Abstract
Objective
This study was to investigate the single nucleotide polymorphism (SNP) in the interferon regulatory factor 6 (IRF6) gene in healthy residents of Guangdong Province, China, for further analysis of their associations with the development of cleft lip with or without palate (CL/P).
Methodology
DNA was extracted from blood samples of 13 healthy residents. IRF6 genes were sequenced and analyzed by alignment to the reference sequences in GenBank.
Results
The IRF6 genes containing 45.21% GC were 25016–25046 bp in length, and had 2215-bp exons and a 1404-bp coding region. There were 65 SNPs, including 58 SNPs in the 5′-untranslated region or introns and 7 SNPs in the exons. These sites had two alleles, including 39 transition sites and 26 transversion sites. Five novel SNPs were identified. c. 459G>T and c. 820G>A in the coding region represented silent and Val274Ile mutations, respectively. The SNPs made two sequences (GenBank HQ875393 and JF346417).
Conclusions
The sequences and SNP profiles of the IRF6 gene in healthy residents of Guangdong Province are consistent with the one in GenBank but with slight variations. Our findings form a basis for further investigation of the association between IRF6 gene polymorphisms and CL/P in residents of Guangdong Province.
1 Introduction
The interferon regulatory factor 6 (IRF6) gene is located on chromosome 1q32-41 and contains 10 exons. The coding region of IRF6 includes exons 3–8 and part of exon 9 [1]. The IRF6 protein belongs to the IRF transcription factor family. It contains two conserved regions, a helix-turn-helix DNA-binding domain and a Smad interferon regulatory (SMIR) factor protein binding domain [2].
The IRF6 gene is the most studied candidate that causes cleft lip with or without palate (CL/P), and the IRF6 protein has been considered a key element for oral and maxillofacial development [3]. In the process of embryonic palate fusion, the levels of IRF6 expression at the edge of the palatal midline are high [1, 4]. In addition, high levels of IRF6 expression can be found in hair follicles, palate wrinkles, teeth, the thyroglossal duct, external genitalia, and the body skin [1]. It has been shown that an IRF6 mutation can cause Van der Woude syndrome (VWS) and popliteal pterygium syndrome (PPS) [1]. Rahimov et al. compared the conserved sequences of the IRF6 gene across multiple species and have shown the presence of the single nucleotide polymorphism (SNP) rs642961, G>A on an enhancer of the IRF6 gene. This SNP can alter the binding of TFAP2A to the IRF6 gene and is associated with the incidence of a simple cleft lip [5]. In addition, some scholars believe that this IRF6 gene polymorphism is significantly correlated with the development of nonsyndromic cleft lip with or without palate (NSCL/P). Variation of the IRF6 gene accounts for 12% of the genetic factors that cause CL/P. The probability of recurrence of CL/P in a family with the IRF6 gene mutation is three times more likely than in normal people [2].
A study by Pan et al. has shown that the rs642961 A and rs2235371 C alleles of the IRF6 gene increase the risk for the development of nonsyndromic orofacial clefts [6]. In contrast, a study by Paranaiba et al. has shown that these IRF6 gene polymorphisms (rs642961 and rs2235371) are not associated with NSCL/P in a Brazilian population [7]. The IRF6 rs642961 polymorphism gene locus is associated with NSCL +/- P in a northern Chinese population [8], Asians, and Caucasians [9]. It can even be considered as a marker of CL/P severity [10]. However, the IRF6 rs642961 polymorphism is not a risk factor for NSCL/P in an Iranian population [11]. These studies suggest that the association of IRF6 genetic variation with the development of CL/P depends on ethnicity. CL/P is an incomplete dominant disease with genetic heterogeneity. Guangdong Province is representative of the southern provinces of China. Its unique geography and climate result in distinct characteristics in many diseases, including CL/P in this region. The IRF6 gene sequences of healthy residents of Guangdong Province can be used to study the association between IRF6 gene variation and CL/P in people from this region. Therefore, the aim of this study was to investigate the presence of sequence and SNP variation in the IRF6 gene in healthy residents of Guangdong Province. This is the first study to investigate the sequences and SNP characteristics of the IRF6 gene in healthy residents of Guangdong Province.
2 Methods
2.1 Ethics statement
This study was conducted in accordance with the principles stated in the Declaration of Helsinki and was approved by the Guangzhou Women and Children’s Medical Center Ethics Committee. Written informed consent was obtained from each subject.
2.2 Sample Collection
Peripheral venous blood samples (3 mL) were collected from 13 healthy people, including 7 males and 6 females, who had fasted overnight. The criteria for selecting healthy subjects for this study were as follows: (1) All subjects belonged to the Han ethnic group and were native to Guangdong Province. There was no kinship among them. (2) No family history of CL/P. (3) No other congenital disease or malformation, or major organ disease.
Blood samples were stored at 4°C in tubes containing the anticoagulant EDTA-K2 (Huasheng Biotechnology Co., Ltd., Beijing). Genomic DNA was extracted from white blood cells using a kit for whole blood genomic DNA extraction (Xinjie Biotechnology Co., Ltd. Hangzhou), according to the manufacturer’s protocol.
2.3 Primer design and IRF gene amplification
A total of 26 pairs of primers (Table 1) for IRF gene sequencing were designed using Primer 5.0, according to the IRF6 gene sequence in GenBank (ACCESSION: NG_007081 VERSION: NG_007081.1, total 25218 bp). A 400–600-bp DNA region could be effectively sequenced using these primers. A total of seven primer pairs (Table 2) for sequencing the coding region were designed. The primers were synthesized by Shanghai Sangon Company.
PCR primers for amplification of the IRF6 genomic sequence.
| No. | Primers | Primer Positions (GenBank: NG 007081.1) | Amplicon (bp) |
|---|---|---|---|
| 1 | F: TCTGAAATCGACACATTGTC | 44–63 | 1304 |
| R: GTGGGACGTACATTTCAGCA | 1328–1347 | ||
| 2 | F: CTTTGGGAGGAGTCCCAGTT | 1121–1140 | 1298 |
| R: AGAGTCAAGTGCCCTGGATG | 2399–2418 | ||
| 3 | F: GGCCGCAGTTTTCTCATCTA | 2221–2240 | 675 |
| R: GCCTCTGTCTCAACCACTGC | 2876–2895 | ||
| 4 | F: CCTTACCCCTCATCCCTCTC | 2706–2725 | 1204 |
| R: AGCTTTTTAGGCCCTGGTGT | 3890–3909 | ||
| 5 | F: TACTGTTGCACAGCCTCCTG | 3587–3606 | 1120 |
| R: CTTCACGGCCCATAATTACC | 4687–4706 | ||
| 6 | F: GATGAGGAAACTGAAACCAAGA | 4450–4471 | 1280 |
| R: TAGCTCCGCTCCTTGGAAAT | 5710–5729 | ||
| 7 | F: CGTCAACAACCCCACTATCC | 5463–5482 | 1298 |
| R: TCTGCGCACCAAGAAAGTAA | 6741–6760 | ||
| 8 | F: GACACAGGCCTTACATTTTGC | 6524–6544 | 1280 |
| R: CTGTGTTTGGGCCTGTCAGA | 7785–7804 | ||
| 9 | F: TCCTTCCATGGGGCTGAG | 7536–7553 | 1280 |
| R: GAGCCAGCATGGGTAAAGTC | 8796–8815 | ||
| 10 | F: TTCCAGGCATCTCATTCTCA | 8591–8610 | 1289 |
| R: TCCTCTTCTTGTTGAGGGCTA | 9859–9879 | ||
| 11 | F: GAAAGGTGGCTGGGAAGG | 9642–9659 | 1252 |
| R: C AC AAC AAGCCTGTCCACCT | 10874–10893 | ||
| 12 | F: GTGATCCTCCCTCCTTAGCC | 10597–10616 | 1295 |
| R: GCCAGGTCTTCCAGTTCTTG | 11872–11891 | ||
| 13 | F: ACCAAGCTCTGATGGCCTTT | 11615–11634 | 1295 |
| R: GCCTAACTGCTGCTCCATTC | 12890–12909 | ||
| 14 | F: CTCTGGTTCTGGGTTGGTCT | 12630–12649 | 602 |
| R: CAACGTTTTCAAGCCAACAG | 13212–13231 | ||
| 15 | F: AAATGGAGGGACCCAAATCT | 12979–12998 | 1259 |
| R: GCACCAAAGTGCAGATGGTA | 14218–14237 | ||
| 16 | F: CTGCACCCAGGTTACAATTC | 14033–14052 | 1194 |
| R: AAATAGGAAAGCTGAGATCCCATA | 15203–15226 | ||
| 17 | F: GCCTCTTTTGGGATTCAAGA | 15044–15063 | 1325 |
| R: GCTGGATTCAAAAGCCCATA | 16349–16368 | ||
| 18 | F: TTTTCGAGTTACGGGCAAAC | 16184–16203 | 1218 |
| R: AAGGAGGCCCCAGTGTCTAT | 17382–17404 | ||
| 19 | F: CCATCAATGGTGGACTGGAT | 17157–17176 | 1236 |
| R: TTGGCCCCAGCAATTAAATA | 18373–18392 | ||
| 20 | F: TTCTTTCCCTGGAGGAGTCA | 18204–18223 | 1213 |
| R: TAACCTGGAGCTGGAGCTGT | 19397–19416 | ||
| 21 | F: CCCATGGTCCCTGTTACTTG | 1920–619225 | 1215 |
| R: CTCAGGACCTGGGAATTT | 20403–20420 | ||
| 22 | F: ACAGGGATGAACAGGCAGAG | 20188–20207 | 1239 |
| R: CCATTCTTCCCCAAAGCATA | 21407–21426 | ||
| 23 | F: GCACATGTGGCTAGTGGCTA | 21244–21263 | 1358 |
| R: ATCCTTGATGTCTGGGGTTG | 22582–22601 | ||
| 24 | F: CAGAATGGGGTCTTCCTCAG | 22376–22395 | 1392 |
| R: TTCGTGCTAGGTGAGCCTTT | 23748–23767 | ||
| 25 | F: GGAGAGGGGGTAGGTGACAG | 23525–23544 | 1249 |
| R: TTGGCACTTTTCCAATACCC | 24754–24773 | ||
| 26 | F: CCAGCTTCGAGGAAAAGTTG | 24474–24493 | 729 |
| R: CCAACACTTGCTCTCCCAGT | 25183–25202 |
PCR primers for amplification of the IRF6 coding sequence.
| Exon | Primers(5′→3′) | Primer positions (GenBank: NG_007081.1 | Amplicon (bp) |
|---|---|---|---|
| 3 | E3F: GAAAGGTGGCTGGGAAGG | 9642–9659 | 1252 |
| E3R: CACAACAAGCCTGTCCACCT | 10874–10893 | ||
| 4 | E4F: CTGCACCCAGGTTACAATTC | 14033–14052 | 1194 |
| E4R: AAATAGGAAAGCTGAGATCCCATA | 15203–15226 | ||
| 5 | E5F: GCCTCTTTTGGGATTCAAGA | 15044–15063 | 1325 |
| E5R: GCTGGATTCAAAAGCCCATA | 16349–16368 | ||
| 6 | E6F: TTCTTTCCCTGGAGGAGTCA | 18204–18223 | 1213 |
| E6R: TAACCTGGAGCTGGAGCTGT | 19397–19416 | ||
| 7 | E7F:ACAGGGATGAACAGGCAGAG | 20188–20207 | 1239 |
| E7R: CCATTCTTCCCCAAAGCATA | 21407–21426 | ||
| 8 | E8F: GCACATGTGGCTAGTGGCTA | 21244–21263 | 1358 |
| E8R: ATCCTTGATGTCTGGGGTTG | 22582–22601 | ||
| 9 | E9F: CAGAATGGGGTCTTCCTCAG | 22376–22395 | 1392 |
| E9R: TTCGTGCTAGGTGAGCCTTT | 23748–23767 |
PCR amplification for genomic DNA and coding region sequencing were carried out in 20 μL and 25 μL volumes, respectively, containing 2 x LA Taq buffer (5 mM Mg2+), 2.5 mM dNTP mixture, 2.5 units of LA Taq DNA polymerase (TaKaRa), and 100 ng of genomic DNA. The final concentrations of each primer were 10 μM and 1 μM, respectively. The PCR was performed using a T GRADIENT PCR instrument (Biometra, Germany) with the following program: 94°C for 5 min; 35 cycles of 94°C for 40 s, 58–60°C for 50 s, and 72°C for 90 s; and a final extension at 72°C for 10 min.
2.4 IRF gene sequencing and analysis
The PCR product (3 μL) was taken and analyzed using 0.8% agarose gel electrophoresis on DYY-4C and DYCP-31DN systems (Beijing, China). The target products were purified with a GeneJETTM Gel Extraction Kit (Fermentas, Thermo Scientific, USA), according to the manufacturer’s protocol, and then used for sequencing on an ABI3730XL instrument (USA) at Shanghai Megiddo Biological Pharmaceutical Co., Ltd. The sequencing results were aligned with the IRF6 gene sequence in GenBank (ACCESSION: NG_007081 VERSION: NG_007081.1) (http://www.ncbi.nlm.nih.gov/nuccore/160358359). CExpress software was used to assemble the IRF6 gene sequences. On-line analysis software NetGene2 (http://www.cbs.dtu.dk/services/NetGene2/) and Splice View (http://zeus2.itb.cnr.it/~webgene/wwwspliceview_ex.html) were used to analyze the sequencing results, including splice site analysis and coding region prediction (initiation codon ATG, and termination codons TAG, TGA, and TAA).
2.5 Analysis of IRF6 SNP
Pertinent IRF6 SNP profiles were obtained through http://www.ncbi.nlm.nih.gov/SNP/. SNPs of the IRF gene sequences in healthy residents of Guangdong Province were analyzed using MegAlign software (DNAstar V7.10).
3 Results
3.1 Make-up of IRF6 genes
To determine the presence of sequence and SNP variation in the IRF6 gene in healthy residents of Guangdong Province, we sequenced the genomic and the coding regions of the IRF6 gene in white blood cell samples. The results showed that the IRF6 gene was 25016–25046 bp in length, with 45.21% GC content. The exons were 2215 bp in length, and the coding region was 1404 bp. There was a slight difference in the length of the introns due to the distinct length of the DNA repeat (Fig. 1).
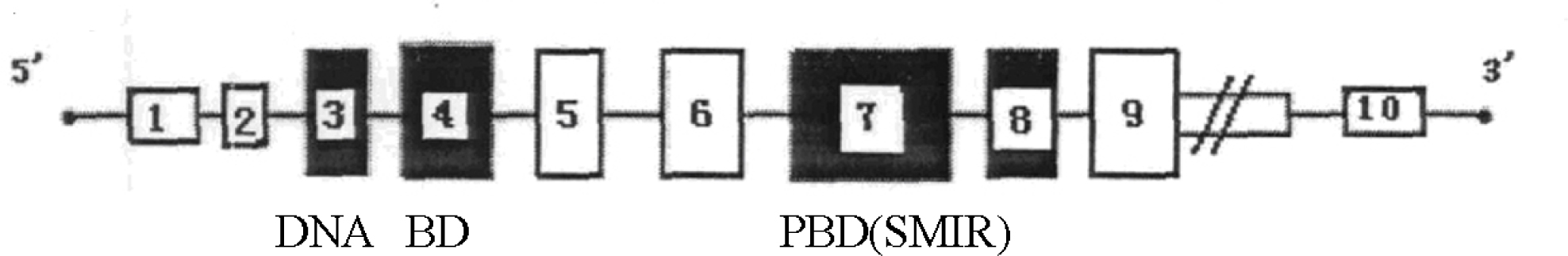
Exons and introns in the IRF6 gene. Box and number in it: exon and its position in the mRNA coding sequence. Solid line: intron. Double vertical lines: the downstream DNA was not sequenced. DNA BD: DNA-binding domain. PBD: protein binding domain. SMIR: Smad interferon regulatory factor. This figure is drawn according to the information on the IRF6 gene in GenBank (Accession Numbers: HQ875393 and JF346417) and our sequencing results in this study.
3.2 SNP profiles of the IRF6 gene
We analyzed the SNPs of the IRF6 gene sequences in healthy residents of Guangdong Province by alignment using MegAlign software (DNAstar V7. 10). The results showed the presence of 65 SNPs in the IRF6 genes of 13 healthy residents of Guangdong Province (Table 3), including 18 SNPs in 5′UTR and 10, 14, 1, 5, 3, 2, and 5 SNPs in introns 1, 3, 4, 5, 6, 7, and 8, respectively. There were a total of 7 SNPs in exons 2, 5, 7, 9, and 10. Two SNPs were found in the coding region. All variable sites had two alleles, including 39 transition sites (22 C/T and 17 G/A) and 26 transversion sites (7 C/A, 6 G/T, 10 C/G, and 3 A/T).
SNPs in healthy residents of Guangdong Province.
| No. | SNP ID | Position | Location (NG_007081) | Allele | ||
|---|---|---|---|---|---|---|
| a1 | a2 | a1:a2 | ||||
| 1 | rs2069068 | 5′UTR | g. 467 | C | A | 9/4 |
| 2 | rs11119347 | 5′UTR | g. 564 | A | G | 11/2 |
| 3 | rs11119346 | 5′UTR | g. 580 | G | A | 11/2 |
| 4 | rs764093 | 5′UTR | g. 1149 | T | C | 10/3 |
| 5 | rs17015259 | 5′UTR | g. 1557 | C | T | 10/3 |
| 6 | rs17015255 | 5′UTR | g. 1742 | T | C | 11/2 |
| 7 | rs632746 | 5′UTR | g. 1965 | G | A | 8/5 |
| 8 | rs6659367 | 5′UTR | g. 2072 | C | A | 11/2 |
| 9 | rs6696825 | 5′UTR | g. 2108 | T | C | 11/2 |
| 10 | rs630984 | 5′UTR | g. 2187 | C | T | 8/5 |
| 11 | rs6540560 | 5′UTR | g. 2407 | G | A | 11/2 |
| 12 | rs6540559 | 5′UTR | g. 2455 | C | T | 8/5 |
| 13 | rs1005287 | 5′UTR | g. 3723 | C | T | 8/5 |
| 14 | rs2357229 | 5′UTR | g. 3991 | C | A | 8/5 |
| 15 | rs6659549 | 5′UTR | g. 4325 | A | C | 9/4 |
| 16 | rs846808 | 5′UTR | g. 4380 | A | G | 12/1 |
| 17 | rs7545542 | 5′UTR | g. 4845 | G | A | 9/4 |
| 18 | rs7545538 | 5′UTR | g. 4867 | G | C | 11/2 |
| 19 | rs12403599 | Intron 1 | g. 5466 | C | G | 7/6 |
| 20 | rs682461 | Intron 1 | g. 5527 | G | C | 0/13 |
| 21 | rs17015250 | Intron 1 | g. 5703 | A | C | 10/3 |
| 22 | rs3753517 | Intron 1 | g. 6382 | G | A | 12/1 |
| 23 | rs3753518 | Intron 1 | g. 6516 | C | G | 11/2 |
| 24 | rs12405750 | Intron 1 | g. 6636 | G | A | 11/2 |
| 25 | rs861020 | Intron 1 | g. 7369 | T | C | 3/10 |
| 26 | rs2073487 | Intron 1 | g. 7834 | A | G | 11/2 |
| 27 | rs2073486 | Intron 1 | g. 8265 | C | T | 7/6 |
| 28 | rs2235377 | Intron 1 | g. 9088 | A | G | 11/2 |
| 29 | rs861019 | Exon 2 | g. 9094 | T | C | 8/5 |
| 30 | rs622832 | Intron 3 | g. 10248 | T | G | 3/10 |
| 31 | rs1160411 | Intron 3 | g. 10558 | A | G | 6/7 |
| 32 | rs2294408 | Intron 3 | g. 10931 | C | T | 7/6 |
| 33 | rs4844494 | Intron 3 | g. 12282 | C | T | 12/1 |
| 34 | rs2005982 | Intron 3 | g. 12405 | C | T | 4/9 |
| 35 | rs2236909 | Intron 3 | g. 12825 | T | C | 7/6 |
| 36 | rs2236908 | Intron 3 | g. 12840 | C | G | 7/6 |
| 37 | rs2236907 | Intron 3 | g. 12852 | G | T | 7/6 |
| 38 | rs2236906 | Intron 3 | g. 12995 | A | C | 7/6 |
| 39 | rs58637469 | Intron 3 | g. 13000 | A | T | 11/2 |
| 40 | rs59043219 | Intron 3 | g. 13870 | C | T | 7/6 |
| 41 | rs7555285 | Intron 3 | g. 14125 | C | G | 4/9 |
| 42 | rs17015226 | Intron 3 | g. 14384 | G | C | 10/2 |
| 43 | rs7552506 | Intron 3 | g. 14578 | C | G | 11/2 |
| 44 | rs17015224 | Intron 4 | g. 15562 | C | T | 12/1 |
| 45 | rs2013162 | Exon 5 | g. 15796 | G | T | 7/6 |
| 46 | rs6685182 | Intron 5 | g. 16161 | T | G | 2/11 |
| 47 | rs79307481 | Intron 5 | g. 16440 | G | A | 10/3 |
| 48 | rs75594643 | Intron 5 | g. 16829 | T | G | 10/3 |
| 49 | rs2179254 | Intron 5 | g. 17100 | A | G | 3/10 |
| 50 | rs595918 | Intron 5 | g. 17637 | T | C | 8/5 |
| 51 | rs17015217 | Intron 5 | g. 17851 | C | T | 12/1 |
| 52 | rs2235375 | Intron 6 | g. 18893 | C | G | 7/6 |
| 53 | rs926348 | Intron 6 | g. 19197 | C | G | 12/1 |
| 54 | rs674433 | Intron 6 | g. 19605 | C | A | 4/9 |
| 55 | rs2235371 | Exon 7 | g. 20400 | G | A | 9/4 |
| 56 | rs2235373 | Intron 7 | g. 20677 | C | T | 7/6 |
| 57 | rs2073485 | Intron 8 | g. 21686 | C | T | 12/1 |
| 58 | rs7522250 | Intron 8 | g. 21941 | A | G | 10/3 |
| 59 | rs614662 | Intron 8 | g. 22061 | G | A | 8/5 |
| 60 | rs926346 | Intron 8 | g. 22276 | T | A | 10/2 |
| 61 | rs742215 | Intron 8 | g. 23457 | A | T | 12/1 |
| 62 | rs742214 | Exon 9 | g. 23555 | A | G | 7/6 |
| 63 | rs599021 | Exon 9 | g. 23558 | T | G | 6/7 |
| 64 | rs2235372 | Exon 9 | g. 24044 | C | T | 7/6 |
| 65 | rs680331 | Exon 10 | g. 24608 | C | T | 3/10 |
Five SNPs that had not been reported previously were observed. At site g. 3420 located in 5′ UTR, the sequence TACAAA was missing in eight samples, aligned to the GenBank sequence that was found in five samples (Fig. 2). At site g. 4475 located in 5′ UTR, the sequence CAAGAAGC was missing in four samples, aligned to the GenBank sequence that was found in nine samples (Fig. 3). At site g. 5527 located in intron 1, -76+339G>C (rs682461) was found in 13 samples aligned to the GenBank sequence (Fig. 4). At site g. 12951–12965 located in intron 3, the sequence TCAAACTTCTTCATT was replaced with C in one sample, aligned to the GenBank sequence with g. 12951–12965 TCAAACTTCTTCATT in seven samples (Fig. 5A and 5B). No clearly separated peak downstream of T was observed in five samples (Fig. 5C). At site g. 21778 located in intron 8, the sequence CACACA was missing in five samples, aligned to the GenBank sequence that was found in eight samples (Fig. 6).

SNP at site g. 3420 located in 5′UTR. (A) The GenBank sequence (ACCESSION: NG_007081 VERSION: NG_007081.1) found in five samples. (B) The sequence TACAAA is missing in eight samples. DNA samples were extracted from white blood cell samples. The target region in the IRF6 gene was amplified using PCR. The PCR product was sequenced by Shanghai Megiddo Biological Pharmaceutical Co., Ltd.
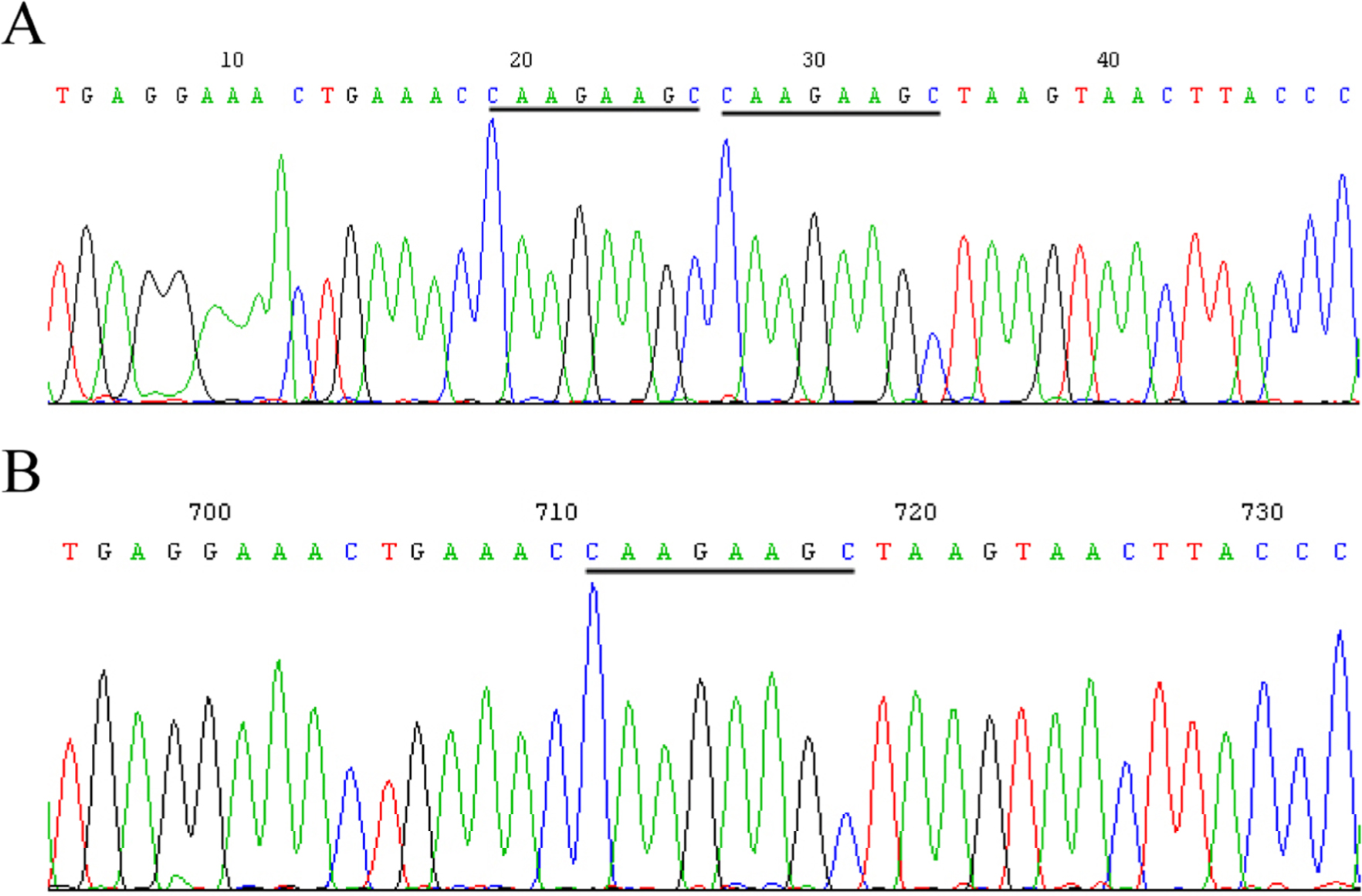
SNP at site g. 4475 located in 5′UTR.(A) The GenBank sequence (ACCESSION: NG_007081 VERSION: NG_007081.1) found in nine samples. (B) The sequence CAAGAAGC is missing in four samples. DNA samples were extracted from white blood cell samples. The target region in the IRF6 gene was amplified using PCR. The PCR product was sequenced by Shanghai Megiddo Biological Pharmaceutical Co., Ltd.

SNP at site g. 5527 located in intron 1. Aligned with GenBank sequence (ACCESSION: NG_007081 VERSION: NG_007081.1), the mutation-76+339G>C (rs682461) was found in 13 samples. DNA samples were extracted from white blood cell samples. The target region in the IRF6 gene was amplified using PCR. The PCR product was sequenced by Shanghai Megiddo Biological Pharmaceutical Co., Ltd.
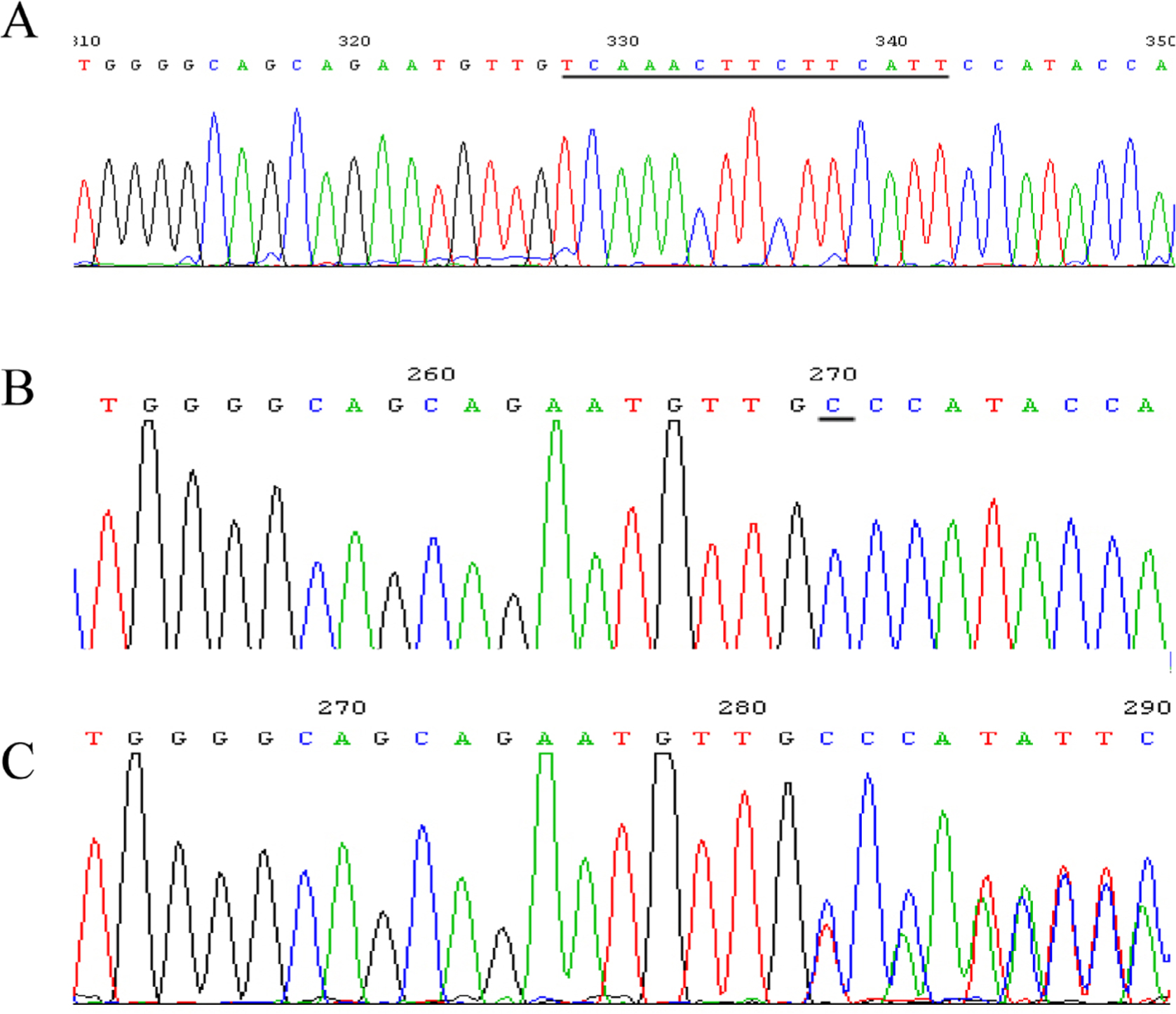
SNP at site g. 12951–12965 located in intron 3.(A) The GenBank sequence (ACCESSION: NG_007081 VERSION: NG_007081.1) with g. 12951-12965 TCAAACTTCTTCATT in seven samples. (B) Replacement of TCAAACTTCTTCATT with C in one sample. (C) No clearly separated peak downstream of T was observed in five samples. DNA samples were extracted from white blood cell samples. The target region in the IRF6 gene was amplified using PCR. The PCR product was sequenced by Shanghai Megiddo Biological Pharmaceutical Co., Ltd.
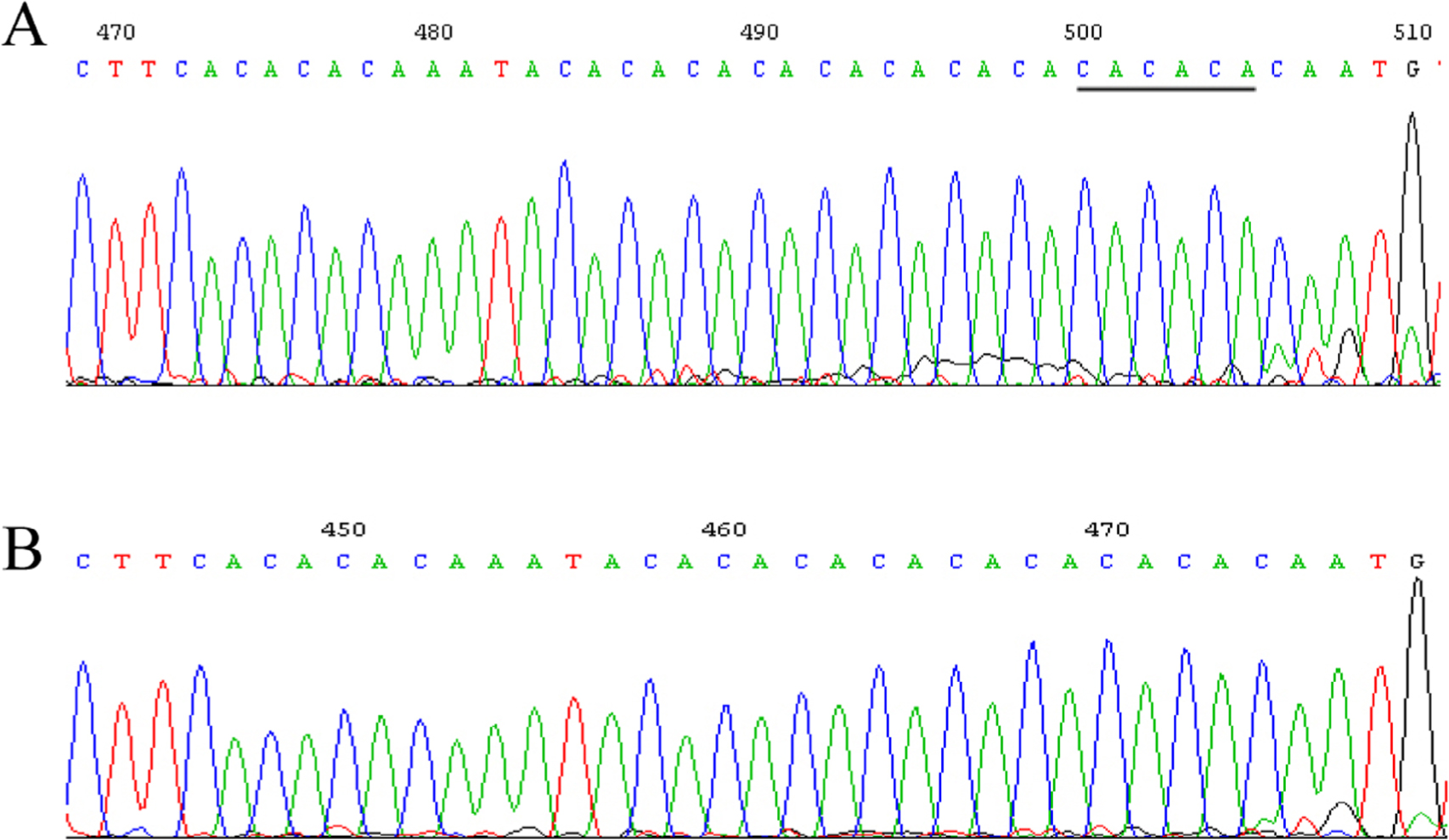
SNP at site g. 21778 located in intron 8. (A) The GenBank sequence (ACCESSION: NG_007081 VERSION: NG_007081.1) found in eight samples. (B) The sequence CACACA is missing in five samples. DNA samples were extracted from white blood cell samples. The target region in the IRF6 gene was amplified using PCR. The PCR product was sequenced by Shanghai Megiddo Biological Pharmaceutical Co., Ltd.
The results also showed the presence of two different sequences through the alignment of the SNPs in the IRF6 genes of 13 samples. Most of the SNP sites (48/65) were located in these two different sequences. They were submitted to GenBank with access numbers HQ875393 (25046 bp) and JF346417 (25016 bp).
3.3 SNP sites in the coding region of the IRF6 gene
Two mutation sites in the coding region were identified to be c. 459G>T and c. 820G>A. Transversion c. 459G>T occurred in the third nucleotide of the codon, e. g., TCG → TCT. Both TCG and TCT encode serine. Therefore, it is a silent mutation and there was no change in the protein sequence. However, the transition c. 820G>A occurred in the first nucleotide of the codon, e. g., GTC → ATC, leading to replacement of valine by an isoleucine (p. Val274Ile), which is a missense mutation.
4 Discussion
In order to study the association between IRF6 gene variation and CL/P in Guangdong Province, in the current study, we investigated the presence of sequence and SNP variation in IRF6 genes in healthy residents of Guangdong Province. We found that the IRF6 gene is 25016–25046 bp in length, with 45.21% GC content. The exons are 2215 bp in length, and the coding region is 1404 bp. There is a slight difference in the length of the introns due to the distinct length of the DNA repeat. The lengths of these sequences are consistent with the IRF6 sequence (ACCESSION: NG_007081 VERSION: NG_007081.1, 25218 bp) in GenBank. Moreover, our results are consistent with those reported by Ben et al. [12].
This is the first study to investigate the sequences and SNP characteristics of the IRF6 gene in healthy residents of Guangdong Province. A total of 65 SNPs were found in the IRF6 gene. Most of them (48/65) are located in two different sequences. Furthermore, there is a slight difference in the length of the introns due to the distinct length of the DNA repeat. It is likely that there are two distinct IRF6 gene sequences (HQ875393 and JF346417, GenBank) in healthy residents of Guangdong Province. Whether there is any difference in the sequences and functions of mRNA and protein from these two distinct IRF6 gene sequences, and whether they are associated with NSCL/P remain to be investigated.
Most SNPs found in the IRF6 gene sequences of healthy residents of Guangdong Province in the current study are NCBI dbSNP (http://www.ncbi.nlm.nih.gov/SNP/snpblastByChr.html). While some well-studied SNPs such as rs642961 were not found in the healthy residents of Guangdong Province in the current study, some of the identified 65 SNPs have been reported to be predictors for risk of NSCL/P. For example, rs2235371 (g. 20400) AA and GA have lower risks of NSCL/P than GG genotypes in Asians, but these genotypes are not significant in Caucasians. rs2013162 (g. 15796) AA carriers have a significantly lower risk than CC carriers for Caucasians but not for Asians [9]. The IRF6 rs2235371 G/A haplotype increases the risk for oral cleft in a Brazilian population [13]. Genetic polymorphisms rs7545538 (g. 4867), rs2235377 (g. 9088), rs2235371 (g. 20400), and rs2235373 (g. 20677) of the IRF6 gene are associated with an increased risk of nsCLP in a Xinjiang Uyghur population [14]. In addition to the known SNPs, five SNPs in healthy residents of Guangdong Province, at sites g. 3420, g. 4475, g. 5527, g. 12951–12965, and g. 21778, have not been reported previously. These SNPs are located in 5′ UTR (2 SNPs) and introns 1, 3, and 8, respectively (Fig. 2–6). All variable sites have two alleles. Whether these newly found SNPs are related to CL/P remains to be investigated in a future study.
There are two SNPs in the coding region of the IRF6 gene in healthy residents of Guangdong Province: c. 459G>T, a silent mutation, and c. 820G>A. These mutations are different from the mutations c. 145C>T (p. Q49X), c. 171T>G (p. F57L), and 1306C>G (p. L436V), which were found in a Thai family with VWS [15]; the mutations c. 251G>T (R84L) and c. 1271C>T (S424L), which were discovered in a Japanese family with PPS [16]; and the frameshift mutations c. 568delG, c. 295G>A, and c. 1219T>C, which were observed in a Pakistani family with VWS [17]. However, the c. 820G>A mutation found in the residents of Guangdong Province results in the replacement of a valine residue with an isoleucine residue. This site is located in the SMIR domain and is commonly seen in 3% of healthy European and 22% of healthy Asian populations [1]. In addition, this mutation has been shown to be associated with NSCL/P in a western Chinese population [18, 19]. We speculate that this missense mutation is probably related to nonsyndromic CL/P in Guangdong Province.
A limitation of this study is the sample size, which may cause a biased result for the population of Guangdong Province. The precise frequency of genotypes, haplotypes, and association with nonsyndromic CL/P should be further investigated by including more samples.
5 Conclusion
The sequences and SNP profiles of the IRF6 gene in healthy residents of Guangdong Province are consistent with the one in GenBank but with slight variations. Our findings form a basis for further investigation of the association between IRF6 gene polymorphisms and CL/P in residents of Guangdong Province.
List of abbreviations
- SNP
single nucleotide polymorphism
- UTR
5′-untranslated region (UTR)
- IRF6
The interferon regulatory factor 6
- SMIR
Smad interferon regulatory
Conflict of interest statement: Authors declare no conflicts of interest.
Author contributions: Hongtao Wang designed the study, conducted all searches, appraised all potential studies, and wrote and revised the draft manuscript and subsequent manuscripts. Wenli Wu and Liang Hua revised the draft manuscript and subsequent manuscripts. Jiansuo Hao and Yiyang Chen assisted with the presentation of findings and assisted with drafting and revising the manuscript. Fan Li and Jiayu Liu conceived and designed the study, assisted with searches, appraised relevant studies, and assisted with drafting and revising the manuscript. All authors read and approved the final manuscript.
References
[1] Kondo S., Schutte B.C., Richardson R.J., Bjork B.C., Knight A.S., Watanabe Y., et al., Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes, Nat. Genet., 2002, 32, 285-910.1038/ng985Search in Google Scholar PubMed PubMed Central
[2] Zucchero T.M., Cooper M.E., Maher B.S., Daack-Hirsch S., Nepomuceno B., Ribeiro L., et al., Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate, N. Engl. J. Med., 2004, 351, 769-8010.1056/NEJMoa032909Search in Google Scholar PubMed
[3] Bailey C.M., Hendrix M.J., IRF6 in development and disease: a mediator of quiescence and differentiation, Cell Cycle, 2008, 7, 1925-3010.4161/cc.7.13.6221Search in Google Scholar PubMed PubMed Central
[4] Knight A.S., Schutte B.C., Jiang R., Dixon M.J., Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome, Dev. Dyn., 2006, 235, 1441-710.1002/dvdy.20598Search in Google Scholar PubMed
[5] Rahimov F., Marazita M.L., Visel A., Cooper M.E., Hitchler M.J., Rubini M., et al., Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip, Nat. Genet., 2008, 40, 1341-710.1038/ng.242Search in Google Scholar PubMed PubMed Central
[6] Pan Y., Ma J., Zhang W., Du Y., Niu Y., Wang M., et al., IRF6 polymorphisms are associated with nonsyndromic orofacial clefts in a Chinese Han population, Am. J. Med. Genet. A, 2010, 152A, 2505-1110.1002/ajmg.a.33624Search in Google Scholar PubMed
[7] Paranaiba L.M., Bufalino A., Martelli-Junior H., de Barros L.M., Graner E., Coletta R.D., Lack of association between IRF6 polymorphisms (rs2235371 and rs642961) and non-syndromic cleft lip and/or palate in a Brazilian population, Oral Dis., 2010, 16, 193-710.1111/j.1601-0825.2009.01627.xSearch in Google Scholar PubMed
[8] Tian J., Wei Y., Wang L., Guo J., Zhou Q., Zhu W., [Relationship between genetic polymorphisms of IRF6 rs642961 and nonsysdromic cleft lip with or without cleft palate], Wei Sheng Yan Jiu, 2015, 44, 543-8 (in Chinese)Search in Google Scholar
[9] Wattanawong K., Rattanasiri S., McEvoy M., Attia J., Thakkinstian A., Association between IRF6 and 8q24 polymorphisms and nonsyndromic cleft lip with or without cleft palate: Systematic review and meta-analysis, Birth Defects Res. A Clin. Mol. Teratol., 2016, 106 (9), 773-8810.1002/bdra.23540Search in Google Scholar PubMed PubMed Central
[10] Kerameddin S., Namipashaki A., Ebrahimi S., Ansari-Pour N., IRF6 Is a Marker of Severity in Nonsyndromic Cleft Lip/Palate, J. Dent. Res., 2015, 94, 226S-32S10.1177/0022034515581013Search in Google Scholar PubMed
[11] Nouri N., Memarzadeh M., Carinci F., Cura F., Scapoli L., Jafary F., et al., Family-based association analysis between nonsyndromic cleft lip with or without cleft palate and IRF6 polymorphism in an Iranian population, Clin. Oral. Investig., 2015, 19, 891-410.1007/s00784-014-1305-3Search in Google Scholar PubMed
[12] Ben J., Jabs E.W., Chong S.S., Genomic, cDNA and embryonic expression analysis of zebrafish IRF6, the gene mutated in the human oral clefting disorders Van der Woude and popliteal pterygium syndromes, Gene Expr. Patterns, 2005, 5, 629-3810.1016/j.modgep.2005.03.002Search in Google Scholar PubMed
[13] de Souza L.T., Kowalski T.W., Ferrari J., Monlleo I.L., Ribeiro E.M., de Souza J., et al., Study of IRF6 and 8q24 region in non-syndromic oral clefts in the Brazilian population, Oral Dis., 2016, 22, 241-510.1111/odi.12432Search in Google Scholar PubMed
[14] Mijiti A., Ling W., Guli, Moming A., Association of single-nucleotide polymorphisms in the IRF6 gene with non-syndromic cleft lip with or without cleft palate in the Xinjiang Uyghur population, Br. J. Oral Maxillofac. Surg., 2015, 53, 268-7410.1016/j.bjoms.2014.12.008Search in Google Scholar PubMed
[15] Yeetong P., Mahatumarat C., Siriwan P., Rojvachiranonda N., Suphapeetiporn K., Shotelersuk V., Three novel mutations of the IRF6 gene with one associated with an unusual feature in Van der Woude syndrome, Am. J. Med. Genet. A, 2009, 149A, 2489-9210.1002/ajmg.a.33048Search in Google Scholar PubMed
[16] Matsuzawa N., Kondo S., Shimozato K., Nagao T., Nakano M., Tsuda M., et al., Two missense mutations of the IRF6 gene in two Japanese families with popliteal pterygium syndrome, Am. J. Med. Genet. A, 2010, 152A, 2262-710.1002/ajmg.a.33338Search in Google Scholar PubMed
[17] Malik S., Kakar N., Hasnain S., Ahmad J., Wilcox E.R., Naz S., Epidemiology of Van der Woude syndrome from mutational analyses in affected patients from Pakistan, Clin. Genet., 2010, 78, 247-5610.1111/j.1399-0004.2010.01375.xSearch in Google Scholar PubMed
[18] Tang W., Du X., Feng F., Long J., Lin Y., Li P., et al., Association analysis between the IRF6 G820A polymorphism and nonsyndromic cleft lip and/or cleft palate in a Chinese population, Cleft Palate Craniofac. J., 2009, 46, 89-9210.1597/07-131.1Search in Google Scholar PubMed
[19] Huang Y., Wu J., Ma J., Beaty T.H., Sull J.W., Zhu L., et al., Association between IRF6 SNPs and oral clefts in West China, J. Dent. Res,. 2009, 88, 715-810.1177/0022034509341040Search in Google Scholar PubMed PubMed Central
© 2016 Wenli Wu et al.
This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 3.0 License.
Articles in the same Issue
- Regular article
- Purification of polyclonal IgG specific for Camelid’s antibodies and their recombinant nanobodies
- Regular article
- Antioxidative defense mechanism of the ruderal Verbascum olympicum Boiss. against copper (Cu)-induced stress
- Regular article
- Polyherbal EMSA ERITIN Promotes Erythroid Lineages and Lymphocyte Migration in Irradiated Mice
- Regular article
- Expression and characterization of a cutinase (AnCUT2) from Aspergillus niger
- Regular article
- The Lytic SA Phage Demonstrate Bactericidal Activity against Mastitis Causing Staphylococcus aureus
- Regular article
- MafB, a target of microRNA-155, regulates dendritic cell maturation
- Regular article
- Plant regeneration from protoplasts of Gentiana straminea Maxim
- Regular article
- The effect of radiation of LED modules on the growth of dill (Anethum graveolens L.)
- Regular article
- ELF-EMF exposure decreases the peroxidase catalytic efficiency in vitro
- Regular article
- Cold hardening protects cereals from oxidative stress and necrotrophic fungal pathogenesis
- Regular article
- MC1R gene variants involvement in human OCA phenotype
- Regular article
- Chondrogenic potential of canine articular cartilage derived cells (cACCs)
- Regular article
- Cloning, expression, purification and characterization of Leishmania tropica PDI-2 protein
- Regular article
- High potential of sub-Mediterranean dry grasslands for sheep epizoochory
- Regular article
- Identification of drought, cadmium and root-lesion nematode infection stress-responsive transcription factors in ramie
- Regular article
- Herbal supplement formula of Elephantopus scaber and Sauropus androgynus promotes IL-2 cytokine production of CD4+T cells in pregnant mice with typhoid fever
- Regular article
- Caffeine effects on AdoR mRNA expression in Drosophila melanogaster
- Regular article
- Effects of MgCl2 supplementation on blood parameters and kidney injury of rats exposed to CCl4
- Regular article
- Effective onion leaf fleck management and variability of storage pathogens
- Regular article
- Improving aeration for efficient oxygenation in sea bass sea cages. Blood, brain and gill histology
- Regular article
- Biogenic amines and hygienic quality of lucerne silage
- Regular article
- Isolation and characterization of lytic phages TSE1-3 against Enterobacter cloacae
- Regular article
- Effects of pH on antioxidant and prooxidant properties of common medicinal herbs
- Regular article
- Relationship between cytokines and running economy in marathon runners
- Regular article
- Anti-leukemic activity of DNA methyltransferase inhibitor procaine targeted on human leukaemia cells
- Regular article
- Research Progress in Oncology. Highlighting and Exploiting the Roles of Several Strategic Proteins in Understanding Cancer Biology
- Regular article
- Ectomycorrhizal communities in a Tuber aestivum Vittad. orchard in Poland
- Regular article
- Impact of HLA-G 14 bp polymorphism and soluble HLA-G level on kidney graft outcome
- Regular article
- In-silico analysis of non-synonymous-SNPs of STEAP2: To provoke the progression of prostate cancer
- Regular article
- Presence of sequence and SNP variation in the IRF6 gene in healthy residents of Guangdong Province
- Regular article
- Environmental and economic aspects of Triticum aestivum L. and Avena sativa growing
- Regular article
- A molecular survey of Echinococcus granulosus sensu lato in central-eastern Europe
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Molecular genetics related to non-Hodgkin lymphoma
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Roles of long noncoding RNAs in Hepatocellular Carcinoma
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Advancement of Wnt signal pathway and the target of breast cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- A tumor suppressive role of lncRNA GAS5 in human colorectal cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- The role of E-cadherin - 160C/A polymorphism in breast cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- The proceedings of brain metastases from lung cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Newly-presented potential targeted drugs in the treatment of renal cell cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Decreased expression of miR-132 in CRC tissues and its inhibitory function on tumor progression
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- The unusual yin-yang fashion of RIZ1/RIZ2 contributes to the progression of esophageal squamous cell carcinoma
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Human papillomavirus infection mechanism and vaccine of vulva carcinoma
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Abnormal expressed long non-coding RNA IRAIN inhibits tumor progression in human renal cell carcinoma cells
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- UCA1, a long noncoding RNA, promotes the proliferation of CRC cells via p53/p21 signaling
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Forkhead box 1 expression is upregulatedin non-small cell lung cancer and correlateswith pathological parameters
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- The development of potential targets in the treatment of non-small cell lung cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Low expression of miR-192 in NSCLC and its tumor suppressor functions in metastasis via targeting ZEB2
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Downregulation of long non-coding RNA MALAT1 induces tumor progression of human breast cancer through regulating CCND1 expression
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Post-translational modifications of EMT transcriptional factors in cancer metastasis
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- EZH2 Expression and its Correlation with Clinicopathological Features in Patients with Colorectal Carcinoma
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- The association between EGFR expression and clinical pathology characteristics in gastric cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- The peiminine stimulating autophagy in human colorectal carcinoma cells via AMPK pathway by SQSTM1
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Activating transcription factor 3 is downregulated in hepatocellular carcinoma and functions as a tumor suppressor by regulating cyclin D1
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Progress toward resistance mechanism to epidermal growth factor receptor tyrosine kinase inhibitor
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Effect of miRNAs in lung cancer suppression and oncogenesis
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Role and inhibition of Src signaling in the progression of liver cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- The antitumor effects of mitochondria-targeted 6-(nicotinamide) methyl coumarin
- Special Issue on CleanWAS 2015
- Characterization of particle shape, zeta potential, loading efficiency and outdoor stability for chitosan-ricinoleic acid loaded with rotenone
- Special Issue on CleanWAS 2015
- Genetic diversity and population structure of ginseng in China based on RAPD analysis
- Special Issue on CleanWAS 2015
- Optimizing the extraction of antibacterial compounds from pineapple leaf fiber
- Special Issue on CleanWAS 2015
- Identification of residual non-biodegradable organic compounds in wastewater effluent after two-stage biochemical treatment
- Special Issue on CleanWAS 2015
- Remediation of deltamethrin contaminated cotton fields: residual and adsorption assessment
- Special Issue on CleanWAS 2015
- A best-fit probability distribution for the estimation of rainfall in northern regions of Pakistan
- Special Issue on CleanWAS 2015
- Artificial Plant Root System Growth for Distributed Optimization: Models and Emergent Behaviors
- Special Issue on CleanWAS 2015
- The complete mitochondrial genomes of two weevils, Eucryptorrhynchus chinensis and E. brandti: conserved genome arrangement in Curculionidae and deficiency of tRNA-Ile gene
- Special Issue on CleanWAS 2015
- Characteristics and coordination of source-sink relationships in super hybrid rice
- Special Issue on CleanWAS 2015
- Construction of a Genetic Linkage Map and QTL Analysis of Fruit-related Traits in an F1 Red Fuji x Hongrou Apple Hybrid
- Special Issue on CleanWAS 2015
- Effects of the Traditional Chinese Medicine Dilong on Airway Remodeling in Rats with OVA-induced-Asthma
- Special Issue on CleanWAS 2015
- The effect of sewage sludge application on the growth and absorption rates of Pb and As in water spinach
- Special Issue on CleanWAS 2015
- Effectiveness of mesenchymal stems cells cultured by hanging drop vs. conventional culturing on the repair of hypoxic-ischemic-damaged mouse brains, measured by stemness gene expression
Articles in the same Issue
- Regular article
- Purification of polyclonal IgG specific for Camelid’s antibodies and their recombinant nanobodies
- Regular article
- Antioxidative defense mechanism of the ruderal Verbascum olympicum Boiss. against copper (Cu)-induced stress
- Regular article
- Polyherbal EMSA ERITIN Promotes Erythroid Lineages and Lymphocyte Migration in Irradiated Mice
- Regular article
- Expression and characterization of a cutinase (AnCUT2) from Aspergillus niger
- Regular article
- The Lytic SA Phage Demonstrate Bactericidal Activity against Mastitis Causing Staphylococcus aureus
- Regular article
- MafB, a target of microRNA-155, regulates dendritic cell maturation
- Regular article
- Plant regeneration from protoplasts of Gentiana straminea Maxim
- Regular article
- The effect of radiation of LED modules on the growth of dill (Anethum graveolens L.)
- Regular article
- ELF-EMF exposure decreases the peroxidase catalytic efficiency in vitro
- Regular article
- Cold hardening protects cereals from oxidative stress and necrotrophic fungal pathogenesis
- Regular article
- MC1R gene variants involvement in human OCA phenotype
- Regular article
- Chondrogenic potential of canine articular cartilage derived cells (cACCs)
- Regular article
- Cloning, expression, purification and characterization of Leishmania tropica PDI-2 protein
- Regular article
- High potential of sub-Mediterranean dry grasslands for sheep epizoochory
- Regular article
- Identification of drought, cadmium and root-lesion nematode infection stress-responsive transcription factors in ramie
- Regular article
- Herbal supplement formula of Elephantopus scaber and Sauropus androgynus promotes IL-2 cytokine production of CD4+T cells in pregnant mice with typhoid fever
- Regular article
- Caffeine effects on AdoR mRNA expression in Drosophila melanogaster
- Regular article
- Effects of MgCl2 supplementation on blood parameters and kidney injury of rats exposed to CCl4
- Regular article
- Effective onion leaf fleck management and variability of storage pathogens
- Regular article
- Improving aeration for efficient oxygenation in sea bass sea cages. Blood, brain and gill histology
- Regular article
- Biogenic amines and hygienic quality of lucerne silage
- Regular article
- Isolation and characterization of lytic phages TSE1-3 against Enterobacter cloacae
- Regular article
- Effects of pH on antioxidant and prooxidant properties of common medicinal herbs
- Regular article
- Relationship between cytokines and running economy in marathon runners
- Regular article
- Anti-leukemic activity of DNA methyltransferase inhibitor procaine targeted on human leukaemia cells
- Regular article
- Research Progress in Oncology. Highlighting and Exploiting the Roles of Several Strategic Proteins in Understanding Cancer Biology
- Regular article
- Ectomycorrhizal communities in a Tuber aestivum Vittad. orchard in Poland
- Regular article
- Impact of HLA-G 14 bp polymorphism and soluble HLA-G level on kidney graft outcome
- Regular article
- In-silico analysis of non-synonymous-SNPs of STEAP2: To provoke the progression of prostate cancer
- Regular article
- Presence of sequence and SNP variation in the IRF6 gene in healthy residents of Guangdong Province
- Regular article
- Environmental and economic aspects of Triticum aestivum L. and Avena sativa growing
- Regular article
- A molecular survey of Echinococcus granulosus sensu lato in central-eastern Europe
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Molecular genetics related to non-Hodgkin lymphoma
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Roles of long noncoding RNAs in Hepatocellular Carcinoma
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Advancement of Wnt signal pathway and the target of breast cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- A tumor suppressive role of lncRNA GAS5 in human colorectal cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- The role of E-cadherin - 160C/A polymorphism in breast cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- The proceedings of brain metastases from lung cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Newly-presented potential targeted drugs in the treatment of renal cell cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Decreased expression of miR-132 in CRC tissues and its inhibitory function on tumor progression
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- The unusual yin-yang fashion of RIZ1/RIZ2 contributes to the progression of esophageal squamous cell carcinoma
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Human papillomavirus infection mechanism and vaccine of vulva carcinoma
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Abnormal expressed long non-coding RNA IRAIN inhibits tumor progression in human renal cell carcinoma cells
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- UCA1, a long noncoding RNA, promotes the proliferation of CRC cells via p53/p21 signaling
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Forkhead box 1 expression is upregulatedin non-small cell lung cancer and correlateswith pathological parameters
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- The development of potential targets in the treatment of non-small cell lung cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Low expression of miR-192 in NSCLC and its tumor suppressor functions in metastasis via targeting ZEB2
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Downregulation of long non-coding RNA MALAT1 induces tumor progression of human breast cancer through regulating CCND1 expression
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Post-translational modifications of EMT transcriptional factors in cancer metastasis
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- EZH2 Expression and its Correlation with Clinicopathological Features in Patients with Colorectal Carcinoma
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- The association between EGFR expression and clinical pathology characteristics in gastric cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- The peiminine stimulating autophagy in human colorectal carcinoma cells via AMPK pathway by SQSTM1
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Activating transcription factor 3 is downregulated in hepatocellular carcinoma and functions as a tumor suppressor by regulating cyclin D1
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Progress toward resistance mechanism to epidermal growth factor receptor tyrosine kinase inhibitor
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Effect of miRNAs in lung cancer suppression and oncogenesis
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- Role and inhibition of Src signaling in the progression of liver cancer
- Topical Issue on Cancer Signaling, Metastasis and Target Therapy
- The antitumor effects of mitochondria-targeted 6-(nicotinamide) methyl coumarin
- Special Issue on CleanWAS 2015
- Characterization of particle shape, zeta potential, loading efficiency and outdoor stability for chitosan-ricinoleic acid loaded with rotenone
- Special Issue on CleanWAS 2015
- Genetic diversity and population structure of ginseng in China based on RAPD analysis
- Special Issue on CleanWAS 2015
- Optimizing the extraction of antibacterial compounds from pineapple leaf fiber
- Special Issue on CleanWAS 2015
- Identification of residual non-biodegradable organic compounds in wastewater effluent after two-stage biochemical treatment
- Special Issue on CleanWAS 2015
- Remediation of deltamethrin contaminated cotton fields: residual and adsorption assessment
- Special Issue on CleanWAS 2015
- A best-fit probability distribution for the estimation of rainfall in northern regions of Pakistan
- Special Issue on CleanWAS 2015
- Artificial Plant Root System Growth for Distributed Optimization: Models and Emergent Behaviors
- Special Issue on CleanWAS 2015
- The complete mitochondrial genomes of two weevils, Eucryptorrhynchus chinensis and E. brandti: conserved genome arrangement in Curculionidae and deficiency of tRNA-Ile gene
- Special Issue on CleanWAS 2015
- Characteristics and coordination of source-sink relationships in super hybrid rice
- Special Issue on CleanWAS 2015
- Construction of a Genetic Linkage Map and QTL Analysis of Fruit-related Traits in an F1 Red Fuji x Hongrou Apple Hybrid
- Special Issue on CleanWAS 2015
- Effects of the Traditional Chinese Medicine Dilong on Airway Remodeling in Rats with OVA-induced-Asthma
- Special Issue on CleanWAS 2015
- The effect of sewage sludge application on the growth and absorption rates of Pb and As in water spinach
- Special Issue on CleanWAS 2015
- Effectiveness of mesenchymal stems cells cultured by hanging drop vs. conventional culturing on the repair of hypoxic-ischemic-damaged mouse brains, measured by stemness gene expression

