Abstract
Statisticians show growing interest in estimating and analyzing heterogeneity in causal effects in observational studies. However, there usually exists a trade-off between accuracy and interpretability for developing a desirable estimator for treatment effects, especially in the case when there are a large number of features in estimation. To make efforts to address the issue, we propose a score-based framework for estimating the conditional average treatment effect (CATE) function in this article. The framework integrates two components: (i) leverage the joint use of propensity and prognostic scores in a matching algorithm to obtain a proxy of the heterogeneous treatment effects for each observation and (ii) utilize nonparametric regression trees to construct an estimator for the CATE function conditioning on the two scores. The method naturally stratifies treatment effects into subgroups over a 2d grid whose axis are the propensity and prognostic scores. We conduct benchmark experiments on multiple simulated data and demonstrate clear advantages of the proposed estimator over state-of-the-art methods. We also evaluate empirical performance in real-life settings, using two observational data from a clinical trial and a complex social survey, and interpret policy implications following the numerical results.
1 Introduction
The questions that motivate many scientific studies in disciplines such as economics, epidemiology, medicine, and political science are not associational but causal in nature. In the study of causal inference, many researchers are interested in inferring average treatment effects, which provide a good sense of whether treatment is likely to deliver more benefit than the control among a whole community. However, the same treatment may affect different individuals very differently. Therefore, a substantial amount of works focus on analyzing heterogeneity in treatment effects, of which the term refers to variation in the effects of treatment across individuals. This variation may provide theoretical insights, revealing how the effect of interventions depends on participants’ characteristics or how varying features of a treatment alters the effect of an intervention.
In this article, we follow the potential outcome framework for causal inference [1,2], where each unit is assigned into either the treatment or the control group. Each unit has an observed outcome variable with a set of covariates. In randomized experiments and observational studies, it is desirable to replicate a sample as closely as possible by obtaining subjects from the treatment and control groups with similar covariate distributions when estimating causal effects. However, it is almost impossible to match observations exactly the same in both treatment and control groups in observational studies. To address this problem, it is usually preferred to define prespecified subgroups under certain conditions and estimate the treatment effects varying among subgroups. Accordingly, the conditional average treatment effect (CATE [3]) function is designed to capture heterogeneity of a treatment effect across subpopulations. The function is often conditioned on some component(s) of the covariates or a single statistic, like propensity score [4] and prognostic score [5]. Propensity scores are the probabilities of receiving the treatment of interest; prognostic scores model the potential outcome under a control group assignment. To understand treatment effect heterogeneity in terms of propensity and prognostic scores, we assume that equal or similar treatment effects are observed along some intervals of the two scores.
We target at constructing an estimator for treatment effects that conditions on only two quantities, propensity and prognostic scores, and we assume a piecewise constant structure in treatment effects. We take a step further from score-based matching algorithms and propose a data-driven approach that integrates the joint use of propensity and prognostic scores in a matching algorithm and a partition over the entire population via a nonparametric regression tree. In the first step, we estimate propensity scores and prognostic scores for each observed unit in the data. Secondly, we perform a
The complementary nature of propensity and prognostic score methods supports that conditioning on both the propensity and prognostic scores has the potential to reduce bias and improve the precision of treatment effect estimates, and it is affirmed in the simulation studies by Leacy and Stuart [6] and Antonelli et al. [7]. We also demonstrate such advantage for our proposed estimator across almost all scenarios examined in the simulation experiments.
Besides high precision in estimation, our proposed estimator demonstrates its superiority over the state-of-arts methods with a few attractive properties as follows:
The estimator is computationally efficient. Propensity and prognostic scores can be easily estimated through simple regression techniques. Our matching algorithm based on the two scores largely reduces dimensionality compared to full matching on the complete covariates. Moreover, growing a single regression tree saves much time over other tree-based estimation methods, such as Bayesian additive regression tree models (BART) [8] and random forests [9, 10].
Many previous works in subgroup analysis, such as studies by Assmann et al. [11] and Abadie et al. [12], set stratification on the sample with a fixed number of subgroups before estimating treatment effects. These approaches require a pre-determination on the number of subgroups contained in the data, and they inevitably introduce arbitrariness into the causal inference. In comparison, our proposed method simultaneously identifies the underlying subgroups in observations through binary split according to propensity and prognostic scores and provides a consequential estimation of treatment effects on each subgroups.
Although random forests-based methods [9, 10] achieve great performance in minimizing bias in estimating treatment effects, these ensemble methods are often referred to as “black boxes.” It is hard to capture the underlying reason why the collective decision with the high number of operations involved is made in their estimation process, especially in the case when there are many features considered in the model. On the contrary, our proposed method provides a straightforward and low-dimensional summary of treatment effects simply based on two scores that are relatively easy to estimate. As a result, given the covariates of an observation, one can easily deduce the positiveness and magnitude of its treatment effect according to its probability of treatment receipt and potential outcome following the structure of the regression tree without carrying out feature selection process.
Instead of relying on a single feature or a subset of features, identifying subgroups with the joint distribution of all features will be of particular interest to policymakers seeking to target policies on those most likely to benefit. Therefore, many existing researches focus on subgroup stratification based on a single index that combines baseline characteristics. For instance, Abadie et al. [12] utilized potential outcomes without treatment (prognostic scores) for endogenous stratification, and Kent and Hayward [13] stratified experimental subjects based on their predicted probability of certain risks (propensity scores). Our method can be considered as a natural extension of the two previous works, which allows researchers and policy makers to merge a large number of baseline characteristics into simply two indices and identify the subgroup who are most in need of help in a population.
We review relevant literature on matching algorithms and estimation of heterogeneous treatment effects in Section 2. In Section 3, we provide the theoretical framework and preliminaries for the causal inference model. We propose our method for estimation and prediction in Section 4. Section 5 lists the results of numerical experiments on multiple simulated data sets and two real-world data sets, following with the comparison with state-of-the-art methods in existing literature and the discussion on policy implications under different realistic scenarios.
2 Relevant literature
Statistical analysis of causality can be dated back to Neyman [1]. Causal inference can be viewed as an identification problem [14], for which statisticians are dedicated to learn the true causality behind the data. In reality, however, we do not have enough information to determine the true value due to a limited number of observations for analysis. This problem is also summarized as a “missing data” problem [15], which stems from the fundamental problem of causal inference [16], that is, for each unit at most one of the potential outcomes is observed. Importantly, the causal effect identification problem, especially for estimating treatment effects, can only be resolved through assumptions. Several key theoretical frameworks have been proposed over the past decades. The potential outcome framework by Rubin [2], often referred to as the Rubin causal model [16], is a common model of causality in statistics at the moment. Dawid [17] developed a decision theoretic approach to causality that rejects counterfactuals. Pearl [18,19] advocated for a model of causality based on non parametric structural equations and path diagrams.
2.1 Matching
To tackle the “missing data” problem when estimating treatment effects in randomized experiments in practice, matching serves as a very powerful tool. The main goal of matching is to find matched groups with similar or balanced observed covariate distributions [20]. The exact
2.2 Subclassification
To understand heterogeneity of treatment effects in the data, subclassification, first used by Cochran [33], is another important research problem. The key idea is to form subgroups over the entire population based on characteristics that are either immutable or observed before randomization. Rosenbaum and Rubin [4,34], and Lunceford and Davidian [35] examined how creating a fixed number of subclasses according to propensity scores removes the bias in the estimated treatment effects, and Yang et al. [36] developed a similar methodology in settings with more than two treatment levels. Full matching [37–39] is a more sophisticated form of subclassification that selects the number of subclasses automatically by creating a series of matched sets. Schou and Marschner [40] presented three measures derived using the theory of order statistics to claim heterogeneity of treatment effect across subgroups. Su et al. [41] pioneered in exploiting standard regression tree methods [42] in subgroup treatment effect analysis. Further, Athey and Imbens [43] derived a recursive partition of the population according to treatment effect heterogeneity. Hill [44] was the first work to advocate for the use of BART [45] for estimating heterogeneous treatment effects, followed by a significant number of research papers focusing on the seminal methodology, including Green and Kern [46], Hill and Su [47], and Hahn et al. [8]. Abadie et al. [12] introduced endogenous stratification to estimate subgroup effects for a fixed number of subgroups based on certain quantiles of the prognostic score. More recently, Padilla et al. [48] combined the fused lasso estimator with score matching methods to lead to a data-adaptive subgroup effects estimator.
2.3 Machine learning for causal inference
For the goal of analyzing treatment effect heterogeneity, supervised machine learning methods play an important role. One of the more common ways for accurate estimation with experimental and observational data is to apply regression [49] or tree-based methods [50]. From a Bayesian perspective, Heckman et al. [51] provided a principled way of adding priors to regression models, and Taddy et al. [52] developed Bayesian nonparametric approaches for both linear regression and tree models. The recent breakthrough work by Wager and Athey [9] proposed the causal forest estimator arising from random forests from Breiman [53]. More recently, Athey et al. [10] took a step forward and enhanced the previous estimator based on generalized random forests. Imai and Ratkovic [54] adapted an estimator from the support vector machine classifier with hinge loss [55]. Bloniarz et al. [56] studied treatment effect estimators with lasso regularization [57] when the number of covariates is large, and Koch et al. [58] applied group lasso for simultaneous covariate selection and robust estimation of causal effects. In the meantime, a series of papers including Qian and Murphy [59], Kü nzel et al. [60], and Syrgkanis et al. [61] focused on developing meta-learners for heterogeneous treatment effects that can take advantage of various machine learning algorithms and data structures.
2.4 Applied work
On the application side, the estimation of heterogeneous treatment effects is particularly an intriguing topic in causal inference with broad applications in scientific research. Gaines and Kuklinski [62] estimated heterogeneous treatment effects in randomized experiments in the context of political science. Dehejia and Wahba [63] explored the use of propensity score matching for nonexperimental causal studies with application in economics. Dahabreh et al. [64] investigated heterogeneous treatment effects to provide the evidence base for precision medicine and patient-centred care. Zhang et al. [65] proposed the survival causal tree method to discover patient subgroups with heterogeneous treatment effects from censored observational data. Rekkas et al. [66] examined three classes of approaches to identify heterogeneity of treatment effect within a randomized clinical trial, and Tanniou et al. [67] rendered a subgroup treatment estimate for drug trials.
3 Preliminaries
Before we introduce our method, we need to provide some mathematical background for treatment effect estimation. We follow Rubin’s framework on causal inference [2] and assume a superpopulation or distribution
One important and commonly used measure of causality in a binary treatment model is the average treatment effect (ATE; [30]), that is, the mean outcome difference between the treatment and control groups. Formally, we write the ATE as
With the
Then, an unbiased estimate of the ATE is the sample average treatment effect
However, we cannot observe
To analyze heterogeneous treatment effects, it is natural to divide the data into subgroups (e.g., by gender, or by race) and investigate if the average treatment effects are different across subgroups. Therefore, instead of estimating the ATE or the ITE directly, statisticians seek to estimate the conditional average treatment effect (CATE), defined by
The CATE can be viewed as an ATE in a subpopulation defined by
We also recall the propensity score [4], denoted by
Thus,
and in this article, we use the conventional definition as the predicted outcome under the control condition [31]:
We are interested in constructing a 2d summary of treatment effects based on propensity and prognostic scores. Instead of conditioning on the entire covariates or a subset of it in the CATE function, we express our estimand, named as scored-based subgroup CATE, by conditioning on the two scores:
For interpretability, we assume that treatment effects are piecewise constant over a 2d grid of propensity and prognostic scores. Specifically, there exists a partition of intervals
where
Moreover, our estimation of treatment effects relies on the following assumptions:
Assumption 1
Throughout the article, we maintain the stable unit treatment value assumption (SUTVA [49]), which consists of two components: no interference and no hidden variations of treatment. Mathematically, for unit
Thus, the SUTVA requires that the potential outcomes of one unit should be unaffected by the particular assignment of treatments to the other units. Furthermore, for each unit, there are no different forms or versions of each treatment level, which lead to different potential outcomes.
Assumption 2
The assumption of probabilistic assignment holds. This requires the assignment mechanism to imply a nonzero probability for each treatment value, for every unit. For the given covariates
almost surely.
This condition, regarding the joint distribution of treatments and covariates, is also known as overlap in some literature (see Assumption 2.2 in the studies by Imbens [30] and D’Amour et al. [68]), and it is necessary for estimating treatment effects everywhere in the defined covariate space. Note that
Assumption 3
We make the assumption that
holds.
This assumption is an implication of the usual unconfoundedness assumption:
Combining Assumption 2 and that in Equation (3), the conditions are typically referred as strong ignorability defined in Rosenbaum and Rubin [4]. Strong ignorability states which outcomes are observed or missing is independent of the missing data conditional on the observed data. It allows statisticians to address the challenge that the “ground truth” for the causal effect is not observed for any individual unit. We rewrite the conventional assumption by replacing the vector of covariates
Provided that Assumptions 1–3 hold, it follows that
and thus our estimand (2) is equivalent to
Thus, in this article, we focus on estimating (4), which is equivalent to (2) if the aforementioned assumptions hold, but might be different if Assumption 3 is violated.
4 Methodology
We now formally introduce our proposal of a three-step method for estimating heterogeneous treatment effects and the estimation rule for a given new observation. We assume a sample of size
4.1 Step 1
We first estimate propensity and prognostic scores for all observations in the sample. For propensity score, we apply a logistic regression on the entire covariate
where
For prognostic score, we restrict to the controlled group and regress the outcome variable
and we estimate prognostic scores as
4.2 Step 2
Next, we perform a nearest-neighbour matching based on the two estimated scores from the previous step. We adapt the notation from Abadie and Imbens [28], and use the Mahalanobis distance norm as the distance metric in the matching algorithm. Mahalanobis distance matching, first proposed by Hansen [69], has been widely used in matching algorithms for causal inference [6,31]. It has been found to perform well when the dimension of covariates are relatively low [20], and thus, it is more suitable than using standard Euclidean distance in our case when we have only propensity scores and prognostic scores as covariates.
Formally, for the units
where
Let
where
We then compute
Intuitively, the construction of
4.3 Step 3
The last step involves denoising of the point estimates of the individual treatment effects
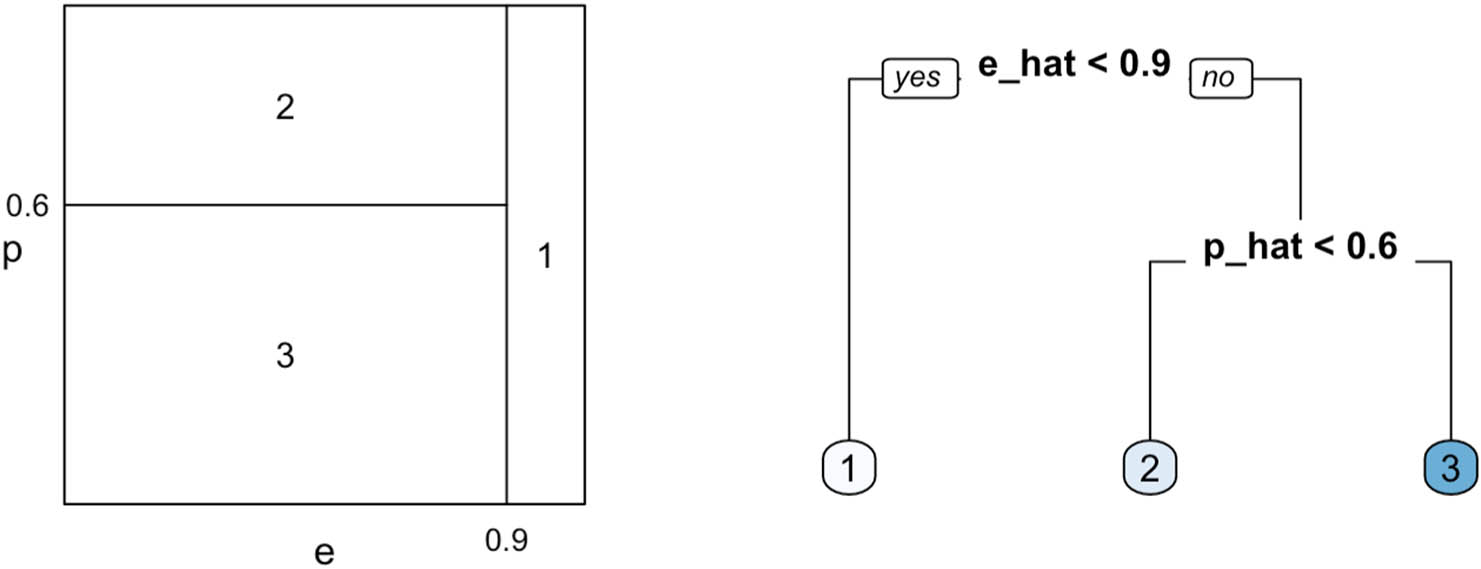
Left: a hypothetical partition over the 2d space of propensity and prognostic scores with the true values of piecewise constant treatment effects; Right: a sample regression tree
To perform such stratification, we grow a regression tree on
The final tree
4.4 Estimation on a new unit
After we obtain the regression tree model
We first compute the estimated propensity and prognostic scores for the new observation by
where
4.5 High-dimensional estimator
In a high-dimensional setting where the covariate dimension
The selection of the tuning parameters
We extend the aforementioned estimator with our proposal of applying a regression tree on the estimated propensity and prognostic scores. The procedure for estimation of subgroup heterogeneous treatment effects and estimation on a new unit remains the same as Step 3 for low-dimensional setups.
Remark
The choice of the number of nearest neighbours is a challenging problem. Updating distance metrics for every observation is computationally expensive, and choosing a value that is too small leads to a higher influence of noise on estimation. With regards to the application of nearest neighbour matching in causal inference, Abadie and Imbens [28] derived large sample properties of matching estimators of average treatment effects with a fixed number of nearest neighbours, but the authors did not provide any details on how to select the exact number of neighbours. Conventional settings on the number of nearest neighbours in current literature is to set
In Appendix A, we conduct a simulation study following one of the generative models from Section 5 to show how sensitive estimation accuracy is to the number of nearest neighbours selected and setting
4.6 Computational complexity
Our method is composed of three steps as introduced before. We first need to implement a logistic regression for estimating propensity score for a sample with size
The overall computational complexity of our method depends on the comparison between the order of
5 Experiments
In this section, we will examine the performance of our proposed estimator (PP) in a variety of simulated and real data sets. The baseline estimators we compete against are leave-one-out endogenous stratification (ST; [12]), causal forest (CF [9]), single-score matching including PSM, and prognostic-score matching. Note that in the original research by Abadie et al. [12], the authors restricted their attention to randomized experiments, because this is the setting where endogenous stratification is typically used. However, they mentioned the possibility of applying the method on observational studies. We take this into consideration and make their method as one of our competitors.
We implement our methods in R, using the packages “MatchIt” for
We evaluate the performance of each method according to two aspects, accuracy and uncertainty quantification. The results for single-score matching algorithms are not reported in this article because of very poor performance throughout all scenarios.
5.1 Simulated data
We first examine on the following simulated data sets under six different data generation mechanisms. We obtain insights from the simulation study by Leacy and Stuart [6] for the models considered in Scenarios 1–3. The propensity score and outcome (prognosis) models in Scenarios 1 and 3 are characterized by additivity and linearity (main effects only), but with different piecewise constant structures in the true treatment effects over a 2d grid of the two scores. We add nonadditivity and nonlinear terms to both propensity and prognosis models in Scenarios 2. In other words, both propensity and prognostic scores are expected to be misspecified in these two models if we apply generalized linear models directly in estimation. Scenario 4 comes from Abadie et al. [12], with a constant treatment effect over all observations. Scenario 5 is modified from the study by Wager and Athey [9] (see Equation 27 there), in which the propensity model follows a continuous distribution instead of a linear structure. In addition, the true treatment effect is not a function of propensity and prognostic scores. In Scenario 6, we break the mold assumption of nicely delineated subgroups in treatment effects over 2d space of the two scores, and use a smooth function instead. A high-dimensional setting (
We first introduce some notations used in the experiments: the sample size
Throughout all the models we consider, we maintain the unconfoundedness assumption discussed in Section 3, generate the covariate
We evaluate the accuracy of an estimator
We record the averaged mean squared error (MSE) over 1,000 Monte Carlo trials for each scenario. For each Monte Carlo trial, we use train-test split to evaluate model performance. In detail, we first obtain
Scenario 1. With
where
Scenario 2. We now add some interaction and nonlinear terms to the propensity and prognostic models in Scenario 1, while keeping the setups of the covariate
Again for each simulation,
Scenario 3. In this case, we define the true treatment effect with a more complicated piecewise constant structure over the 2d grid, under the same model used in Scenario 1, with
Scenario 4. Setting
where
Scenario 5. The data satisfy
where
Scenario 6. For this scenario, we examine the performance when the treatment effect takes on a smooth interval of propensity and prognostic scores. We use the same model in Scenario 1 for generating the two scores with
Scenario 7. In the last case, we study the performance of different estimators on a high-dimensional data. The data model follows
We select
The boxplots that depict the distribution of MSEs obtained under all scenarios are presented in Figure 2. We can see that for Scenario 1, our proposed estimator achieves better accuracy when the sample size

Comparison of mean squared errors over 1,000 Monte Carlo simulations under different generative models for our proposed method (PP), endogenous stratification (ST), and causal forest (CF).
We now take a careful look at the visualization comparison between the true treatment effect and the predictions obtained from our method for Scenarios 1, 2, 3, 6, and 7. We confine both the true signal and the predictive model in a 2d grid scaled on the true propensity and prognostic scores, as shown in Figure 3. It is not surprising that our proposed estimators provide a descent recovery of the piecewise constant partition in the true treatment effects over the 2d grid as in Scenarios 1, 2, 3, and 7, with only a small difference in the magnitude of treatment effects. In the setting when treatment effects are smoothly distributed across the grid of the two scores as in Scenario 6, our estimation does not exactly capture the soft boundary of the shape, but still provides us with a simplified delineation that help us understand general heterogeneity in treatment effects over the 2d space.

One instance comparison between the true treatment effects and the estimates from the score-based method for Scenarios 1, 2, 3, 6, and 7 (from the top to the bottom). Plots on the left column depict the true treatment effect on the 2d grid of propensity and prognostic scores, and the right plots demonstrate the estimated treatment effects from our proposed method over the same grid.
With regard to uncertainty quantification, we examine coverage rates with a target confidence level of 0.95 for each method under different scenarios, and the corresponding results, along with the within iteration standard error estimates, are recorded in Table 1. It is quite clear that our proposed method achieves nominal coverage over the other two methods in almost all scenarios. It is worth to notice that although our estimator achieves a better coverage rate than the benchmarks, it suffers from large variation due to the nature of nearest-neighbour matching, especially in the case with small sample size. In general, our method that incorporates both propensity and prognostic scores in identifying subgroups and estimating treatment effects show its advantage in reducing bias, but the resulting large variance is worrisome and it remains space for future study to solve this issue.
Reported coverage rates with a target confidence level of 0.95 and within iteration standard error estimates (in parentheses)
| Scenario |
|
|
PP | ST | CF |
|---|---|---|---|---|---|
| 1 | 1,000 | 2 | 0.996 (0.177) | 0.748 (0.158) | 0.969 (0.128) |
| 5,000 | 10 | 0.987 (0.116) | 0.731 (0.062) | 0.736 (0.061) | |
| 2 | 1,000 | 10 | 0.994 (0.230) | 0.753 (0.152) | 0.797 (0.157) |
| 5,000 | 10 | 0.956 (0.141) | 0.619 (0.064) | 0.829 (0.224) | |
| 3 | 1,000 | 10 | 0.989 (0.382) | 0.420 (0.163) | 0.769 (0.297) |
| 5,000 | 10 | 0.949 (0.164) | 0.307 (0.075) | 0.750 (0.252) | |
| 4 | 1,000 | 10 | 1.000 (28.621) | 0.933 (16.578) | 0.993 (18.067) |
| 5,000 | 10 | 1.000 (6.693) | 0.998(6.049) | 0.973 (5.957) | |
| 5 | 1,000 | 10 | 1.000 (0.524) | 0.950 (0.353) | 0.978 (0.429) |
| 5,000 | 10 | 0.845 (0.336) | 0.622 (0.287) | 0.716 (0.321) | |
| 6 | 1,000 | 10 | 0.995 (0.472) | 0.495 (0.161) | 0.865 (0.474) |
| 5,000 | 10 | 0.999 (0.179) | 0.578 (0.089) | 0.821 (0.259) |
We also compare the computational cost of our proposed scored-based method and causal forest, with simulated data from Scenario 1. Endogenous stratification is not considered in this comparison because the method exerts an arbitrary subclassification into a predetermined number of subgroups, and it does not involve a cross-validation process. We record the averaged time consumed to obtain the estimate over 1,000 simulations for both methods. Figure 4 demonstrates that our proposed method (PP) is comparably time-efficient over its competitor, and CF can be very expensive in operational time for large-size problems.

A plot of time per simulation in seconds for our proposed method (PP) and CF against problem size
These results highlight the promise of our method for accurate estimation of subgroups in heterogeneous treatment effects, all while emphasizing avenues for further work. An immediate challenge is to control the bias in estimation of propensity and prognostic scores. Using more powerful models instead of simple linear regression in estimation is a good way to reduce bias by enabling the trees to focus more closely on the coordinates with the greatest signal. The study of splitting rules for trees designed to estimate causal effects is still in its infancy and improvements may be possible.
5.2 Real data analysis
To illustrate the behaviour of our estimator, we apply our method on the two real-world data sets, one from a clinical study and the other from a complex social survey. Propensity score-based methods are frequently used for confounding adjustment in observational studies, where baseline characteristics can affect the outcome of policy interventions. Therefore, the results from our method are expected to provide meaningful implications for these real data sets. However, one potential limitation of our proposed tree-based approach is that it imposes arbitrary dichotomization with sharp thresholds and thus may obscure potential continuous gradient over the space. For real data analysis in general, our method provides a reasonable simplification that is easy to interpret for the audience and gives some framework for thinking about when such a simplification would be appropriate.
5.2.1 Right heart catheterization (RHC) analysis
While randomized control trials are widely encouraged as the ideal methodology for causal inference in clinical and medical research, the lack of randomized data due to high costs and potential high risks leads to the studies based on observational data. In this section, we are interested in examining the association between the use of RHC during the first 24 h of care in the intensive care unit (ICU) and the short-term survival conditions of the patients. RHC is a procedure for directly measuring how well the heart is pumping blood to the lungs. RHC is often applied to critically ill patients for directing immediate and subsequent treatment. However, RHC imposes a small risk of causing serious complications when administering the procedure. Therefore, the use of RHC is controversial among practitioners, and scientists want to statistically validate the causal effects of RHC treatments. The causal study using observational data can be dated back to Connors et al. [80], where the authors implemented propensity score matching and concluded that RHC treatment lead to lower survival than not performing the treatment. Later, Hirano and Imbens [81] proposed a more efficient propensity-score-based method, and the recent study by Loh and Vansteelandt [82] using a modified propensity score model suggested that RHC significantly affected mortality rate in a short-term period.
A dataset for analysis was first used in the study by Connors et al. [80], and it is suitable for the purpose of applying our method because of its extremely well-balanced distribution of confounders across levels of the treatment [83]. The treatment variable
The result of the prediction model from our proposed method is reported in Figure 5. We observe that the sign of estimated treatment effects varies depending on the value of the propensity score and prognostic score. This particular pattern implies that RHC procedures indeed offer both benefits and risks in affecting patients’ short-term survival conditions. Specifically, we are interested in the occurrence of large positive treatment effects (increase in chance of death) from the estimation. An estimated treatment effect of 0.17 (with a 95% confidence interval between

Prediction model for RHC data. Left: a tree diagram of predictions on each split based on the two scores, with 95% confidence intervals provided on the bottom. Right: a 2d plot of prediction stratification on the intervals of propensity and prognostic scores, with a colour scale on magnitude of treatment effects.
5.2.2 National medical expenditure survey (NMES)
For the next experiment, we analyze a complex social survey data. In many complex surveys, data are not usually well balanced due to potential biased sampling procedure. To incorporate score-based methods with complex survey data requires an appropriate estimation on propensity and prognostic scores. DuGoff et al. [84] suggested that combining a propensity score method and survey weighting is necessary to achieve unbiased treatment effect estimates that are generalizable to the original survey target population. Austin et al. [85] conducted numerical experiments and showed that greater balance in measured baseline covariates and decreased bias is observed when natural retained weights are used in propensity score matching. Therefore, we include sampling weight as an baseline covariate when estimating propensity and prognostic scores in our analysis.
In this study, we aim to answer the research question: how smoking habit affects medical expenditure over lifetime, and we use the same data set as given in the study by Johnson et al. [86], which is originally extracted from the 1987 NMES. The NMES included detailed information about frequency and duration of smoking with a large nationally representative data base of nearly 30,000 adults, and that 1987 medical costs are verified by multiple interviews and additional data from clinicians and hospitals. A large amount of literature focus on applying various statistical methods to analyze the causal effects of smoking on medical expenditures using the NMES data. In the original study by Johnson et al. [86], the authors first estimated the effects of smoking on certain diseases and then examined how much those diseases increased medical costs. In contrast, Rubin [87], Imai and van Dyk [88], and Zhao et al. [89] proposed to directly estimate the effects of smoking on medical expenditures using propensity-score-based matching and subclassification. Hahn et al. [8] applied Bayesian regression tree models to assess heterogeneous treatment effects.
For our analysis, we explore the effects of extensive exposure to cigarettes on medical expenditures, and we use pack-year as a measurement of cigarette measurement, the same as given in the studies by Imai and van Dyk [88] and Hahn et al. [8]. Pack-year is a clinical quantification of cigarette smoking used to measure a person’s exposure to tobacco, defined by
Following that, we determine the treatment indicator
The subject-level covariates
The prediction model obtained from our method, as shown in Figure 6, is simple and easy to interpret. We derive a positive treatment effect across the entire sample, and the effect becomes significant when the predicted potential outcome is relatively low (less than 5.7). These results indicate that more reliance on smoking will deteriorate one’s health condition, especially for those who currently do not have a large amount of medical expenditure. Moreover, we observe a significant positive treatment effect of 1.4 (with a 95% confidence interval between 0.56 and 2.79), and in other words, a certain and substantial increase in medical expenditure for the subgroup with propensity score less than 0.17. It is intuitive to assume that a smaller possibility of engaging in excessive tobacco exposure is associated with healthier living styles. This phenomenon is another evidence that individuals who are more likely to stay healthy may suffer more from excessive exposure to tobacco products. In all, these results support policymakers and social activists who advocate for nationwide smoking ban.

Prediction model for NMES data. Left: a tree diagram of predictions on each split based on the two scores, with 95% confidence intervals provided on the bottom. Right: a 2d plot of prediction stratification on the intervals of propensity and prognostic scores, with a colour scale on magnitude of treatment effects.
6 Discussion
Our method is different from existing methods on estimating heterogeneous treatment effects in a way that we incorporate both matching algorithms and nonparametric regression trees in estimation, and the final estimate can be regarded as a 2d summary on treatment effects. Moreover, our method exercises a simultaneous stratification across the entire population into subgroups with the same treatment effects. Subgroup treatment effect analysis is an important but challenging research topic in observational studies, and our method can be served as an efficient tool to reach a reasonable partition.
Our numerical experiments on various simulated and real-life data lay out empirical evidence of the superiority of our estimator over state-of-the-art methods in accuracy and our proposal provides insightful interpretation on heterogeneity in treatment effects across different subgroups. We also discovered that our method is powerful in investigating subpopulations with significant treatment effects. Identifying representative subpopulations that receive extreme results after treatment is a paramount task in many practical contexts. Through empirical experiments on two real-world datasets from observational studies, our method demonstrates its ability in identifying these significant effects.
Although our method shows its outstanding performance in estimating treatments effects under the piecewise constant structure assumption, it remains meaningful and requires further study to develop more accurate recovery of such structure. For example, a potential shortcoming of using conventional regression trees for subclassification is that the binary partition over the true signals is not necessarily unique. Using some variants of CART, like optimal trees [90] and dyadic regression trees [91], would be more appropriate for estimation under additional assumptions. Another particular concern with tree-based methods is the curse of dimensionality. Applying other nonparametric regression techniques, such as
Moreover, we acknowledge that fitting propensity score and prognostic scores normally would demand more than main terms linear and logistic regressions as used in the current method to have hope of eliminating bias in estimating the two scores. The key takeaway of our proposal is not to estimate the two scores more accurately, but to provide a lower-dimensional summary on heterogeneous treatment effects. If more foreknowledge on the structure of the underlying functions of the two scores can be incorporated, we can certainly select a more ideal algorithm or machine learning model for estimation. To compromise the lack of such information under a usual scenario, a SuperLearner ensemble estimator, proposed by van der Laan et al. [93], may offer a more promising solution for estimating the two scores by taking a weighted average of multiple prediction models. Yet, an important challenge for future work is to design rules that can automatically choose the best model to fit the data.
Another question to consider is the source of heterogeneity in causal effects. Our proposal that utilizes two scores can be seen as a reasonable starting point to analyze heterogeneity from a low-dimensional perspective, but it cannot comprehensively capture the sources of such effects. For instance, it is seemingly possible to exist a covariate that leads to substantial heterogeneous treatment effects on the response but does not itself have a large effect on propensity or prognostic scores. There is no particular guiding theories about this issue in current literature, and thus, it remains huge space for further investigation.
Acknowledgments
The authors would like to thank three anonymous referees and the Editor for their constructive comments that improved the quality of this article.
-
Funding information: Steven Siwei Ye was partially funded with the UCLA 2022–23 Faculty Career Development Award awarded to Oscar Madrid Padilla.
-
Author contributions: Steven Siwei Ye worked on all the different parts of the project, Oscar Madrid Padilla, and Yanzhen Chen worked in the methodology and applications.
-
Conflict of interest: The authors state no conflict of interest.
-
Data availability statement: The data and R codes for implementing the method introduced in the article are publicly available on one of the author’s Github page (https://github.com/stevenysw/causal_pp).
Appendix A Study on the choice of the number of nearest neighbours
In this section, we examine how the number of nearest neighbours in the matching algorithm affects the estimation accuracy. Recall in Step 2 of our proposed method, we implement a
We follow the same generative model in Scenario 1 from Section 5 and compute the averaged mean squared error over 1,000 Monte Carlo simulations for

The plot of averaged MSE against the number of nearest neighbours.
Appendix B A summary table of simulation studies
In this appendix, we include a table to provide a text summary of all simulation models considered in Section 5.1. We add some notes to help the audience understand similarities, differences, and sources of these data generative models.
| Scenario | Propensity model | Prognostic model | Treatment effect model | Note |
|---|---|---|---|---|
| 1 | Linearity in covariates | Linearity in covariates | A sharp delineation over the 2d grid of propensity and prognostic scores with two subgroups | |
| 2 | Linear model with additional interactions and nonlinear terms | Linear model with additional interactions and nonlinear terms | A sharp delineation over the 2d grid of propensity and prognostic scoreswith two subgroups | The same treatment effect model as in Scenario 1; both propensity and prognostic scores are misspecified with linear model |
| 3 | Linearity in covariates | Linearity in covariates | A sharp delineation over the 2d grid of propensity and prognostic scores with four subgroups | The same propensity and prognostic models as in Scenario 1; more subgroups in treatment effects |
| 4 | Random with a fixed number of treated units | Linearity in covariates with an intercept | Constant zero | The same setting from Abadie et al. [12] |
| 5 | A smooth function of single covariate | Proportional to single covariate | A function of the covariates | Modified from Equation 27 of Wager and Athey [9] |
| 6 | Linearity in covariates | Linearity in covariates | A smooth function of the two scores | Modified from Equation 28 of Wager and Athey [9] |
| 7 | Linearity in a few covariates with sparsity in others | Linearity in a few covariates with sparsity in others | A sharp delineation over the 2d grid of propensity and prognostic scores with two subgroups | A high-dimensional setting corresponding to Scenario 1 |
Appendix C Non-parametric bootstrap in simulation studies
In Section 5, we use nonparametric bootstrap to construct confidence intervals for endogenous stratification and our proposed method. We use these bootstrap samples to compute coverage rates with respect to a target level of 95% as a measurement of uncertainty. The bootstrap method, introduced by [94], is a simple but powerful tool to obtain a robust nonparametric estimate of the confidence intervals through sampling from the empirical distribution function of the observed data. It is worth noting that causal forests or generalized random forests can compute confidence intervals quickly without requiring a bootstrap approach on which our method relies.
In this appendix, we introduce the details on how we implement nonparametric bootstrap for the purpose of computing coverage rates in the simulation experiments. For each scenario in Section 5, we start with generating a sample following the defined data generation model with a sample size
References
[1] Neyman J. On the application of probability theory to agricultural experiments. Ann Agricult Sci. 1923;10:1–51. Search in Google Scholar
[2] Rubin DB. Estimating causal effects of treatments in randomized and nonrandomized studies. J Educ Psychol. 1974;66(5):688–701. 10.1037/h0037350Search in Google Scholar
[3] Hahn J. On the role of the propensity score in efficient semiparametric estimation of average treatment effects. Econometrica. 1998;66(2):315–31. 10.2307/2998560Search in Google Scholar
[4] Rosenbaum PR, Rubin DB. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70(1):41–55. 10.1093/biomet/70.1.41Search in Google Scholar
[5] Hansen BB. The prognostic analogue of the propensity score. Biometrika. 2008;95(2):481–8. 10.1093/biomet/asn004Search in Google Scholar
[6] Leacy FP, Stuart EA. On the joint use of propensity and prognostic scores in estimation of the average treatment effect on the treated: a simulation study. Stat Med. 2014;33(20):3488–508. 10.1002/sim.6030Search in Google Scholar PubMed PubMed Central
[7] Antonelli J, Cefalu M, Palmer N, Agniel D. Doubly robust matching estimators for high dimensional confounding adjustment. Biometrics. 2018;74(4):1171–9. 10.1111/biom.12887Search in Google Scholar PubMed PubMed Central
[8] Hahn PR, Murray JS, Carvalho CM. Bayesian regression tree models for causal inference: regularization, confounding, and heterogeneous effects. Bayesian Anal. 2020;15(3):965–1056. 10.1214/19-BA1195Search in Google Scholar
[9] Wager S, Athey S. Estimation and inference of heterogeneous treatment effects using random forests. J Amer Stat Assoc. 2018;113(523):1228–42. 10.1080/01621459.2017.1319839Search in Google Scholar
[10] Athey S, Tibshirani J, Wager S. Generalized random forests. Ann Stat. 2019;47(2):1148–78. 10.1214/18-AOS1709Search in Google Scholar
[11] Assmann SF, Pocock SJ, Enos LE, Kasten LE. Subgroup analysis and other (mis)uses of baseline data in clinical trials. Lancet. 2000;355(9209):1064–9. 10.1016/S0140-6736(00)02039-0Search in Google Scholar PubMed
[12] Abadie A, Chingos MM, West MR. Endogenous stratification in randomized experiments. Rev Econ Stat. 2018;C(4):567–80. 10.1162/rest_a_00732Search in Google Scholar
[13] Kent DM, Hayward RA. Limitations of applying summary results of clinical trials to individual patients: the need for risk stratification. J Amer Med Assoc. 2007;298(10):1209–12. 10.1001/jama.298.10.1209Search in Google Scholar PubMed
[14] Keele L. The statistics of causal inference: a view from political methodology. Political Anal. 2015;23(3):313–35. 10.1093/pan/mpv007Search in Google Scholar
[15] Ding P, Li F. Causal inference: a missing data perspective. Stat Sci 2018;33(2):214–37. 10.1214/18-STS645Search in Google Scholar
[16] Holland PW. Statistics and causal Inference. J Amer Stat Assoc. 1986;81(396):945–60. 10.1080/01621459.1986.10478354Search in Google Scholar
[17] Dawid AP. Causal inference without counterfactuals. J Amer Stat Assoc. 2000;95(450):407–24. 10.1080/01621459.2000.10474210Search in Google Scholar
[18] Pearl J. Causal diagrams for empirical research. Biometrika 1995;82(4):669–710. 10.1093/biomet/82.4.702Search in Google Scholar
[19] Pearl J. Causality: models, reasoning, and inference. Cambridge, UK: Cambridge University Press; 2009. 10.1017/CBO9780511803161Search in Google Scholar
[20] Stuart EA. Matching methods for causal inference: a review and a look forward. Stat Sci Rev J Inst Math Stat. 2010;25(1):1–21. 10.1214/09-STS313Search in Google Scholar PubMed PubMed Central
[21] Smith H. Matching with multiple controls to estimate treatment effects in observational studies. Sociol Methodol. 1997;27(1):325–53. 10.1111/1467-9531.271030Search in Google Scholar
[22] Rubin DB, Thomas N. Combining propensity score matching with additional adjustments for prognostic covariates. J Amer Stat Assoc. 2000;95(450):573–85. 10.1080/01621459.2000.10474233Search in Google Scholar
[23] Ming K, Rosenbaum PR. A note on optimal matching with variable controls using the assignment algorithm. J Comput Graph Stat. 2001;10(3):455–63. 10.1198/106186001317114938Search in Google Scholar
[24] Rosenbaum PR. Optimal matching for observational studies. J Amer Stat Assoc.1989;84(408):1024–32. 10.1080/01621459.1989.10478868Search in Google Scholar
[25] Gu XS, Rosenbaum PR. Comparison of multivariate matching methods: structures, distances, and algorithms. J Comput Graph Stat. 1993;2(4):405–20. 10.1080/10618600.1993.10474623Search in Google Scholar
[26] Zubizarreta JR. Using mixed integer programming for matching in an observational study of kidney failure after surgery. J Amer Stat Assoc. 2012107(500):1360–71. 10.1080/01621459.2012.703874Search in Google Scholar
[27] Zubizarreta JR, Keele L. Optimal multilevel matching in clustered observational studies: a case study of the effectiveness of private schools under a large-scale voucher system. J Amer Stat Assoc. 2017;112(518):547–60. 10.1080/01621459.2016.1240683Search in Google Scholar
[28] Abadie A, Imbens GW. Large sample properties of matching estimators for average treatment effects. Econometrica. 2006;74(1):235–67. 10.1111/j.1468-0262.2006.00655.xSearch in Google Scholar
[29] Rubin DB, Thomas N. Matching using estimated propensity scores: relating theory to practice. Biometrics. 1996;52(1):249–64. 10.2307/2533160Search in Google Scholar
[30] Imbens GW. Nonparametric estimation of average treatment effects under exogeneity: a review. Rev Econ Stat. 2004;86:4–29. 10.1162/003465304323023651Search in Google Scholar
[31] Aikens RC, Greaves D, Baiocchi M. A pilot design for observational studies: using abundant data thoughtfully. Stat Med. 2020;39(30):4821–40. 10.1002/sim.8754Search in Google Scholar PubMed
[32] Aikens RC, Baiocchi M. Assignment-control plots: a visual companion for causal inference study design. Amer Stat. 2023;77(1):72–84. 10.1080/00031305.2022.2051605Search in Google Scholar PubMed PubMed Central
[33] Cochran WG. The effectiveness of adjustment by subclassification in removing bias in observational studies. Biometrics. 1968;24(2):295–313. 10.2307/2528036Search in Google Scholar
[34] Rosenbaum PR, Rubin DB. Constructing a control group using multivariate matched sampling methods that incorporate the propensity score. Amer Stat. 1985;39(1):33–38. 10.1080/00031305.1985.10479383Search in Google Scholar
[35] Lunceford JK, Davidian M. Stratification and weighting via the propensity score in estimation of causal treatment effects: a comparative study. Stat Med. 2004;23(19):2937–60. 10.1002/sim.1903Search in Google Scholar PubMed
[36] Yang S, Imbens GW, Cui Z, Faries DE, Kadziola Z. Propensity score matching and subclassification in observational studies with multi-level treatments. Biometrics. 2016;72(4):1055–65. 10.1111/biom.12505Search in Google Scholar PubMed
[37] Rosenbaum PR. A characterization of optimal designs for observational studies. J R Stat Soc Ser B (Stat Methodol). 1991;53(3):597–610. 10.1111/j.2517-6161.1991.tb01848.xSearch in Google Scholar
[38] Hansen BB. Full matching in an observational study of coaching for the SAT. J Amer Stat Assoc. 2004;99(467):609–18. 10.1198/016214504000000647Search in Google Scholar
[39] Stuart EA, Green KM. Using full matching to estimate causal effects in non-experimental studies: examining the relationship between adolescent marijuana use and adult outcomes. Development Psychol. 2008;44(2):395–406. 10.1037/0012-1649.44.2.395Search in Google Scholar PubMed PubMed Central
[40] Schou IM, Marschner IC. Methods for exploring treatment effect heterogeneity in subgroup analysis: an application to global clinical trials. Pharmaceut Stat. 2015;14(1):44–55. 10.1002/pst.1656Search in Google Scholar PubMed
[41] Su X, Tsai C, Wang H, Nickerson DM, Li B. Subgroup analysis via recursive partitioning. J Machine Learn Res. 2009;10:141–58. 10.2139/ssrn.1341380Search in Google Scholar
[42] Breiman L, Friedman J, Stone CJ, Olshen RA. Classification and regression trees. New York: Taylor & Francis; 1984. Search in Google Scholar
[43] Athey S, Imbens GW. Recursive partitioning for heterogeneous causal effects. Proc Nat Acad Sci. 2016;113(27):7353–60. 10.1073/pnas.1510489113Search in Google Scholar PubMed PubMed Central
[44] Hill JL. Bayesian nonparametric modeling for causal inference. J Comput Graph Stat. 2011;20(1):217–40. 10.1198/jcgs.2010.08162Search in Google Scholar
[45] Chipman HA, George EI, McCulloch RE. BART: Bayesian additive regression trees. Ann Appl Stat. 2010;4(1):266–98. 10.1214/09-AOAS285Search in Google Scholar
[46] Green DP, Kern HL. Modeling heterogeneous treatment effects in survey experiments with Bayesian additive regression trees. Public Opinion Quarter. 2012;76(3):491–511. 10.1093/poq/nfs036Search in Google Scholar
[47] Hill JL, Su Y. Assessing lack of common support in causal inference using Bayesian nonparametrics: implications for evaluating the effect of breastfeeding on childrenas cognitive outcomes. Ann Appl Stat. 2013;7(3):1386–420. 10.1214/13-AOAS630Search in Google Scholar
[48] Padilla OHM, Ding P, Chen Y, Ruiz G. A causal fused lasso for interpretable heterogeneous treatment effects estimation. 2021. arXiv: 2110.00901. Search in Google Scholar
[49] Imbens GW, Rubin DB. Causal inference for statistics, social, and biomedical sciences: an introduction. Cambridge, UK: Cambridge University Press; 2015. 10.1017/CBO9781139025751Search in Google Scholar
[50] Imai K, Strauss A. Estimation of heterogeneous treatment effects from randomized experiments, with application to the optimal planning of the get-out-the-vote campaign. Politic Anal. 2011;19:1–19. 10.1093/pan/mpq035Search in Google Scholar
[51] Heckman JJ, Lopes HF, Piatek R. Treatment effects: a Bayesian perspective. Econom Rev. 2014;33(1–4):36–67. 10.1080/07474938.2013.807103Search in Google Scholar PubMed PubMed Central
[52] Taddy M, Gardner M, Chen L, Draper D. A nonparametric Bayesian analysis of heterogenous treatment effects in digital experimentation. J Business Econom Stat. 2016;34(4):661–72. 10.1080/07350015.2016.1172013Search in Google Scholar
[53] Breiman L. Random forests. Machine Learning. 2001;45:5–32. 10.1023/A:1010933404324Search in Google Scholar
[54] Imai K, Ratkovic M. Estimating treatment effect heterogeneity in randomized program evaluation. Ann Appl Stat. 2013;7(1):443–70. 10.1214/12-AOAS593Search in Google Scholar
[55] Wahba G. Soft and hard classification by reproducing kernel Hilbert space methods. Proc Nat Acad Sci. 2002;99(26):16524–30. 10.1073/pnas.242574899Search in Google Scholar PubMed PubMed Central
[56] Bloniarz A, Liu H, Zhang C, Sekhon JS, Yu B. Lasso adjustments of treatment effect estimates in randomized experiments. Proc Nat Acad Sci. 2016;113(27):7383–90. 10.1073/pnas.1510506113Search in Google Scholar PubMed PubMed Central
[57] Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Ser B (Stat Meth) 1996;58(1):267–88. 10.1111/j.2517-6161.1996.tb02080.xSearch in Google Scholar
[58] Koch B, Vock DM, Wolfson J. Covariate selection with group lasso and doubly robust estimation of causal effects. Biometrics. 2018;74(1):8–17. 10.1111/biom.12736Search in Google Scholar PubMed PubMed Central
[59] Qian M, Murphy SA. Performance guarantees for individualized treatment rules. Ann Stat 2011;39(2):1180–210. 10.1214/10-AOS864Search in Google Scholar PubMed PubMed Central
[60] Kü nzel SR, Sekhon JS, Bickel PJ, Yu B. Metalearners for estimating heterogeneous treatment effects using machine learning. Proc Nat Acad Sci. 2019;116(10):4156–65. 10.1073/pnas.1804597116Search in Google Scholar PubMed PubMed Central
[61] Syrgkanis V, Lei V, Oprescu M, Hei M, Oprescu M, Battocchi K, et al. Machine learning estimation of heterogeneous treatment effects with instruments. Advances in Neural Information Processing Systems 32: Annual Conference on Neural Information Processing Systems 2019, NeurIPS 2019, December 8-14, 2019, Vancouver, BC, Canada. p. 15167–76. Search in Google Scholar
[62] Gaines B, Kuklinski J. Estimation of heterogeneous treatment effects related to self-selection. Amer J Politic Sci. 2011;55(3):724–36. 10.1111/j.1540-5907.2011.00518.xSearch in Google Scholar
[63] Dehejia RH, Wahba S. Propensity score matching methods for nonexperimental causal studies. Rev Econ Stat. 2002;84(1):151–61. 10.1162/003465302317331982Search in Google Scholar
[64] Dahabreh IJ, Hayward R, Kent DM. Using group data to treat individuals: understanding heterogeneous treatment effects in the age of precision medicine and patient-centred evidence. Int J Epidemiol. 2016;45(6):2184–93. 10.1093/ije/dyw125Search in Google Scholar PubMed PubMed Central
[65] Zhang W, Le TD, Liu L, Zhou Z, Li J. Mining heterogeneous causal effects for personalized cancer treatment. Bioinformatics. 2017;33(15):2372–8. 10.1093/bioinformatics/btx174Search in Google Scholar PubMed
[66] Rekkas A, Paulus JK, Raman G, Wong JB, Steyerberg EW, Rijnbeek PR, et al. Predictive approaches to heterogeneous treatment effects: a scoping review. BMC Med Res Methodol. 2020;20(1):264. 10.1186/s12874-020-01145-1Search in Google Scholar PubMed PubMed Central
[67] Tanniou J, van der Tweel I, Teerenstra S, Roes KCB. Estimates of subgroup treatment effects in overall nonsignificant trials: to what extent should we believe in them? Pharmaceut Stat. 2017;16(4):280–95. 10.1002/pst.1810Search in Google Scholar PubMed
[68] D’Amour A, Ding P, Feller A, Lei L, Sekhon J. Overlap in observational studies with high-dimensional covariates. J Econ. 2021;221:644–54. 10.1016/j.jeconom.2019.10.014Search in Google Scholar
[69] Hansen BB. Bias reduction in observational studies via prognosis scores. Statistics Department, University of Michigan; Ann Arbor, Michigan: Technical Report. 2006. 441. Search in Google Scholar
[70] Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference, and prediction. New York: Springer; 2001. 10.1007/978-0-387-21606-5Search in Google Scholar
[71] Austin PC, Schuster T. The performance of different propensity score methods for estimating absolute effects of treatments on survival outcomes: a simulation study. Stat Meth Med Res. 2016;25(5):2214–37. 10.1177/0962280213519716Search in Google Scholar PubMed PubMed Central
[72] Ming K, Rosenbaum PR. Substantial gains in bias reduction from matching with a variable number of controls. Biometrics. 2001;56(1):118–24. 10.1111/j.0006-341X.2000.00118.xSearch in Google Scholar
[73] Brito MR, Chávez EL, Quiroz AJ, Yukich JE. Connectivity of the mutual k-nearest-neighbour graph in clustering and outlier detection. Stat Probability Lett. 1997;35(1):33–42. 10.1016/S0167-7152(96)00213-1Search in Google Scholar
[74] Yuan G, Ho C, Lin C. An improved GLMNET for l1-regularized logistic regression. J Machine Learn Res. 2012;13:1999–2030. 10.1145/2020408.2020421Search in Google Scholar
[75] Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. 10.18637/jss.v033.i01Search in Google Scholar
[76] von Luxburg U. A tutorial on spectral clustering. Stat Comput. 2007;17(4):395–416. 10.1007/s11222-007-9033-zSearch in Google Scholar
[77] Athey S, Wager S. Estimating treatment effects with causal forests: an application. Observ Stud. 2019;19(5):37–51. 10.1353/obs.2019.0001Search in Google Scholar
[78] Nie X, Wager S. Quasi-oracle estimation of heterogeneous treatment effects. Biometrika. 2021;108(2):299–319. 10.1093/biomet/asaa076Search in Google Scholar
[79] Karoui NE, Purdom E. Can we trust the bootstrap in high-dimensions? The case of linear models. J Machine Learn Res. 2018;19(5):1–66. Search in Google Scholar
[80] Connors AF, Speroff T, Dawson NV, Thomas C, Harrell FE, Wagner D, et al. The effectiveness of right heart catheterization in the initial care of critically ill patients. J Amer Med Assoc. 1996;276(11):889–97. 10.1001/jama.276.11.889Search in Google Scholar PubMed
[81] Hirano K, Imbens GW. Estimation of causal effects using propensity Score weighting: an application to data on Right Heart Catheterization. Health Services Outcomes Res Methodol. 2001;2(3):259–78. 10.1023/A:1020371312283Search in Google Scholar
[82] Loh WW, Vansteelandt S. Confounder selection strategies targeting stable treatment effect estimators. Stat Med. 2021;40(3):607–30. 10.1002/sim.8792Search in Google Scholar PubMed
[83] Smith MJ, Mansournia MA, Maringe C, Zivich PN, Cole SR, Leyrat C, et al. Introduction to computational causal inference using reproducible Stata, R, and Python code: A tutorial. Stat Med. 2021;41(2):407–32. 10.1002/sim.9234Search in Google Scholar PubMed
[84] DuGoff EH, Schuler M, Stuart EA. Generalizing observational study results: applying propensity score methods to complex surveys. Health Service Res. 2014;49(1):284–303. 10.1111/1475-6773.12090Search in Google Scholar PubMed PubMed Central
[85] Austin PC, Jembere N, Chiu M. Propensity score matching and complex surveys. Stat Meth Med Res. 2018;27(4):1240–57. 10.1177/0962280216658920Search in Google Scholar PubMed PubMed Central
[86] Johnson E, Dominici F, Griswold M, Zeger SL. Disease cases and their medical costs attributable to smoking: an analysis of the national medical expenditure survey. J Econometr. 2003;112(1):135–51. 10.1016/S0304-4076(02)00157-4Search in Google Scholar
[87] Rubin DB. Using propensity scores to help design observational studies: application to the tobacco litigation. Health Services Outcomes Res Methodol. 2001;2:169–88. 10.1023/A:1020363010465Search in Google Scholar
[88] Imai K, van Dyk DA. Causal inference with general treatment regimes. J Amer Stat Assoc. 2004;99(467):854–66. 10.1198/016214504000001187Search in Google Scholar
[89] Zhao S, van Dyk DA, Imai K. Propensity score-based methods for causal inference in observational studies with non-binary treatments. Stat Meth Med Res 2020;29(3):709–27. 10.1177/0962280219888745Search in Google Scholar PubMed
[90] Bertsimas D, Dunn J. Optimal classification trees. Machine Learn. 2017;106(7):1039–82. 10.1007/s10994-017-5633-9Search in Google Scholar
[91] Donoho DL. CART and best-ortho-basis: a connection. Ann Stat. 1997;25(5):1870–911. 10.1214/aos/1069362377Search in Google Scholar
[92] Padilla OHM, Sharpnack J, Chen Y, Witten D. Adaptive non-parametric regression with the K-NN fused lasso. Biometrika. 2020;107(2):293–310. 10.1093/biomet/asz071Search in Google Scholar PubMed PubMed Central
[93] van der Laan MJ, Polley EC, Hubbard AE. Super learner. Stat Appl Genetic Mol Biol. 2007;6(1):1–23. 10.2202/1544-6115.1309Search in Google Scholar PubMed
[94] Efron B. Bootstrap methods: another look at the Jackknife. Ann Stat. 1979;7(1):1–26. 10.1214/aos/1176344552Search in Google Scholar
© 2023 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Research Articles
- Adaptive normalization for IPW estimation
- Matched design for marginal causal effect on restricted mean survival time in observational studies
- Robust inference for matching under rolling enrollment
- Attributable fraction and related measures: Conceptual relations in the counterfactual framework
- Causality and independence in perfectly adapted dynamical systems
- Sensitivity analysis for causal decomposition analysis: Assessing robustness toward omitted variable bias
- Instrumental variable regression via kernel maximum moment loss
- Randomization-based, Bayesian inference of causal effects
- On the pitfalls of Gaussian likelihood scoring for causal discovery
- Double machine learning and automated confounder selection: A cautionary tale
- Randomized graph cluster randomization
- Efficient and flexible mediation analysis with time-varying mediators, treatments, and confounders
- Minimally capturing heterogeneous complier effect of endogenous treatment for any outcome variable
- Quantitative probing: Validating causal models with quantitative domain knowledge
- On the dimensional indeterminacy of one-wave factor analysis under causal effects
- Heterogeneous interventional effects with multiple mediators: Semiparametric and nonparametric approaches
- Exploiting neighborhood interference with low-order interactions under unit randomized design
- Robust variance estimation and inference for causal effect estimation
- Bounding the probabilities of benefit and harm through sensitivity parameters and proxies
- Potential outcome and decision theoretic foundations for statistical causality
- 2D score-based estimation of heterogeneous treatment effects
- Identification of in-sample positivity violations using regression trees: The PoRT algorithm
- Model-based regression adjustment with model-free covariates for network interference
- All models are wrong, but which are useful? Comparing parametric and nonparametric estimation of causal effects in finite samples
- Confidence in causal inference under structure uncertainty in linear causal models with equal variances
- Special Issue on Integration of observational studies with randomized trials - Part II
- Personalized decision making – A conceptual introduction
- Precise unbiased estimation in randomized experiments using auxiliary observational data
- Conditional average treatment effect estimation with marginally constrained models
- Testing for treatment effect twice using internal and external controls in clinical trials
Articles in the same Issue
- Research Articles
- Adaptive normalization for IPW estimation
- Matched design for marginal causal effect on restricted mean survival time in observational studies
- Robust inference for matching under rolling enrollment
- Attributable fraction and related measures: Conceptual relations in the counterfactual framework
- Causality and independence in perfectly adapted dynamical systems
- Sensitivity analysis for causal decomposition analysis: Assessing robustness toward omitted variable bias
- Instrumental variable regression via kernel maximum moment loss
- Randomization-based, Bayesian inference of causal effects
- On the pitfalls of Gaussian likelihood scoring for causal discovery
- Double machine learning and automated confounder selection: A cautionary tale
- Randomized graph cluster randomization
- Efficient and flexible mediation analysis with time-varying mediators, treatments, and confounders
- Minimally capturing heterogeneous complier effect of endogenous treatment for any outcome variable
- Quantitative probing: Validating causal models with quantitative domain knowledge
- On the dimensional indeterminacy of one-wave factor analysis under causal effects
- Heterogeneous interventional effects with multiple mediators: Semiparametric and nonparametric approaches
- Exploiting neighborhood interference with low-order interactions under unit randomized design
- Robust variance estimation and inference for causal effect estimation
- Bounding the probabilities of benefit and harm through sensitivity parameters and proxies
- Potential outcome and decision theoretic foundations for statistical causality
- 2D score-based estimation of heterogeneous treatment effects
- Identification of in-sample positivity violations using regression trees: The PoRT algorithm
- Model-based regression adjustment with model-free covariates for network interference
- All models are wrong, but which are useful? Comparing parametric and nonparametric estimation of causal effects in finite samples
- Confidence in causal inference under structure uncertainty in linear causal models with equal variances
- Special Issue on Integration of observational studies with randomized trials - Part II
- Personalized decision making – A conceptual introduction
- Precise unbiased estimation in randomized experiments using auxiliary observational data
- Conditional average treatment effect estimation with marginally constrained models
- Testing for treatment effect twice using internal and external controls in clinical trials

