresses opsins at higher levels in the absence of a normal day–night cycle. A long period of darkness represents a stressful and dangerous condition for the sessile Hydra polyp as its behavior strictly depends on light radiation and activities slowdown in the darkness. In addition, the absence of a normal cycle would not allow the maintenance of opsin levels as Hydra’s cells continuously proliferate and are frequently lost by sloughing or budding. The overexpression of opsins in the continuous absence of external light stimuli would allow the animal to find itself fully reconstituted once it emerges from the unsafe darkening condition and the opsins themselves could play a role in this process. From literature, it appears clear that, depending on the context and tissue, opsins could act not only as photosensors but also as critical regulators of many biological processes thanks to their ability to interact with a plethora of molecules as polymodal sensory receptors. A major discovery is that invertebrate opsins could have sensory roles that are light independent. They can constitutively bind their downstream G-protein partner to switch or accelerate a pathway signaling [54] or alternatively work as thermosensor [55]. In this case, a significant drop in temperature could be the switch needed to detect a prolonged darkening condition and to induce overexpression.
Finally, the effect of the light regulation on behavioral patterns could depend also on the spatial distribution of the photoreceptors’ expression levels, although currently the opsin expression seems to be not segregated in single animal parts but appears diffused along the whole ectodermal sheet [19,25]. Further approaches with technologies like mRNA in situ hybridization and antibody staining could be useful to verify possible spatial localization of expression changes.
These hints open a window on an intriguing phylogenetic scenario and should be the object of future study, as no biological clocks have as yet been molecularly identified in Hydra (although expression levels for 380 genes underlying diel, i.e. diurnal, behavior have been found [46]). Conversely, clock genes have been characterized in other phylogenetically close cnidarian species, the sea anemone Nematostella [56,57], and the coral Acropora [58]. Likewise, opsin photopigments have been proven to be involved in circadian clock entrainment and synchronization in Drosophila [59].
Furthermore, we report the first evidence of light regulation of non-visual opsin transcripts, melanopsin, and peropsin, in a lower invertebrate. Melanopsin is a vertebrate non-visual opsin even if resembles the invertebrate ones in various aspects, therefore classified as belonging to the rhabdomeric type [60]. Early identified in the inner retina of amphibian [61] and then in the fish [62], lizard [63], bird [64], mouse, and human [65] and currently widely identified in vertebrate species including humans [66], melanopsin is considered a key molecule in circadian response to light [13,67]. In the same way, peropsin is a non-visual opsin first identified from human ocular tissue and is nowadays considered a regulatory molecule of the visual physiology [68,69]. In Hydra, a possible role of peropsin as photoisomerase could be not excluded, supported by the presence of a similar kind of photoprotein in the jellyfish ocelli [70].
Light and circadian regulation of melanopsin has been mainly reported for vertebrate inner retina [32,62,71,72,73] while accounts for invertebrate extra-ocular systems are substantially lacking. Although very recently long-wavelength opsins have been found in extraocular regions of the blind shrimp Creaseria, they are strictly related to the synchronization of biological processes and diurnal vertical migration [74]. Therefore, our data could represent the first account of the expression of a light-regulated opsin gene similar to melanopsin in a primitive invertebrate. According to our results, the presence and the regulation of these transcripts in the lower cnidarian Hydra suggest that the so-called non-visual modality evolved early to allow the detection of the quantity and of the quality of the changes of light for adaptive advantage. Moreover, in the frame of the current controversial debate on cnidarian opsin phylogeny, the coexistence of visual-opsin transcripts in an eyeless cnidarian suggests that orthologs of visual opsin could have evolved prior to the evolution of animal visual systems [75]. Additionally, regarding non-visual opsins, the coexistence of non-visual and ocular expression of opsins has been already reported [76] and, for melanopsin in particular, correlation has been proved between its expression and the onset of several diseases [77,78], including those caused by the dysregulation of temporal physiological processes [79,80].
Conclusions
Overall, our study in Hydra can be summarized according to these key points:
the irradiance regulates the diurnal expression of visual and non-visual opsins, while its absence in the circadian cycle could induce an upregulation of their expression;
a putative, still unidentified, circadian system autonomously drives opsin expression; and
the gain of circadian transcriptional profiles is consistent with no irradiance detection.
To our knowledge, in this study, we report the first evidence of the light-regulated expression of visual and non-visual opsin transcripts in a lower invertebrate. Therefore, we could state that the irradiance signaling system has evolved early in the Cnidaria, hypothesizing that orthologs of visual opsin would have evolved prior to the evolution of animal visual systems. In conclusion, the discovery of non-visual – extra-ocular – photoreceptor proteins (including above all opsins) and the study of their expression and of the sensory ambient modality that regulates them raise the NVP to a light-sensing signaling complementary to vision, rather than an evolutive residual modality [13,14].
Finally, in the wake of the integrated view of a visual system mutually interacting with non-visual and temporal mechanisms and processes [6], our study strongly supports the suitability of Hydra as an appropriate model for identifying ancestral precursors of what the current literature defines as visual and non-visual photosensitive modalities (including their irradiance signaling system and the relative phototransduction pathways), and for phylogenetic and comparative studies on the evolution of visual and circadian systems.
Acknowledgments
SS and CM are profoundly indebted to Cloe Taddei-Ferretti (former CNR Institute of Cybernetics, Pozzuoli, Naples, Italy) who introduced them to Hydra and its “magic” research field. We are most grateful to Pierangelo Orlando (ICB-CNR, Pozzuoli, Naples, Italy) for masterful RT-PCR analysis and data curation and for incisive comments on the final manuscript. We are very grateful to Michael Whalen (IBF-CNR, Trento, I) for the accurate revision of the English and useful hints on the final manuscript. We thank Elena Gerola (IBF-CNR Trento, I) for administrative support and fund management. This work was carried out in the frame of the ISASI-CNR@Pozzuoli – IBF-CNR@Trento 2022-2025 Scientific Collaboration Operating Agreement (Convenzione Operativa di Collaborazione Scientifica) with SS and CM responsible, respectively.
-
Conflict of interest: Carlo Musio is a member of Biomolecular concepts’ Editorial Board. The other authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Terakita A. The opsins. Genome Biol. 2002;6:213. https://genomebiology.biomedcentral.com/articles/. 10.1186/gb-2005-6-3-213.Search in Google Scholar PubMed PubMed Central
[2] Nagata T, Inoue K. Rhodopsins at a glance. J Cell Sci. 2021;134(22):jcs258989.10.1242/jcs.258989Search in Google Scholar PubMed
[3] Yau KW, Hardie RC. Phototransduction motifs and variations. Cell. 2009;139(2):246–64.10.1016/j.cell.2009.09.029Search in Google Scholar PubMed PubMed Central
[4] Fain GL, Hardie R, Laughlin SB. Phototransduction and the evolution of photoreceptors. Curr Biol. 2010;20(3):R114–24.10.1016/j.cub.2009.12.006Search in Google Scholar PubMed PubMed Central
[5] Shichida Y, Matsuyama T. Evolution of opsins and phototransduction. Philos Trans R Soc Lond B Biol Sci. 2009;364(1531):2881–95.10.1098/rstb.2009.0051Search in Google Scholar PubMed PubMed Central
[6] Gehring WJ. The evolution of vision. Wiley Interdiscip Rev Dev Biol. 2014;3(1):1–40.10.1002/wdev.96Search in Google Scholar PubMed
[7] Bentrop J, Paulsen R. Invertebrate rhodopsins. In: Batschauer A, editor. Photoreceptors and Light Signalling (Comprehensive Series in Photochemical). Vol. 3. 1st edn. Royal Society Chemistry; 2003. p. 40–76.10.1039/9781847551665-00040Search in Google Scholar
[8] de Grip WJ, Ganapathy S. Rhodopsins: An excitingly versatile protein species for research, development and creative engineering. Front Chem. Vol. 10; 2022. p. 87960910.3389/fchem.2022.879609Search in Google Scholar PubMed PubMed Central
[9] Yoshida M. Extraocular photoreception. In: Autrum H, editor. Handbook of Sensory Physiology, Vol. 7/6A. Vision in invertebrates, A: Invertebrate photoreceptors. Berlin: Springer-Verlag; 1979. p. 581–640.10.1007/978-3-642-66999-6_10Search in Google Scholar
[10] Musio C. Extraocular photosensitivity in invertebrates. In: Taddei-Ferretti C editor. Biophysics of photoreception: Molecular and phototransductive events. Singapore: World Scientific; 1997. p. 245–62.Search in Google Scholar
[11] Musio C, Santillo S. Non-visual photoreception in invertebrates. In: Smith KC editor. Photobiological Sciences Online. American Society for Photobiology; 2009. http://photobiology.info. NLM ID 101673292.Search in Google Scholar
[12] Foster RG, Hankins MW. Circadian vision. Curr Biol. 2007;17(17):R746–51.10.1016/j.cub.2007.07.007Search in Google Scholar PubMed
[13] Musio C, Santillo S. Nonvisual photosensitivity and circadian vision. In: Griesbeck A, Oelgemöller M, Ghetti F. CRC Handb. of organic photochemistry and photobiology. 3rd edn. Boca Raton: CRC Press; 2012. p. 1185–200.10.1201/b12252-51Search in Google Scholar
[14] Andrabi M, Upton BA, Lang RA, Vemaraju S. An expanding role for nonvisual opsins in extraocular light sensing physiology. Annu Rev Vis Sci. 2023;9:245–67.10.1146/annurev-vision-100820-094018Search in Google Scholar PubMed
[15] Santillo S, Orlando P, De Petrocellis L, Cristino L, Guglielmotti V, Musio C. Evolving visual pigments: hints from the opsin-based proteins in a phylogenetically old eyeless invertebrate. BioSystems. 2006;86(1–3):3–17.10.1016/j.biosystems.2006.03.008Search in Google Scholar PubMed
[16] Plachetzki DC, Degnan BM, Oakley TH. The origins of novel protein interactions during animal opsin evolution. PLoS One. 2007;2(10):e1054.10.1371/journal.pone.0001054Search in Google Scholar PubMed PubMed Central
[17] Suga H, Schmid V, Gehring WJ. Evolution and functional diversity of jellyfish opsins. Curr Biol. 2008;18(1):51–5.10.1016/j.cub.2007.11.059Search in Google Scholar PubMed
[18] Macias-Munõz A, Murad R, Mortazavi A. Molecular evolution and expression of opsin genes in Hydra vulgaris. BMC Genomics. 2019;20(1):1–19. https://bmcgenomics.biomedcentral.com/articles 10.1186/s12864-019-6349-y Search in Google Scholar PubMed PubMed Central
[19] Plachetzki DC, Fong CR, Oakley TH. Cnidocyte discharge is regulated by light and opsin-mediated phototransduction. BMC Biol. 2012;10:17.10.1186/1741-7007-10-17Search in Google Scholar PubMed PubMed Central
[20] Vöcking O, Macias-Muñoz A, Jaeger SJ, Oakley TH. Deep diversity: Extensive variation in the components of complex visual systems across animals. Cells. 2022;11(24):3966.10.3390/cells11243966Search in Google Scholar PubMed PubMed Central
[21] Passano LM, McCullough CB. The light response and the rhythmic potentials of Hydra. Proc Natl Acad Sci USA. 1962;48(8):1376–82.10.1073/pnas.48.8.1376Search in Google Scholar PubMed PubMed Central
[22] Taddei-Ferretti C, Di Maio V, Musio C, Cotugno A. Modulation of Hydra attenuata rhythmic activity. VI. Combined effects of background and pulse light wavelength. J Photochem Photobiol, B: Biol. 1992;15:307–15.10.1016/1011-1344(92)85137-JSearch in Google Scholar
[23] Taddei-Ferretti C, Musio C. Photobehaviour of Hydra (Cnidaria, Hydrozoa) and correlated mechanisms: a case of extraocular photosensitivity. J Photochem Photobiol B. 2000;55(2–3):88–101.10.1016/S1011-1344(00)00041-5Search in Google Scholar
[24] Taddei-Ferretti C, Musio C, Santillo S, Cotugno A. The photobiology of Hydra’s periodic activity. Hydrobiologia. 2004;530/531:129–34.10.1007/s10750-004-2680-6Search in Google Scholar
[25] Musio C, Santillo S, Taddei-Ferretti C, Robles LJ, Vismara R, Barsanti L, Gualtieri P. First identification and localization of a visual pigment in Hydra (Cnidaria, Hydrozoa). J Comp Physiol A. 2001;187(1):79–81.10.1007/s003590100180Search in Google Scholar PubMed
[26] GenomeWiki (Genome Browser Wiki). Opsin evolution: key critters (Cnidaria). Santa Cruz, USA: UCSC University of California Genome Browser Group; 2010. http://genomewiki.ucsc.edu/index.php? title = Opsin_evolution:_key_critters_(cnidaria).Search in Google Scholar
[27] Katti C, Kempler K, Porter ML, Legg A, Gonzalez R, Garcia-Rivera E, et al. Opsin co-expression in Limulus photoreceptors: differential regulation by light and a circadian clock. J Exp Biol. 2010;213(15):2589–601.10.1242/jeb.043869Search in Google Scholar PubMed PubMed Central
[28] Battelle BA. Opsins and their expression patterns in the Xiphosuran Limulus polyphemus. Biol Bull. 2017;233(1):3–20.10.1086/693730Search in Google Scholar PubMed
[29] Sasagawa H, Narita R, Kitagawa Y, Kadowaki T. The expression of genes encoding visual components is regulated by a circadian clock, light environment and age in the honeybee (Apis mellifera). Eur J Neurosci. 2003;17(5):963–70.10.1046/j.1460-9568.2003.02528.xSearch in Google Scholar PubMed
[30] Korenbrot JI, Fernald RD. Circadian rhythm and light regulate opsin mRNA in rod photoreceptors. Nature. 1989;337(6206):454–7.10.1038/337454a0Search in Google Scholar PubMed
[31] Halstenberg S, Lindgren KM, Samagh SP, Nadal-Vicens M, Balt S, Fernald RD. Diurnal rhythm of cone opsin expression in the teleost fish Haplochromis burtoni. Vis Neurosci. 2005;22(2):135–41.10.1017/S0952523805222022Search in Google Scholar PubMed
[32] Salgado D, Mariluz BR, Araujo M, Lorena J, Perez LN, Ribeiro RL, et al. Light-induced shifts in opsin gene expression in the four-eyed fish Anableps. Front Neurosci. 2022;16:995469.10.3389/fnins.2022.995469Search in Google Scholar PubMed PubMed Central
[33] Li P, Temple S, Gao Y, Haimberger TJ, Hawryshyn CW, Li L. Circadian rhythms of behavioral cone sensitivity and long wavelength opsin mRNA expression: a correlation study in zebrafish. J Exp Biol. 2005;208(3):497–504.10.1242/jeb.01424Search in Google Scholar PubMed
[34] Grimaldi P, Orlando P, Di Siena S, Lolicato F, Petrosino S, Bisogno T, et al. The endocannabinoid system and pivotal role of the CB2 receptor in mouse spermatogenesis. Proc Natl Acad Sci U S A. 2009;106(27):11131–6.10.1073/pnas.0812789106Search in Google Scholar PubMed PubMed Central
[35] Aviello G, Romano B, Borrelli F, Capasso R, Gallo L, Piscitelli F, et al. Chemopreventive effect of the non-psychotropic phytocannabinoid cannabidiol on experimental colon cancer. J Mol Med (Berl). 2012;90(8):925–34.10.1007/s00109-011-0856-xSearch in Google Scholar PubMed
[36] Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36.10.1093/nar/30.9.e36Search in Google Scholar PubMed PubMed Central
[37] Panda S. The arrival of circadian medicine. Nat Rev Endocrinol. 2019;15(2):67–9.10.1038/s41574-018-0142-xSearch in Google Scholar PubMed
[38] Comitato A, Di Salvo MT, Turchiano G, Montanari M, Sakami S, Palczewski K, et al. Dominant and recessive mutations in rhodopsin activate different cell death pathways. Hum Mol Genet. 2016;25(13):2801–12.10.1093/hmg/ddw137Search in Google Scholar PubMed
[39] Nash TR, Chow ES, Law AD, Fu SD, Fuszara E, Bilska A, et al. Daily blue-light exposure shortens lifespan and causes brain neurodegeneration in Drosophila. NPJ Aging Mech Dis. 2019;5:8.10.1038/s41514-019-0038-6Search in Google Scholar PubMed PubMed Central
[40] Lichtenstein L, Grübel K, Spaethe J. Opsin expression patterns coincide with photoreceptor development during pupal development in the honey bee, Apis mellifera. BMC Dev Biol. 2018;18(1):1. https://bmcdevbiol.biomedcentral.com/articles/10.1186/s12861-018-0162-8 Search in Google Scholar PubMed PubMed Central
[41] Souto-Neto JA, David DD, Zanetti G, Sua-Cespedes C, Freret-Meurer NV, Moraes MN, et al. Light-specific wavelengths differentially affect the exploration rate, opercular beat, skin color change, opsin transcripts, and the oxi-redox system of the longsnout seahorse Hippocampus reidi. Comp Biochem Physiol A Mol Integr Physiol. 2023;288:111551.10.1016/j.cbpa.2023.111551Search in Google Scholar PubMed
[42] David CN, Hager G. Formation of a primitive nervous system: nerve cell differentiation in the polyp hydra. Perspect Dev Neurobiol. 1994;2(2):135–40.Search in Google Scholar
[43] Dalal JS, Jinks RN, Cacciatore C, Greenberg RM, Battelle BA. Limulus opsins: diurnal regulation of expression. Vis Neurosci. 2003;20(5):523–34.10.1017/S095252380320506XSearch in Google Scholar PubMed
[44] Jessop AL, Ogawa Y, Bagheri ZM, Partridge JC, Hemmi JM. Photoreceptors and diurnal variation in spectral sensitivity in the fiddler crab Gelasimus dampieri. J Exp Biol. 2020;223(23):jeb230979.10.1242/jeb.230979Search in Google Scholar PubMed
[45] Collantes-Alegre JM, Mattenberger F, Barberà M, Martínez-Torres D. Characterisation, analysis of expression and localisation of the opsin gene repertoire from the perspective of photoperiodism in the aphid Acyrthosiphon pisum. J Insect Physiol. 2018;104:48–59.10.1016/j.jinsphys.2017.11.009Search in Google Scholar PubMed
[46] Yan S, Zhu J, Zhu W, Zhang X, Li Z, Liu X, et al. The expression of three opsin genes from the compound eye of Helicoverpa armigera (Lepidoptera: Noctuidae) is regulated by a circadian clock, light conditions and nutritional status. PLoS One. 2014;9(10):e111683.10.1371/journal.pone.0111683Search in Google Scholar PubMed PubMed Central
[47] Garm A, Bielecki J, Petie R, Nilsson DE. Opposite patterns of diurnal activity in the box jellyfish Tripedalia cystophora and Copula sivickisi. Biol Bull. 2012;222(1):35–45.10.1086/BBLv222n1p35Search in Google Scholar PubMed
[48] Kanaya HJ, Kobayakawa Y, Itoh TQ. Hydra vulgaris exhibits day-night variation in behavior and gene expression levels. Zoological Lett. 2019;5:10.10.1186/s40851-019-0127-1Search in Google Scholar PubMed PubMed Central
[49] Flensburg SB, Garm A, Funch P. The contraction-expansion behaviour in the demosponge Tethya wilhelma is light controlled and follows a diurnal rhythm. J Exp Biol. 2022;225(24):jeb244751.10.1242/jeb.244751Search in Google Scholar PubMed
[50] Xu M, Li J, Guo B, Qi P, Ye Y, Yan X. Identification and characterization of light-responsive genes in pre-settlement eyed veligers of Mytilus coruscus. Aquacult Rep. 2023;33:101768.10.1016/j.aqrep.2023.101768Search in Google Scholar
[51] Passano LM, McCullough CB. Co-ordinating systems and behaviour in Hydra. I. Pacemaker system of the periodic contractions. J Exp Biol. 1964;41:643–64.10.1242/jeb.41.3.643Search in Google Scholar
[52] Badhiwala KN, Gonzales DL, Vercosa DG, Avants BW, Robinson JT. Microfluidics for electrophysiology, imaging, and behavioral analysis of Hydra. Lab Chip. 2018;18(17):2523–39.10.1039/C8LC00475GSearch in Google Scholar PubMed
[53] Tommasini G, De Simone M, Santillo S, Dufil G, Iencharelli M, Mantione D, et al. In vivo neuromodulation of animal behavior with organic semiconducting oligomers. Sci Adv. 2023;9(42):eadi5488.10.1126/sciadv.adi5488Search in Google Scholar PubMed PubMed Central
[54] van Wyk M, Kleinlogel S, van Wyk M, Kleinlogel S. A visual opsin from jellyfish enables precise temporal control of G protein signalling. Nat Commun. 2023;14(1):2450.10.1038/s41467-023-38231-zSearch in Google Scholar PubMed PubMed Central
[55] Leung NY, Montell C. Unconventional roles of opsins. Annu Rev Cell Dev Biol. 2017;33:241–64.10.1146/annurev-cellbio-100616-060432Search in Google Scholar PubMed PubMed Central
[56] Oren M, Tarrant AM, Alon S, Simon-Blecher N, Elbaz I, Appelbaum L, et al. Profiling molecular and behavioral circadian rhythms in the non-symbiotic sea anemone Nematostella vectensis. Sci Rep. 2015;5:11418.10.1038/srep11418Search in Google Scholar PubMed PubMed Central
[57] Leach WB, Macrander J, Peres R, Reitzel AM. Transcriptome-wide analysis of differential gene expression in response to light:dark cycles in a model cnidarian. Comp Biochem Physiol Part D Genomics Proteomics. 2018;26:40–9.10.1016/j.cbd.2018.03.004Search in Google Scholar PubMed PubMed Central
[58] Shoguchi E, Tanaka M, Shinzato C, Kawashima T, Satoh N. A genome-wide survey of photoreceptor and circadian genes in the coral, Acropora digitifera. Gene. 2013;515(2):426–31.10.1016/j.gene.2012.12.038Search in Google Scholar PubMed
[59] Klarsfeld A, Picot M, Vias C, Chélot E, Rouyer F. Identifying specific light inputs for each subgroup of brain clock neurons in Drosophila larvae. J Neurosci. 2011;31(48):17406–15.10.1523/JNEUROSCI.5159-10.2011Search in Google Scholar PubMed PubMed Central
[60] Koyanagi M, Kubokawa K, Tsukamoto H, Shichida Y, Terakita A. Cephalochordate melanopsin: evolutionary linkage between invertebrate visual cells and vertebrate photosensitive retinal ganglion cells. Curr Biol. 2005;15(11):1065–9.10.1016/j.cub.2005.04.063Search in Google Scholar PubMed
[61] Provencio I, Jiang G, De Grip WJ, Hayes WP, Rollag MD. Melanopsin: An opsin in melanophores, brain, and eye. Proc Natl Acad Sci U S A. 1998;95(1):340–5.10.1073/pnas.95.1.340Search in Google Scholar PubMed PubMed Central
[62] Bellingham J, Whitmore D, Philp AR, Wells DJ, Foster RG. Zebrafish melanopsin: isolation, tissue localisation and phylogenetic position. Brain Res Mol Brain Res. 2002;107(2):128–36.10.1016/S0169-328X(02)00454-0Search in Google Scholar PubMed
[63] Frigato E, Vallone D, Bertolucci C, Foulkes NS. Isolation and characterization of melanopsin and pinopsin expression within photoreceptive sites of reptiles. Naturwissenschaften. 2006;93(8):379–85.10.1007/s00114-006-0119-9Search in Google Scholar PubMed
[64] Chaurasia SS, Rollag MD, Jiang G, Hayes WP, Haque R, Natesan A, et al. Molecular cloning, localization and circadian expression of chicken melanopsin (Opn4): differential regulation of expression in pineal and retinal cell types. J Neurochem. 2005;92(1):158–70.10.1111/j.1471-4159.2004.02874.xSearch in Google Scholar PubMed
[65] Provencio I, Rodriguez IR, Jiang G, Hayes WP, Moreira EF, Rollag MD. A novel human opsin in the inner retina. J Neurosci. 2000;20(2):600–5.10.1523/JNEUROSCI.20-02-00600.2000Search in Google Scholar PubMed PubMed Central
[66] Contreras E, Nobleman AP, Robinson PR, Schmidt TM. Melanopsin phototransduction: beyond canonical cascades. J Exp Biol. 2021;224(23):jeb226522.10.1242/jeb.226522Search in Google Scholar PubMed PubMed Central
[67] Foster RG, Hankins MW. Non-rod, non-cone photoreception in the vertebrates. Prog Retin Eye Res. 2002;21(6):507–27.10.1016/S1350-9462(02)00036-8Search in Google Scholar
[68] Sun H, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. Peropsin, a novel visual pigment-like protein located in the apical microvilli of the retinal pigment epithelium. Proc Natl Acad Sci U S A. 1997;94(18):9893–8.10.1073/pnas.94.18.9893Search in Google Scholar PubMed PubMed Central
[69] Cook JD, Ng SY, Lloyd M, Eddington S, Sun H, Nathans J, et al. Peropsin modulates transit of vitamin A from retina to retinal pigment epithelium. J Biol Chem. 2017;292(52):21407–16.10.1074/jbc.M117.812701Search in Google Scholar PubMed PubMed Central
[70] Garm A, Svaerke JE, Pontieri D, Oakley TH. Expression of opsins of the box jellyfish Tripedalia cystophora reveals the first photopigment in cnidarian ocelli and supports the presence of photoisomerases. Front Neuroanat. 2022;16:916510.10.3389/fnana.2022.916510Search in Google Scholar PubMed PubMed Central
[71] Poletini MO, Ramos BC, Moraes MN, Castrucci AM. Nonvisual opsins and the regulation of peripheral clocks by light and hormones. Photochem Photobiol. 2015;91(5):1046–55.10.1111/php.12494Search in Google Scholar PubMed
[72] Tomonari S, Takagi A, Noji S, Ohuchi H. Expression pattern of the melanopsin-like (cOpn4m) and VA opsin-like genes in the developing chicken retina and neural tissues. Gene Expr Patterns. 2007;7(7):746–53.10.1016/j.modgep.2007.06.001Search in Google Scholar PubMed
[73] Grone BP, Zhao S, Chen CC, Fernald RD. Localization and diurnal expression of melanopsin, vertebrate ancient opsin, and pituitary adenylate cyclase-activating peptide mRNA in a teleost retina. J Biol Rhythms. 2007;22(6):558–61.10.1177/0748730407308285Search in Google Scholar PubMed PubMed Central
[74] Pérez-Calderón JR, Pérez-León JA, Simões N, Aguirre-Ramírez M, Malpica-Calderon R, Botello A. Extravisual opsins in the blind shrimp Creaseria morleyi: presence and expression. Nauplius. 2023;31:e2023024. 10.1590/2358-2936e2023024 Search in Google Scholar
[75] Birch S, Picciani N, Oakley T, Plachetzki D. Cnidarians: Diversity and evolution of cnidarian visual systems. In: Buschbeck E, Bok M, editors. Distributed Vision: From simple sensor to sophisticated combination eyes. Springer Series in Vision Research. London, Berlin: Springer; 2023. p. 21–47.10.1007/978-3-031-23216-9_2Search in Google Scholar
[76] Bielecki J, Zaharoff AK, Leung NY, Garm A, Oakley TH. Ocular and extraocular expression of opsins in the rhopalium of Tripedalia cystophora (Cnidaria: Cubozoa). PLoS One. 2014;9(6):e98870.10.1371/journal.pone.0098870Search in Google Scholar PubMed PubMed Central
[77] Ksendzovsky A, Pomeraniec IJ, Zaghloul KA, Provencio JJ, Provencio I. Clinical implications of the melanopsin-based non-image-forming visual system. Neurology. 2017;88(13):1282–90.10.1212/WNL.0000000000003761Search in Google Scholar PubMed PubMed Central
[78] Esquiva G, Hannibal J. Melanopsin-expressing retinal ganglion cells in aging and disease. Histol Histopathol. 2019;34(12):1299–1311.Search in Google Scholar
[79] Sulli G, Lam MTY, Panda S. Interplay between circadian clock and cancer: new frontiers for cancer treatment. Trends Cancer. 2019;5(8):475–94.10.1016/j.trecan.2019.07.002Search in Google Scholar PubMed PubMed Central
[80] Nassan M, Videnovic A. Circadian rhythms in neurodegenerative disorders. Nat Rev Neurol. 2022;18(1):7–24.10.1038/s41582-021-00577-7Search in Google Scholar PubMed
© 2024 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Research Articles
- Antitumor activity of 5-hydroxy-3′,4′,6,7-tetramethoxyflavone in glioblastoma cell lines and its antagonism with radiotherapy
- Digital methylation-specific PCR: New applications for liquid biopsy
- Synergistic effects of essential oils and phenolic extracts on antimicrobial activities using blends of Artemisia campestris, Artemisia herba alba, and Citrus aurantium
- β-Amyloid peptide modulates peripheral immune responses and neuroinflammation in rats
- A novel approach for protein secondary structure prediction using encoder–decoder with attention mechanism model
- Diurnal and circadian regulation of opsin-like transcripts in the eyeless cnidarian Hydra
- Withaferin A alters the expression of microRNAs 146a-5p and 34a-5p and associated hub genes in MDA-MB-231 cells
- Toxicity of bisphenol A and p-nitrophenol on tomato plants: Morpho-physiological, ionomic profile, and antioxidants/defense-related gene expression studies
- Review Articles
- Polycystic ovary syndrome and its management: In view of oxidative stress
- Senescent adipocytes and type 2 diabetes – current knowledge and perspective concepts
- Seeing beyond the blot: A critical look at assumptions and raw data interpretation in Western blotting
- Biochemical dynamics during postharvest: Highlighting the interplay of stress during storage and maturation of fresh produce
- A comprehensive review of the interaction between COVID-19 spike proteins with mammalian small and major heat shock proteins
- Exploring cardiovascular implications in systemic lupus erythematosus: A holistic analysis of complications, diagnostic criteria, and therapeutic modalities, encompassing pharmacological and adjuvant approaches
Articles in the same Issue
- Research Articles
- Antitumor activity of 5-hydroxy-3′,4′,6,7-tetramethoxyflavone in glioblastoma cell lines and its antagonism with radiotherapy
- Digital methylation-specific PCR: New applications for liquid biopsy
- Synergistic effects of essential oils and phenolic extracts on antimicrobial activities using blends of Artemisia campestris, Artemisia herba alba, and Citrus aurantium
- β-Amyloid peptide modulates peripheral immune responses and neuroinflammation in rats
- A novel approach for protein secondary structure prediction using encoder–decoder with attention mechanism model
- Diurnal and circadian regulation of opsin-like transcripts in the eyeless cnidarian Hydra
- Withaferin A alters the expression of microRNAs 146a-5p and 34a-5p and associated hub genes in MDA-MB-231 cells
- Toxicity of bisphenol A and p-nitrophenol on tomato plants: Morpho-physiological, ionomic profile, and antioxidants/defense-related gene expression studies
- Review Articles
- Polycystic ovary syndrome and its management: In view of oxidative stress
- Senescent adipocytes and type 2 diabetes – current knowledge and perspective concepts
- Seeing beyond the blot: A critical look at assumptions and raw data interpretation in Western blotting
- Biochemical dynamics during postharvest: Highlighting the interplay of stress during storage and maturation of fresh produce
- A comprehensive review of the interaction between COVID-19 spike proteins with mammalian small and major heat shock proteins
- Exploring cardiovascular implications in systemic lupus erythematosus: A holistic analysis of complications, diagnostic criteria, and therapeutic modalities, encompassing pharmacological and adjuvant approaches

