Abstract
Objectives
Breast cancer (BC) is a malignant tumor characterized by high heterogeneity. The current study aims to examine the process underlying the LINC00578/miR-495-3p/RNF8 regulatory pathway in BC progression, aiming to discover new therapeutic targets.
Methods
The levels of LINC00578, miR-495-3p, and RNF8 were quantified. The prognostic significance of LINC00578 was assessed through the utilization of Kaplan-Meier survival curves along with Cox regression analysis. The proliferation, migration, and invasion abilities of BC cells were assessed. The targeting relationships between LINC00578 and miR-495-3p, along with between miR-495-3p and RNF8, were verified using a dual luciferase reporter assay.
Results
LINC00578 and RNF8 were significantly elevated in BC tissues and cells, while miR-495-3p was lowly expressed. Compared to BC patients with high expression, those with low expression of LINC00578 exhibit substantially higher 5-year overall survival rate. LINC00578 expression, Lymph Node Metastasis (LNM), and tumour, node, and metastasis (TNM) stage were independent prognostic indicators. LINC00578 targeted and regulated miR-495-3p, and knockdown of miR-495-3p overturned the inhibitory action of LINC00578 interference on the proliferation, invasion, and migration of BC cells. Furthermore, miR-495-3p targeted and regulated RNF8, and knocking down RNF8 reversed the stimulatory effects of miR-495-3p interference on RNF8 expression.
Conclusions
The upregulation of LINC00578 is associated with the deterioration of BC and indicates a poor prognosis. LINC00578 expression, LNM, and TNM stage were independent prognostic indicators. LINC00578 influences the advancement of BC by targeting and regulating the miR-495-3p/RNF8 axis.
Introduction
Breast cancer (BC) is among the typical malignancies among women in China, with its incidence and mortality showing a significant upward trend. Due to its insidious and rapid onset, as well as the low screening rate, timely diagnosis and treatment are often hindered, resulting in poor patient prognosis [1], [2], [3]. After scientific and standardized treatment, the survival rate at 5 years for patients with early triple-negative BC is 85 %, while that of patients with other types of early BC is 94–99 % [4]. However, there is still a considerable number of patients with intermediate-stage and advanced-stage BC whose conditions have been delayed. When they seek medical treatment, the tumor has already invaded other important tissues and organs, resulting in poor prognosis and high mortality rates [5]. In recent years, a plethora of new targeted drugs for BC have emerged, significantly improving the outlook for patients with early BC. Nevertheless, the outlook for patients with locally advanced or metastatic BC continues to be unfavorable. The specific mechanisms driving the invasion and metastasis of BC cells are still not fully understood, highlighting the urgency to delve into the regulatory pathways of BC progression and identify novel therapeutic targets.
Long non-coding RNA (lncRNA) is abnormally expressed in various cancers including lung cancer, and BC [6]. As a novel lncRNA, LINC00578 has been relatively understudied, and its specific functions and mechanisms of action in BC are not fully understood. Some studies have found that the risk model of LINC00578 has important prognostic value for BC and may become a therapeutic target related to autophagy in clinical practice [7], [8], [9]. By studying the expression, regulatory network, and function of LINC00578 in BC cells, its unique function in the initiation and progression of BC can be unveiled, offering a fresh insight into the molecular mechanisms underlying BC.
MicroRNAs (miRNAs) often exhibiting a competitive binding interaction with lncRNAs. They can participate in various physiological processes such as cell differentiation, and apoptosis, and are also important targets in cancer treatment [10]. miRNAs can affect mRNA stability or influence the activity of target gene expression. After screening the potential downstream miRNAs of LINC00578 using the lncRNASNP database, it was found that miR-495-3p is downregulated in most tumors. Studies have shown that the downregulation of miR-495-3p in gastric cancer patients is associated with the malignant phenotype of gastric cancer [11]. NEAT1 can promote the development of colon cancer by targeting miR-495-3p [12]. Lin et al. [13] discovered that miR-495-3p is involved in the regulation of BC and can predict poor prognosis in BC patients. Therefore, it is conceivable that LINC00578 may play a part in the advancement of BC by modulating the levels of miR-495-3p. Additionally, studies have shown that overexpressed miR-495-3p can inhibit the proliferation, invasion, and migration of osteosarcoma cells by targeting CTRP3 [14]. Given the complex relationship between miR-495-3p and various types of tumors, as well as its potential association with LINC00578, it is speculated that LINC00578 may participate in the progression of BC by regulating miR-495-3p.
Through the TargetScan Human database, RNF8 has been recognized as a downstream target gene of miR-495-3p. RNF8 is a ubiquitin E3 ligase containing 484 amino acids [15], 16]. Recent studies have shown that RNF8 plays a significant role in BC. RNF8 is highly expressed in clinical BC samples, and upregulation of RNF8 may facilitate BC progression by enabling enhanced estrogen receptor α (ERα)-induced transactivation, which promotes the expression of ERα target genes [17]. Additionally, RNF8 is regulated by miRNAs. For instance, overexpression of miR-214 inhibits the proliferation and invasion of BC cells and regulates the epithelial-mesenchymal transition (EMT) process by downregulating RNF8 [18]. Therefore, it can be speculated that miR-495-3p contributes to the regulation of BC by targeting RNF8.
This study preliminarily investigates the clinical significance of LINC00578 in BC, delves into the molecular mechanisms of the LINC00578/miR-495-3p axis in BC cell proliferation, invasion, and migration, and further explores the regulatory effects of the LINC00578/miR-495-3p/RNF8 axis on BC, holding promise to provide novel theoretical insights for targeted BC therapies.
Materials and methods
Patients and sample collection
This study has been approved by the Ethics Committee of Cangzhou Central Hospital, and all patients signed written informed consent forms.
This study selected 116 BC patients who underwent surgical intervention at Cangzhou Central Hospital from January 2017 to December 2018. Among them, there were 108 cases of invasive carcinoma (including 105 cases of invasive ductal carcinoma and 3 cases of invasive lobular carcinoma), and 8 cases of ductal carcinoma in situ. TNM staging: There were 23 cases in stage I, 54 cases in stage II, and 39 cases in stage III. Inclusion criteria: (1) Patients with a clear postoperative pathological diagnosis; (2) All patients had not undergone preoperative radiotherapy or chemotherapy; (3) In good mental condition with stable vital signs and able to cooperate with researchers. Exclusion criteria: Patients with other concurrent malignant tumors, severe cardiovascular and cerebrovascular diseases, liver and kidney dysfunction, endocrine disorders, and other such conditions. This study has been approved by the hospital ethics committee and have signed a written informed consent form. During surgical procedures, samples of BC tissues as well as adjacent normal tissues were obtained. The tissues were promptly frozen to prepare for subsequent experimental procedures. Survival data for the patients was collected via telephone follow-up over a 5-year period.
Cell culture
Human normal breast epithelial cells (MCF-10A) and human BC cell lines (SK-BR-3, MCF-7, MDA-MB-468, MDA-MB-231) were purchased from American Type Culture Collection (Manassas, MA, USA). After thawing the cryopreservation vial, centrifuge to remove the supernatant. Add the corresponding complete medium according to the cell type: MCF-10A cells are cultured in MCF10A-specific medium, while MCF-7 and SK-BR-3 cells are cultured in DMEM complete medium. MDA-MB-468 and MDA-MB-231 cells are cultured in L-15 complete medium. Carry out routine culturing in a cell incubator maintained at 37 °C, 5 % CO2, and saturated humidity. The cells were passaged in a timely manner, and the logarithmic phase cells were taken after 3 passages for experiments.
Cell transfection
Cells were plated into 6-well dishes (at a concentration of 1 × 106 cells/mL) to ensure a cell confluence of over 50 % at the time of transient transfection. According to the protocol of the Lipofectamine 3,000 reagent kit (Thermo Fisher Scientific, USA), cells were divided into four groups based on the transfection vectors: the si-NC group (transfected with si-NC), the si-LINC group (transfected with si-LINC00578), the si+miR-NC group (co-transfected with si-LINC00578 and miR-NC), and the si+miR-inhibitor group (co-transfected with si-LINC00578 and miR-inhibitor). And the constructed wild-type(WT) and mutant plasmids(MUT) were co-transfected with miR-NC, miR-mimic, or miR-inhibitor, respectively. After 48 h of transfection, and subsequent experiments were conducted. Transfection sequence:si-LINCO0578 5′-GGATGCCTGTGTTGTTGTTTG-3′, miR-mimic 5′-AAACAAACAUGGUGCACUUCUU-3′, miR-inhibitor 5′, AAAGUACGUACCAUGAAAACAUUA-3′, si-RNF: TGATGATGTGAGGGATAGGGCGTAG. The negative control was purchased from Wuhan Sanying Biotechnology Co., Ltd.
qRT-PCR
Total RNA was isolated from cells and tissues utilizing the TRIzol reagent (supplied by ThermoFisher Scientific, Waltham, MA, USA), following the established protocol. Reverse transcription was performed using reverse transcriptase (TOYOBO, Japan) to synthesize cDNA, which was stored at −20 °C. Using SYBR Green Premix (TOYOBO, Japan) in the PCR System (Applied Biosystems, USA), miRNA is standardized using U6 as the reference, while RNF8 is standardized using GAPDH as the reference. The 2−ΔΔCt method was employed to assess and quantify gene expression levels.
Dual-luciferase reporter assay
Wild-type plasmids (LINC00578-WT and RNF8-WT) and mutant plasmids (LINC00578-MUT and RNF8-MUT) for LINC00578 and RNF8 were constructed. The mutant versions of LINC00578 and RNF8 were established through point mutations at their binding sites. The plasmid vector used was pMIR-REPORT (Ambion, USA). The constructed wild-type and mutant plasmids were co-transfected into cells with miR-NC, miR-mimic, or miR-inhibitor, respectively, using the Lipofectamine 3,000 reagent kit (Thermo Fisher Scientific, USA). Using Renilla luciferase as an internal control, luciferase activity was measured after 48 h. Each experiment was repeated three times.
CCK-8 assay
After cell transfection, the cells were plated into 96-well plates at a density of 5 × 103 cells per well. After 0 h, 24 h, 48 h, and 72 h, CCK-8 reagent (sourced from Dojindo, Kumamoto, Japan) was added to each well, and the cells were incubated under conditions of 37 °C and 5 % CO2 for 4 h. The absorbance (OD) at 450 nm was measured using a microplate reader.
Transwell assay
The Matrigel was dissolved overnight at 4 °C and then diluted at a ratio of 1:7 with serum-free medium on ice. Twenty-five μL of the diluted Matrigel were added to the upper chamber of the Transwell insert and kept sterile at 37 °C overnight to ensure complete polymerization of the Matrigel. Note that the Matrigel was not used for the Transwell cell migration assay. The transfected cells were collected and resuspended in serum-free medium to adjust the cell density to 5 × 104 cells/mL. Five hundred microliters of cell culture medium containing serum were added to the lower chamber of the Transwell as a chemotactic agent for migration. The cell suspension was then added to the upper chamber of the Transwell, which was placed in a 24-well plate for 24 h of incubation. The cells on the upper surface of the membrane were gently wiped off with a cotton swab. After washing with PBS, the cells were stained with crystal violet. The cells were observed and photographed under an inverted phase-contrast microscope at × 200 magnification. Five random fields were observed and photographed for cell counting. The experiment was repeated three times.
Statistical analysis
The data were evaluated utilizing SPSS 23.0 software, with results reported as mean±SD Group comparisons were conducted using the independent-samples t-test. Statistical significance was determined by p<0.05.
Results
Expression levels of LINC00578, miR-495-3p, and RNF8
LINC00578 and RNF8 in BC tissues were notably elevated compared to neighboring non-cancerous breast tissues (p<0.01), while the level of miR-495-3p in BC tissues was significantly decreased compared to adjacent normal breast tissues (p<0.01, Figure 1A). Compared to MCF-10A cells, LINC00578 and RNF8 were significantly elevated in other cells (p<0.01), while miR-495-3p was markedly decreased (p<0.01, Figure 1B). Among these cell lines, MCF-7 and SK-BR-3 exhibited the most pronounced differences in the expression of these three factors, hence they were selected for upcoming experiments.

Expression levels of LINC00578, miR-495-3p, and RNF8. A) Expression levels of LINC00578, miR-495-3p, and RNF8 in BC tissues and adjacent normal tissues B) expression levels of LINC00578, miR-495-3p, and RNF8 in four BC cell lines compared to normal cells. Note: Compared with MCF-10A, ***p<0.01.
Correlation of LINC00578 expression with patients’ clinical characteristics
Based on the expression levels of LINC00578 in BC patients, an average value of 1.35 was calculated. Using this average value, 116 BC patients were divided into two groups: high and low. LINC00578 exhibited a notable correlation with the TNM stage (p=0.022) and LNM (p=0.034) of BC patients (p<0.05, Table 1). A lack of significant correlation was noted between other characteristics, including age, menopause, HER-2, PR, tumor size, and differentiation (p>0.05).
Correlation between LINC00578 expression levels and clinical features of BC patients.
| Variables | BC patients (n=116) | LINC00578 expression | p-Value | |
|---|---|---|---|---|
| Low (n=56) | High (n=60) | |||
| Age | 0.181 | |||
| <50 | 63 | 34 | 29 | |
| ≥50 | 53 | 22 | 31 | |
| Menopause | 0.71 | |||
| No | 58 | 29 | 29 | |
| Yes | 58 | 27 | 31 | |
| HER-2 | 0.309 | |||
| Negative | 69 | 36 | 33 | |
| Positive | 47 | 20 | 27 | |
| PR | 0.432 | |||
| Negative | 64 | 33 | 31 | |
| Positive | 52 | 23 | 29 | |
| ER | 0.472 | |||
| Negative | 62 | 28 | 34 | |
| Positive | 54 | 28 | 26 | |
| Tumor size | 0.349 | |||
| <5 | 59 | 31 | 28 | |
| ≥ 5 | 57 | 25 | 32 | |
| Differentiation | 0.366 | |||
| Well-moderate | 76 | 39 | 37 | |
| Poor | 40 | 17 | 23 | |
| TNM stage | ||||
| I-II | 77 | 43 | 34 | 0.022 |
| III | 39 | 13 | 26 | |
| LNM | 0.034 | |||
| No | 78 | 43 | 35 | |
| Yes | 38 | 13 | 25 | |
-
HER2, human epidermal growth factor receptor 2; PR, progesterone receptor; ER, estrogen receptor; TNM, tumor node metastasis; LNM, lymph node metastasis.
Prognostic analysis of BC patients
A Kaplan-Meier survival curve was constructed based on LINC00578 over a five-year follow-up period (Figure 2). The Kaplan-Meier curve confirmed that BC patients with lower LINC00578 expression levels had markedly higher five-year overall survival rates compared to those with higher LINC00578 expression levels (p=0.016). Cox regression analysis revealed that LINC00578 (HR=2.932, 95 %CI=1.235–6.964, p=0.015), TNM stage (HR=2.967, 95 %CI=1.2–7.336, p=0.019), and LNM (HR=2.41, 95 %CI=1.098–5.292, p=0.028) serve as independent predictors of BC outcome (Table 2).

Kaplan-Meier curves for BC patients based on the expression of LINC00578.
Cox regression analysis to evaluate clinical characteristic indicators.
| Variables | HR | 95 % CI | p-Value |
|---|---|---|---|
| LINC00578 | 2.932 | 1.235–6.964 | 0.015 |
| Menopause | 0.602 | 0.2–1.815 | 0.367 |
| Age | 2.047 | 0.697–6.017 | 0.193 |
| Tumor size | 1.78 | 0.799–3.967 | 0.158 |
| Differentiation | 1.146 | 0.496–2.652 | 0.749 |
| TNM | 2.967 | 1.2–7.336 | 0.019 |
| LNM | 2.41 | 1.098–5.292 | 0.028 |
| HER-2 | 1.592 | 0.735–3.448 | 0.239 |
| PR | 1.905 | 0.793–4.572 | 0.149 |
| ER | 0.474 | 0.202–1.111 | 0.086 |
-
T, tumour size; N, lymph node; M, distant metastasis; TNM, tumour, node, and metastasis; LNM, lymph node metastasis; HER-2, human epidermal growth factor receptor-2; PR, progesterone receptor; ER, estrogen receptor.
Targeting relationship between LINC00578 and miR-495-3p
Through the lncRNASNP database, miR-495-3p was predicted to bind with LINC00578. The locations where LINC00578 interacts with miR-495-3p are illustrated in Figure 3A. miR-495-3p negatively regulates the luciferase activity of LINC00578-WT in both MCF-7 (Figure 3B) and SK-BR-3 (Figure 3C) cell lines (p<0.01). No notable disparity was observed in luciferase activity of LINC00578-MUT in either cell line (p>0.05). After knocking down LINC00578, the expression levels of LINC00578 in the MCF-7 and SK-BR-3 cell lines were significantly reduced (p<0.01, Figure 3D), while the expression levels of miR-495-3p were significantly increased (p<0.01, Figure 3E). However, compared with si+miR-NC, knocking down miR-495-3p did not significantly alter the expression of LINC00578 (p>0.05). Silencing miR-495-3p could reverse the enhancing effect of LINC00578 knockdown on miR-495-3p expression (p<0.01). These results indicate that LINC00578 targets and regulates the expression of miR-495-3p.

Verification of the targeting relationship between LINC00578 and miR-495-3p using luciferase reporter gene assays. A) Predicted targeting sites between LINC00578 and miR-495-3p. B) Luciferase reporter assay for LINC00578 in MCR-7 cells. C) Luciferase reporter assay for LINC00578 in SK-BR-3 cells. D) Expression levels of LINC00578 in cell lines after transfection. E) Expression levels of miR-495-3p in cell lines after transfection. ***p<0.01.
Effects of the LINC00578/miR-495-3p axis on BC cell proliferation, invasion, and migration
After knocking down LINC00578, the proliferation ability of MCF-7 (Figure 4A) and SK-BR-3 (Figure 4B) cell lines was significantly reduced (p<0.01). Conversely, knocking down miR-495-3p can overcome the inhibitory action of LINC00578 interference on the MCF-7 and SK-BR-3 cell lines (p<0.01). Similarly, silencing LINC00578 markedly decreased the number of migrated (Figure 4C and D) and invaded (Figure 4E and F) cells (p<0.01). In contrast, knocking down miR-495-3p can reverse the impact of disrupting LINC00578 on cellular migratory and invasive capabilities (p<0.01).
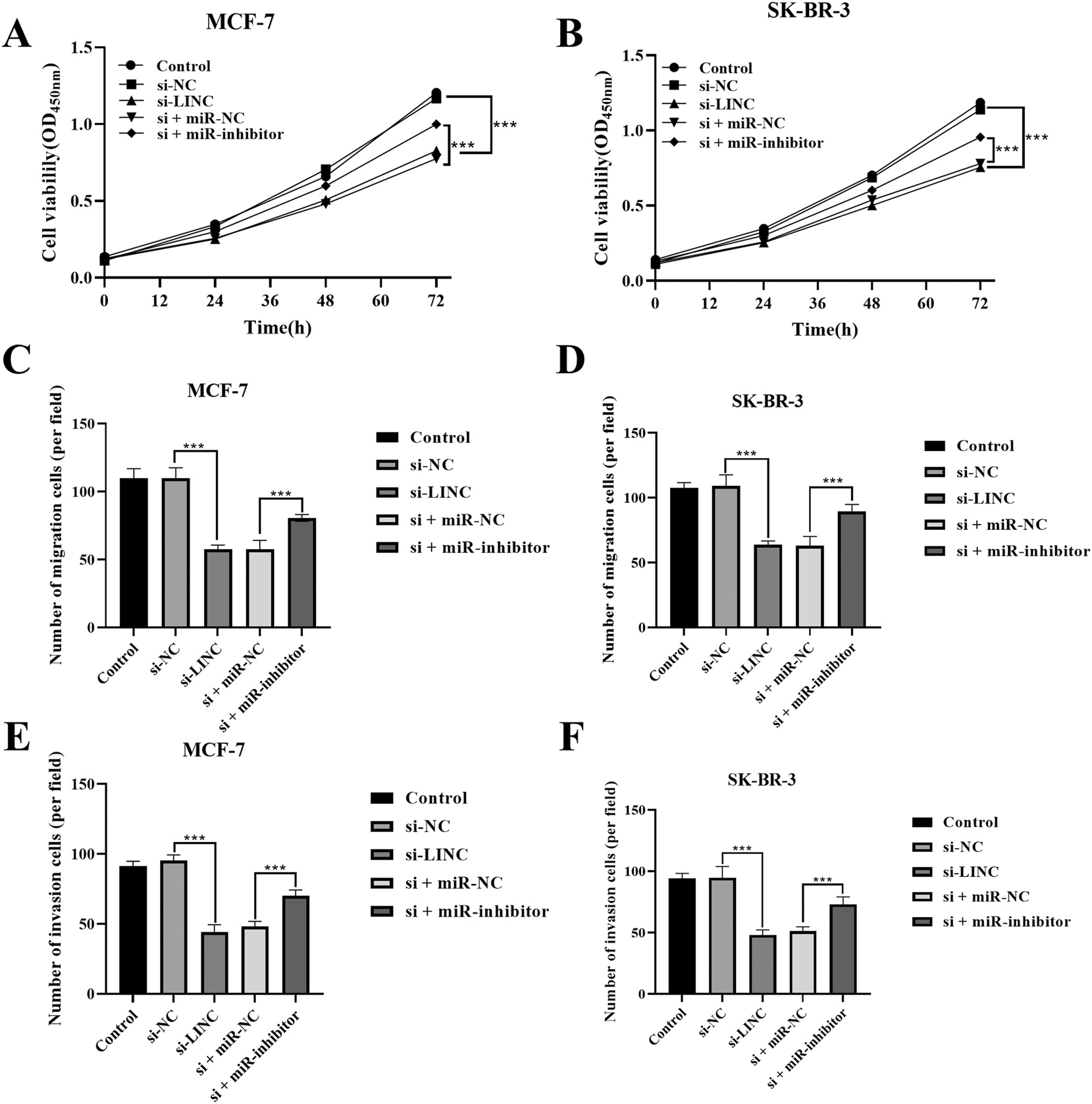
Proliferation, migration, and invasion capabilities of BC cells. A) Proliferation capability of MCR-7. B) Proliferation capability of SK-BR-3. C) Migration capability of MCR-7. D) Migration capability of SK-BR-3. E) Invasion capability of MCR-7. F) Invasion capability of SK-BR-3. ***p<0.01.
Targeting relationship between miR-495-3p and RNF8
Through the TargetScan database, RNF8 was identified as a downstream target gene of miR-495-3p. The interaction points between miR-495-3p and RNF8 are shown in Figure 5A. miR-495-3p negatively regulates the luciferase activity of RNF8-WT in both MCF-7 (Figure 5B) and SK-BR-3 (Figure 5C) cells (p<0.01). A notable disparity was absent in the luciferase activity of RNF8-MUT in either cell line (p>0.05). Knocking down miR-495-3p markedly increased RNF8 in both SK-BR-3 and MCF-7 cell lines (p<0.01). After inhibiting miR-495-3p, the expression levels of RNF8 in the MCF-7 and SK-BR-3 cell lines were significantly increased (p<0.01, Figure 5D), while the expression levels of miR-495-3p were significantly decreased (p<0.01, Figure 5E). However, knocking down RNF8 had no significant effect on the expression of miR-495-3p but could reverse the stimulatory effect of miR-495-3p interference on RNF8. These results suggest that miR-495-3p regulates RNF8 in a targeted manner.

Verification of the targeting relationship between miR-495-3p and RNF8 using luciferase reporter gene assays. A) Predicted targeting sites between miR-495-3p and RNF8. B) Luciferase reporter assay for RNF8 in MCR-7 cells. C) Luciferase reporter assay for RNF8 in SK-BR-3 cells. D) Expression levels of RNF8 in cell lines after transfection. E) Expression levels of miR-495-3p in cell lines after transfection. ***p<0.01.
Discussion
BC is a prevalent malignancy among women, distinguished by its significant heterogeneity, propensity for recurrence and metastasis, as well as its rapid progression [19]. The treatment effect and prognosis of BC are gradually improving, but the prognosis of patients in the middle and late stages is still unsatisfactory [20]. Hence, discovering novel therapeutic targets and approaches for BC holds immense importance in enhancing the prognosis of BC patients. lncRNAs are capable of regulating diverse biological behaviors of tumor cells and contributing to tumor development [21].
Abnormal expression of LINC00578 has been confirmed in numerous kinds of solid tumors. Specifically, LINC00578 is highly expressed in both pancreatic cancer tissues and serum, and its expression level correlates with clinical staging [22]. LINC00578 is expressed at high levels in lung cancer, and it is tightly linked to the metastasis of lung adenocarcinoma and poor patient survival. [9], 23]. The findings of this study further revealed a notable upregulation of LINC00578 in BC tissues and cells. Prior research has suggested that LINC00578 expression is elevated in pancreatic cancer tissues, with its expression level increasing as the tumor stage advances. Furthermore, elevated expression levels of LINC00578 are strongly associated with adverse prognosis outcomes, indicating its possible use as a predictive indicator for pancreatic cancer [22]. In BC, LINC00578 has been identified as a lncRNA associated with autophagy, cellular stemness, and immune regulation. Elevated expression levels of LINC00578 are tightly linked to unfavorable prognosis among BC patients [7], 8], 24]. This aligns with the findings of our research, which found that LINC00578 is significantly correlated with TNM stage and LNM. Furthermore, LINC00578 expression, TNM stage, and LNM are independent risk factors for the prognosis of BC, and BC patients with high LINC00578 expression have a worse prognosis. These findings imply that LINC00578 is intimately linked to tumor progression and disease severity in BC patients.
Through the lncRNA SNP database, it was discovered that LINC00578 harbors a binding site for miR-495-3p. miR-495-3p expression has been found to decrease in multiple types of cancer, including BC [25], 26], leukemia [27], and esophageal cancer [28]. This indicates that miR-495-3p may play a role in inhibiting tumor development in the context of cancer pathogenesis. This study found that miR-495-3p was significantly reduced in BC tissues and cells. Additionally, studies have shown that upregulation of miR-495-3p inhibits invasion, migration, and proliferation of pancreatic cancer (PC) cells, while enhancing apoptosis [29]. The findings of this study suggest that LINC00578 targets and regulates miR-495-3p. Silencing miR-495-3p reversed the inhibitory effects of LINC00578 interference on the proliferation, invasion, and migration of BC cells. This implies that LINC00578 may play be involved in regulating the progression of BC by modulating miR-495-3p.
Furthermore, the bioinformatics prediction in the present study also revealed a binding site between miR-495-3p and RNF8. RNF8 can promote BC metastasis. [30]. RNF8 has been implicated in playing an oncogenic role in BC, where it can promote the EMT process in BC cells, thereby inducing BC metastasis [16], 31]. This indicates that RNF8 is essential for the metastasis of BC. Kuang J and colleagues discovered that RNF8 is abundantly expressed in BC cell lines and correlates with an unfavorable prognosis. They also discovered that RNF8 can induce EMT in BC cells, thereby promoting BC metastasis [16]. The results of this study showed that RNF8 can be found in high abundance in BC tissues and cells. Furthermore, knocking down RNF8 reversed the stimulatory effect of miR-495-3p interference on RNF8, indicating that miR-495-3p targets and regulates RNF8.
The patients included in this study are those with ER/PR-positive and HER2-positive status, excluding the TNBC subtype. The sample size in this study is relatively small, potentially unable to adequately represent the characteristics of the entire study population and susceptible to the influence of chance factors. Future research will employ a larger sample size and incorporate more subtypes, including TNBC, to validate and consolidate the current findings, thereby enhancing the reliability and reproducibility of the study. In summary, LINC00578 is highly expressed in BC, and its low expression can inhibit the development and progression of malignancy in BC by targeting the miR-495-3p/RNF8 axis. This study provides scientific evidence and reference to further investigate the mechanisms driving BC development and the development of potential therapeutic targets. However, further validation of the relevant mechanisms is needed.
-
Research ethics: The study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Cangzhou Central Hospital before the study began.
-
Informed consent: Informed consent was obtained from all individuals included in this study.
-
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and approved its submission. Conceptualization, H.L. J.Y. and J.L.; Data curation, H.L. J.Y. and J.L.; Formal analysis, J.Y. and J.L.; Funding acquisition, H.L.; Investigation, J.Y. and J.L.; Methodology, H.L., J.Y. and J.L.; Project administration, H.L.; Resources, J.Y. and J.L.; Software, J.Y. and J.L.; Supervision, H.L.; Validation, J.Y. and J.L.; Visualization, H.L.; Roles/Writing – original draft, H.L.; Writing – review & editing, H.L.
-
Use of Large Language Models, AI and Machine Learning Tools: None declared.
-
Conflict of interests: Authors state no conflict of interest.
-
Research funding: None declared.
-
Data availability: The data that support the findings of this study are available from the corresponding author, H.L., upon reasonable request.
References
1. Criscitiello, CCC. Breast cancer genetics: diagnostics and treatment. Genes (Basel) 2022;13. https://doi.org/10.3390/genes13091593.Search in Google Scholar PubMed PubMed Central
2. Katsura, C, Ogunmwonyi, I, Kankam, HK, Saha, S. Breast cancer: presentation, investigation and management. Br J Hosp Med (Lond). 2022;83:1–7. https://doi.org/10.12968/hmed.2021.0459.Search in Google Scholar PubMed
3. Tsang, JYS, Tse, GM. Molecular classification of breast cancer. Adv Anat Pathol 2020;27:27–35. https://doi.org/10.1097/pap.0000000000000232.Search in Google Scholar PubMed
4. Waks, AG, Winer, EP. Breast cancer treatment: a review. Jama 2019;321:288–300. https://doi.org/10.1001/jama.2018.19323.Search in Google Scholar PubMed
5. Franzoi, MA, Saúde-Conde, R, Ferreira, SC, Eiger, D, Awada, A, de Azambuja, E. Clinical outcomes of platinum-based chemotherapy in patients with advanced breast cancer: an 11-year single institutional experience. Breast 2021:5786–94. https://doi.org/10.1016/j.breast.2021.03.002.Search in Google Scholar PubMed PubMed Central
6. Chen, S, Shen, X. Long noncoding RNAs: functions and mechanisms in colon cancer. Mol Cancer 2020;19:167. https://doi.org/10.1186/s12943-020-01287-2.Search in Google Scholar PubMed PubMed Central
7. Li, X, Li, Y, Yu, X, Jin, F. Identification and validation of stemness-related lncRNA prognostic signature for breast cancer. J Transl Med 2020;18:331. https://doi.org/10.1186/s12967-020-02497-4.Search in Google Scholar PubMed PubMed Central
8. Ma, W, Zhao, F, Yu, X, Guan, S, Suo, H, Tao, Z, et al.. Immune-related lncRNAs as predictors of survival in breast cancer: a prognostic signature. J Transl Med 2020;18:442. https://doi.org/10.1186/s12967-020-02522-6.Search in Google Scholar PubMed PubMed Central
9. Wang, L, Zhao, H, Xu, Y, Li, J, Deng, C, Deng, Y, et al.. Systematic identification of lincRNA-based prognostic biomarkers by integrating lincRNA expression and copy number variation in lung adenocarcinoma. Int J Cancer 2019;144:1723–34. https://doi.org/10.1002/ijc.31865.Search in Google Scholar PubMed
10. Hill, MTN. miRNA interplay: mechanisms and consequences in cancer. Dis Model Mech 2021;14. https://doi.org/10.1242/dmm.047662.Search in Google Scholar PubMed PubMed Central
11. Chen, S, Wu, J, Jiao, K, Wu, Q, Ma, J, Chen, D, et al.. MicroRNA-495-3p inhibits multidrug resistance by modulating autophagy through GRP78/mTOR axis in gastric cancer. Cell Death Dis 2018;9:1070. https://doi.org/10.1038/s41419-018-0950-x.Search in Google Scholar PubMed PubMed Central
12. He, Z, Dang, J, Song, A, Cui, X, Ma, Z, Zhang, Z. NEAT1 promotes colon cancer progression through sponging miR-495-3p and activating CDK6 in vitro and in vivo. J Cell Physiol 2019;234:19582–91. https://doi.org/10.1002/jcp.28557.Search in Google Scholar PubMed
13. Lin, S, Zhao, M, Lv, Y, Mao, G, Ding, S, Peng, F. The lncRNA GATA3-AS1/miR-495-3p/CENPU axis predicts poor prognosis of breast cancer via the PLK1 signaling pathway. Aging (Albany NY) 2021;13:13663–79. https://doi.org/10.18632/aging.202909.Search in Google Scholar PubMed PubMed Central
14. Zhao, G, Zhang, L, Qian, D, Sun, Y, Liu, W. miR-495-3p inhibits the cell proliferation, invasion and migration of osteosarcoma by targeting C1q/TNF-related protein 3. OncoTargets Ther 2019:126133–43. https://doi.org/10.2147/ott.s193937.Search in Google Scholar
15. Fang, S, Weissman, AM. A field guide to ubiquitylation. Cell Mol Life Sci 2004;61:1546–61. https://doi.org/10.1007/s00018-004-4129-5.Search in Google Scholar PubMed PubMed Central
16. Kuang, J, Li, L, Guo, L, Su, Y, Wang, Y, Xu, Y, et al.. RNF8 promotes epithelial-mesenchymal transition of breast cancer cells. J Exp Clin Cancer Res 2016;35:88. https://doi.org/10.1186/s13046-016-0363-6.Search in Google Scholar PubMed PubMed Central
17. Wang, S, Luo, H, Wang, C, Sun, H, Sun, G, Sun, N, et al.. RNF8 identified as a co-activator of estrogen receptor α promotes cell growth in breast cancer. Biochim Biophys Acta, Mol Basis Dis 2017;1863:1615–28. https://doi.org/10.1016/j.bbadis.2017.02.011.Search in Google Scholar PubMed
18. Min, L, Liu, C, Kuang, J, Wu, X, Zhu, L. miR-214 inhibits epithelial-mesenchymal transition of breast cancer cells via downregulation of RNF8. Acta Biochim Biophys Sin (Shanghai) 2019;51:791–8. https://doi.org/10.1093/abbs/gmz067.Search in Google Scholar PubMed
19. Wilkinson, L, Gathani, T. Understanding breast cancer as a global health concern. Br J Radiol 2022;95:20211033. https://doi.org/10.1259/bjr.20211033.Search in Google Scholar PubMed PubMed Central
20. Coughlin, SS. Epidemiology of breast cancer in women. Adv Exp Med Biol 2019:11529–9.10.1007/978-3-030-20301-6_2Search in Google Scholar PubMed
21. Cao, HL, Liu, ZJ, Huang, PL, Yue, YL, Xi, JN. lncRNA-RMRP promotes proliferation, migration and invasion of bladder cancer via miR-206. Eur Rev Med Pharmacol Sci 2019;23:1012–21. https://doi.org/10.26355/eurrev_201902_16988.Search in Google Scholar PubMed
22. Zhang, B, Li, C, Sun, Z. Long non-coding RNA LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 are novel prognostic markers for pancreatic cancer. Am J Transl Res 2018;10:2648–58.Search in Google Scholar
23. Zhao, B, Xu, H, Ai, X, Adalat, Y, Tong, Y, Zhang, J, et al.. Expression profiles of long noncoding RNAs in lung adenocarcinoma. OncoTargets Ther 2018:115383–90. https://doi.org/10.2147/ott.s167633.Search in Google Scholar
24. Li, X, Jin, F, Li, Y. A novel autophagy-related lncRNA prognostic risk model for breast cancer. J Cell Mol Med 2021;25:4–14. https://doi.org/10.1111/jcmm.15980.Search in Google Scholar PubMed PubMed Central
25. Liu, M, Yang, J, Lv, W, Wang, S, Du, T, Zhang, K, et al.. Down-regulating GRP78 reverses pirarubicin resistance of triple negative breast cancer by miR-495-3p mimics and involves the p-AKT/mTOR pathway. Biosci Rep 2022;42. https://doi.org/10.1042/bsr20210245.Search in Google Scholar PubMed PubMed Central
26. Guan, YX, Zhang, MZ, Chen, XZ, Zhang, Q, Liu, SZ, Zhang, YL. Lnc RNA SNHG20 participated in proliferation, invasion, and migration of breast cancer cells via miR-495. J Cell Biochem 2018;119:7971–81. https://doi.org/10.1002/jcb.26588.Search in Google Scholar PubMed
27. Rittavee, Y, Artus, J, Desterke, C, Simanic, I, de Souza, LEB, Riccaldi, S, et al.. miR-495-3p sensitizes BCR-ABL1-expressing leukemic cells to tyrosine kinase inhibitors by targeting multidrug resistance 1 gene in T315I mutated cells. Exp Hematol 2023:11840–52. https://doi.org/10.1016/j.exphem.2022.12.003.Search in Google Scholar PubMed
28. Huang, GM, Zang, HL, Geng, YX, Li, YH. LncRNA FAM83A-AS1 aggravates the malignant development of esophageal cancer by binding to miR-495-3p. Eur Rev Med Pharmacol Sci 2020;24:9408–15. https://doi.org/10.26355/eurrev_202009_23024.Search in Google Scholar PubMed
29. Chen, F, Liu, L, Wang, S. Long non-coding RNA NORAD exhaustion represses prostate cancer progression through inhibiting TRIP13 expression via competitively binding to miR-495-3p. Cancer Cell Int 2020;20323. https://doi.org/10.1186/s12935-020-01371-z.Search in Google Scholar PubMed PubMed Central
30. Kuang, J, Duan, T, Gao, C, Liu, C, Chen, S, Zhu, LY, et al.. RNF8 depletion attenuates hepatocellular carcinoma progression by inhibiting epithelial-mesenchymal transition and enhancing drug sensitivity. Acta Biochim Biophys Sin (Shanghai) 2023;55:661–71. https://doi.org/10.3724/abbs.2023076.Search in Google Scholar PubMed PubMed Central
31. Lee, HJ, Li, CF, Ruan, D, Powers, S, Thompson, PA, Frohman, MA, et al.. The DNA damage transducer RNF8 facilitates cancer chemoresistance and progression through twist activation. Mol Cell 2016;63:1021–33. https://doi.org/10.1016/j.molcel.2016.08.009.Search in Google Scholar PubMed PubMed Central
© 2025 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Review Article
- Unveiling the hidden clinical and economic impact of preanalytical errors
- Research Articles
- To explore the role of hsa_circ_0053004/hsa-miR-646/CBX2 in diabetic retinopathy based on bioinformatics analysis and experimental verification
- Study on the LINC00578/miR-495-3p/RNF8 axis regulating breast cancer progression
- Comparison of two different anti-mullerian hormone measurement methods and evaluation of anti-mullerian hormone in polycystic ovary syndrome
- The evaluation of the relationship between anti angiotensin type I antibodies in hypertensive patients undergoing kidney transplantation
- Evaluation of neopterin, oxidative stress, and immune system in silicosis
- Assessment of lipocalin-1, resistin, cathepsin-D, neurokinin A, agmatine, NGF, and BDNF serum levels in children with Autism Spectrum Disorder
- Regulatory nexus in inflammation, tissue repair and immune modulation in Crimean-Congo hemorrhagic fever: PTX3, FGF2 and TNFAIP6
- Pasteur effect in leukocyte energy metabolism of patients with mild, moderate, and severe COVID-19
- Thiol-disulfide homeostasis and ischemia-modified albumin in patients with sepsis
- Myotonic dystrophy type 1 and oxidative imbalance: evaluation of ischemia-modified albumin and oxidant stress
- Antioxidant and alpha-glucosidase inhibitory activities of flavonoids isolated from fermented leaves of Camellia chrysantha (Hu) Tuyama
- Examination of the apelin signaling pathway in acetaminophen-induced hepatotoxicity in rats
- Integrating network pharmacology, in silico molecular docking and experimental validation to explain the anticancer, apoptotic, and anti-metastatic effects of cosmosiin natural product against human lung carcinoma
- Validation of Protein A chromatography: orthogonal method with size exclusion chromatography validation for mAb titer analysis
- The evaluation of the efficiency of Atellica UAS800 in detecting pathogens (rod, cocci) causing urinary tract infection
- Case Report
- Exploring inherited vitamin B responsive disorders in the Moroccan population: cutting-edge diagnosis via GC-MS profiling
- Letter to the Editor
- Letter to the Editor: “Gene mining, recombinant expression and enzymatic characterization of N-acetylglucosamine deacetylase”
Articles in the same Issue
- Frontmatter
- Review Article
- Unveiling the hidden clinical and economic impact of preanalytical errors
- Research Articles
- To explore the role of hsa_circ_0053004/hsa-miR-646/CBX2 in diabetic retinopathy based on bioinformatics analysis and experimental verification
- Study on the LINC00578/miR-495-3p/RNF8 axis regulating breast cancer progression
- Comparison of two different anti-mullerian hormone measurement methods and evaluation of anti-mullerian hormone in polycystic ovary syndrome
- The evaluation of the relationship between anti angiotensin type I antibodies in hypertensive patients undergoing kidney transplantation
- Evaluation of neopterin, oxidative stress, and immune system in silicosis
- Assessment of lipocalin-1, resistin, cathepsin-D, neurokinin A, agmatine, NGF, and BDNF serum levels in children with Autism Spectrum Disorder
- Regulatory nexus in inflammation, tissue repair and immune modulation in Crimean-Congo hemorrhagic fever: PTX3, FGF2 and TNFAIP6
- Pasteur effect in leukocyte energy metabolism of patients with mild, moderate, and severe COVID-19
- Thiol-disulfide homeostasis and ischemia-modified albumin in patients with sepsis
- Myotonic dystrophy type 1 and oxidative imbalance: evaluation of ischemia-modified albumin and oxidant stress
- Antioxidant and alpha-glucosidase inhibitory activities of flavonoids isolated from fermented leaves of Camellia chrysantha (Hu) Tuyama
- Examination of the apelin signaling pathway in acetaminophen-induced hepatotoxicity in rats
- Integrating network pharmacology, in silico molecular docking and experimental validation to explain the anticancer, apoptotic, and anti-metastatic effects of cosmosiin natural product against human lung carcinoma
- Validation of Protein A chromatography: orthogonal method with size exclusion chromatography validation for mAb titer analysis
- The evaluation of the efficiency of Atellica UAS800 in detecting pathogens (rod, cocci) causing urinary tract infection
- Case Report
- Exploring inherited vitamin B responsive disorders in the Moroccan population: cutting-edge diagnosis via GC-MS profiling
- Letter to the Editor
- Letter to the Editor: “Gene mining, recombinant expression and enzymatic characterization of N-acetylglucosamine deacetylase”


