Abstract
DEAD-box helicases catalyze RNA duplex unwinding in an ATP-dependent reaction. Members of the DEAD-box helicase family consist of a common helicase core formed by two RecA-like domains. According to the current mechanistic model for DEAD-box mediated RNA unwinding, binding of RNA and ATP triggers a conformational change of the helicase core, and leads to formation of a compact, closed state. In the closed conformation, the two parts of the active site for ATP hydrolysis and of the RNA binding site, residing on the two RecA domains, become aligned. Closing of the helicase core is coupled to a deformation of the RNA backbone and destabilization of the RNA duplex, allowing for dissociation of one of the strands. The second strand remains bound to the helicase core until ATP hydrolysis and product release lead to re-opening of the core. The concomitant disruption of the RNA binding site causes dissociation of the second strand. The activity of the helicase core can be modulated by interaction partners, and by flanking N- and C-terminal domains. A number of C-terminal flanking regions have been implicated in RNA binding: RNA recognition motifs (RRM) typically mediate sequence-specific RNA binding, whereas positively charged, unstructured regions provide binding sites for structured RNA, without sequence-specificity. Interaction partners modulate RNA binding to the core, or bind to RNA regions emanating from the core. The functional interplay of the helicase core and ancillary domains or interaction partners in RNA binding and unwinding is not entirely understood. This review summarizes our current knowledge on RNA binding to the DEAD-box helicase core and the roles of ancillary domains and interaction partners in RNA binding and unwinding by DEAD-box proteins.
Introduction: DEAD-box helicases and RNA unwinding
DEAD-box helicases are the largest family of RNA helicases. They locally destabilize RNA duplexes in an ATP-dependent manner and play central roles in all processes that involve RNA. DEAD-box helicases are involved in transcription, translation, mRNA splicing, RNA transport and RNA decay. Malfunction of DEAD-box proteins is frequently associated with cancer and other malignancies (reviewed in Steimer and Klostermeier, 2012).
The DEAD-box helicase core, a ∼400 amino acid region shared by all members of the family, carries a set of conserved motifs that mediate ATP binding and hydrolysis, RNA binding, and local RNA unwinding (Figure 1A,B). The helicase core is formed by two RecA-like domains (RecA_N, RecA_C) that are connected by a flexible linker region with little conservation in sequence. The helicase core is the functional unit that provides RNA-dependent ATPase- and ATP-dependent RNA unwinding activities (Richter et al., 1999; Rogers et al., 1999). The eukaryotic translation initiation factor eIF4A is a minimal DEAD-box helicase that consists of a helicase core only, and is regarded as a prototypical representative of the DEAD-box family (Linder et al., 1989; Rogers et al., 2002; reviewed in Andreou and Klostermeier, 2012a,b). In most DEAD-box proteins, the helicase core is flanked by N- and C-terminal regions (Figure 1A) that modulate the activity of the core.
![Figure 1: DEAD-box helicase architecture and unwinding mechanism.(A) Domain structure of DEAD-box helicases. The eukaryotic translation initiation factor eIF4A is a minimal DEAD-box helicase that consists of a helicase core only, formed by two RecA-like domains (RecA_N, RecA_C). The helicase core contains the conserved motifs Q, I, Ia, Ib, Ic, II, III, IV, IVa, V, Va and VI that mediate ATP binding and hydrolysis, RNA binding, and RNA unwinding. In most DEAD-box proteins, the helicase core is flanked by N- and/or C-terminal regions. In B. subtilis YxiN, the canonical helicase core is followed by an RNA binding domain (RBD). T. thermophilus Hera carries a C-terminal dimerization domain (DD) between the core and the RBD. In the S. cerevisiae splicing helicase Mss116, the helicase core is flanked by an unstructured NTE, and an α-helical CTE, followed by a basic C-tail. (B) Sequence and position of the conserved helicase motifs in the DEAD-box proteins discussed in this work. Dme: Drosophila melanogaster, Hsa: Homo sapiens, Sce: Saccharomyces cerevisiae. (C) Conformational changes in the catalytic cycle of the helicase core lead to RNA unwinding. ATP and RNA binding to the helicase core in an open conformation (dark gray) leads to closure of the cleft between the RecA domains and formation of the closed state of the helicase core (light gray). Cleft closure is coupled to RNA deformation and local duplex destabilization. Rearrangement leads to the formation of a hydrolysis- and unwinding-competent state (star), from which the first strand (red) of the RNA duplex dissociates. The helicase core remains in the closed state during ATP hydrolysis. Re-opening of the inter-domain cleft occurs upon phosphate release (red circle). The disruption of the RNA binding site formed by both RecA domains leads to release of the second strand of the duplex (black). After ADP release, the helicase core is ready for subsequent catalytic cycles. Modified after Harms et al. (2014) [Harms, U., Andreou, A.Z., Gubaev, A., and Klostermeier, D. (2014). eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res. 42 (12), 7911–7922] with permission.](/document/doi/10.1515/hsz-2014-0277/asset/graphic/j_hsz-2014-0277_fig_001.jpg)
DEAD-box helicase architecture and unwinding mechanism.
(A) Domain structure of DEAD-box helicases. The eukaryotic translation initiation factor eIF4A is a minimal DEAD-box helicase that consists of a helicase core only, formed by two RecA-like domains (RecA_N, RecA_C). The helicase core contains the conserved motifs Q, I, Ia, Ib, Ic, II, III, IV, IVa, V, Va and VI that mediate ATP binding and hydrolysis, RNA binding, and RNA unwinding. In most DEAD-box proteins, the helicase core is flanked by N- and/or C-terminal regions. In B. subtilis YxiN, the canonical helicase core is followed by an RNA binding domain (RBD). T. thermophilus Hera carries a C-terminal dimerization domain (DD) between the core and the RBD. In the S. cerevisiae splicing helicase Mss116, the helicase core is flanked by an unstructured NTE, and an α-helical CTE, followed by a basic C-tail. (B) Sequence and position of the conserved helicase motifs in the DEAD-box proteins discussed in this work. Dme: Drosophila melanogaster, Hsa: Homo sapiens, Sce: Saccharomyces cerevisiae. (C) Conformational changes in the catalytic cycle of the helicase core lead to RNA unwinding. ATP and RNA binding to the helicase core in an open conformation (dark gray) leads to closure of the cleft between the RecA domains and formation of the closed state of the helicase core (light gray). Cleft closure is coupled to RNA deformation and local duplex destabilization. Rearrangement leads to the formation of a hydrolysis- and unwinding-competent state (star), from which the first strand (red) of the RNA duplex dissociates. The helicase core remains in the closed state during ATP hydrolysis. Re-opening of the inter-domain cleft occurs upon phosphate release (red circle). The disruption of the RNA binding site formed by both RecA domains leads to release of the second strand of the duplex (black). After ADP release, the helicase core is ready for subsequent catalytic cycles. Modified after Harms et al. (2014) [Harms, U., Andreou, A.Z., Gubaev, A., and Klostermeier, D. (2014). eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res. 42 (12), 7911–7922] with permission.
In the absence of nucleotide or RNA, the two RecA domains of the helicase core are splayed apart and do not contact each other. A large variety of juxtapositions of the two RecA domains has been captured in crystal structures of different DEAD-box helicases cores (Caruthers et al., 2000; Story et al., 2001; Cheng et al., 2005; Andersen et al., 2006; Hogbom et al., 2007; Klostermeier 2013; Mathys et al., 2014; Mohlmann et al., 2014). By contrast, solution studies support a more homogenous, less widely open conformation (Wang et al., 2007; Theissen et al., 2008; Hilbert et al., 2011; Mallam et al., 2011). Binding of ATP and RNA to DEAD-box proteins is accompanied by a conformational change of the helicase core to the closed state (Sengoku et al., 2006; Theissen et al., 2008; Aregger and Klostermeier, 2009; Andreou and Klostermeier, 2014; Harms et al., 2014). In this closed conformation, the two RecA domains form an extensive inter-domain interface. The nucleotide is bound at the interface of the two domains, and contacted mostly by RecA_N, although ATP hydrolysis requires interaction of the nucleotide with both RecA domains. The Q-motif of RecA_N (Tanner et al., 2003) provides specificity for adenine nucleotides by hydrogen bonding to the Watson-Crick face of adenine. Motifs I, II, and VI also contribute to nucleotide binding. An arginine from motif VI acts as an arginine finger (Ahmadian et al., 1997; Sengoku et al., 2006) that is involved ATP hydrolysis (Elles and Uhlenbeck, 2008). The RNA binding site extends over both RecA domains, perpendicular to the inter-domain interface (Figures 1C and 2). The RNA is contacted by amino acids from motifs Ia, Ib, Ic, IV, IVa, and V (Figure 2B). In addition to interactions with the bound nucleotide and RNA, a network of interactions between the conserved helicase motifs is established in the closed conformation (Andersen et al., 2006; Bono et al., 2006; Sengoku et al., 2006; Del Campo and Lambowitz, 2009; von Moeller et al., 2009; Montpetit et al., 2011).
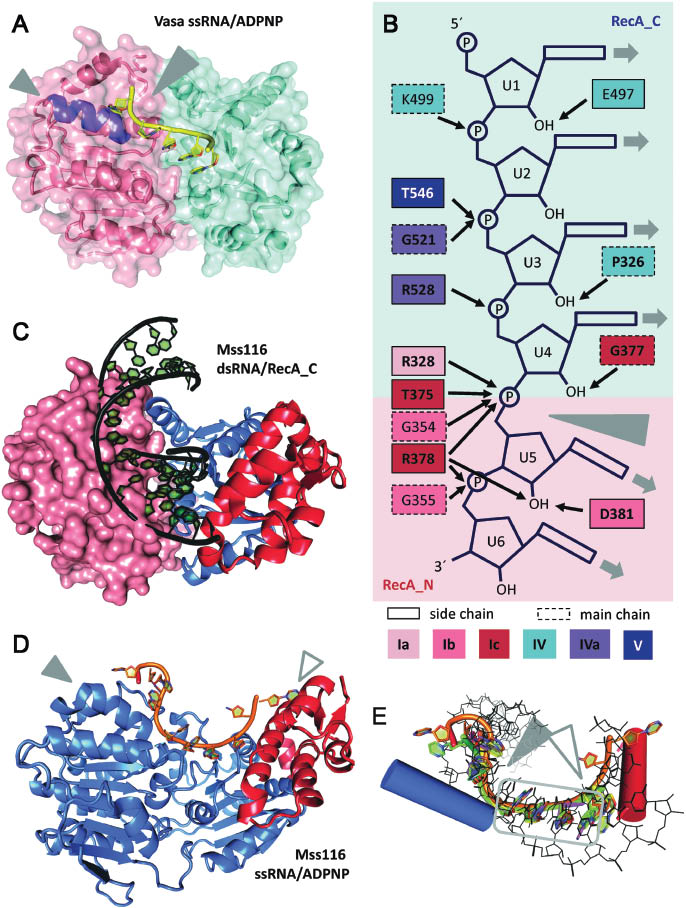
RNA binding to the helicase core.
(A) Structure of the D. melanogaster DEAD-box helicase Vasa in complex with single-stranded RNA and ADPNP (Sengoku et al., 2006; PDB-ID 2db3). The helicase core adopts a closed conformation, with tight interactions between the two RecA domains (Rec_N: salmon, RecA_C: cyan). The bound RNA (yellow) is bent (large gray arrowhead) because of the steric hindrance with the ‘wedge’ helix (purple, small gray arrowhead). The interactions between Vasa and the RNA are detailed in panel B. (B) Schematic depiction of interactions between amino acids from conserved helicase motifs in Vasa and the central six nucleotides of the bound ssRNA. Amino acids are colored according to the corresponding conserved motifs in red (RecA_N) and blue hues (RecA_C). A dotted outline indicates main chain interactions, the other amino acids bind the RNA via side chain interactions. Conserved amino acids are shown in bold. The gray arrowhead marks the kink in the RNA caused by the ‘wedge’ helix. The uridine bases of the bound RNA are solvent accessible, and in principle available for base-pairing (gray arrows). (C) Structure of a Mss116 fragment comprising RecA_C (blue) and the CTE (red) in complex with a 14 bp dsRNA (black; Mallam et al., 2012; PDB-ID 4db2). The modeled position of a RecA_N domain (salmon) in the closed conformation is indicated in surface representation. The dsRNA and RecA_N in the closed helicase core overlap, indicating that structural rearrangements of the RNA have to occur upon closure of the helicase core. The same figure with a surface representation of RecA_C, colored according to the electrostatic potential, is shown in Supplementary Figure S1. (D) Structure of Mss116 (helicase core: blue, CTE: red) in complex with ssRNA (orange) and ADPNP (not shown; Mohr et al., 2011; PDB-ID 3i5). The RNA bound to Mss116 is kinked twice, once caused by the ‘wedge’ helix (filled arrowhead), and a second time because of steric hindrance with the CTE (open arrowhead). (E) Superposition of ssRNA (yellow: Vasa, PDB-ID 2db3; orange: Mss116, PDB-ID 3i5x; magenta: eIF4A-III, PDB-ID 3ex7; green: DDX19, PDB-ID 3g0h) and dsRNA (black, Mss116, PDB-ID 4db2) bound to DEAD-box helicase cores. The blue cylinder marks the position of the ‘wedge’ helix in RecA_N that causes a bend in the RNA backbone (filled arrowhead). The red cylinder indicates the position of the helix in the Mss116 CTE that causes the second kink (open arrowhead). The gray box highlights the four central nucleotides preceding the kink that are bound identically in all DEAD-box structures known to date.
The conformational change of the helicase core is directly linked to RNA unwinding. Single-stranded RNA bound to the closed conformation is bent, which is incompatible with RNA duplex geometry. The introduction of such a bend into one strand of a double-stranded RNA substrate will lead to local destabilization of the duplex (Sengoku et al., 2006). Mutations of residues at the interface of the two RecA domains that interfere with the conformational change of the helicase core uncouple ATP hydrolysis and RNA unwinding (Sengoku et al., 2006; Karow and Klostermeier, 2009).
In the current model of RNA unwinding by DEAD-box helicases, a nucleotide-dependent conformational cycle of the helicase core drives duplex destabilization and strand separation (Figure 1C). As a first step, cooperative binding of ATP and (ds)RNA to the helicase core triggers the transition from an open to the closed state. A structural rearrangement leads to the formation of an active, unwinding-competent state (Talavera and De La Cruz, 2005). The RNA backbone of one of the strands of the RNA duplex is distorted, leading to the disruption of a few base pairs and local destabilization of the duplex. At this stage of the catalytic cycle, the first strand of the RNA duplex dissociates (Sengoku et al., 2006), and is free to engage in interactions with other partners. The second strand of the RNA remains bound to the helicase core and bent, and is thus not available for re-binding. Release of the two strands of the duplex at different times prevents re-annealing and futile cycles. After ATP hydrolysis and phosphate release, re-opening of the helicase core (Theissen et al., 2008; Aregger and Klostermeier, 2009) allows for release of the second RNA strand, completing duplex separation and resetting the helicase for further catalytic cycles (reviewed in Hilbert et al., 2009; Linder and Jankowsky, 2011; Andreou and Klostermeier, 2012a,b; Henn et al., 2012; Putnam and Jankowsky, 2013).
RNA binding to the helicase core
The first glimpse on RNA binding to the DEAD-box helicase core came from the crystal structure of the Drosophila DEAD-box protein Vasa, bound to a single-stranded U10 RNA and the non-hydrolyzable ATP-analog 2′/3′-O-(N-methyl-anthraniloyl)-adenosine-5′-[(β,γ)-imido] triphosphate (ADPNP; Sengoku et al., 2006). Vasa is a positive translational regulator for genes involved in embryonic patterning and germ line differentiation (reviewed in Raz, 2000). With the structure of the Vasa/ADPNP/RNA complex, the closed state of the DEAD-box helicase core has been visualized for the first time. The structure reveals the binding sites for nucleotide and RNA on the helicase core, the polarity of the bound RNA, and the molecular details of nucleotide and ssRNA recognition (Figure 2A,B). It rationalizes the specificity of DEAD-box proteins for RNA, but the lack of sequence-specificity, and reveals a network of interactions that inter-connects the conserved motifs. Subsequently reported structures of the DEAD-box proteins eIF4A-III (Andersen et al., 2006; Bono et al., 2006), DDX19/Dbp5 (Collins et al., 2009; von Moeller et al., 2009; Montpetit et al., 2011), and Mss116 (Del Campo and Lambowitz, 2009) in complex with single-stranded RNAs and non-hydrolyzable ATP analogs (summarized in Table 1) confirmed these common denominators in RNA and nucleotide recognition by DEAD-box proteins (Figure 2D,E). eIF4A-III is a component of the exon junction complex involved in mRNA transport, translation and quality control (Andersen et al., 2006; Bono et al., 2006; reviewed in Tange et al., 2004), DDX19/Dbp5 is involved in nuclear export of mRNAs (Collins et al., 2009; von Moeller et al., 2009; Montpetit et al., 2011), and Mss116 is a general RNA chaperone involved in group I and II intron splicing (Del Campo and Lambowitz, 2009). Strikingly, the closed state of the prototypical DEAD-box helicase eIF4A has not been captured until recently. According to single molecule FRET data, the closed state of eIF4A is less stabilized by RNA and ATP binding than for other DEAD-box proteins, and its population requires the additional presence of other translation initiation factors (Andreou and Klostermeier, 2014; Harms et al., 2014). In this section, we will first summarize our current knowledge on RNA binding to the helicase core. The contributions of flanking domains and interaction partners on RNA binding to DEAD-box proteins, and the implications for RNA unwinding are discussed in the subsequent sections.
Crystal structures of the closed state of the DEAD-box protein helicase core in complex with ssRNA and a non-hydrolyzable nucleotide analog.
| DEAD-box protein | nucleotide | RNAa | PDB-ID/resolution (Å) | Reference |
|---|---|---|---|---|
| Vasa | ADPNP | U10 (U7) | 2db3/2.2 | Sengoku et al. (2006) |
| eIF4A-III | ADPNP | U15 (U6) | 2j0sb/2.21 | Bono et al. (2006) |
| ADPNP | poly-Uc (U6) | 2hyib/2.3 | Andersen et al. (2006) | |
| ADPNP | U15 (U8) | 2xb2d/3.4 | Buchwald et al. (2010) | |
| ADP·AlFx | poly-Uc (U6) | 3ex7b/2.3 | Nielsen et al. (2008) | |
| Dbp5/DDX19 | ADPNP | U10 (U6) | 3fht/2.2 | von Moeller et al. (2009) |
| ADPNP | U10 (U7) | 3g0h/2.7 | Collins et al. (2009) | |
| ADP·BeFx | U6 (U5) | 3pey/1.4 | Montpetit et al. 2011) | |
| Mss116 | ADPNP | U10 (U10) | 3i5xe/1.9, 3sqwf/1.91, 3sqxg/2.11 | Del Campo and Lambowitz (2009) |
| ADP·AlFx | U10 (U10) | 3i62e/1.95 | Del Campo and Lambowitz (2009) | |
| ADP·BeFx | U10 (U10) | 3i61e/2.1 | Del Campo and Lambowitz (2009) |
aThe number in brackets refers to nucleotides included in the final model.
beIF4A-III in complex with Mago/Y14, Btz (MLN51).
cInitial complexes formed with poly-U RNA, trimmed by RNase A treatment.
deIF4A-III in complex with Mago/Y14, Btz (MLN51), Ufp3b.
e3ix5, 3i61, 3i62: Mss116 (aa 37–597), lacking the mitochondrial targeting sequence and the basic C-tail.
f3sqw: Mss116 deletion construct (aa 88–664) lacking the mitochondrial targeting sequence and the NTE.
g3sqx: Mss116 deletion construct (aa 88–597) lacking the mitochondrial targeting sequence, the NTE and the basic C-tail).
Binding of ssRNA by the helicase core: an enzyme-product complex?
In the structure of Vasa in complex with ssRNA and ADPNP, seven nucleotides (U1–U7) of the U10 RNA used for crystallization are visible in the electron density (Sengoku et al., 2006). The RNA binding site follows a path perpendicular to the inter-domain cleft. The 5′-region of the RNA is bound by RecA_C, the 3′-end contacts RecA_N. Nucleotides U1–U5 interact with motifs IV, IVa, and V of RecA_C, and U6 and U7 form contacts with residues from motifs Ia, Ib, and Ic of RecA_N (Figure 2B). Notably, the central nucleotides U4 and U5 interact with both RecA domains. Four 2′-OH groups (U2, U4-U6) are engaged in hydrogen bonds to the protein (Figure 2B), rationalizing the specificity of DEAD-box proteins for RNA (Rogers et al., 2001). The phosphate groups are bound by a number of polar interactions. All bases of the single-stranded RNA are facing the solvent, and are thus in principle accessible for base-pairing and duplex formation. The bases of U1–U5 are continuously stacked. Stacking is interrupted between U5 and U6, and the conformation of the phosphate-ribose backbone in this region causes a sharp bend between U5 and U6. The bend is imposed by a ‘wedge’ helix (helix α7) in RecA_N that contains the conserved motif Ib and blocks a continuous path of the RNA backbone, diverting the RNA backbone downstream of the kink (Figure 2A,D,E). The bases U6 and U7 are again stacked in the Vasa complex (although not in all structures of DEAD-box proteins bound to ssRNA, Figure 2E).
The bending point is extensively contacted by main chain atoms of Gly354 (motif Ib) and Arg378 (motif Ic), and by the side chains of Arg328 (motif Ia) and Thr375 (motif Ic). An inter-domain interaction network involving Gln525 and Arg528 (motif IVa), and Arg328 and Glu329 (motif Ia) may help stabilize the closed conformation and the position of the bend. The bend in the RNA backbone is not compatible with continuous base-pairing within an A-form RNA duplex. Thus, the base pairs located 5′ or 3′ of the bend will be disrupted when one strand of an RNA duplex is bound to a DEAD-box protein. New interactions of the bound RNA strand with residues of the helicase core would compensate for the energetically unfavorable disruption of base pairs (Sengoku et al., 2006).
Except for Arg403 (interacting with U3 in Vasa, but not in all DEAD-box proteins) and Glu497 (interacting with the 2′-OH of U1 in Vasa, Figure 2B), all residues contacting the RNA are conserved among DEAD-box proteins. A similar set of contacts between residues of the helicase core and single-stranded RNA has been observed in the crystal structures of the DEAD-box proteins eIF4A-III (Andersen et al., 2006; Bono et al., 2006), DDX19/Dbp5 (Collins et al., 2009; von Moeller et al., 2009; Montpetit et al., 2011), and Mss116 (Del Campo and Lambowitz, 2009, Figure 2B). The bend imposed by the ‘wedge’ helix is present in the same position in all of these DEAD-box proteins (Figure 2E; between U4 and U5 in eIF4A-III and DDX19/Dbp5, U6 and U7 in Mss116). The position of the ‘wedge’ helix is also conserved among DEAD-box proteins, but does not superimpose with the corresponding helix in other helicase families (Sengoku et al., 2006). The ‘wedge’ helix therefore seems to be a specific and mechanistically important element of DEAD-box proteins. The common structural features of the DEAD-box helicase core imply that members of this family couple conformational changes to duplex destabilization and strand separation by a common mechanism.
ATP hydrolysis is not necessary for duplex separation (Liu et al., 2008). Structural studies of DEAD-box proteins in complex with ssRNA and the non-hydrolyzable ATP analogs ADPNP (Andersen et al., 2006; Bono et al., 2006; Theissen et al., 2008; Aregger and Klostermeier, 2009; Collins et al., 2009; Del Campo and Lambowitz, 2009; von Moeller et al., 2009; Montpetit et al., 2011), and ADP·BeFx (Theissen et al., 2008; Aregger and Klostermeier, 2009; Del Campo and Lambowitz, 2009; Montpetit et al., 2011), and with the transition state analogs ADP·AlFx (Nielsen et al., 2008; Del Campo and Lambowitz, 2009) and ADP·MgFx (Theissen et al., 2008; Aregger and Klostermeier, 2009) have revealed similar conformations and interaction patterns with bound ssRNA. The structures of DEAD-box helicase cores with non-hydrolyzable analogs and ssRNA therefore most likely reflect the product state, after dissociation of the first strand of the duplex, but before ATP hydrolysis and product dissociation.
Binding of dsRNA to the helicase core: an enzyme-substrate complex
The ssRNA bound to the DEAD-box helicase core could in principle form a duplex by base-pairing with a second RNA strand before or after the kink, rationalizing the capacity of the helicase core to bind dsRNA. The only structural information on binding of dsRNA to DEAD-box proteins at high resolution comes from a crystal structure of the isolated RecA_C and C-terminal extension (CTE) domains of Mss116 in complex with a 14 bp dsRNA (Mallam et al., 2012; Figure 2C). The structure reveals a positively charged binding pocket for the RNA duplex on RecA_C (Supplementary Figure S1). Amino acids from motifs IV, IVa, V, and Va form extensive interactions with one strand of the bound duplex (Mallam et al., 2012; Figure 2C). In contrast, the second RNA strand is not contacted by the helicase core, but is only engaged in a few hydrogen bonds between hydroxyl groups and residues from the CTE. The nucleobases are base-paired, and not available for interactions with the helicase core. Mss116 forms similar interactions with an RNA/RNA duplex and with the DNA strand of an A-form RNA/DNA hybrid (Mallam et al., 2012). DEAD-box helicases can unwind DNA duplexes that contain only a few consecutive ribonucleotides (Yang et al., 2007), suggesting that they recognize the A-form-specific backbone geometry of their substrates.
It has been proposed that the structure of RecA_C bound to the RNA duplex reflects the initial complex upon binding of duplex RNA to the open helicase core (Mallam et al., 2012). The overall structure of RecA_C in complex with an RNA duplex (Mallam et al., 2012) and as part of the helicase core bound to single-stranded RNA and ADPNP (Del Campo and Lambowitz, 2009) is identical. However, the path of the RNA duplex emanating from RecA_C is sterically not compatible with the position of the RecA_N domain in the closed state of Mss116 (Mallam et al., 2012; Figure 2C). The movement of RecA_N towards RecA_C upon closing of the helicase core would cause a deformation of the duplex bound to RecA_C, and lead to duplex destabilization and disruption (Mallam et al., 2012).
Coupling of nucleotide and RNA binding with conformational changes causes duplex separation
The active site for ATP hydrolysis and the RNA binding site are formed by both RecA domains of the helicase core, rationalizing the positive thermodynamic linkage between nucleotide and RNA binding, and the RNA-dependent ATPase activity of DEAD-box proteins. Positive thermodynamic linkage of (ss)RNA and nucleotide binding has been reported for a number of DEAD-box proteins, although the degree of cooperativity varies (reviewed in Hilbert et al., 2009; Henn et al., 2012; Putnam and Jankowsky, 2013). Nevertheless, the isolated RecA_N binds nucleotides independently (Mallam et al., 2012; Samatanga and Klostermeier, 2014), exemplified by several crystal structures of isolated RecA_N domains in complex with nucleotide (Rudolph et al., 2006; Hogbom et al., 2007; Schutz et al., 2010; Strohmeier et al., 2011), and by structures of the helicase core in the open conformation with nucleotide (AMP, ADP or ADPNP) bound exclusively to RecA_N via the Q-motif and motifs I and II (Hogbom et al., 2007; Schutz et al., 2008; Collins et al., 2009; Klostermeier, 2013; Jacewicz et al., 2014). The structure of the Mss116 RecA_C/CTE with dsRNA shows that the isolated RecA_C can independently bind to RNA duplexes. Strikingly, its affinity for dsRNA is identical to the affinity of the complete helicase core, and it has been suggested that RecA_C is the dedicated duplex recognition module of the DEAD-box helicase core (Mallam et al., 2012). However, different pictures have emerged for initial duplex recognition by helicase cores from other DEAD-box proteins (Samatanga and Klostermeier, 2014): studies of Bacillus subtilis YxiN have shown that neither of the individual RecA domains show significant affinity for dsRNA (Samatanga and Klostermeier, 2014). The YxiN helicase core also shows no detectable RNA affinity in the absence of nucleotide (Kd>100 μm), but binds RNA with high affinity in the presence of nucleotide (Samatanga and Klostermeier, 2014), pointing to sequential binding of nucleotide and RNA. The isolated RecA domains of Thermus thermophilus Hera, conversely, both bind to dsRNA individually (Samatanga and Klostermeier, 2014). The Hera helicase core binds RNA with higher affinity than each of the isolated domains (Samatanga and Klostermeier, 2014), indicating cooperativity between the RecA domains in RNA binding. Taken together, these studies point to a wide spectrum of initial RNA recognition modes (Samatanga and Klostermeier, 2014). Duplex destabilization therefore does not always have to be caused by the incursion of RecA_N towards the RNA-bound RecA_C as for Mss116 (Mallam et al., 2012), but may alternatively result from a coordinated movement of both RecA domains relative to the bound RNA and towards each other (e.g. for Hera). Eventually, these different scenarios lead to the same closed conformation of the helicase core, and lead to local destabilization of the duplex, followed by strand separation.
In the closed conformation, interactions between the conserved motifs mediate coupling of ATP hydrolysis to RNA binding and unwinding. Compared to the open state, the nucleotide is now also contacted by residues from motifs V and VI, completing the active site for ATP hydrolysis and triggering the hydrolysis event (Sengoku et al., 2006). The contiguous motifs V/Va have been ascribed a central role in coupling because they are the only motifs that interact with both nucleotide and RNA in the closed conformation of the helicase core (Sengoku et al., 2006). Mutations of motif Va impede closing of the cleft between the RecA domains, reduce the positive thermodynamic linkage of ADPNP and ssRNA binding, and uncouple ATPase and unwinding activities (Cheng et al., 2005; Sengoku et al., 2006; Karow and Klostermeier, 2009). In Mss116, motif Va interacts with the duplex bound to RecA_C, but contributes to formation of the ATPase site in the closed state (Mallam et al., 2012). It has been suggested that this movement of motif Va upon closure of the helicase core might trigger ATP hydrolysis (Mallam et al., 2012). Another major player in coupling is motif III, the only motif that contacts neither the nucleotide nor the RNA. Motif III interacts with motif II (DEAD-box) in the isolated RecA_N and in the open state of the DEAD-box helicase core, and is additionally connected to motif VI in the closed state. Mutations in motif III lead to various degrees of uncoupling of ATP hydrolysis and RNA unwinding (Pause and Sonenberg, 1992; Banroques et al., 2009; Karow and Klostermeier, 2009). These findings implicate motif III in inter-domain communication, but its precise role has not been defined.
RNA binding by domains flanking the helicase core
As discussed in the previous section, the helicase core is the functional unit that provides non-specific RNA binding and unwinding activities. Flanking domains confer RNA specificity and recruit the helicase to its cellular substrate, or they mediate unspecific interactions with RNA and flexibly tether the helicase core to its target.
Sequence-specific binding of single-stranded RNA by a C-terminal RRM
The Escherichia coli DEAD-box helicase DbpA and its B. subtilis ortholog YxiN contain a C-terminal RNA binding domain (RBD). The RBD mediates specific binding to hairpin 92 of 23S ribosomal RNA with high affinity (Diges and Uhlenbeck, 2001; Kossen et al., 2002; Karginov et al., 2005). Assembly of 50S ribosomal subunits is impaired in a dominant negative DbpA mutant, implicating DbpA (and YxiN) in ribosome biogenesis (Sharpe Elles et al., 2009).
The structure of the isolated YxiN RBD has been determined in the apo form (Wang et al., 2006) and in complex with a 73-nucleotide fragment of 23S rRNA, comprising helices 90 and 91, and hairpin 92 (Hardin et al., 2010; Figure 3A). The RBD contains an RRM that folds into a four-stranded antiparallel β-sheet with two α-helices on one side, similar to eukaryotic RRMs (Wang et al., 2006). In the classical RNA binding mode of RRMs, single-stranded RNA is bound across the central β-sheet, involving two conserved aromatic residues on its surface (Oubridge et al., 1994). The corresponding aromatic residues are not critical for the interaction of the YxiN RRM with RNA (Wang et al., 2006), however, in agreement with a different mode of RNA binding (see below). In the structure of the YxiN RBD in complex with the 23S RNA fragment, the three RNA helices form a three-way junction in which helices 90 and 91 are coaxially stacked. The RBD is inserted at the junction between helices 90 and the stem of hairpin 92 (Hardin et al., 2010). The overall structure of the RRM does not change upon RNA binding, but two lysine-rich loops that were not identified in the apo structure are now well-ordered and interact with the RNA. The RRM binds nucleotides in the loop of hairpin 92 across α-helix α1 (Figure 3A). The first uridine of the loop, Ura2552, is not contacted. The Watson-Crick face of the subsequent guanine, Gua2553, forms three hydrogen bonds with the RRM (Figure 3B), leading to sequence-specificity in this position. The bases of Ura2554 and Ura2555 each form one hydrogen bond with the RRM via their O2. The O4 of both uridines is not specifically contacted, and a cytosine could also occupy these positions. According to the interaction pattern in the structure, the YxiN RRM binds to GYYX stretches (Y=pyrimidine, X=purine/pyrimidine). An arginine is inserted into the loop of hairpin 92, and stacks on the ultimate G-C base pair of its stem (Hardin et al., 2010; Supplementary Figure S2). In addition to binding the loop of hairpin 92, the RRM also engages in interactions with the single-stranded nucleotides in the bulge of helix 90 over its distal side (Supplementary Figure S2). Overall, the YxiN RRM thus mediates tight and specific binding to the loop region of hairpin 92 in the structural context of 23S rRNA, and presumably anchors the helicase core on the RNA for unwinding of nearby duplex regions (see below).
![Figure 3: RNA binding to domains flanking the helicase core.(A) Structure of the B. subtilis YxiN RBD bound to the single-stranded region of hairpin 92 in 23S ribosomal RNA (Hardin et al., 2010; PDB-ID 3moj). The RNA lies across the first α-helix of the RRM (see also panel E). The RBD interacts with the nucleotides Gua2553, Ura2554 and Ura2555 that are located in the loop region of hairpin 92. The interaction pattern is consistent with binding to GYYX stretches. (B) Interactions of YxiN with the individual nucleotides of the loop in hairpin 92. Interactions of the RRM with other nucleotides of the bound RNA are shown in Supplementary Figure S2. (C) Structure of the T. thermophilus Hera RBD in complex with a 4mer GGGC RNA oligonucleotide (Steimer et al., 2013; PDB-ID 4i67). As in YxiN, the RNA straddles the first α-helix. The N-terminal subdomain (highlighted in gray) specific to Hera creates a guanine-specific binding pocket and contacts the first nucleotide, Gua1. Reprinted with permission from Steimer et al. (2013) [Steimer, L., Wurm, J.P., Linden, M.H., Rudolph, M.G., Wohnert, J., and Klostermeier, D. (2013). Recognition of two distinct elements in the RNA substrate by the RNA binding domain of the T. thermophilus DEAD box helicase Hera. Nucleic Acids Res. 41 (12), 6259–6272]. (D) Interactions of Hera with individual nucleotides. Gua1 and Gua2 are recognized base-specifically. The recognition pattern for Gua3 is also compatible with other bases in this position. At position 4, a uracil could be accommodated. The Hera RBD thus binds to GGXY sequences. Reprinted from from Steimer et al. (2013) with permission. (E) Superposition of the YxiN (cyan, RNA in green) and Hera (magenta, RNA in yellow) RBDs (stereo figure). In both RBDs, the bound RNA straddles the same α-helix. Owing to subtle differences in the RNA binding at the 5′- and 3′-ends, the RNA entering and emanating from the RBD follows different paths for both proteins, and may take different paths towards the helicase core (arrows; magenta: Hera, cyan: YxiN). Upper panels: front view, bottom panels: top view (relative to the depiction in panel A). Modified with permission from Steimer et al. (2013) [Steimer, L., Wurm, J.P., Linden, M.H., Rudolph, M.G., Wohnert, J., and Klostermeier, D. (2013). Recognition of two distinct elements in the RNA substrate by the RNA binding domain of the T. thermophilus DEAD box helicase Hera. Nucleic Acids Res. 41 (12), 6259–6272].](/document/doi/10.1515/hsz-2014-0277/asset/graphic/j_hsz-2014-0277_fig_003.jpg)
RNA binding to domains flanking the helicase core.
(A) Structure of the B. subtilis YxiN RBD bound to the single-stranded region of hairpin 92 in 23S ribosomal RNA (Hardin et al., 2010; PDB-ID 3moj). The RNA lies across the first α-helix of the RRM (see also panel E). The RBD interacts with the nucleotides Gua2553, Ura2554 and Ura2555 that are located in the loop region of hairpin 92. The interaction pattern is consistent with binding to GYYX stretches. (B) Interactions of YxiN with the individual nucleotides of the loop in hairpin 92. Interactions of the RRM with other nucleotides of the bound RNA are shown in Supplementary Figure S2. (C) Structure of the T. thermophilus Hera RBD in complex with a 4mer GGGC RNA oligonucleotide (Steimer et al., 2013; PDB-ID 4i67). As in YxiN, the RNA straddles the first α-helix. The N-terminal subdomain (highlighted in gray) specific to Hera creates a guanine-specific binding pocket and contacts the first nucleotide, Gua1. Reprinted with permission from Steimer et al. (2013) [Steimer, L., Wurm, J.P., Linden, M.H., Rudolph, M.G., Wohnert, J., and Klostermeier, D. (2013). Recognition of two distinct elements in the RNA substrate by the RNA binding domain of the T. thermophilus DEAD box helicase Hera. Nucleic Acids Res. 41 (12), 6259–6272]. (D) Interactions of Hera with individual nucleotides. Gua1 and Gua2 are recognized base-specifically. The recognition pattern for Gua3 is also compatible with other bases in this position. At position 4, a uracil could be accommodated. The Hera RBD thus binds to GGXY sequences. Reprinted from from Steimer et al. (2013) with permission. (E) Superposition of the YxiN (cyan, RNA in green) and Hera (magenta, RNA in yellow) RBDs (stereo figure). In both RBDs, the bound RNA straddles the same α-helix. Owing to subtle differences in the RNA binding at the 5′- and 3′-ends, the RNA entering and emanating from the RBD follows different paths for both proteins, and may take different paths towards the helicase core (arrows; magenta: Hera, cyan: YxiN). Upper panels: front view, bottom panels: top view (relative to the depiction in panel A). Modified with permission from Steimer et al. (2013) [Steimer, L., Wurm, J.P., Linden, M.H., Rudolph, M.G., Wohnert, J., and Klostermeier, D. (2013). Recognition of two distinct elements in the RNA substrate by the RNA binding domain of the T. thermophilus DEAD box helicase Hera. Nucleic Acids Res. 41 (12), 6259–6272].
Sequence-specific RNA binding by a C-terminal RRM and structure recognition by a short basic tail
A more complex example of a C-terminal flanking region is found in the T. thermophilus DEAD-box protein Hera (Morlang et al., 1999), a DEAD-box protein of unknown function (Klostermeier, 2013). The Hera C-terminal region is subdivided into a dimerization domain (DD) and an RBD (Klostermeier and Rudolph, 2009). The Hera RBD adopts an RRM fold (Rudolph and Klostermeier, 2009; Figure 3C) with similar topology to the YxiN RRM, despite a lack of sequence homology. In contrast to YxiN, where an α-helix connects β-strands β3 and β4, these strands are linked by an extended, well-defined loop in Hera. At the N-terminus of the Hera RRM, a small globular subdomain is formed by residues following the linker connecting the DD to the RBD. This subdomain attaches to the central β-sheet of the RRM (Rudolph and Klostermeier, 2009; Figure 3C) and is responsible for some of the sequence-specificity of the Hera RRM (see below).
The Hera C-terminal flanking region contributes to binding of ribosomal RNA and of RNAse P RNA (Linden et al., 2008). The structure of the Hera RBD in complex with a RNA tetranucleotide reveals recognition of a GGXY stretch by the RRM (Steimer et al., 2013). The tetranucleotide straddles helix α1, and binds to the same side of the RRM as does the loop region of hairpin 92 to the YxiN RBD (Figure 3C,E). Gua1 is bound base-specifically by residues from the small N-terminal subdomain that precedes the Hera RRM. Gua2 is also bound base-specifically, whereas Gua3 and Cyt4 are not recognized in a base-specific manner (Figure 3D). The overall fold of the RRM is unchanged in the RNA-bound form, with the exception of the flexible C-tail that was disordered in the apo structure, but becomes ordered in the RNA complex. The C-tail is important for high affinity RNA binding, and most likely interacts with duplex regions in the vicinity of the single-stranded region recognized by the RRM (Steimer et al., 2013). Thus, the Hera RBD appears to recognize two distinct elements in the Hera RNA substrate, a single-stranded GGXY stretch, and a nearby duplex. Possibly, the Hera RBD mediates binding to single-stranded regions within structured RNA molecules, and presents duplexes in the vicinity to the helicase core for unwinding.
Both the YxiN and the Hera RRM bind RNA on top of helix α1, and provide guanine-specific recognition (Gua2 or Gua2553). Yet, both RRMs recognize different sequences. In addition, the different interactions of both RRMs with the nucleotides at the ends of the bound segment lead to different directions of the RNA leaving the RRM (Figure 3E). At the 5′-end of the RNA, YxiN does not contact the first uridine (Ura2552 of the loop), whereas Hera binds the corresponding guanine (Gua1) in a binding pocket created by its N-terminal subdomain. As a result, the position of these bases differs by 15 Å. Similarly, Cyt4 (Hera) and Ura2555 (YxiN) at the 3′-end are bound differently because of different sizes of the contacting side chains, and Cyt4 in Hera is 3 Å more distant from the RRM than Ura2555 in YxiN (Figure 3E). These differences in geometry of the RNA emanating from the RRMs may impact on the presentation of adjacent RNA duplexes to their respective helicase cores (see below).
Non-specific RNA binding by basic C-tails
A very different type of a C-terminal flanking region is found in the Saccharomyces cerevisiae DEAD-box protein Mss116. The Mss116 helicase core is followed by a domain dubbed CTE (Figure 1) that forms a four-helix bundle flanked by an α-helix and a two-stranded β-sheet (Figure 2C). The CTE packs tightly against the RecA_C domain, and the two domains form a compact structural unit (Del Campo and Lambowitz, 2009), which is different from the loose association of the RBDs in Hera or YxiN with the helicase core. The CTE has been ascribed a role in stabilization of the Mss116 helicase core (Del Campo and Lambowitz, 2009). Residues from the CTE interact with single-stranded RNA emanating from the helicase core, and steric hindrance by the CTE causes a second bend in the RNA backbone (Figure 2D,E). Such a ‘crimping’ of RNA by inducing two bends may also be employed by other DEAD-box proteins, possibly through domains outside the core or by contributions from other binding partners (Del Campo and Lambowitz, 2009). Following the structured CTE, Mss116 contains an unstructured, basic C-terminal tail (C-tail) in a position to interact with RNA regions further upstream. The C-tails of Mss116 and of the closely related Neurospora crassa DEAD-box protein Cyt-19 have been implicated in binding to structured RNAs (Tijerina et al., 2006; Grohman et al., 2007; Mallam et al., 2011).
Anchoring DEAD-box proteins on their physiological substrate: RNA binding to complete DEAD-box proteins
Despite the advances in our understanding of RNA binding, nucleotide binding, and ATP-dependent duplex destabilization by the DEAD-box helicase core, structural information on complete DEAD-box helicases in complex with RNA is still very limited. In the following, three examples for structural models of full-length DEAD-box helicases will be presented, and their implications for RNA binding and unwinding will be discussed.
YxiN: cooperation of a helicase core with a C-terminal RRM
The crystal structures of the isolated YxiN RBD, both alone and in complex with RNA (Wang et al., 2006; Hardin et al., 2010) and of the RecA_C domain (Caruthers et al., 2006) have been determined. A high-resolution structure of B. subtilis YxiN is still lacking, but SAXS data on authentic YxiN have pointed to a loose attachment of the helicase core to the RBD, and to a distended, elongated shape of the full-length protein (Wang et al., 2007). A structural model for full-length YxiN in solution has been constructed using distance restraints from FRET experiments, introducing donor and acceptor fluorophores either on RecA_N and the RBD, or on RecA_C and the RBD (Karow and Klostermeier, 2010; Figure 4A). The model places the RBD close to RecA_C, above a surface region that is formed by several loops (Karow and Klostermeier, 2010). The corresponding region is also involved in interactions of eIF4A-III with Upf3 (up-frameshift suppressor 3), a protein involved in nonsense-mediated decay of RNA (Buchwald et al., 2010, 2013), of eIF4A with the translation initiation factor eIF4G (Schutz et al., 2008), and of Dbp5 with its regulator Gle1 (Montpetit et al., 2011). This region on the helicase core may provide an adaptive interface for the interaction of the DEAD-helicase core with other domains or interaction partners. When the YxiN model is superimposed with the structure of the YxiN RBD/RNA complex (Hardin et al., 2010; Figure 4A), there are no clashes of the helicase core with the RNA bound to the RBD, suggesting that this conformation may be representative of the YxiN/RNA complex in the absence of nucleotide. Strikingly, helix 91 of the ribosomal RNA that is unwound by YxiN is positioned far away from the helicase core (Figure 4A). Large-scale conformational changes of YxiN, with a movement of the RBD relative to the helicase core, would be required to bring the target RNA duplex to the helicase core for unwinding.
![Figure 4: Binding of RNA to complete DEAD-box proteins.(A) Homology model of full-length YxiN bound to ribosomal RNA. Superposition (right) of a model for full-length YxiN (Karow and Klostermeier, 2010) and the crystal structure of the RBD/RNA complex (Hardin et al., 2010; PDB-ID 3moj; left) on the RBD. The YxiN model was obtained by mapping the position of the RBD relative to the helicase core with distance restraints from smFRET (Karow and Klostermeier, 2010). The helix 91 that is unwound by YxiN using model RNA substrates, highlighted by the red box, is pointing away from the RNA binding site of the helicase core (arrow). (B) Model for the full-length Hera dimer (Klostermeier and Rudolph, 2009; Klostermeier, 2013), showing the RNA bound to the helicase core and to the RRM of the RBD (Steimer et al., 2013). The model was constructed from the structures comprised of RecA_C and the DD (Klostermeier and Rudolph, 2009; PDB-ID 3eas), RecA_C, the DD, and the RBD (Rudolph and Klostermeier, 2009; PDB-ID 3i32), the RBD in complex with RNA (Steimer et al., 2013; PDB-ID 4i67), and a homology model for the closed helicase core (RecA_N, RecA_C), using the Vasa structure in complex with RNA and ADPNP (Sengoku et al., 2006; PDB-ID 2db3) as a template. One protomer is shown in gray, the second is color-coded according to the electrostatic surface potential. The two RNA binding sites are connected by a positively charged patch, but the 5′-ends are facing towards each other, arguing against binding to a contiguous strand of RNA in larger substrates. The right panel shows the dimer rotated upwards around the horizontal axis by about 60°, with one protomer in gray, one in orange, and the bound RNA in yellow. The RNAs bound to the helicase cores of the dimer are facing towards each other, suggesting that the two cores might act concertedly on different regions of the same large RNA molecule. Reprinted from Klostermeier (2013) [Klostermeier, D. (2013). Rearranging RNA structures at 75°C? towards the molecular mechanism and physiological function of the Thermus thermophilus DEAD-box helicase hera. Biopolymers 99 (12), 1137–1146] with permission. (C) Model for RNA binding to Mss116. Left: the structure of Mss116, comprising the helicase core (blue), the CTE (red) and the basic C-tail (gray), as determined from SAXS studies (Mallam et al. 2012). Bound RNA is depicted in orange, according to its position in the crystal structure. The C-tail is flexible, and two alternative conformations are depicted to illustrate the conformational space covered relative to the helicase core. Right: model for a complex of Mss116, bound to the group I intron (yellow). The flexibility of the C-tail may allow for presentation of different regions of the large RNA substrate to the helicase core for unwinding (arrows). Modified after Mallam et al. (2011) [Mallam, A.L., Jarmoskaite, I., Tijerina, P., Del Campo, M., Seifert, S., Guo, L., Russell, R., and Lambowitz, A.M. (2011). Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc. Natl. Acad. Sci. USA 108, 12254–12259], reprinted with permission.](/document/doi/10.1515/hsz-2014-0277/asset/graphic/j_hsz-2014-0277_fig_004.jpg)
Binding of RNA to complete DEAD-box proteins.
(A) Homology model of full-length YxiN bound to ribosomal RNA. Superposition (right) of a model for full-length YxiN (Karow and Klostermeier, 2010) and the crystal structure of the RBD/RNA complex (Hardin et al., 2010; PDB-ID 3moj; left) on the RBD. The YxiN model was obtained by mapping the position of the RBD relative to the helicase core with distance restraints from smFRET (Karow and Klostermeier, 2010). The helix 91 that is unwound by YxiN using model RNA substrates, highlighted by the red box, is pointing away from the RNA binding site of the helicase core (arrow). (B) Model for the full-length Hera dimer (Klostermeier and Rudolph, 2009; Klostermeier, 2013), showing the RNA bound to the helicase core and to the RRM of the RBD (Steimer et al., 2013). The model was constructed from the structures comprised of RecA_C and the DD (Klostermeier and Rudolph, 2009; PDB-ID 3eas), RecA_C, the DD, and the RBD (Rudolph and Klostermeier, 2009; PDB-ID 3i32), the RBD in complex with RNA (Steimer et al., 2013; PDB-ID 4i67), and a homology model for the closed helicase core (RecA_N, RecA_C), using the Vasa structure in complex with RNA and ADPNP (Sengoku et al., 2006; PDB-ID 2db3) as a template. One protomer is shown in gray, the second is color-coded according to the electrostatic surface potential. The two RNA binding sites are connected by a positively charged patch, but the 5′-ends are facing towards each other, arguing against binding to a contiguous strand of RNA in larger substrates. The right panel shows the dimer rotated upwards around the horizontal axis by about 60°, with one protomer in gray, one in orange, and the bound RNA in yellow. The RNAs bound to the helicase cores of the dimer are facing towards each other, suggesting that the two cores might act concertedly on different regions of the same large RNA molecule. Reprinted from Klostermeier (2013) [Klostermeier, D. (2013). Rearranging RNA structures at 75°C? towards the molecular mechanism and physiological function of the Thermus thermophilus DEAD-box helicase hera. Biopolymers 99 (12), 1137–1146] with permission. (C) Model for RNA binding to Mss116. Left: the structure of Mss116, comprising the helicase core (blue), the CTE (red) and the basic C-tail (gray), as determined from SAXS studies (Mallam et al. 2012). Bound RNA is depicted in orange, according to its position in the crystal structure. The C-tail is flexible, and two alternative conformations are depicted to illustrate the conformational space covered relative to the helicase core. Right: model for a complex of Mss116, bound to the group I intron (yellow). The flexibility of the C-tail may allow for presentation of different regions of the large RNA substrate to the helicase core for unwinding (arrows). Modified after Mallam et al. (2011) [Mallam, A.L., Jarmoskaite, I., Tijerina, P., Del Campo, M., Seifert, S., Guo, L., Russell, R., and Lambowitz, A.M. (2011). Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc. Natl. Acad. Sci. USA 108, 12254–12259], reprinted with permission.
Hera: cooperation of a core with a C-terminal RRM and a basic tail in context of a dimer
Hera consists of a helicase core followed by a DD and an RBD, and forms a stable dimer in solution (Klostermeier and Rudolph, 2009). Again, a high-resolution structure of the full-length protein has not been reported to date. Crystal structures of overlapping constructs, i.e. RecA_C and the DD (PDB-ID 3eas), and RecA_C, DD, and the RBD (PDB-ID 3i32; Klostermeier and Rudolph, 2009; Rudolph and Klostermeier, 2009; Klostermeier, 2013), in conjunction with a homology model based on the structure of Vasa (Sengoku et al., 2006; PDB-ID 2db3) have allowed construction of a model for the dimeric T. thermophilus DEAD-box protein Hera (Figure 4B). In this model, the DD of one protomer distends from RecA_C of the helicase core to connect with the DD from the second protomer, and then loops back to RecA_C from its own protomer. As a consequence, the RBD of each Hera protomer is located adjacent to RecA_C of the same protomer (Figure 4B), positioned at the end of the RNA binding cleft on the surface of the helicase core. Overall, the domain arrangement is therefore similar to the relative configuration in the monomeric YxiN that lacks the DD, although the RBDs in YxiN and Hera lie on different sides of RecA_C (Supplementary Figure S3). Owing to the structural plasticity of the DD in Hera and its flexible linkages to the core, the two helicase cores of the Hera dimer cover a large conformational space in the absence of RNA (Klostermeier, 2013). Upon RNA and ATP binding to the helicase cores, and their conformational change to the closed state, this conformational freedom is substantially reduced (Klostermeier, 2013; Figure 4B).
From a superposition of the Hera model with structures of the helicase core of Vasa (Sengoku et al., 2006) and of the RBD bound to RNA (Steimer et al., 2013), it is evident that the RNA bound to the RBD and the core have opposite polarities, with their 5′-ends facing towards each other (Figure 4B). Although both RNA binding sites are connected by an elongated patch of positive electrostatic potential, it is clear from this polarity that they do not simply bind to one contiguous strand of a larger RNA. Binding of the Hera RBD to short single-stranded RNA stretches next to an exposed duplex may anchor Hera on a structured RNA substrate. Duplexes in the neighborhood are then unwound by the Hera helicase core.
Strikingly, the two RNA binding sites of the helicase cores in the Hera dimer are facing each other (Figure 4B). It is possible that both Hera cores can act on two duplexes of the same RNA molecule, and can concertedly unwind different regions of hitherto unidentified physiological RNA substrates.
Mss116: presentation of RNA to the core by a flexible tether
The only structural information on RNA binding by full-length DEAD-box proteins, including N- and C-terminal regions flanking the helicase core, stems from SAXS experiments on S. cerevisiae Mss116 and Neurospora crassa Cyt 19 (Mallam et al., 2011). The structural model of full-length Mss116 is consistent with the NTE and the basic C-tail being unstructured extensions. The NTE extends from RecA_N. The CTE forms one compact unit with RecA_C. It binds ssRNA emanating from the binding site on the helicase core (Del Campo and Lambowitz, 2009), and contacts the second strand of duplex RNA bound to RecA_C (Mallam et al., 2012). The basic C-tail following the CTE extends outward from RecA_C/CTE, and thus away from the helicase core. The C-tail is flexible, and can cover a large conformational space relative to the helicase core (Mallam et al., 2011). Despite its lack of regular structure, SAXS data argue against a fully extended conformation of the C-tail. Instead, they are consistent with favored conformations of the C-tail close to RecA_C and the CTE, in a position to contact the RNA extending from the helicase core (Mallam et al., 2011). Overall, the C-tail appears to serve as a functional tether that interacts electrostatically with exposed helices (Pan et al., 2014) and anchors Mss116 on its large RNA substrate (Mallam et al., 2011). The helicase core then unwinds duplexes within reach from this anchoring site.
In all three DEAD-box proteins discussed here, the flanking domains act as anchors on a large RNA substrate, and determine which duplex regions are accessible for unwinding by the helicase core. The anchoring can be achieved with limited sequence-specificity by RRMs (YxiN, Hera), and non-sequence-specifically by structured domains (Mss116) and unstructured basic C-tails (Hera, Mss116). It is possible that the RNA binding modules remain attached to the RNA during multiple rounds of RNA unwinding by the helicase core. Despite the apparent modular architecture of DEAD-box proteins, attempts to generate functional chimera by attaching an RBD to a different helicase core have been largely unsuccessful (Rogers et al., 2002; Banroques et al., 2011), suggesting that we have not yet fully uncovered the molecular basis for functional cooperation of the helicase core with extra domains. The function of individual elements also appears to be context-dependent. An example is the C-tail of Mss116 and Cyt19. Despite its common role as a functional tether (Tijerina et al., 2006; Mallam et al., 2011; Jarmoskaite et al., 2014), deletion of the C-tail has different effects in the two proteins: Deleting the C-tail is deleterious for Cyt19 activity (Grohman et al., 2007), but causes only a moderate reduction in Mss116 activity (Mohr et al., 2008). The individual domains of DEAD-box helicases thus do not appear to be functionally independent modules. More work is needed to define the mode of cooperation of helicase cores with flanking domains, and to identify the role of the linkers connecting these elements.
Positive and negative contributions of interaction partners to RNA binding
Contributions to RNA binding by a set of interaction partners
Although structural information on RNA binding to DEAD-box proteins is limited to binding of short oligonucleotides that do not extend beyond the helicase core, it has already become clear that interaction partners can contribute to binding of RNA regions flanking the stretch that is bound by the helicase core. A first glimpse of RNA binding by interaction partners is provided by the structure of the exon junction complex. The RNA bound to eIF4A-III is contacted at the 5′-end by the complex component Btz/MLN51 (Andersen et al., 2006; Bono et al., 2006), pointing to a joint binding of exon junction complex components to the mRNA. Coordinated binding of mRNA by a helicase and its interaction partners is also found in translation initiation, where the translation initiation factor eIF4A interacts with the 5′-UTR of mRNAs. The translation initiation factors eIF4G, eIF4E and eIF4B/eIF4H are also RNA binding proteins, and together with eIF4A interact with different elements of the mRNA. The available data point to a network of protein/RNA- and protein/protein interactions in the translation initiation complex, but the molecular basis for the cooperation of these translation factors with eIF4A in mRNA binding and ribosome scanning is currently not well-understood.
Modulation of RNA binding to the helicase core
Binding partners can also show a negative effect on RNA binding to DEAD-box proteins. eIF4G binds to RecA_N and RecA_C of eIF4A, stabilizing it in a half-open conformation (Schutz et al., 2008; Hilbert et al., 2011). In the eIF4A-eIF4G complex, the contiguous RNA binding site normally formed by RecA_N and RecA_C is not aligned, and eIF4G therefore stimulates RNA release from eIF4A (Montpetit et al., 2011). Gle1, an interaction partner of the yeast DEAD-box protein Dbp5, stimulates RNA release from Dbp5 by the same mechanism (Montpetit et al., 2011). Dbp5 (DDX19 in humans) is a DEAD-box protein involved in mRNA export from the nucleus. Dbp5 localizes to the nuclear pore complex by binding to the cytoplasmic nucleoporin Nup159 (NUP214 in humans). The RNA binding site on the helicase core of DDX19/Dbp5 overlaps with the binding site for the nucleoporin NUP214/Nup159, rendering RNA and NUP214/Nup159 binding mutually exclusive (von Moeller et al., 2009; Montpetit et al., 2011). In addition to occluding the RNA binding site, NUP214/Nup159 prevents formation of the closed state of the DDX19/Dbp5 helicase core, and inhibits its RNA-stimulated ATPase activity (von Moeller et al., 2009; Montpetit et al., 2011). During mRNA export, Nup159 binding may either trigger release of the transported mRNA from Dbp5 or binding of the mRNA releases Dbp5 from Nup159, and thus from the nuclear pore (von Moeller et al., 2009). The Dbp5 interaction partner Gle1, also located at the cytoplasmic side of the nuclear pore, displaces an N-terminal helix of DDX19/Dbp5 that functions as an autoinhibitory element for RNA binding (Collins et al., 2009) and alleviates this inhibitory effect (Montpetit et al., 2011). Although the order of events during nuclear export of RNA is not yet entirely clear, Gle1 and NUP214/Nup159 undoubtedly modulate RNA binding to the DDX19/Dbp5 helicase core in multiple ways to provide spatial and temporal regulation of DDX19/Dbp5 activity.
The two examples illustrate that interaction partners can modulate RNA binding to the helicase core itself, or can provide RNA binding sites for additional interactions with the RNA substrate outside the region contacted by the helicase core. Modulation of RNA binding to DEAD-box proteins by interaction partners allows for the spatial and temporal regulation of their activity.
Emerging principles and perspective
From the structural insight into the interaction of the helicase core with ssRNA and of the RecA_C domain with dsRNA, a coherent picture for RNA binding and unwinding by DEAD-box helicase cores has emerged. An RNA duplex binds to RecA_C (or to both RecA domains) of the open conformation of the helicase core. Nucleotide binding then triggers the transition to the closed state of the helicase core. The two RecA domains move towards each other, and residues from both domains are aligned to form a contiguous RNA binding site. The concomitant approach of the ‘wedge’ helix causes a distortion of the RNA backbone, leading to the local disruption of a few base pairs in the bound RNA duplex. While six nucleotides of one RNA strand are bound in a sequence-independent manner by the helicase core, the second strand can dissociate from the destabilized duplex. ATP hydrolysis, phosphate release, re-opening of the helicase core, and dissociation of the second RNA strand complete the catalytic cycle.
RNA binding to the helicase core can be modulated by interaction partners that occlude the RNA binding site or affect the orientation of the two parts of the RNA binding site. Interaction partners can also bind to RNA regions not contacted by the DEAD-box helicase core. Domains flanking the helicase core also contribute to RNA binding. RRMs provide sequence-specific binding to single-stranded RNA regions in the vicinity of the unwound duplex, outside the region contacted by the helicase core, and confer substrate specificity. In contrast to RRMs, basic regions act as sequence-unspecific tethers that anchor DEAD-box proteins near structured regions of larger RNA molecules.
The molecular basis of the functional interplay between the helicase core and additional domains or interaction partners in RNA binding and unwinding is not yet completely understood. The available data point to relative movements of the helicase core and flanking domains involved in RNA binding. Conformational dynamics is ensured by connecting the helicase core to RBDs with flexible linkers, or by the intrinsic flexibility of unstructured tails. Binding of flanking domains to a specific site of the RNA substrate determines which duplex regions are within reach of the helicase core and can be unwound. In the few structural models available to date, flanking domains that contribute to RNA binding are in different positions relative to the helicase core, indicating that a wide spectrum of solutions is realized among different DEAD-box proteins. To arrive at a comprehensive understanding of conformational changes that allow for the functional cooperation of RBDs and the helicase core, future studies should address the dynamics of DEAD-box protein/RNA complexes. While most mechanistic studies have been limited to isolated model duplexes as unwinding substrates, recent work has moved towards understanding the action of DEAD-box proteins on duplexes within larger, structured RNAs. For many DEAD-box proteins, the in vivo RNA targets are still unknown. The current efforts in many laboratories to identify physiological substrates of DEAD-box proteins will be the key towards the identification of their in vivo function. At the same time, the identification of in vivo substrates will also provide an impetus for future mechanistic studies to dissect contributions of domains outside the helicase core and their role for unwinding of physiological substrates by the helicase core. In the long run, mechanistic studies of DEAD-box helicases in the context of their functional interaction networks will lead to a comprehensive understanding of DEAD-box protein function in the cellular environment.
Acknowledgments
We thank Anna Mallam and Alan Lambowitz for sharing the structural model of Mss116 (Figure 4C), and previous and current members of the Klostermeier group and collaborators for their contributions. Funding: Work in the Klostermeier laboratory was supported by the Swiss National Science Foundation (31003A_125437) and the Deutsche Forschungsgemeinschaft (KL 1153/7-1).
References
Ahmadian, M.R., Stege, P., Scheffzek, K., and Wittinghofer, A. (1997). Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 4, 686–689.10.1038/nsb0997-686Suche in Google Scholar PubMed
Andersen, C.B., Ballut, L., Johansen, J.S., Chamieh, H., Nielsen, K.H., Oliveira, C.L., Pedersen, J.S., Seraphin, B., Le Hir, H., and Andersen, G.R. (2006). Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313, 1968–1972.10.1126/science.1131981Suche in Google Scholar PubMed
Andreou, A.Z. and Klostermeier, D. (2012a). Conformational changes of DEAD-box helicases monitored by single molecule fluorescence resonance energy transfer. Methods Enzymol. 511, 75–109.10.1016/B978-0-12-396546-2.00004-8Suche in Google Scholar PubMed
Andreou, A.Z. and Klostermeier, D. (2012b). The DEAD-box helicase eIF4A: paradigm or the odd one out? RNA Biol. 1, 19–32.10.4161/rna.21966Suche in Google Scholar PubMed PubMed Central
Andreou, A.Z. and Klostermeier, D. (2014). eIF4B and eIF4G jointly stimulate eIF4A ATPase and unwinding activities by modulation of the eIF4A conformational cycle. J. Mol. Biol. 426, 51–61.10.1016/j.jmb.2013.09.027Suche in Google Scholar PubMed
Aregger, R. and Klostermeier, D. (2009). The DEAD-box helicase YxiN maintains a closed conformation during ATP hydrolysis. Biochemistry 48, 10679–10681.10.1021/bi901278pSuche in Google Scholar PubMed
Banroques, J., Doere, M., Dreyfus, M., Linder, P., and Tanner, N.K. (2009). Motif III in superfamily 2 “helicases” helps convert the binding energy of ATP into a high-affinity RNA binding site in the yeast DEAD-box protein Ded1. J. Mol. Biol. 396, 949–966.10.1016/j.jmb.2009.12.025Suche in Google Scholar PubMed
Banroques, J., Cordin, O., Doere, M., Linder, P., and Tanner, N.K. (2011). Analyses of the functional regions of DEAD-box RNA “helicases” with deletion and chimera constructs tested in vivo and in vitro. J. Mol. Biol. 413, 451–472.10.1016/j.jmb.2011.08.032Suche in Google Scholar PubMed
Bono, F., Ebert, J., Lorentzen, E., and Conti, E. (2006). The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126, 713–725.10.1016/j.cell.2006.08.006Suche in Google Scholar PubMed
Buchwald, G., Ebert, J., Basquin, C., Sauliere, J., Jayachandran, U., Bono, F., Le Hir, H., and Conti, E. (2010). Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proc. Natl. Acad. Sci. USA 107, 10050–10055.10.1073/pnas.1000993107Suche in Google Scholar PubMed PubMed Central
Buchwald, G., Schussler, S., Basquin, C., Le Hir, H., and Conti, E. (2013). Crystal structure of the human eIF4AIII-CWC22 complex shows how a DEAD-box protein is inhibited by a MIF4G domain. Proc. Natl. Acad. Sci. USA 110, E4611–E4608.10.1073/pnas.1314684110Suche in Google Scholar PubMed PubMed Central
Caruthers, J.M., Johnson, E.R., and McKay, D.B. (2000). Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. USA 97, 13080–13085.10.1073/pnas.97.24.13080Suche in Google Scholar PubMed PubMed Central
Caruthers, J.M., Hu, Y., and McKay, D.B. (2006). Structure of the second domain of the Bacillus subtilis DEAD-box RNA helicase YxiN. Acta Crystallogr. Sect. F 62, 1191–1195.10.1107/S1744309106044642Suche in Google Scholar PubMed PubMed Central
Cheng, Z., Coller, J., Parker, R., and Song, H. (2005). Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA 11, 1258–1270.10.1261/rna.2920905Suche in Google Scholar PubMed PubMed Central
Collins, R., Karlberg, T., Lehtio, L., Schutz, P., van den Berg, S., Dahlgren, L.G., Hammarstrom, M., Weigelt, J., and Schuler, H. (2009). The DExD/H-box RNA helicase DDX19 is regulated by an α-helical switch. J. Biol. Chem. 284, 10296–10300.10.1074/jbc.C900018200Suche in Google Scholar PubMed PubMed Central
Del Campo, M. and Lambowitz, A.M. (2009). Structure of theyeast DEAD-box protein Mss116p reveals two wedges that crimp RNA. Mol. Cell 35, 598–609.10.1016/j.molcel.2009.07.032Suche in Google Scholar PubMed PubMed Central
Diges, C.M. and Uhlenbeck, O.C. (2001). Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 20, 5503–5512.10.1093/emboj/20.19.5503Suche in Google Scholar PubMed PubMed Central
Elles, L.M. and Uhlenbeck, O.C. (2008). Mutation of the arginine finger in the active site of Escherichia coli DbpA abolishes ATPase and helicase activity and confers a dominant slow growth phenotype. Nucleic Acids Res. 36, 41–50.10.1093/nar/gkm926Suche in Google Scholar PubMed PubMed Central
Grohman, J.K., Campo, M.D., Bhaskaran, H., Tijerina, P., Lambowitz, A.M., and Russell, R. (2007). Probing the mechanisms of DEAD-box proteins as general RNA chaperones: the C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry 46, 3013–3022.10.1021/bi0619472Suche in Google Scholar PubMed PubMed Central
Hardin, J.W., Hu, Y.X., and McKay, D.B. (2010). Structure of the RNA Binding Domain of a DEAD-Box Helicase Bound to Its Ribosomal RNA Target Reveals a Novel Mode of Recognition by an RNA Recognition Motif. J. Mol. Biol. 402, 412–427.10.1016/j.jmb.2010.07.040Suche in Google Scholar PubMed PubMed Central
Harms, U., Andreou, A.Z., Gubaev, A., and Klostermeier, D. (2014). eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res. 42, 7911–7922.10.1093/nar/gku440Suche in Google Scholar PubMed PubMed Central
Henn, A., Bradley, M.J., and De La Cruz, E.M. (2012). ATP Utilization and RNA Conformational Rearrangement by DEAD-Box Proteins. Annu. Rev. Biophys. 41, 247–267.10.1146/annurev-biophys-050511-102243Suche in Google Scholar PubMed PubMed Central
Hilbert, M., Karow, A.R., and Klostermeier, D. (2009). The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol. Chem. 390, 1237–1250.10.1515/BC.2009.135Suche in Google Scholar PubMed
Hilbert, M., Kebbel, F., Gubaev, A., and Klostermeier, D. (2011). eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res. 39, 2260–2270.10.1093/nar/gkq1127Suche in Google Scholar PubMed PubMed Central
Hogbom, M., Collins, R., van den Berg, S., Jenvert, R.M., Karlberg, T., Kotenyova, T., Flores, A., Hedestam, G.B., and Schiavone, L.H. (2007). Crystal Structure of Conserved Domains 1 and 2 of the Human DEAD-box Helicase DDX3X in Complex with the Mononucleotide AMP. J. Mol. Biol. 372, 150–159.10.1016/j.jmb.2007.06.050Suche in Google Scholar PubMed
Jacewicz, A., Schwer, B., Smith, P., and Shuman, S. (2014). Crystal structure, mutational analysis and RNA-dependent ATPase activity of the yeast DEAD-box pre-mRNA splicing factor Prp28. Nucleic Acids Res. 42, 12885–12898.10.1093/nar/gku930Suche in Google Scholar PubMed PubMed Central
Jarmoskaite, I., Bhaskaran, H., Seifert, S., and Russell, R. (2014). DEAD-box protein CYT-19 is activated by exposed helices in a group I intron RNA. Proc. Natl. Acad. Sci. USA 111, E2928–E2936.10.1073/pnas.1404307111Suche in Google Scholar PubMed PubMed Central
Karginov, F.V., Caruthers, J.M., Hu, Y., McKay, D.B., and Uhlenbeck, O.C. (2005). YxiN is a modular protein combining a DEx(D/H) core and a specific RNA-binding domain. J. Biol. Chem. 280, 35499–35505.10.1074/jbc.M506815200Suche in Google Scholar PubMed
Karow, A.R. and Klostermeier, D. (2009). A conformational change in the helicase core is necessary but not sufficient for RNA unwinding by the DEAD box helicase YxiN. Nucleic Acids Res. 37, 4464–4471.10.1093/nar/gkp397Suche in Google Scholar PubMed PubMed Central
Karow, A.R. and Klostermeier, D. (2010). A structural model for the DEAD box helicase YxiN in solution: localization of the RNA-binding domain. J. Mol. Biol. 402, 629–637.10.1016/j.jmb.2010.07.049Suche in Google Scholar PubMed
Klostermeier, D. (2013). Rearranging RNA structures at 75°C? towards the molecular mechanism and physiological function of the Thermus thermophilus DEAD-box helicase hera. Biopolymers 99, 1137–1146.Suche in Google Scholar
Klostermeier, D. and Rudolph, M.G. (2009). A novel dimerization motif in the C-terminal domain of the Thermus thermophilus DEAD box helicase Hera confers substantial flexibility. Nucleic Acids Res. 37, 421–430.10.1093/nar/gkn947Suche in Google Scholar
Kossen, K., Karginov, F.V., and Uhlenbeck, O.C. (2002). The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J. Mol. Biol. 324, 625–636.10.1016/S0022-2836(02)01140-3Suche in Google Scholar
Linden, M.H., Hartmann, R.K., and Klostermeier, D. (2008). The putative RNase P motif in the DEAD box helicase Hera is dispensable for efficient interaction with RNA and helicase activity. Nucleic Acids Res. 36, 5800–5811.10.1093/nar/gkn581Suche in Google Scholar PubMed PubMed Central
Linder, P. and Jankowsky, E. (2011). From unwinding to clamping – the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12, 505–516.10.1038/nrm3154Suche in Google Scholar PubMed
Linder, P., Lasko, P.F., Ashburner, M., Leroy, P., Nielsen, P.J., Nishi, K., Schnier, J., and Slonimski, P.P. (1989). Birth of the D-E-A-D box. [erratum appears in Nature (1989) 20;340:246]. Nature 337, 121–122.10.1038/337121a0Suche in Google Scholar PubMed
Liu, F., Putnam, A., and Jankowsky, E. (2008). ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl. Acad. Sci. USA 51, 20209–20214.10.1073/pnas.0811115106Suche in Google Scholar PubMed PubMed Central
Mallam, A.L., Jarmoskaite, I., Tijerina, P., Del Campo, M., Seifert, S., Guo, L., Russell, R., and Lambowitz, A.M. (2011). Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc. Natl. Acad. Sci. USA 108, 12254–12259.10.1073/pnas.1109566108Suche in Google Scholar PubMed PubMed Central
Mallam, A.L., Del Campo, M., Gilman, B., Sidote, D.J., and Lambowitz, A.M. (2012). Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p. Nature 490, 121–125.10.1038/nature11402Suche in Google Scholar PubMed PubMed Central
Mathys, H., Basquin, J., Ozgur, S., Czarnocki-Cieciura, M., Bonneau, F., Aartse, A., Dziembowski, A., Nowotny, M., Conti, E., and Filipowicz, W. (2014). Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol. Cell 54, 751–765.10.1016/j.molcel.2014.03.036Suche in Google Scholar PubMed
Mohlmann, S., Mathew, R., Neumann, P., Schmitt, A., Luhrmann, R., and Ficner, R. (2014). Structural and functional analysis of the human spliceosomal DEAD-box helicase Prp28. Acta Crystallogr. D Biol. Crystallogr. 70, 1622–1630.10.1107/S1399004714006439Suche in Google Scholar PubMed PubMed Central
Mohr, G., Del Campo, M., Mohr, S., Yang, Q., Jia, H., Jankowsky, E., and Lambowitz, A.M. (2008). Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J. Mol. Biol. 375, 1344–1364.10.1016/j.jmb.2007.11.041Suche in Google Scholar PubMed PubMed Central
Mohr, G., Del Campo, M., Turner, K.G., Gilman, B., Wolf, R.Z., and Lambowitz, A.M. (2011). High-throughput genetic identification of functionally important regions of the yeast DEAD-box protein Mss116p. J. Mol. Biol. 413, 952–972.10.1016/j.jmb.2011.09.015Suche in Google Scholar PubMed PubMed Central
Montpetit, B., Thomsen, N.D., Helmke, K.J., Seeliger, M.A., Berger, J.M., and Weis, K. (2011). A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472, 238–242.10.1038/nature09862Suche in Google Scholar PubMed PubMed Central
Morlang, S., Weglohner, W., and Franceschi, F. (1999). Hera from Thermus thermophilus: the first thermostable DEAD-box helicase with an RNase P protein motif. J. Mol. Biol. 294, 795–805.10.1006/jmbi.1999.3282Suche in Google Scholar PubMed
Nielsen, K.H., Chamieh, H., Andersen, C.B., Fredslund, F., Hamborg, K., Le Hir, H., and Andersen, G.R. (2008). Mechanism of ATP turnover inhibition in the EJC. RNA 15, 67–75.10.1261/rna.1283109Suche in Google Scholar PubMed PubMed Central
Oubridge, C., Ito, N., Evans, P.R., Teo, C.H., and Nagai, K. (1994). Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372, 432–438.10.1038/372432a0Suche in Google Scholar PubMed
Pan, C., Potratz, J.P., Cannon, B., Simpson, Z.B., Ziehr, J.L., Tijerina, P., and Russell, R. (2014). DEAD-box helicase proteins disrupt RNA tertiary structure through helix capture. PLoS Biol 12, e1001981.10.1371/journal.pbio.1001981Suche in Google Scholar PubMed PubMed Central
Pause, A. and Sonenberg, N. (1992). Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 11, 2643–2654.10.1002/j.1460-2075.1992.tb05330.xSuche in Google Scholar PubMed PubMed Central
Putnam, A.A. and Jankowsky, E. (2013). DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim. Biophys. Acta 1829, 884–893.10.1016/j.bbagrm.2013.02.002Suche in Google Scholar PubMed PubMed Central
Raz, E. (2000). The function and regulation of vasa-like genes in germ-cell development. Genome Biol. 1, reviews1017.1–reviews1017.6.10.1186/gb-2000-1-3-reviews1017Suche in Google Scholar PubMed PubMed Central
Richter, N.J., Rogers, G.W., Jr., Hensold, J.O., and Merrick, W.C. (1999). Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H. J. Biol. Chem. 274, 35415–35424.10.1074/jbc.274.50.35415Suche in Google Scholar
Rogers, G.W. Jr., Richter, N.J., and Merrick, W.C. (1999). Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274, 12236–12244.10.1074/jbc.274.18.12236Suche in Google Scholar
Rogers, G.W., Jr., Lima, W.F., and Merrick, W.C. (2001). Further characterization of the helicase activity of eIF4A. Substrate specificity. J. Biol. Chem. 276, 12598–12608.10.1074/jbc.M007560200Suche in Google Scholar
Rogers, G.W., Jr., Komar, A.A., and Merrick, W.C. (2002). eIF4A: the godfather of the DEAD box helicases. Prog. Nucleic Acid Res. Mol. Biol. 72, 307–331.10.1016/S0079-6603(02)72073-4Suche in Google Scholar
Rudolph, M.G. and Klostermeier, D. (2009). The Thermus thermophilus DEAD box helicase Hera contains a modified RNA recognition motif domain loosely connected to the helicase core. RNA 15, 1993–2001.10.1261/rna.1820009Suche in Google Scholar PubMed PubMed Central
Rudolph, M.G., Heissmann, R., Wittmann, J.G., and Klostermeier, D. (2006). Crystal Structure and Nucleotide Binding of the Thermus thermophilus RNA Helicase Hera N-terminal Domain. J. Mol. Biol. 361, 731–743.10.1016/j.jmb.2006.06.065Suche in Google Scholar PubMed
Samatanga, B. and Klostermeier, D. (2014). DEAD-box RNA helicase domains exhibit a continuum between complete functional independence and high thermodynamic coupling in nucleotide and RNA duplex recognition. Nucleic Acids Res. 42, 10644–10654.10.1093/nar/gku747Suche in Google Scholar PubMed PubMed Central
Schutz, P., Bumann, M., Oberholzer, A.E., Bieniossek, C., Trachsel, H., Altmann, M., and Baumann, U. (2008). Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc. Natl. Acad. Sci. USA 105, 9564–9569.10.1073/pnas.0800418105Suche in Google Scholar PubMed PubMed Central
Schutz, P., Karlberg, T., van den Berg, S., Collins, R., Lehtio, L., Hogbom, M., Holmberg-Schiavone, L., Tempel, W., Park, H.W., Hammarstrom, M., et al. (2010). Comparative structural analysis of human DEAD-box RNA helicases. PLoS One 5, e12791.10.1371/journal.pone.0012791Suche in Google Scholar PubMed PubMed Central
Sengoku, T., Nureki, O., Nakamura, A., Kobayashi, S., and Yokoyama, S. (2006). Structural Basis for RNA Unwinding by the DEAD-Box Protein Drosophila Vasa. Cell 125, 287–300.10.1016/j.cell.2006.01.054Suche in Google Scholar PubMed
Sharpe Elles, L.M., Sykes, M.T., Williamson, J.R., and Uhlenbeck, O.C. (2009). A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res. 37, 6503–6514.10.1093/nar/gkp711Suche in Google Scholar
Steimer, L. and Klostermeier, D. (2012). RNA helicases in infection and disease. RNA Biol. 9, 751–771.10.4161/rna.20090Suche in Google Scholar
Steimer, L., Wurm, J.P., Linden, M.H., Rudolph, M.G., Wohnert, J., and Klostermeier, D. (2013). Recognition of two distinct elements in the RNA substrate by the RNA binding domain of the T. thermophilus DEAD box helicase Hera. Nucleic Acids Res. 41, 6259–6272.10.1093/nar/gkt323Suche in Google Scholar
Story, R.M., Li, H., and Abelson, J.N. (2001). Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc. Natl. Acad. Sci. USA 98, 1465–1470.10.1073/pnas.98.4.1465Suche in Google Scholar
Strohmeier, J., Hertel, I., Diederichsen, U., Rudolph, M.G., and Klostermeier, D. (2011). Changing nucleotide specificity of the DEAD-box helicase Hera abrogates communication between the Q-motif and the P-loop. Biol. Chem. 392, 357–369.10.1515/bc.2011.034Suche in Google Scholar
Talavera, M.A. and De La Cruz, E.M. (2005). Equilibrium and kinetic analysis of nucleotide binding to the DEAD-box RNA helicase DbpA. Biochemistry 44, 959–970.10.1021/bi048253iSuche in Google Scholar
Tange, T.O., Nott, A., and Moore, M.J. (2004). The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 16, 279–284.10.1016/j.ceb.2004.03.012Suche in Google Scholar
Tanner, N.K., Cordin, O., Banroques, J., Doere, M., and Linder, P. (2003). The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell 11, 127–138.10.1016/S1097-2765(03)00006-6Suche in Google Scholar
Theissen, B., Karow, A.R., Kohler, J., Gubaev, A., and Klostermeier, D. (2008). Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc. Natl. Acad. Sci. USA 105, 548–553.10.1073/pnas.0705488105Suche in Google Scholar PubMed PubMed Central
Tijerina, P., Bhaskaran, H., and Russell, R. (2006). Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc. Natl. Acad. Sci. USA 103, 16698–16703.10.1073/pnas.0603127103Suche in Google Scholar PubMed PubMed Central
von Moeller, H., Basquin, C., and Conti, E. (2009). The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat. Struct. Mol. Biol. 16, 247–254.10.1038/nsmb.1561Suche in Google Scholar PubMed
Wang, S., Hu, Y., Overgaard, M.T., Karginov, F.V., Uhlenbeck, O.C., and McKay, D.B. (2006). The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold. RNA 12, 959–967.10.1261/rna.5906Suche in Google Scholar PubMed PubMed Central
Wang, S., Overgaard, M.T., Hu, Y., and McKay, D.B. (2007). The Bacillus subtilis RNA Helicase YxiN is Distended in Solution. Biophys. J. 94, L01–L03.Suche in Google Scholar
Yang, Q., Del Campo, M., Lambowitz, A.M., and Jankowsky, E. (2007). DEAD-box proteins unwind duplexes by local strand separation. Mol. Cell 28, 253–263.10.1016/j.molcel.2007.08.016Suche in Google Scholar PubMed
Supplemental Material
The online version of this article (DOI: 10.1515/hsz-2014-0277) offers supplementary material, available to authorized users.
©2015 by De Gruyter
Artikel in diesem Heft
- Frontmatter
- Reviews
- Ras activation revisited: role of GEF and GAP systems
- When core competence is not enough: functional interplay of the DEAD-box helicase core with ancillary domains and auxiliary factors in RNA binding and unwinding
- Cathepsin S: therapeutic, diagnostic, and prognostic potential
- Minireview
- Overview of the roles of Sox2 in stem cell and development
- Research Articles/Short Communications
- Genes and Nucleic Acids
- Transcriptional and translational mechanisms contribute to regulate the expression of Discs Large 1 protein during different biological processes
- Membranes, Lipids, Glycobiology
- Rapid transfer of overexpressed integral membrane protein from the host membrane into soluble lipid nanodiscs without previous purification
- Molecular Medicine
- Characterization of a new dual-targeting fully human antibody with potent antitumor activity against nasopharyngeal carcinoma
- Cell Biology and Signaling
- Lithium chloride improves the efficiency of induced pluripotent stem cell-derived neurospheres
- SIRT2 suppresses non-small cell lung cancer growth by targeting JMJD2A
- Troglitazone suppresses glutamine metabolism through a PPAR-independent mechanism
Artikel in diesem Heft
- Frontmatter
- Reviews
- Ras activation revisited: role of GEF and GAP systems
- When core competence is not enough: functional interplay of the DEAD-box helicase core with ancillary domains and auxiliary factors in RNA binding and unwinding
- Cathepsin S: therapeutic, diagnostic, and prognostic potential
- Minireview
- Overview of the roles of Sox2 in stem cell and development
- Research Articles/Short Communications
- Genes and Nucleic Acids
- Transcriptional and translational mechanisms contribute to regulate the expression of Discs Large 1 protein during different biological processes
- Membranes, Lipids, Glycobiology
- Rapid transfer of overexpressed integral membrane protein from the host membrane into soluble lipid nanodiscs without previous purification
- Molecular Medicine
- Characterization of a new dual-targeting fully human antibody with potent antitumor activity against nasopharyngeal carcinoma
- Cell Biology and Signaling
- Lithium chloride improves the efficiency of induced pluripotent stem cell-derived neurospheres
- SIRT2 suppresses non-small cell lung cancer growth by targeting JMJD2A
- Troglitazone suppresses glutamine metabolism through a PPAR-independent mechanism


