Loss of respiratory complex I subunit NDUFB10 affects complex I assembly and supercomplex formation
-
Tasnim Arroum
Abstract
The orchestrated activity of the mitochondrial respiratory or electron transport chain (ETC) and ATP synthase convert reduction power (NADH, FADH2) into ATP, the cell’s energy currency in a process named oxidative phosphorylation (OXPHOS). Three out of the four ETC complexes are found in supramolecular assemblies: complex I, III, and IV form the respiratory supercomplexes (SC). The plasticity model suggests that SC formation is a form of adaptation to changing conditions such as energy supply, redox state, and stress. Complex I, the NADH-dehydrogenase, is part of the largest supercomplex (CI + CIII2 + CIVn). Here, we demonstrate the role of NDUFB10, a subunit of the membrane arm of complex I, in complex I and supercomplex assembly on the one hand and bioenergetics function on the other. NDUFB10 knockout was correlated with a decrease of SCAF1, a supercomplex assembly factor, and a reduction of respiration and mitochondrial membrane potential. This likely is due to loss of proton pumping since the CI P P -module is downregulated and the P D -module is completely abolished in NDUFB10 knock outs.
1 Introduction
The mitochondrial respiratory chain (RC) is comprised of four complexes (C), complex I to complex IV (CI to CIV). CI is the NADH-ubiquinone-oxidoreductase, CII is the succinate-dehydrogenase, CIII is the ubiquinol-cytochrome c oxidoreductase and CIV is the cytochrome c oxidase. Three of the four complexes, CI, CIII and CIV, assemble into multimeric units, so-called supercomplexes (SC). Originally, the N-respirasome (SC I + III2 + IV) and the Q-respirasome (SC III2 + IV) have been discovered by the analysis of protein complexes separated by native gel electrophoresis (Schagger and Pfeiffer 2000). Several compositions have been found in different tissues, with different stoichiometries of CI, CIII and CIV, including SC without CI. Off note, different detergents yield distinct higher molecular weight band patterns in native gels, so the question remained for some time, whether the different SCs were in fact artifacts generated by the extraction method. However, the cryo-electron tomography (cryo-TEM) structure of respirasome’s provided structural evidence for SC assembly in situ (Gu et al. 2016; Letts et al. 2016, 2019; Vercellino and Sazanov 2021) (mammals), (Guo et al. 2017; Letts et al. 2019) (human), (Hartley et al. 2019, 2020; Rathore et al. 2019) (yeast), (Maldonado et al. 2021) (plants) (Cogliati et al. 2016; Guo et al. 2017; Melber and Winge 2016), supported by functional data (Acin-Perez et al. 2008; Lapuente-Brun et al. 2013). SC formation primarily provides structural stability for the contributing complexes. Several studies suggest that SC formation has a functional advantage, i.e., in reducing electron leak and enhancing the efficiency of the electron transport chain (ETC) (Acin-Perez and Enriquez 2014; Barrientos and Ugalde 2013; Blaza et al. 2014), or minimizing mitochondrial ROS production (Lopez-Fabuel et al. 2016), however, this is still under debate (Milenkovic et al. 2017). Likewise, SC could play a role in metabolic regulation (Guaras et al. 2016). Interestingly, the stoichiometry of respiratory complexes CI, CIII, and CIV is not equal and the mobile electron carriers are in excess (Schwerzmann et al. 1986). Thus, under which circumstances and to which extend, if at all, mitochondrial respirasome formation contributes to functional ETC optimization is still not well understood and needs further elucidation.
An indirect indication for the physiological relevance of SC in vivo is the existence of the SC assembly factor 1 (SCAF1, also known as COX7A2L). In particular, SCAF1 is required for SC III2 + IV assembly (Cogliati et al. 2016). It first binds to III2 and recruits CIV into the Q-respirasome (III2 + IV) (Lobo-Jarne et al. 2018; Perez-Perez et al. 2016). SCAF1 is relevant for SC formation and OXPHOS modulation in mouse (Cogliati et al. 2016), zebrafish (Garcia-Poyatos et al. 2020) and ovine (Letts et al. 2016). In B57BL/6 mice, a mutated SCAF1 variant (111 amino acids) lacking the ability to stabilize Q-respirasome, resulted in impaired exercise (Calvo et al. 2020). Other studies have suggested that SCAF1 is not necessary for N-respirasome formation (Calvo et al. 2020; Fernandez-Vizarra et al. 2021). This might be related to the fact that distinct N-respirasomes can be formed, either with SCAF1 or COX7A2. For instance, kidney SCs contained the isoform COX7A2 (Zong et al. 2018). The presence of SCAF1 seems to alter the structural conformation of the N-respirasome; two distinct respirasome species were reported (Calvo et al. 2020). SCAF1 respirasomes were found in the tight structure and led to increased NADH-dependent respiration and decreased ROS production. Furthermore, it was suggested that two different Coenzyme Q-pool exist that regulate NADH oxidation by complex I depending on complex I assembly status (free complex I or attached to III2). Thus the preferential expression of COX7A subunit isoforms, COX7A2 and SCAF1 (COX7A2L) leads to a different mitochondrial respiratory complex make-up: COX7A2 or SCAF1 (COX7A2L) based respirasomes. Interestingly, the two forms of N-respirasome were bioenergetically different and produced different leakage of reactive oxygen species (ROS) (Calvo et al. 2020). Moreover, in zebrafish, the absence of SCAF1 causes the loss of metabolic efficiency (Garcia-Poyatos et al. 2020). The presence of SCAF1 or COX7A2L in the N-respirasome could be detected by Blue native PAGE (BNGE) because of their differential migration: slower for I + III2 + IVSCAF1 with respect to I + III2 + IVCOX7A2. How SCAF1 specifically affects N-respirasome structural conformation in different tissues needs to be further investigated.
The role of complex I in supercomplex formation and activity is not yet satisfactorily clarified. As suggested in a recent study, the distal part of the membrane arm of complex I (PD-a module) may be sufficient to build a scaffold for the assembly of complexes III and IV (CIII2 + CIV) to form a respirasome subcomplex (Fang et al. 2021). The role of CIII is less understood, it was reported that the depletion of CIII prevented CI assembly by blocking the integration of the N module in 43B-TK osteosarcoma-derived cybrid cells supporting the interconnected assembly model (Protasoni et al. 2020), but in another report cells with complete loss of CIII did assemble a functional CI (Guaras et al. 2016). Further, the impeding of CI function and the diminishing of its interaction with CIV in NDUFAF4 deficient patients with fatal early encephalopathy caused the abnormal accumulation of CIV subunits together with CIII in SC III2 + IV (Protasoni et al. 2020). On the other hand, COX-deficient Caenorhabditis elegans showed impaired CI activity, suggesting a feedback loop between CIV function and CI function (Suthammarak et al. 2009).
Mature CI is comprised of a matrix arm and a membrane-embedded arm, both arranged to form a boot-like structure. The matrix arm consists of the N- and Q-modules and the membrane arm consists of the P-module. The P-module has a proximal (PP) and a distal (PD) part, further divided into PD-a, PD-b, PP-a and PP-b (Brandt 2006; Guerrero-Castillo et al. 2017) (Figure 1Aa). The N-part is the NADH-Dehydrogenase, the Q-module, the ubiquinone oxidoreductase, transfers electrons to ubiquinone. The P-arm comprises the proton pumps, whereby the exact proton translocation sites are still under debate (Kravchuk et al. 2022). Interestingly, all mitochondrially encoded subunits are found in the membrane arm of CI, i.e., the hydrophobic region. NDUFB10 is an accessory subunit in the PD-a module (Figure 1Ab), which is essential for CI formation (Stroud et al. 2016).

Loss of NDUFB10 disrupts complex I and supercomplex assembly. (A) Structure of human complex I. a: modules are indicated in color, N-module, Q-module, P-submodules: distal PD; proximal PP; b: localization of NDUFB10 in the PD-a module. PDB structure (5XTD). (B) BNGE followed by immunoblotting with anti-NDUFB10 (CI), anti-UQCRC2 (CIII) and anti-COXIV (CIV) antibodies; WT, HAP1 WT; KO, HAP1 NDUFB10 knockout; RS, KO stably rescued with NDUFB10-MYC-FLAG. The colored lines indicate from which semiquantitative analyses were performed. (C) In-gel activity assay for NADH-Dehydrogenase activity of complex I (CI-IGA, left panel), Coomassie staining of the gel (right panel) with molecular weight markers. (D) Immunodetection of subunit NDUFS3 (Q-module subunit) after separation by BNGE. Supercomplex SCI + III2 + IV; respiratory complex RC III2 + IV and comigrating III2, IV2; #, unspecific; *, subcomplexes of complex I containing the Q-module subunit NDUFS3. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, Bonforri test.
Complex I is composed of 44 subunits in mammals with a molecular weight of about 1 MDa (Stroud et al. 2016; Zickermann et al. 2003). Its coordinated biogenesis and assembly is a challenge due to the fact that numerous supernumerary subunits are nuclear-encoded and have to be imported into mitochondria, where they have to be assembled in a well-orchestrated manner with their mitochondrially encoded counterparts (Guerrero-Castillo et al. 2017). N-module subunits are found to be added at the end of the assembly pathway. Different assembly factors are required to successfully attach the subunits to their appropriate subcomplex, as validated by knockout studies (Stroud et al. 2016). The assembly factors detach from CI before the assembly is complete (Guerrero-Castillo et al. 2017). Free CI is usually not detected in OXPHOS-dependent tissues like neurons but is found exclusively in SCs. In contrast, in astrocytes that are mostly glycolytic also free CI was found. Free CI was associated with higher reactive oxygen species (ROS) (Lopez-Fabuel et al. 2016). CI subunit NDUFS1 plays a vital role in directing CI assembly into SC: The more elevated the expression of NDUFS1, the less free CI was observed, such as in neurons versus astrocytes (Lopez-Fabuel et al. 2016). Thus, CI subunit composition directly contributes to SC regulation and ROS homeostasis. In addition, CI subunit NDUFV3 is the first identified tissue-specific subunit in mammals (Bridges et al. 2017), similar to CIV, for which tissue-specific isoforms have been found. Here, we concentrate on the central role of the accessory subunit NDUFB10 for CI and SC assembly. NDUFB10 is a subunit of the PD-a module and likely has a role in stabilizing the PD-module. We here determined the effect of NDUFB10 knockout on CI, SC formation and respiratory activity. We found that knockout of NDUFB10 resulted in depletion of CI and SC I1III2IV, and pertubation of respiratory activity in the absence of complex II substrate.
2 Results
2.1 Knockout of NDUFB10 results in loss of complex I and respiratory supercomplexes
Wildtype HAP1 and HAP1 NDUFB10 knock out cell lines were purchased from Horizon Discovery (Horizon Genomics GmbH). A rescue cell line was generated by introducing NDUFB10-MYCFLAG into the AAVS1 (HDR) safe harbor site using CRISPR/CAS9 technology. Crude mitochondria from wildtype HAP1, NDUFB10KO, and rescued NDUFB10KO with NDUFB10-MYCFLAG tag were isolated and membrane complexes were solubilized with digitonin under native conditions. The proteins were separated by blue native gel electrophoresis in BNGE (Wittig et al. 2006). To resolve SC and single complexes, immunoblotting was conducted against complex I subunit (NADH:ubiquinone oxidoreductase subunit B10, NDUFB10), complex III subunit (ubiquinol-cytochrome C reductase core protein III, UQCRC2), complex IV (Cytochrome oxidase subunit IV, COXIV) (Figure 1B). In the wild type cell line, plotting against NDUFB10 revealed that most of the subunit was found in supercomplexes of the composition I1 + III2 + IV and I1 + III2 (Figure 1B). In addition, CIII was found in respirasome complex RC III2 + IV. CIV assembled in SCs and RCs, but in addition there was a considerable amount of free CIV (Figure 1B). In the NDUFB10KO line, hardly NDUFB10 could be detected and no supercomplexes with complex I were found, when the membrane was blotted against complex IV (COXIV) and complex III (UQCRC2). Respirasome complexes containing CIII and CIV were still present indicating that respiratory complexes CIII and CIV could assemble in the absence or knockdown of CI. We validated the importance of the accessory subunit NDUFB10 in complex I assembly and SCs formation by rescuing the HAP1 NDUFB10KO cells via CRISPR/CAS9 mediated insertion of NDUFB10 cDNA into the genomic safe harbor locus site (AAVS1). In the rescue cell line, we observed the full recovery of CI and supercomplexes SC I + III2 + IV and SC I + III2 (Figure 1B). Consequently, the P-module-NDUFB10 subunit is crucial for complex I assembly.
In CI-depleted conditions, we found higher levels of RC III2 + IV as well as free III2, IV and IV2, while in rescued cells due to the formation of SC, free CIII2 and CIV decreased again because CIII and CIV re-assembled into SC I1 + III2 + IVn (Figure 1B). This suggests that loss of CI did not reduce the biogenesis and assembly of CIII and CIV per se. This was further explored by generating expression and protein profiles.
2.2 NDUFB10 is essential for the assembly of the N- and Q-module into full CI complex
Since complex I biogenesis is a progressive process, where modules are sequentially assembled (Guerrero-Castillo et al. 2017), we next asked, how knockout of NDUFB10 would affect the different assembly steps and activity of the modules. For example, in some knockout cell lines for complex I nuclear genes (NDUFS6, NDUFS4) the N module can be assembled but cannot be mounted on the Q/P 830 kDa complex (Kahlhöfer et al. 2021). The NDUFB10 is part of the PD-a module, which provides part of the assembly platform for the Q- and N-module (Guerrero-Castillo et al. 2017). We determined the activity of the N-module, which is the last module added during complex I biogenesis. Therefore, we tested for NADH:ubiquinone reductase enzyme activity by an in-gel activity assay (IGA) (Figure 1C). While the SC protein band in control cells displayed NADH oxidizing activity, we found no NADH oxidizing function in NDUFB10KO at the level of SC. However, we found NADH oxidizing activity in a subcomplex (# in Figure 1C), which is likely attributed to a partial N-module/Q-module.
Next, we probed the assembly of the Q-module by staining NDUFS3, a subunit that is attributed to the Q-module. The wild type mitochondrial extract showed the presence of supercomplexes and several subcomplexes of lower molecular weight that were positive for NDUFS3 (Figure 1D, marked as # and *). In the NDUFB10KO, no signal in the height of SC assembly was detected, but the subcomplexes detected in the wildtype (#, *) plus an additional subcomoplex with MW<480 were detected (Figure 1D, marked as **). In particular, the subcomplex of MW ∼500 kDa accumulated. Probably, diverse subcomplexes accumulated since they could not be further processed to full complex I respectively supercomplexes, likely due to the lack of the anchoring platform PD (Guerrero-Castillo et al. 2017). The assembly intermediates were further characterized in Section 3.5.
2.3 Loss of intact complex I and supercomplexes results in perturbation of the respiratory activity and decrease of ΔΨm
Next, we asked how the loss of supercomplexes and perturbance of complex I would affect the electron transport in the respiratory chain. We determined the bioenergetic profile of the NDUFB10 deficient cells by monitoring oxygen consumption rates (OCR) with a Seahorse XF96 Extracellular Flux Analyzer. Different OXPHOS complex inhibitors were sequentially added to determine basal respiration, ATP synthase-linked respiration, maximal respiratory capacity and non-respiratory oxygen consumption (for details see material and methods). We tested HAP wt cells, NDUFB10KO and rescued NDUFB10KO cells. The wt and rescued cells exhibited similar responses to the inhibitors, namely decrease of OCR after oligomycin application, increase of OCR after FCCP application, and shut down of ETC-related respiration after inhibition of rotenone and antimycin addition (Figure 2A). The rescue was not complete, though, since for example full capacity was not reached. Basal respiration (wt: OCRbasal = 1.29 ± 0.25 [SD] pmol min−1 1000 cells−1 and rescued: OCRbasal = 1.39 ± 0.14 [SD] pmol min−1 1000 cells−1) and ATP synthesis related respiration (wt: OCRATP = 0.87 ± 0.17 [SD] pmol min−1 1000 cells−1 and rescued: OCRATP = 0.98 ± 0.15 [SD] pmol min−1 1000 cells−1) were comparable in wt and rescued cells (Figure 2A). Interestingly, in both cell lines the uncoupler FCCP restored an OCR slightly lower than the respective basal OCR but did not further stimulate respiration (wtspare capacity = 0.96 ± 0.36%, rescuedspare capacity = 0.79 ± 0.48%). This can have several reasons: lack of sufficient substrate to reach maximal OCR, or no spare capacity in wt and rescued cells. Contrary to control and rescued cells, NDUFB10KO cells showed a significant reduction of mitochondrial respiration and no response to ETC inhibitors and uncouplers (Figure 2A, Supplementary Figure S1).
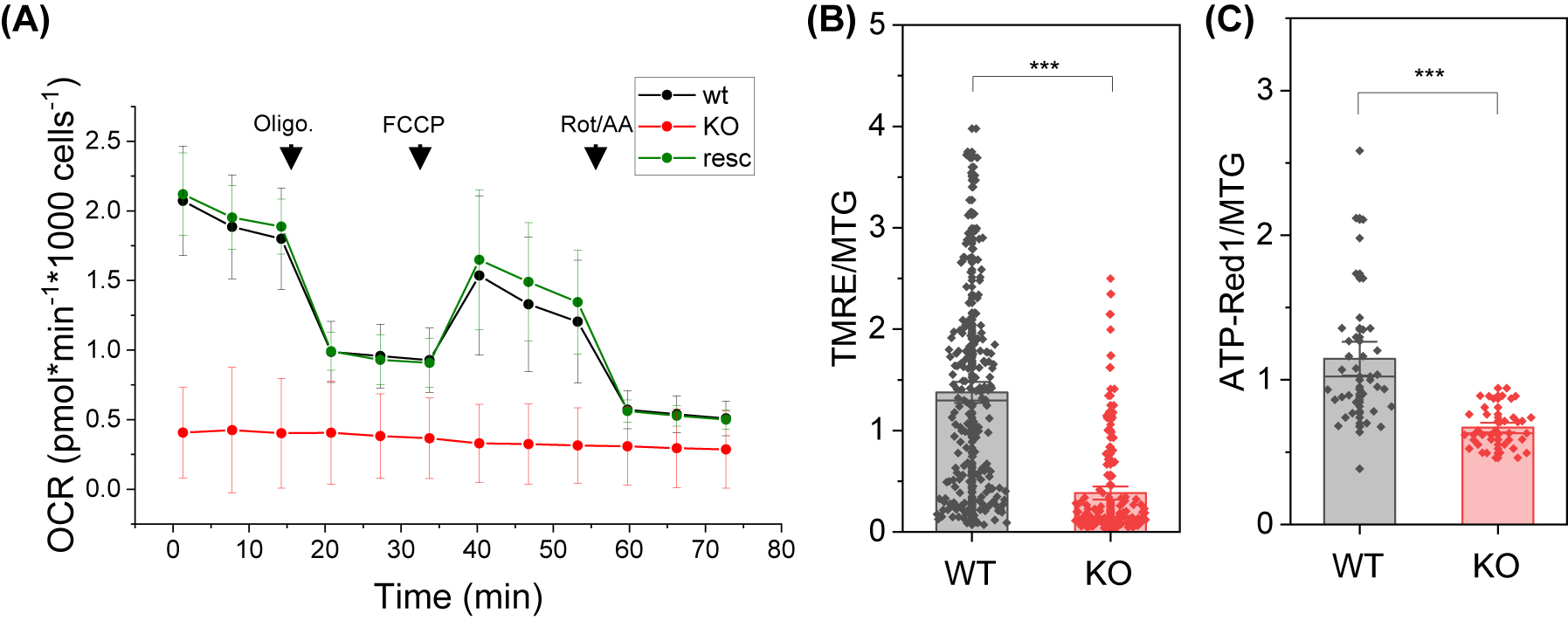
Loss of complex I and supercomplex assembly results in pertubation of mitochondrial respiration. (A) Respiration measurement of wild type cells, NDUFB10KO and NDUFB10KO rescued. Seahorse Mito Stress test showing oxygen consumption rate (OCR; mean and standard deviation) under high glucose conditions; 1, oligomycin injection (2 µM); 2, FCCP injection (0.5 µM); 3, rotenone and antimycin A injection (0.5 µM); a representative experiment is shown. (B) Mitochondrial membrane potential measurements with TMRE (25 nM) from five independent experiments and 140 cells. Normalization on MTG (200 nM). (C) Relative mitochondrial ATP levels were determined by ATP-Red1™ (5 µM); measurements from three independent experiments and 40 cells. WT, HAP1 wildtype; KO, HAP1 NDUFB10 knockout; two-sided t-test *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. Charts: The error bars denote the standard error (SE); the charts represent 50 percentiles. The vertical lines in the boxes represent the mean values. Technical replicate #1. Biological replicates: N = 28 for wt, N = 30 for KO and n=13 for rescue.
Reduction of the respiratory activity will likely affect the ΔΨm, which is build by the activity of the primary proton pumps CI, CIII and CIV. Missing CI activity reduces the net proton pumping by 8 H+/O2 (N-pathway CI/CIII2/CIV: 20 H+/O2; CIII2/CIV: 12 H+/O2).
To validate the loss of function of the ETC, we determined the mitochondrial membrane potential (ΔΨm), which is directly linked to OXPHOS activity.
The relative ΔΨm was determined with the ΔΨm-sensitive dye Tetramethylrhodamine ethyl ester (TMRE). TMRE accumulates in active polarized mitochondria in dependence on the ΔΨm and thus its fluorescence intensity is an indicator for ΔΨm. The TMRE fluorescence intensity was normalized to MitoTracker™Green (MTG) fluorescence. The experiment revealed a significant reduction of ΔΨm in NDUFB10KO cells (Figure 2B). This data is in accordance with the reduced respiration and thus proton pumping into the intra cristae space in NDUFB10KO. Eventually, a reduced proton motive force will result in less ATP production. To test this, we determined relative mitochondrial ATP levels with the dye ATP-Red1™. The fluorescence of ATP-Red1 is proportional to mitochondrial ATP levels. To account for mitochondrial mass, the fluorescence was normalized to MTG. As anticipated, the NDUFB10KO showed significantly reduced ATP levels (Figure 2C).
2.4 Knockout of NDUFB10 does not affect the expression of other ETC complexes
Loss of complex I related to NDUFB10 knockout and its effects on OXPHOS activity could be regulated on the level of gene expression and/or protein level. To check this, relative expression levels of OXPHOS complexes were determined by quantitative PCR. Complex I expression was probed by its subunit NDUFA9, complex II by subunit SDHA, complex III by subunit UQCRC1, complex IV by subunit Cox8a and ATP synthase by subunit ATP5A. Moreover, we tested for the expression of SCAF1, a chaperone involved in supercomplex assembly. All values were normalize on β-tubulin. Compared to HAP1 WT cells, NDUFB10KO cells showed no significant difference in NDUFA9 mRNA levels, a subunit that resides in the interface of the N- and P-module and is not assigned to a specific complex I module (Stroud et al. 2016) (Figure 3A). This does not necessarily mean that NDUFA9 was also unchanged on the protein level, because likely most of complex I regulation happens at the protein level (Stroud et al. 2016). In contrast, complex II subunit SDHA trended to an increased expression. For CIII and CIV, the mRNA levels of subunits UQCRC1 and Cox8a decreased significantly, while CV expression was apparently not altered in NDUFB0KO as the expression of ATPFA was unchanged (Figure 3A). Expression profiling also revealed that supercomplex assembly factor 1 (SCAF1) was reduced in NDUFB10KO cells. SCAF1 is an essential factor in SC formation. In particular the interaction between CIII and CIV is modulated by SCAF1 (also known as COX7A2L) (Cogliati et al. 2016; Maekawa et al. 2020) (Figure 3A). This suggests, that disrupted CI biogenesis might compromise III(2) + IV assembly/interaction due to loss of SCAF1.

NDUFB10 effects on the expression of respiratory complexes and ATP synthase. (A) Expression of OXPHOS subunits NDUFA9 (CI), SDHA (CII), UQCRC1 (CIII), Cox8a (CIV), ATP5A (CV) and the supercomplex assembly factor SCAF1. mRNA levels were determined using RT-qPCR analysis, normalized on beta-tubulin. Statistics: N = 3 independent experiments, n = 3 biological samples and four technical replicates each (ΔΔCt). (B) Immunoblot of complex I subunit NDUFS3. WT, wildtype; MW, molecular weight marker. (C) Immunoblot of complex II subunit SDHA. (D) Immunoblot of respiratory complex subunits as indicated. a: lower, b: higher exposure of the same blot. Antibodies against NDUFB8, SDHB, UQCRC2, MTCO1, and ATP5A. (E) Quantification of protein levels normalized on Tom20.
Following, subunits of different OXPHOS complexes were probed on the protein level (Figure 3B–D). We found a significant decrease in the level of complex I subunit NDUFS3, a subunit assigned to the Q-module (Figure 3B and E). Complex II subunit SDHA was not changed in NDUFB10KO (Figure 3C). Next, we analyzed protein subunits of all OXPHOS complexes: NDUFB8, SDHB, UQCRC2, MTCO1, and ATP5A. Protein levels were normalized on Tom20 levels, that did not change (Figure 3E), (Supplementary Figure S2). SDHA protein was not changed in NDUFB10KO, although mRNA indicated an increase in expression (Figure 3A and E), but SDHB protein levels were significantly increased. For complex III, we observed a slight increase in the UQCRC2 in NDUFB10KO (Figure 3E). The mitochondrial encoded complex IV subunit MTCO1 was not affected in NDUFB10KO. Finally, we assessed complex V subunit α (ATP5A) protein levels. The NDUFB10KO exhibited the same levels as the WT cells. This was matched by the expression levels of the same subunit, which also displayed no difference between WT and NDUFB10KO (Figure 3A and E). For complex I, we found a reduction of NDUFS3 and an almost complete loss of subunit NDUFB8, part of the ND5 module, in NDUFB10KO cells (Figure 3E). From expression and protein analysis data, a tendency towards an increase of complex II can be deduced. The protein data for complex III and complex IV, however, do not indicate a difference between wildtype and NDUFB10KO. Finally, complex V was also not affected by complex I depletion.
2.5 NDUFB10KO results in incomplete complex I assembly
To get a more comprehensive understanding of NDUFB10KO effects on assembly of the different modules of complex I, mass spectrometry analysis was performed. We asked which other subunits of CI were affected by NDUFB10 downregulation. According to quantitative PCR analysis of mRNA, NDUFA9 (Q-module) expression was not changed in NDUFB10KO cells (Supplementary Figure S3). The expression of mitochondrial encoded subunit ND1, which is part of the P P -module, in NDUFB10KO was only 66% of the level in WT, which is in principle a knock down (KD). For several other subunits, expression levels also were lowered, however, this was not significant. In two of the biological replicates of NDUFB10KO cells no ND1 was detected. It has to mentioned, though, that CI mt-DNA encoded subunits are always very hard to find due their high hydrophobicity. ND1 together with TIMMDC1 provides the anchoring platform for the Q-module in the membrane at an early step during CI biogenesis (Fang et al. 2021; Guerrero-Castillo et al. 2017). Starting from this building block, CI continues to be assembled from both the N-, Q- and P-modules as more N, Q, and P subunits are added (Guerrero-Castillo et al. 2017). The P D -module, which contains NDUFB10, is added several steps later. Therefore, the loss or downregulation of ND1 is critical for the assembly of CI. Although we could not assess ND1 levels, subunits NDUFA3 and NDUFA13, part of the ND1 (PP-a) module were not decreased in NDUFB10KO, while subunits NDUFA2 and NDUFA7, assigned to the N-module, were downregulated (Figure 4B). In addition, the abundance of NDUFA10, part of ND2-(PP-b)module, was lower in NDUFB10KO. From this data it appears that the assembly of the N-module and the PP-b-module, is affected in NDUFB10KO, while the ND1 (PP-a) module is not concerned. The subunits ND1, NDUFA3, NDUFA8 and NDUFA13 are part of an intermediate Q/PP-a complex (Guerrero-Castillo et al. 2017) and indeed, we found no changes in Q-subunit NDUFS2, NDUFS3, NDUFS7 and NDUFS8 levels in NDUFB10KO (Figure 4B and C). In contrast, the abundance of N-(NDUFS1, NDUFS4, NDUFS6, NDUFV1, NDUFV2) and ND2/PP-b-(NDUFC1, NDUFC2, NDUFS5) module were less abundant in NDUFB10KO cells. Also, when looking at the subunits of the PD-b and PD-a modules, it is obvious, that the abundance of these sub-modules is decreased in NDUFB10KO cells (Figure 4D).
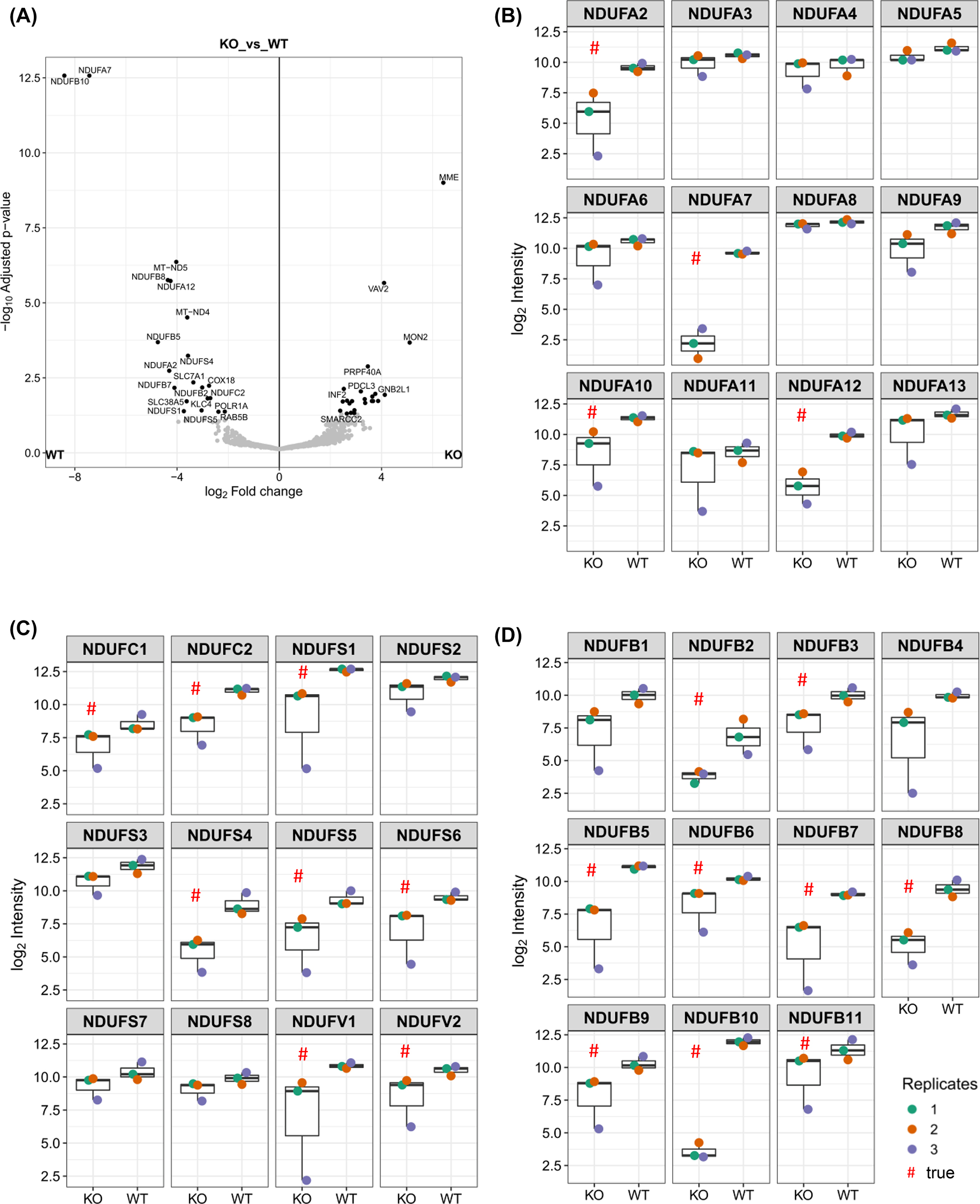
Changes in the abundance of complex I subunits in NDUFB10KO. (A) Volcano plot showing log2 changes in NDUFB10KO cells in comparison to the wildtype. (B) Abundance of CI subunits of the N-, Q- and ND1/ND2 modules. (C) Abundance of N-(NDUFS1, NDUFS4, NDUFS6, NDUFV1, NDUFV2), ND2/PP-b-(NDUFC1, NDUFC2, NDUFS5), and Q-(NDUFS2, NDUFS3, NDUFS7, NDUFS8) module subunits in WT and NDUFB10KO cells. (D) Abundance of subunits of the PD-a (NDUFB1, NDUFB5, NDUFB6, NDUFB10, NDUFB11) and PD-b modules (NDFUB7, NDUFB8, NDUFB9, NDUFB3, NDUFB2).
Together, the CI-proteome data suggest that in NDUFB10KO cells no fully assembled CI but only subcomplexes containing Q-module subunits, ND1 and partial ND2/PP-b subunits are built, while N-, PD-b and PD-a module subunits are reduced. The immune-staining of NDUFS3, a Q-module subunit, was mildly decreased and NDUFB8, a PD-b subunit was strongly reduced. These observations are in line with the MS-data. Together, the data suggests that the assembly of CI is halted at an early stage. Likely, this is just the last step before ND1 and TIMMDC1 anchor the Q-subunits to the inner mitochondrial membrane (Guerrero-Castillo et al. 2017). Thus, neither the N-module nor the PD-arm will be assembled.
2.6 Loss of CI and respiratory supercomplexes affects mitochondrial morphology
Complex I and SC are mainly found in lamellar cristae sheets (Davies et al. 2011, 2018). Mitochondria are highly dynamic organelles whose architectural variations are closely linked to function (Hackenbrock 1968). Therefore, one can expect an impact of loss of CI and respiration on mitochondrial architecture. Transmission electron micrographs show tubular and more roundish mitochondria. Cristae were rather sparse (Figure 5A). It has to be mentioned, however, that cells had to be kept in high glucose conditions (25 mM Glucose) because NDUFB10KO cells were exclusively relying on glycolysis and were not able to survive with low glucose supply (5.6 mM glucose). For accurate quantification of subtle changes in mitochondrial morphological parameters, the analyze particles function in ImageJ software was used. After analyzing about 200 mitochondria for each condition (WT and KO cell lines), we found no difference in mitochondrial perimeter, while the circularity of mitochondria decreased in NDUFB10KO conditions (Figure 5B). Statistical analysis further revealed a significant decrease in mitochondrial width but no change in mitochondrial length for NDUFB10KO mitochondria compared to WT (Figure 5C). Consequently, the calculated aspect ratio (AR; length-to-width ratio) (Krebiehl et al. 2010) was significantly increased in NDUFB10KO, without an effect on the mitochondrial area. The cristae architecture was also affected in NDUFB10KO conditions (Figure 5D). The number and length of cristae per mitochondrion decreased, while the cristae width increased, resulting in an altered aspect ratio for cristae architecture. In sum, we found moderate changes of mitochondrial morphology and ultrastructure in NDUFB10KO cells, whereby cristae widening was the most prominent outcome.
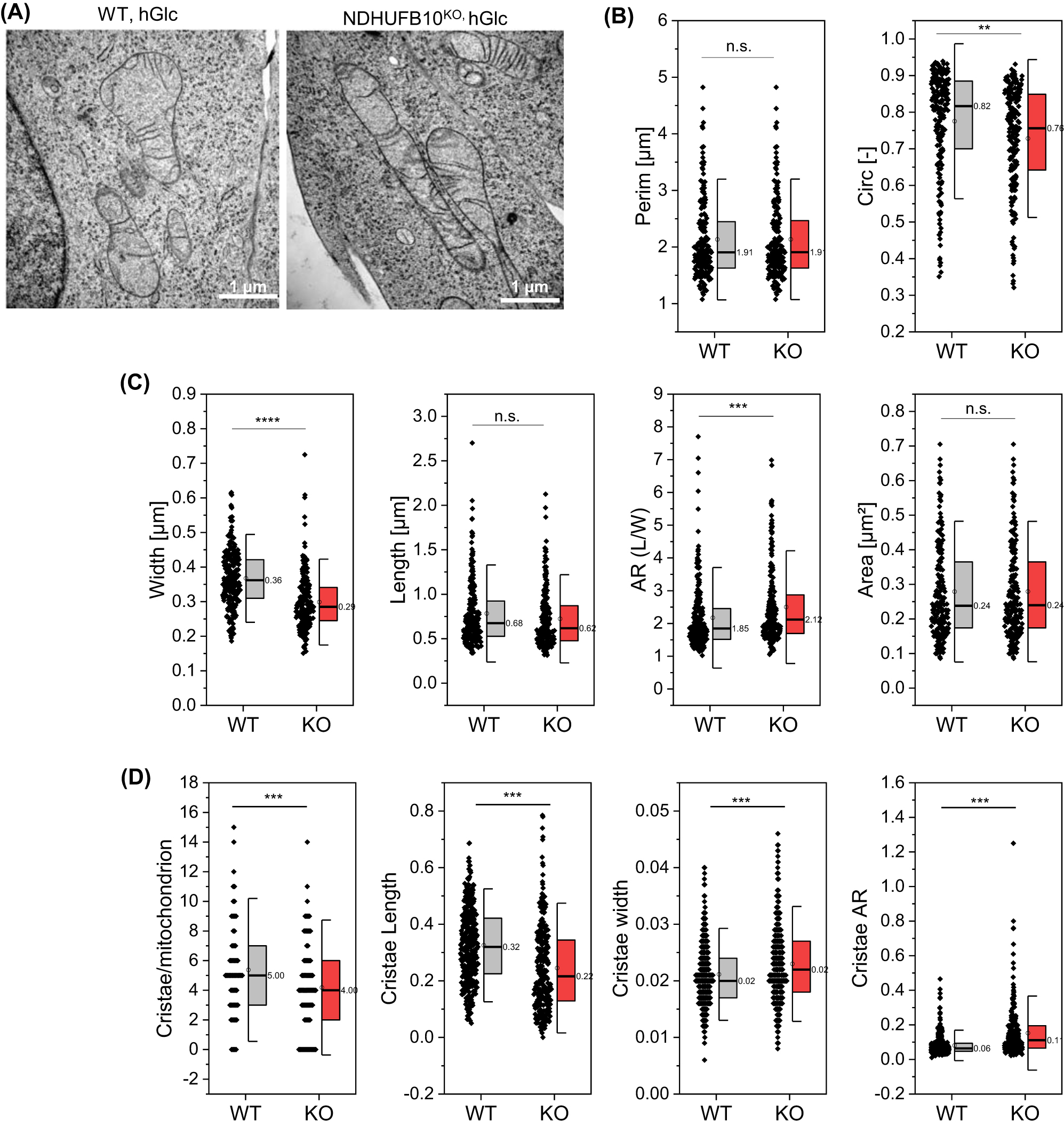
Mitochondrial morphology changes in NDUFB10KO cells. (A) Transmission electron micrographs of mitochondria in wild-type and NDUFB10KO cells. Images are representative of 190 (WT), respectively 197 (KO) imaged mitochondria. (B) Quantification of mitochondrial shape parameters. Mitochondrial perimeter and circularity. (C) Mitochondrial width, length and resulting aspect ratio (AR), which is the ratio between the minor and major axes of the ellipse equivalent to the object – that represents mitochondrial elongation and reflects the “width-to-length ratio”. Also, mitochondrial area is shown. (D) Characterization of cristae. Two-sided t-test, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 10−10; error bars indicate standard deviation HGlc, high glucose culture.
3 Discussion
The interaction of mitochondrial respiratory complexes to dynamically form supercomplexes is an important, though not well understood phenomenon. Here, we showed that knockout of subunit NDUFB10, which is part of the PD-module of complex I, resulted in a pertubation of NADH-dehydrogenase related respiration and ΔΨm generation, explained by depletion of fully assembled CI (Figure 6). While due to the loss of CI, no SC I + III2 + IV or SC I + III2 was built, III2 and IV are still found in the Q-respirasome (III2 + IV) and as individual complexes III2, IV2, and IV. Protein levels of III2 and IV were not changed as earlier reported for NDUFB10KO HEK293T cells (Stroud et al. 2016).
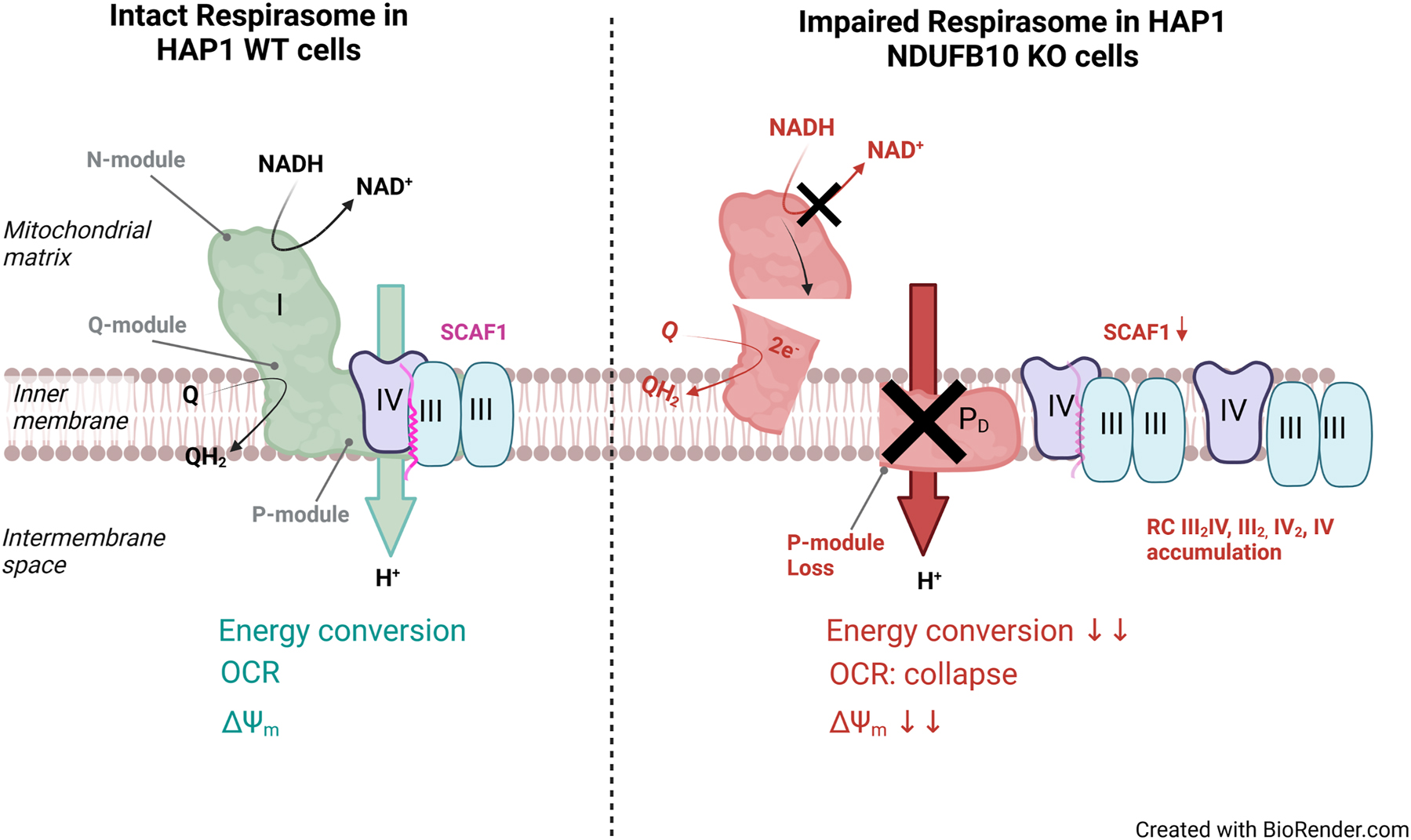
Schematic representation of the processes affected by NDUFB10KO.
The knockout of NDUFB10 resulted in rudimentary modules of complex I, though. Similar, as observed before (Stroud et al. 2016) and in agreement with the biogenesis pathway of complex I (Guerrero-Castillo et al. 2017), assembly of complex I was stalled at intermediate states. While the assembly of the Q-module apparently was unaffected, the N-module was incompletely assembled, as it was the ND2 module. Parts of the P-module of complex I were severely affected in NDUFB10KO. The P-arm is the membrane-embedded part of complex I. It is divided in a proximal PP and a distal PD part. NDUFB10 is assigned to the PD-a-module. While subunits of the PP-module were minorly affected in NDUFB10KO, subunits of the PD-module were massively downregulated. In recent years, it was demonstrated that several subunits attributed either to the proximal or distal P-module are critical for CI assembly. Cells without complex I assembly factor TIMMDC1 or expressing disease-related mutations accumulated subcomplexes, but were unable to build fully assembled CI (Fang et al. 2021). Previously, it was shown that depletion of the C. elegans NDUFA11 homologue NDUF-11 had similar effects on CI assembly and SC formation. NDUF-11 is an integral membrane protein (Gu et al. 2016). Its depletion also resulted in perturbation of respiration and comparably affected mitochondrial morphology as NDUFB10KO (Knapp-Wilson et al. 2021). However, NDUFA11 is later added during CI biogenesis than NDUFB10 (Guerrero-Castillo et al. 2017). Depletion of NDUFB10 or NDUFA11, respectively, hampers SC formation probably by two different mechanisms. NDUFA11 is localized at the interface between CI and CIII and interacts with subunits UQCRB and UQCRQ thereby stabilizing the SC (Letts et al. 2019). NDUFB10, on the other hand, is an accessory subunit that stabilizes the P-arm of CI. Its depletion resulted in the loss of subunits of the N-module and P-module, either by interfering with their assembly or by decreasing the stability of complex I. Since SC formation requires fully assembled CI (Guerrero-Castillo et al. 2017), loss of intact complex I affects SC formation. Because we found no bands of higher molecular weight in BNGE in NDUFB10KO than those indicating III2 + IV, III2, IV2, and IV we conclude that CI-subcomplexes did not assemble with III2 + IV, III2, IV2, and IV other than recently suggested (Fang et al. 2021). Therefore, our data is in line with previous observations that fully assembled CI is required for SC formation (Moreno-Lastres et al. 2012). NDUFB10 depletion resulted in the accumulation of RC III2 + IV in NDUFB10KO cells and no or reduced endogenous respiratory activity when no external substrates were added. Electrons can still feed in complex II, explaining the reduced oxygen consumption. It is also plausible that complex I dysfunction causes NADH accumulation in the mitochondrial matrix, which has synergistic effects on reducing OXPHOS complex activity: Adverse NADH/NAD+ levels reduce mitochondrial SIRT3 deacetylase activity, which activates CI and CII in a NAD+-dependent manner (Cimen et al. 2010; Martinez-Reyes and Chandel 2020).
In line with reduced OCR, we observed a significant reduction of ΔΨm and mitochondrial ATP in the NDUFB10KO cells. The ΔΨm is build by the activity of the primary proton pumps CI, CIII and CIV. Missing CI activity reduces the net proton pumping by 8 H+/O2 (N-pathway CI/CIII2/CIV: 20 H+/O2; CIII2/CIV: 12 H+/O2), which will results in a decreased ΔpH and electrochemical gradient ΔΨm, which constitute the proton motive force pmf. Loss of PMF/ΔΨm is detrimental for protein import (Neupert and Herrmann 2007) and a reduced ΔΨm will likely trigger quality control mechanisms. Indeed, PMF/ΔΨm regulates mitochondrial proteostasis, in particular complex I turnover (Patron et al. 2018). Also, Opa1, the inner membrane fusion and shaping protein, is processed in a PMF/ΔΨm dependent manner (Song et al. 2007), which might explain the variations in the cristae width observed in NDUFB10KO. Cristae alterations also could be indirectly affected by loss of CI. The partial loss of the large membrane-spanning P-arm might alter the protein-lipid ratio, reduce the molecular crowding and thus change the properties of the inner mitochondrial membrane (Lowe et al. 2020), which eventually impact on OXPHOS efficiency (Jezek et al. 2023). Effects of CI deficiency on mitochondrial morphology have been reported earlier: Mitochondrial shape and number varied significantly among 13 different complex I deficient primary fibroblast cells (Koopman et al. 2005) (Opa1-dependent). Cristae modulation is relevant for cellular adaptation to metabolic demands (Gomes et al. 2011a,b; Patten et al. 2014). This relation might explain, why the knockdown of NDUFB10, or likewise the core subunit NDUFS1, significantly reduced the reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) (Skvortsova et al. 2022): likely by preventing the required metabolic shift during reprogramming (Nishimura et al. 2019). Mutations in NDUFB11 located in the PD-a module and encoded from a gene in the X chromosome caused a complex I defect in two male patients (Reinson et al. 2019).
A specific mutation in NDUFB10 that changes its posttranslational modification caused assembly defects of CI which resulted in fatal infantile lactic acidosis and cardiomyopathy (Friederich et al. 2017).
In summary, our results confirm the importance of NDUFB10 for complex I assembly, and extend previous findings that pathogenic NDUFB10 mutations cause isolated complex I deficiency by impairing complex I assembly and compromising its enzymatic function (Friederich et al. 2017). This highlights the importance of the P-module in CI biogenesis and function. Likewise, it is this part of CI that is also indirectly important for the assembly of functional supercomplexes. Thus, the accessory subunits of the P module are crucial for a functional assembly of the ETC in several respects. Finally, it should be mentioned that loss of NDUFB10 resulted in a decrease of SCAF1 expression. SCAF1 is important for stabilizing the CIII-CIV interaction (Cogliati et al. 2016) and loss of SCAF1 will therefore lead to a weaker interaction between CIII and CIV. Whether this has functional and structural consequences needs to be elucidated in further studies.
4 Materials and methods
4.1 Cell culture
HAP1 cells have a near-haploid genotype, thereby facilitating genetic manipulation. In this project, HAP1 parental wildtype (WT) and NDUFB10 knockout cells were purchased from Horizon Discovery (Horizon Genomics GmbH) and used to generate different isogenic cell lines. Under normal culture conditions, HAP cell lines were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) (Gibco, #12440; containing 25 mM glucose) supplemented with 10% FBS at 37 °C and 5% CO2. When cells reached 70–75% confluency, cells were detached with Trypsin/EDTA for 1 min at 37 °C and the desired number of cells was passaged into a cell culture flask or imaging dish.
4.2 Generation of rescued HAP1 NDUFB10KO cell lines
We rescued HAP1 NDUFB10 knockout cells via CRISPR-Cas9 targeted NDUFB10 insertion into the AAVS1 Safe Harbor locus (GSHs). Two plasmids were transfected into the cells, the pAAVS1-EF1a-Puro-DNR (AAVS1-donor plasmid), containing the labeled NDUFB10 sequence with MYC-FLAG (small Tag) and pCas-Guide-AAVS1 plasmid, containing the CAS9 and gRNA sequences. For transfection, Turbofectin 8.0 reagent was mixed 4:1 to total plasmid DNA concentration, whereas AAVS1-donorplasmid and the pCas-Guide-AAVS1 plasmid were prepared in a 1:3 ratio. After 48 h, cells were selected via puromycin resistance (1 μg/mL). Cells were cultivated in the puromycin-containing medium until single cell clones could be observed. Single cells were picked using serial dilution and expanded in 96-well plates. When confluent, cells were transferred to a larger growth surface and frozen for long-term storage.
4.3 Transmission electron microscopy (TEM)
For high-resolution imaging of mitochondrial ultrastructure, HAP1 cells were seeded on coverslips that were previously coated with RGD-grafted poly-L-lysin-graft-(polyethylene glycol) (PLL-PEG-RGD). When the desired confluency was reached, cells were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate for 20 min on ice. Then cells were washed with 0.1 M sodium cacodylate buffer containing 3 μM calcium chloride. Next, a solution containing 1% OsO4, 0.8% potassium ferrocyanide, 3 μM calcium chloride in 0.1 M sodium cacodylate was prepared and added to the cells. After 30 min incubation, cells were washed with double distilled water and stained with 2% uranyl acetate. Next, cells were incubated in EtOH solutions, increasing the concentration gradually (20%, 50%, 70%, 90%, 100%). A mixture of 50% EtOH and 50% Durcupan ACM resin (Sigma Aldrich, #44611) was prepared and added to the cells. Subsequently, cells were incubated 2 × 3 h in 100% Durcupan, which was polymerized for at least 48 h at 60 °C in an oven. For imaging, thin sections of 70 nm were prepared and recorded with a Zeiss EM902 operated at 80 kV (Zeiss) and 300 kV.
4.4 MitoTracker Green FM staining
MitoTracker Green (MTG) is a membrane potential-independent mitochondrial tracker with excitation/emission maxima λ = 490/λ = 516 nm. However, a minimum amount of membrane potential is needed to allow the entrance of the dye. In this project, we used 200 nm MTG (Invitrogen, #M7514) to stain mitochondrial mass, whereby the DMSO concentration was below 1%. Cells were incubated for 30 min at 37 °C and 5% CO2 before medium exchange and subsequent imaging.
4.5 Membrane potential measurements
Tetramethylrhodamine ethyl ester (TMRE) is a cell-permeable, cationic, fluorescent dye with excitation/emission maxima λ = 549/λ = 575 nm. It is used to determine mitochondrial membrane potential ΔΨm since its accumulation in mitochondria is ΔΨm dependent. Cells were stained with 25 nM TMRE (BioMol #ABD-22220) for 30 min. Simultaneously, cells were stained with 200 nM MitoTracker™Green (MTG) as a reference for mitochondrial mass. After 30 min staining, cells were washed twice with PBS solution and imaging medium without phenol red was added.
4.6 Determination of ATP
The BioTracker™ ATP-Red dye (Sigma Aldrich, #SCT045) allows for live-cell imaging for mitochondrial targeting ATP molecules. The dye is not fluorescent until a negatively charged ATP is present that breaks the covalent bonds between boron and ribose, causing ring-opening and fluorescence. The red fluorescent dye has excitation/emission maxima of λ = 510 nm/λ = 570 nm. Cells were incubated with 5 μM ATP-red for 15 min at 37 °C. Cells were simultaneously stained with MTG as mitochondrial mass reference. After staining, cells were washed twice with PBS solution and fresh imaging medium was added.
4.7 Blue native PAGE (BNGE)
For BNGE, cells were collected from two confluent T175 flasks and processed as described previously (Salewskij et al. 2020). Briefly, harvested cells were lysed mechanically using the cell homogenizer. Then mitochondria were enriched via differential centrifugation. Digitonin ∼50% (TLC) (Sigma-Aldrich, #D5628) was used for mitochondrial solubilization in a ratio of 6 g/g as described in (Wittig et al. 2006). 50–100 μg protein were loaded per pocket and separated via a vertical native gel 3–12% SERVAGel™ (Serva, #43251.01). NativeMark™ unstained was used as a protein standard (Thermo Fisher, #LC0725) for molecular weight estimation. However, it should be noted that in gradient gels, native membrane proteins do not move at the same speed as a free protein standard, particularly in the more increased molecular weight range. All buffers were prepared as described in (Wittig et al. 2006).
4.8 In-gel activity assay
The native gel was incubated for 45 min in 20 mL complex I substrate solution until violets bands were clear indicating active complex I. The reaction was stopped by denaturing the native complexes with 10% acetic acid solution. Then the gel was washed with water and documented. Complex I substrate solution is prepared as described in (Jha et al. 2016).
4.9 Immunoblotting
PVDF membranes were blocked with 10% non-fat dry milk in TBS-T (200 mM Tris, 1.37 M NaCl and 0.1% Tween 20). Super Signal West Pico by Thermo Scientific (#34080) was used as a chemiluminescent substrate to visualize protein bands. Immunoblotting was conducted against complex I subunits NDUFB10 and NDUFS3 and NDUFB8, complex II subunit SDHA, complex III subunit UQCRC2, complex IV subunit Cox4L1. Tom20 was used as endogenous control. The secondary antibody was anti-mouse DyLight™ 800 from Rockland (#610-145-002) or anti-rabbit (#611-645-122). Antibodies are listed in Table 1. The signal was detected using the ChemiDOC™MP Infrared Imaging System (BioRad®). The mean density of the detected bands was determined using the analysis tool measure of ImageJ®.
Antibodies used in this study.
| Antibody | Company | Cat. no. |
|---|---|---|
| NDUFB10 | Abcam | ab196019 |
| NDUFS3 | Abcam | ab183733 |
| Cox4L1 | Abcam | ab183629 |
| UQCRC2 | Abcam | ab14745 |
| OxPhos cocktail | Abcam | Ab110413 |
| Tom20 | SantaCruz | sc-17764 |
4.10 Respiration measurements
4.10.1 Seahorse XF Mitostress test
For studying mitochondrial respiration, an automatic flux analyzer XF 96 (Seahorse/Agilent) was used. One day before the experiment, 50.000 HAP cells/well were seeded in a Seahorse XF 96 Cell Culture Microplate and incubated overnight at 37 °C and 5% CO2. In addition, 200 μL of sterile water was added to each Calibration Hydrate Sensor Cartridge well for calibration and incubated at 37°C, environmental CO2. The Seahorse XF 96 Analyzer had to be switched on at least 12 h before the measurement and calibrated at 37 °C. On the day of the experiment, the water in the sensor cartridge was replaced by 200 μL of XF Calibrant/well and incubated for 45–60 min at 37 °C with environmental CO2. Prior to measurement, cells were incubated for 1 h in 180 μL XF Base Assay Medium (25 mM D-glucose, 1 mM pyruvate, 2 mM L-glutamine in XF Base Medium with 25 mM glucose) at 37 °C, environmental CO2. After calibration with the Hydrate Sensor Cartridge, the Seahorse XF 96 Cell Culture Microplate was mounted in the XF 96 Analyzer. First, basal respiration was determined. Then, through sequential inhibitor injections (1_oligomycin injection [2 µM]; 2_ Carbonylcyanid-p-trifluoromethoxyphenylhydrazon [FCCP] injection [0.5 µM]; 3_rotenone and antimycin A injection [0.5 µM each]) the ATP synthesis related respiration, the maximum respiration, the proton leak and the non-mitochondrial respiration were determined at 37 °C. With the final injection, cells were stained with Hoechst 33,342 and cells were counted with the Cytation 1 Cell Imager (Agilent) to allow for normalization to cell number.
4.11 Quantitative polymerase chain reaction
Total RNA of HAP1 cell lines was obtained with the Monarch® Total RNA Miniprep Kit (NEB #T2010), and cDNA was transcribed with the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific #K1621). Quantitative PCR (qPCR) was performed on the StepOnePlus™ Real-Time PCR System (Applied Biosystems). The PCR reaction was prepared from PowerUP™ SYBR™ Green Master Mix (Applied Biosystems # A25741), 30 ng cDNA, and 0.1 nM of each forward and reverse primer per sample. The PCR protocol started with an initial denaturation step of 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. Primers were purchased from Eurofins Genomics. The list of primers used is found in Table 2.
RTq-PCR primer sequences used in this work.
| Primer name | Nucleotide sequence (5′ → 3′) |
|---|---|
| Mitochondrial targets | |
| Complex I | |
| NDUFB10_forward | TAGAGCGGCAGCACGCAAAG |
| NDUFB10_reverse | CTGACAGGCTTTGAGCCGATC |
| ND1_forward | CTACTACAACCCTTCGCTGAC |
| ND1_reverse | GGATTGAGTAAACGGCTAGG |
| ND4_forward | CTAGGCTCACTAAACATTCTA |
| ND4_reverse | CCTAGTTTTAAGAGTACTGCG |
| ND5_forward | TCGAATAATTCTTCTCACCC |
| ND5_reverse | TAGTAATGAGAAATCCTGCG |
| NDUFA5_forward | GAGAAGCTGGCTATGGTTAAAGCG |
| NDUFA5_reverse | CCACTAATGGCTCCCATAGTTTCC |
| NDUFV1_forward | TGAGACGGTGCTGATGGACTTC |
| NDUFV1_reverse | AGGCGGGCGATGGCTTTC |
| NDUFA10_forward | CACCTGCGATTACTGGTTCAG |
| NDUFA10_reverse | GCAGCTCTCTGAACTGATGTA |
| NDUFB11_forward | GGAAAGCGGCCCCCAGAACCGAC |
| NDUFB11_reverse | CCACGCTCTTGGACACCCTGTGC |
| NDUFA9_forward | AGC TTC ATC ATG CCC TCA TGC C |
| NDUFA9_reverse | TCT CGC GTC CCA TTC CAG AAA C |
| Complex II | |
| SDHA_forward | GCA AAA TCA TGC TGC CGT GTT C |
| SDHA_reverse | ATC CGC ACC TTG TAG TCT TCC C |
| Complex III | |
| UQCRC1_forward | GAG CAC CAG CAA CTG TTA GAC C |
| UQCRC1_reverse | CAG TGA AGC GGC ATG GAG TAA G |
| Complex IV | |
| Cox8a_forward | AAG ATC CAT TCG TTG CCG C |
| Cox8a_reverse | TAG GTC TCC AGG TGT GAC AG |
| Complex V | |
| ATP5F1A_forward | CCA AAC CAG GGC TAT GAA GCA G |
| ATP5F1A_reverse | CAC CCG CAT AGA TAA CAG CCA C |
| ATP5F1B_forward | GAA GAC AAG TTG ACC GTG TCC C |
| ATP5F1B_reverse | TCA CGA TGA ATG CTC TTC AGC C |
| SCAF1_forward | TGG TTT CCA CAG AAG CAC CAC |
| SCAF1_reverse | ATC AGG CAG TAG ATG GTC CCT C |
| Metabolic regulators | |
| AMPK | |
| AMPK_forward | ACA GCC GAG AAG CAG AAA CAC |
| AMPK_reverse | GCC CAG TCA ATT CAT GTT TGC C |
| mTOR | |
| mTOR_forward | AGC ATC GGA TGC TTA GGA GTG G |
| mTOR_reverse | CAG CCA GTC ATC TTT GGA GAC C |
| GLUT4_forward | CCA TCC TGA TGA CTG TGG CTC T |
| GLUT4_reverse | GCC ACG ATG AAC CAA GGA ATG G |
| Housekeeping genes | |
| GAPDH | |
| GAPDH_forward | CTG GTA AAG TGG ATA TTG TTG CCA T |
| GAPDH_reverse | TGG AAT CAT ATT GGA ACA TGT AAA CC |
| Actin | |
| beta Actin_forward | CAC CAT TGG CAA TGA GCG GTT C |
| beta Actin_reverse | AGG TCT TTG CGG ATG TCC ACG T |
| Tubulin | |
| beta V Tubulin_forward | CTG GAC CGC ATC TCT GTG TAC T |
| beta V Tubulin_reverse | GCC AAA AGG ACC TGA GCG AAC A |
4.12 Mass spectrometry
4.12.1 Sample preparation
Mitochondria were lysed in 4% (w/v) SDS/1mM PMSF/1mM Benzamidine in 100 mM Tris/HCl, pH8 by heating to 95°C for 5 min in a Thermomixer at 1000 rpm and subsequent sonification for 15 min. Protein concentrations were determined by BCA assay (Thermo Fisher Scientific) using BSA as standard. Tryptic protein digestion was carried out according to the FASP protocol (Wisniewski et al. 2009). Resulting peptides were desalted using in-house made C18-STAGE tips (Rappsilber and Ishihama 2007), then dried by vacuum centrifugation, and resolubilized in 4% acetonitrile/0.05% TFA prior to MS analysis.
4.12.2 Liquid chromatography–mass spectrometry (LC-MS/MS)
Chromatographic separation of peptides was achieved using an Ultimate 3000 RSLCnano System (Dionex, part of Thermo Fisher Scientific). The mobile phases for the loading pump consisted of 0.05% (v/v) TFA in ultrapure water (A) and 80% acetonitrile/0.05% (v/v) TFA in ultrapure water (B). The sample was loaded on a trapping column (C18 PepMap 100, 300 µM × 5 mm, 5 µm particle size, 100 Å pore size; Thermo Scientific) and desalted for 5 min using eluent A at a flow rate of 10 μL/min. Then the trap column was switched to back-flush position in series with the separation column (Acclaim PepMap100 C18, 75 µm × 50 cm, 2 µM particle size, 100 Å pore size, Thermo Scientific). The mobile phases for elution of peptides consisted of 0.1% (v/v) formic acid in ultrapure water (A*) and 80% acetonitrile/0.1% (v/v) formic acid in ultrapure water (B*). Peptides were eluted at a flow rate of 250 nL/min using the following gradient profile: 5% B* over 3 min, 5–24% B* over 120 min, 24–36% B* over 40 min, 36–99% B* over 10 min and 99% B* over 20 min.
The LC system was coupled via a nanospray source to a Q Exactive Plus mass spectrometer (Thermo Scientific). MS full scans (m/z 350–1400) were acquired in positive ion mode at a resolution of 70,000 (FWHM) with internal lock mass calibration on m/z 445.12003 and 50 ms maximum injection time. The mass spectrometer was operated in data-dependent acquisition (DDA) mode, selecting the 12 most intense ions of a full scan for fragmentation by HCD (NCE 27). Fragmentation scans (MS/MS) were recorded at a resolution of 17,500 (FWHM) and a maximum injection time of 80 ms. Automatic gain control (AGC) target values were 3 × 106 and 5 × 104 for MS full and MS/MS scans, respectively. Dynamic exclusion was enabled with an exclusion duration of 60 s. Singly charged ions, ions with unassigned charge states and charge states >4 were rejected.
4.12.3 LC-MS/MS data analysis
LC-MS/MS data was processed with MaxQuant 2.0.3.0 for peptide/protein identification and label-free quantification (LFQ). Raw files were searched against the Uniprot human reference proteome database (UP000005640, downloaded 13/05/2020) and a list of common contaminants. Default search and quantification settings were applied, with activation of “Match between runs” as the only exception. Oxidation of methionine and N-acetylation of protein N-termini were set as variable and carbamidomethylation of cysteine as fixed modifications. Peptides and proteins were filtered to satisfy a false discovery rate (FDR) of 0.01.
For statistical analysis non-normalized intensity data was imported into Perseus (version 1.6.15.0). Contaminant proteins, proteins only identified by site and reverse hits were removed. Intensities were log2 transformed and proteins with less than three valid intensity values in at least one genotype were discarded. To compensate for differences in mitochondria purification efficiencies, intensity data of each sample were normalized to mitochondrial citrate synthase (CS, O75390). Differential expression analysis was carried out with Limma (Ritchie et al. 2015) implemented in LFQ-Analyst (Shah et al. 2020). For imputation of left-censored missing values the MinProb algorithm was used. Log2-fold changes of <−1 or >1 and with an FDR <0.05 were considered as significant. The mass spectrometry proteomics raw experimental data has been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2019) partner repository with the dataset identifier PXD038890 und DOI 10.6019/PXD038890.
Funding source: Human Frontier Science Program
Award Identifier / Grant number: RGP0016/2018
Funding source: GRC, German Research Fundation
Award Identifier / Grant number: CRC944 (INST190/1672 and the z-project)
Acknowledgments
The TEM data was obtained at the iBiOS facility in Osnabrück, Germany. We thank Silke Morris for editorial help.
-
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Research funding: The study was supported by a grant from CRC944 (INST190/1672 and the z-project). Tasnim Arroum was supported by an HFSP doctoral fellowship (RGP0016/2018).
-
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
Acin-Perez, R. and Enriquez, J.A. (2014). The function of the respiratory supercomplexes: the plasticity model. Biochim. Biophys. Acta 1837: 444–450, https://doi.org/10.1016/j.bbabio.2013.12.009.Search in Google Scholar PubMed
Acin-Perez, R., Fernandez-Silva, P., Peleato, M.L., Perez-Martos, A., and Enriquez, J.A. (2008). Respiratory active mitochondrial supercomplexes. Mol. Cell. 32: 529–539, https://doi.org/10.1016/j.molcel.2008.10.021.Search in Google Scholar PubMed
Barrientos, A. and Ugalde, C. (2013). I function, therefore I am: overcoming skepticism about mitochondrial supercomplexes. Cell Metabol. 18: 147–149, https://doi.org/10.1016/j.cmet.2013.07.010.Search in Google Scholar PubMed PubMed Central
Blaza, J.N., Serreli, R., Jones, A.J., Mohammed, K., and Hirst, J. (2014). Kinetic evidence against partitioning of the ubiquinone pool and the catalytic relevance of respiratory-chain supercomplexes. Proc. Natl. Acad. Sci. U.S.A. 111: 15735–15740, https://doi.org/10.1073/pnas.1413855111.Search in Google Scholar PubMed PubMed Central
Brandt, U. (2006). Energy converting NADH: quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75: 69–92, https://doi.org/10.1146/annurev.biochem.75.103004.142539.Search in Google Scholar PubMed
Bridges, H.R., Mohammed, K., Harbour, M.E., and Hirst, J. (2017). Subunit NDUFV3 is present in two distinct isoforms in mammalian complex I. Biochim. Biophys. Acta Bioenerg. 1858: 197–207, https://doi.org/10.1016/j.bbabio.2016.12.001.Search in Google Scholar PubMed PubMed Central
Calvo, E., Cogliati, S., Hernansanz-Agustin, P., Loureiro-Lopez, M., Guaras, A., Casuso, R.A., Garcia-Marques, F., Acin-Perez, r., Marti-Mateos, y., Silla-Castro, J.C., et al.. (2020). Functional role of respiratory supercomplexes in mice: SCAF1 relevance and segmentation of the Q(pool). Sci. Adv. 6: eaba7509, https://doi.org/10.1126/sciadv.aba7509.Search in Google Scholar PubMed PubMed Central
Cimen, H., Han, M.J., Yang, Y., Tong, Q., Koc, H., and Koc, E.C. (2010). Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry 49: 304–311, https://doi.org/10.1021/bi901627u.Search in Google Scholar PubMed PubMed Central
Cogliati, S., Calvo, E., Loureiro, M., Guaras, A.M., Nieto-Arellano, R., Garcia-Poyatos, C., Ezkurdia, I., Mercader, N., Vazquez, J., and Enriquez, J.A. (2016). Mechanism of super-assembly of respiratory complexes III and IV. Nature 539: 579–582, https://doi.org/10.1038/nature20157.Search in Google Scholar PubMed
Davies, K.M., Strauss, M., Daum, B., Kief, J.H., Osiewacz, H.D., Rycovska, A., Zickermann, V., and Kuhlbrandt, W. (2011). Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc. Natl. Acad. Sci. U.S.A. 108: 14121–14126, https://doi.org/10.1073/pnas.1103621108.Search in Google Scholar PubMed PubMed Central
Davies, K.M., Blum, T.B., and Kuhlbrandt, W. (2018). Conserved in situ arrangement of complex I and III2 in mitochondrial respiratory chain supercomplexes of mammals, yeast, and plants. Proc. Natl. Acad. Sci. U.S.A. 115: 3024–3029, https://doi.org/10.1073/pnas.1720702115.Search in Google Scholar PubMed PubMed Central
Fang, H., Ye, X., Xie, J., Li, Y., Li, H., Bao, X., Yang, Y., Lin, Z., Jia, M., Han, Q., et al.. (2021). A membrane arm of mitochondrial complex I sufficient to promote respirasome formation. Cell Rep. 35: 108963, https://doi.org/10.1016/j.celrep.2021.108963.Search in Google Scholar PubMed
Fernandez-Vizarra, E., Lopez-Calcerrada, S., Formosa, L.E., Perez-Perez, R., Ding, S., Fearnley, I.M., Arenas, J., Martin, M.A., Zeviani, M., Ryan, M.T., et al.. (2021). SILAC-based complexome profiling dissects the structural organization of the human respiratory supercomplexes in SCAFI(KO) cells. Biochim. Biophys. Acta Bioenerg. 1862: 148414, https://doi.org/10.1016/j.bbabio.2021.148414.Search in Google Scholar PubMed
Friederich, M.W., Erdogan, A.J., Coughlin, C.R.2nd., Elos, M.T., Jiang, H., O’rourke, C.P., Lovell, M.A., Wartchow, E., Gowan, K., Chatfield, K.C., et al.. (2017). Mutations in the accessory subunit NDUFB10 result in isolated complex I deficiency and illustrate the critical role of intermembrane space import for complex I holoenzyme assembly. Hum. Mol. Genet. 26: 702–716, https://doi.org/10.1093/hmg/ddw431.Search in Google Scholar PubMed PubMed Central
Garcia-Poyatos, C., Cogliati, S., Calvo, E., Hernansanz-Agustin, P., Lagarrigue, S., Magni, R., Botos, M., Langa, X., Amati, F., Vazquez, J., et al.. (2020). Scaf1 promotes respiratory supercomplexes and metabolic efficiency in zebrafish. EMBO Rep. 21: e50287, https://doi.org/10.15252/embr.202050287.Search in Google Scholar PubMed PubMed Central
Gomes, L.C., Di Benedetto, G., and Scorrano, L. (2011a). During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 13: 589–598, https://doi.org/10.1038/ncb2220.Search in Google Scholar PubMed PubMed Central
Gomes, L.C., Di Benedetto, G., and Scorrano, L. (2011b). Essential amino acids and glutamine regulate induction of mitochondrial elongation during autophagy. Cell Cycle 10: 2635–2639, https://doi.org/10.4161/cc.10.16.17002.Search in Google Scholar PubMed
Gu, J., Wu, M., Guo, R., Yan, K., Lei, J., Gao, N., and Yang, M. (2016). The architecture of the mammalian respirasome. Nature 537: 639–643, https://doi.org/10.1038/nature19359.Search in Google Scholar PubMed
Guaras, A., Perales-Clemente, E., Calvo, E., Acin-Perez, R., Loureiro-Lopez, M., Pujol, C., Martinez-Carrascoso, I., Nunez, E., Garcia-Marques, F., Rodriguez-Hernandez, M.A., et al.. (2016). The CoQH2/CoQ ratio serves as a sensor of respiratory chain efficiency. Cell Rep. 15: 197–209, https://doi.org/10.1016/j.celrep.2016.03.009.Search in Google Scholar PubMed
Guerrero-Castillo, S., Baertling, F., Kownatzki, D., Wessels, H.J., Arnold, S., Brandt, U., and Nijtmans, L. (2017). The assembly pathway of mitochondrial respiratory chain complex I. Cell Metabol. 25: 128–139, https://doi.org/10.1016/j.cmet.2016.09.002.Search in Google Scholar PubMed
Guo, R., Zong, S., Wu, M., Gu, J., and Yang, M. (2017). Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell 170: 1247–1257. e12, https://doi.org/10.1016/j.cell.2017.07.050.Search in Google Scholar PubMed
Hackenbrock, C.R. (1968). Ultrastructural bases for metabolically linked mechanical activity in mitochondria. II. Electron transport-linked ultrastructural transformations in mitochondria. J. Cell Biol. 37: 345–369, https://doi.org/10.1083/jcb.37.2.345.Search in Google Scholar PubMed PubMed Central
Hartley, A.M., Lukoyanova, N., Zhang, Y., Cabrera-Orefice, A., Arnold, S., Meunier, B., Pinotsis, N., and Marechal, A. (2019). Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc1. Nat. Struct. Mol. Biol. 26: 78–83, https://doi.org/10.1038/s41594-018-0172-z.Search in Google Scholar PubMed PubMed Central
Hartley, A.M., Meunier, B., Pinotsis, N., and Marechal, A. (2020). Rcf2 revealed in cryo-EM structures of hypoxic isoforms of mature mitochondrial III-IV supercomplexes. Proc. Natl. Acad. Sci. U.S.A. 117: 9329–9337, https://doi.org/10.1073/pnas.1920612117.Search in Google Scholar PubMed PubMed Central
Jezek, P., Jaburek, M., Holendova, B., Engstova, H., and Dlaskova, A. (2023). Mitochondrial cristae morphology reflecting metabolism, superoxide formation, redox homeostasis, and pathology. Antioxid. Redox Signaling. https://doi.org/10.1089/ars.2022.0173 (Epub ahead of print).Search in Google Scholar PubMed
Jha, P., Wang, X., and Auwerx, J. (2016). Analysis of mitochondrial respiratory chain supercomplexes using blue native polyacrylamide gel electrophoresis (BN-PAGE). Curr. Protoc. Mol. Biol. 6: 1–14, https://doi.org/10.1002/9780470942390.mo150182.Search in Google Scholar PubMed PubMed Central
Kahlhöfer, F., Gansen, M., and Zickermann, V. (2021). Accessory subunits of the matrix arm of mitochondrial complex I with a focus on subunit NDUFS4 and its role in complex I function and assembly. Life 11, https://doi.org/10.3390/life11050455.Search in Google Scholar PubMed PubMed Central
knapp-Wilson, A., Pereira, G.C., Buzzard, E., Ford, H.C., Richardson, A., Corey, R.A., Neal, C., Verkade, P., Halestrap, A.P., Gold, V.A.M., et al.. (2021). Maintenance of complex I and its supercomplexes by NDUF-11 is essential for mitochondrial structure, function and health. J. Cell Sci. 134, https://doi.org/10.1242/jcs.258399.Search in Google Scholar PubMed PubMed Central
Koopman, W.J., Visch, H.J., Verkaart, S., van den Heuvel, L.W., Smeitink, J.A., and Willems, P.H. (2005). Mitochondrial network complexity and pathological decrease in complex I activity are tightly correlated in isolated human complex I deficiency. Am. J. Physiol. Cell Physiol. 289: C881–C890, https://doi.org/10.1152/ajpcell.00104.2005.Search in Google Scholar PubMed
Kravchuk, V., Petrova, O., Kampjut, D., Wojciechowska-Bason, A., Breese, Z., and Sazanov, L. (2022). A universal coupling mechanism of respiratory complex I. Nature 609: 808–814, https://doi.org/10.1038/s41586-022-05199-7.Search in Google Scholar PubMed
Krebiehl, G., Ruckerbauer, S., Burbulla, L.F., Kieper, N., Maurer, B., Waak, J., Wolburg, H., Gizatullina, Z., Gellerich, F.N., Woitalla, D., et al.. (2010). Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson’s disease-associated protein DJ-1. PLoS One 5: e9367, https://doi.org/10.1371/journal.pone.0009367.Search in Google Scholar PubMed PubMed Central
Lapuente-Brun, E., Moreno-Loshuertos, R., Acin-PereZ, R., Latorre-Pellicer, A., Colas, C., Balsa, E., Perales-Clemente, E., Quiros, P.M., Calvo, E., Rodriguez-Hernandez, M.A., et al.. (2013). Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340: 1567–1570, https://doi.org/10.1126/science.1230381.Search in Google Scholar PubMed
Letts, J.A., Fiedorczuk, K., and Sazanov, L.A. (2016). The architecture of respiratory supercomplexes. Nature 537: 644–648, https://doi.org/10.1038/nature19774.Search in Google Scholar PubMed
Letts, J.A., Fiedorczuk, K., Degliesposti, G., Skehel, M., and Sazanov, L.A. (2019). Structures of respiratory supercomplex I+III2 reveal functional and conformational crosstalk. Mol. Cell. 75: 1131–1146.e6, https://doi.org/10.1016/j.molcel.2019.07.022.Search in Google Scholar PubMed PubMed Central
Lobo-Jarne, T., Nyvltova, E., Perez-Perez, R., Timon-Gomez, A., Molinie, T., Choi, A., Mourier, A., Fontanesi, F., Ugalde, C., and Barrientos, A. (2018). Human COX7A2L regulates complex III biogenesis and promotes supercomplex organization remodeling without affecting mitochondrial bioenergetics. Cell Rep. 25: 1786–1799.e4, https://doi.org/10.1016/j.celrep.2018.10.058.Search in Google Scholar PubMed PubMed Central
Lopez-Fabuel, I., Le Douce, J., Logan, A., James, A.M., Bonvento, G., Murphy, M.P., Almeida, A., and Bolanos, J.P. (2016). Complex I assembly into supercomplexes determines differential mitochondrial ROS production in neurons and astrocytes. Proc. Natl. Acad. Sci. U.S.A. 113: 13063–13068, https://doi.org/10.1073/pnas.1613701113.Search in Google Scholar PubMed PubMed Central
Lowe, M., Kalacheva, M., Boersma, A.J., and Kedrov, A. (2020). The more the merrier: effects of macromolecular crowding on the structure and dynamics of biological membranes. FEBS J. 287: 5039–5067, https://doi.org/10.1111/febs.15429.Search in Google Scholar PubMed
Maekawa, S., Takada, S., Furihata, T., Fukushima, A., Yokota, T., and Kinugawa, S. (2020). Mitochondrial respiration of complex II is not lower than that of complex I in mouse skeletal muscle. Biochem. Biophys. Rep. 21: 100717, https://doi.org/10.1016/j.bbrep.2019.100717.Search in Google Scholar PubMed PubMed Central
Maldonado, M., Guo, F., and Letts, J.A. (2021). Atomic structures of respiratory complex III(2), complex IV, and supercomplex III(2)-IV from vascular plants. Elife 10, https://doi.org/10.7554/elife.62047.Search in Google Scholar PubMed PubMed Central
Martinez-Reyes, I. and Chandel, N.S. (2020). Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 11: 102, https://doi.org/10.1038/s41467-019-13668-3.Search in Google Scholar PubMed PubMed Central
Melber, A. and Winge, D.R. (2016). Inner secrets of the respirasome. Cell 167: 1450–1452, https://doi.org/10.1016/j.cell.2016.11.025.Search in Google Scholar PubMed PubMed Central
Milenkovic, D., Blaza, J.N., Larsson, N.G., and Hirst, J. (2017). The enigma of the respiratory chain supercomplex. Cell Metabol. 25: 765–776, https://doi.org/10.1016/j.cmet.2017.03.009.Search in Google Scholar PubMed
Moreno-Lastres, D., Fontanesi, F., Garcia-Consuegra, I., Martin, M.A., Arenas, J., Barrientos, A., and Ugalde, C. (2012). Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metabol. 15: 324–335, https://doi.org/10.1016/j.cmet.2012.01.015.Search in Google Scholar PubMed PubMed Central
Neupert, W. and Herrmann, J.M. (2007). Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76: 723–749, https://doi.org/10.1146/annurev.biochem.76.052705.163409.Search in Google Scholar PubMed
Nishimura, K., Fukuda, A., and Hisatake, K. (2019). Mechanisms of the metabolic shift during somatic cell reprogramming. Int. J. Mol. Sci. 20, https://doi.org/10.3390/ijms20092254.Search in Google Scholar PubMed PubMed Central
Patron, M., Sprenger, H.G., and Langer, T. (2018). m-AAA proteases, mitochondrial calcium homeostasis and neurodegeneration. Cell Res. 28: 296–306, https://doi.org/10.1038/cr.2018.17.Search in Google Scholar PubMed PubMed Central
Patten, D.A., Wong, J., Khacho, M., Soubannier, V., Mailloux, R.J., Pilon-Larose, K., Maclaurin, J.G., Park, D.S., Mcbride, H.M., Trinkle-Mulcahy, L., et al.. (2014). OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J. 33: 2676–2691, https://doi.org/10.15252/embj.201488349.Search in Google Scholar PubMed PubMed Central
Perez-Perez, R., Lobo-Jarne, T., Milenkovic, D., Mourier, A., Bratic, A., Garcia-Bartolome, A., Fernandez-Vizarra, E., Cadenas, S., Delmiro, A., Garcia-Consuegra, I., et al.. (2016). COX7A2L is a mitochondrial complex iii binding protein that stabilizes the III2+IV supercomplex without affecting respirasome formation. Cell Rep. 16: 2387–2398, https://doi.org/10.1016/j.celrep.2016.07.081.Search in Google Scholar PubMed PubMed Central
Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D.J., Inuganti, A., Griss, J., Mayer, G., Eisenacher, M., et al.. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47: D442–D450, https://doi.org/10.1093/nar/gky1106.Search in Google Scholar PubMed PubMed Central
Protasoni, M., Perez-Perez, R., Lobo-Jarne, T., Harbour, M.E., Ding, S., Penas, A., Diaz, F., Moraes, C.T., Fearnley, I.M., Zeviani, M., et al.. (2020). Respiratory supercomplexes act as a platform for complex III-mediated maturation of human mitochondrial complexes I and IV. EMBO J. 39: e102817, https://doi.org/10.15252/embj.2019102817.Search in Google Scholar PubMed PubMed Central
Rappsilber, J. and Ishihama, M. (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2: 1896–1906, https://doi.org/10.1038/nprot.2007.261. 17703201.Search in Google Scholar PubMed
Rathore, S., Berndtsson, J., Marin-Buera, L., Conrad, J., Carroni, M., Brzezinski, P., and Ott, M. (2019). Cryo-EM structure of the yeast respiratory supercomplex. Nat. Struct. Mol. Biol. 26: 50–57, https://doi.org/10.1038/s41594-018-0169-7.Search in Google Scholar PubMed
Reinson, K., Kovacs-Nagy, R., Oiglane-Shlik, E., Pajusalu, S., Noukas, M., Wintjes, L.T., Van Den Brandt, F.C.A., Brink, M., Acker, T., Ahting, U., et al.. (2019). Diverse phenotype in patients with complex I deficiency due to mutations in NDUFB11. Eur. J. Med. Genet. 62: 103572, https://doi.org/10.1016/j.ejmg.2018.11.006.Search in Google Scholar PubMed
Ritchie, M.E., Phipson, B., Wu, D., Hu, Y., Law, C.W., Shi, W., and Smyth, G.K. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47, https://doi.org/10.1093/nar/gkv007.Search in Google Scholar PubMed PubMed Central
Salewskij, K., Rieger, B., Hager, F., Arroum, T., Duwe, P., Villalta, J., Colgiati, S., Richter, C.P., Psathaki, O.E., Enriquez, J.A., et al.. (2020). The spatio-temporal organization of mitochondrial F1FO ATP synthase in cristae depends on its activity mode. Biochim. Biophys. Acta Bioenerg. 1861: 148091, https://doi.org/10.1016/j.bbabio.2019.148091.Search in Google Scholar PubMed
Schagger, H. and Pfeiffer, K. (2000). Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19: 1777–1783, https://doi.org/10.1093/emboj/19.8.1777.Search in Google Scholar PubMed PubMed Central
Schwerzmann, K., Cruz-Orive, L.M., Eggman, R., Sanger, A., and Weibel, E.R. (1986). Molecular architecture of the inner membrane of mitochondria from rat liver: a combined biochemical and stereological study. J. Cell Biol. 102: 97–103, https://doi.org/10.1083/jcb.102.1.97.Search in Google Scholar PubMed PubMed Central
Shah, A.D., Goode, R.J.A., Huang, C., Powell, D.R., and Schittenhelm, R.B. (2020). LFQ-analyst: an easy-to-use interactive web platform to analyze and visualize label-free proteomics data preprocessed with MaxQuant. J. Proteome Res. 19: 204–211, https://doi.org/10.1021/acs.jproteome.9b00496.Search in Google Scholar PubMed
Skvortsova, E.V., Nazarov, I.B., Tomilin, A.N., and Sinenko, S.A. (2022). Dual mode of mitochondrial ROS action during reprogramming to pluripotency. Int. J. Mol. Sci. 23, https://doi.org/10.3390/ijms231810924.Search in Google Scholar PubMed PubMed Central
Song, Z., Chen, H., Fiket, M., Alexander, C., and Chan, D.C. (2007). OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 178: 749–755, https://doi.org/10.1083/jcb.200704110.Search in Google Scholar PubMed PubMed Central
Stroud, D.A., Surgenor, E.E., Formosa, L.E., Reljic, B., Frazier, A.E., Dibley, M.G., Osellame, L.D., Stait, T., Beilharz, T.H., Thorburn, D.R., et al.. (2016). Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature 538: 123–126, https://doi.org/10.1038/nature19754.Search in Google Scholar PubMed
Suthammarak, W., Yang, Y.Y., Morgan, P.G., and Sedensky, M.M. (2009). Complex I function is defective in complex IV-deficient Caenorhabditis elegans. J. Biol. Chem. 284: 6425–6435, https://doi.org/10.1074/jbc.m805733200.Search in Google Scholar
Vercellino, I. and Sazanov, L.A. (2021). Structure and assembly of the mammalian mitochondrial supercomplex CIII(2)CIV. Nature 598: 364–367, https://doi.org/10.1038/s41586-021-03927-z.Search in Google Scholar PubMed
Wisniewski, J.R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Method. 6: 359–362, https://doi.org/10.1038/nmeth.1322. 19377485.Search in Google Scholar PubMed
Wittig, I., Carrozzo, R., Santorelli, F.M., and Schagger, H. (2006). Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim. Biophys. Acta 1757: 1066–1072, https://doi.org/10.1016/j.bbabio.2006.05.006.Search in Google Scholar PubMed
Zickermann, V., Bostina, M., Hunte, C., Ruiz, T., Radermacher, M., and Brandt, U. (2003). Functional implications from an unexpected position of the 49-kDa subunit of NADH:ubiquinone oxidoreductase. J. Biol. Chem. 278: 29072–29078, https://doi.org/10.1074/jbc.m302713200.Search in Google Scholar
Zong, S., Wu, M., Gu, J., Liu, T., Guo, R., and Yang, M. (2018). Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 28: 1026–1034, https://doi.org/10.1038/s41422-018-0071-1.Search in Google Scholar PubMed PubMed Central
Supplementary Material
This article contains supplementary material (https://doi.org/10.1515/hsz-2022-0309).
© 2023 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Highlight: Physiology and Dynamics of Cellular Microcompartments
- Highlight: on the past and the future of cellular microcompartments
- Nuclear redox processes in land plant development and stress adaptation
- The readily retrievable pool of synaptic vesicles
- Loss of respiratory complex I subunit NDUFB10 affects complex I assembly and supercomplex formation
- Modulation of self-organizing circuits at deforming membranes by intracellular and extracellular factors
- Computational resolution in single molecule localization – impact of noise level and emitter density
- Setting up a data management infrastructure for bioimaging
- Molecular insights into endolysosomal microcompartment formation and maintenance
- The role of lysosomes in lipid homeostasis
- Membrane damage and repair: a thin line between life and death
- Neuronal stress granules as dynamic microcompartments: current concepts and open questions
- Molecular determinants of protein half-life in chloroplasts with focus on the Clp protease system
- Neprilysin 4: an essential peptidase with multifaceted physiological relevance
- Determinants of synergistic cell-cell interactions in bacteria
- Drosophila collagens in specialised extracellular matrices
Articles in the same Issue
- Frontmatter
- Highlight: Physiology and Dynamics of Cellular Microcompartments
- Highlight: on the past and the future of cellular microcompartments
- Nuclear redox processes in land plant development and stress adaptation
- The readily retrievable pool of synaptic vesicles
- Loss of respiratory complex I subunit NDUFB10 affects complex I assembly and supercomplex formation
- Modulation of self-organizing circuits at deforming membranes by intracellular and extracellular factors
- Computational resolution in single molecule localization – impact of noise level and emitter density
- Setting up a data management infrastructure for bioimaging
- Molecular insights into endolysosomal microcompartment formation and maintenance
- The role of lysosomes in lipid homeostasis
- Membrane damage and repair: a thin line between life and death
- Neuronal stress granules as dynamic microcompartments: current concepts and open questions
- Molecular determinants of protein half-life in chloroplasts with focus on the Clp protease system
- Neprilysin 4: an essential peptidase with multifaceted physiological relevance
- Determinants of synergistic cell-cell interactions in bacteria
- Drosophila collagens in specialised extracellular matrices

