Abstract
Background
TCDD-inducible poly (ADP-ribose) polymerase (TiPARP) is a DNA repair enzyme with functions in energy metabolism, signal transduction, cell differentiation, and other biological processes, which may closely related to lipid metabolism and is highly expressed in adipose tissue. Adipose tissue can be divided into white adipose tissue (WAT) that stores energy and brown adipose tissue (BAT) that releases energy and generates heat. In the present study, we investigated whether TiPARP can affect adipogenesis in adipose tissue and thus participate in the development of obesity.
Methods
BAT primary cells or 3T3-L1 cells infected with adenovirus expressing TiPARP or TiPARP-targeted short hairpin RNA (shTiPARP) were cultured to induce adipogenic differentiation. The expression of TiPARP was detected by real-time PCR and Western blotting. The expression of specific BAT- and WAT-related markers was detected by real-time PCR. The accumulation of lipid droplets in differentiated cells was detected by Oil Red O staining.
Results
TiPARP was highly expressed in both subcutaneous WAT and BAT, and TiPARP mRNA level increased significantly along with adipogenic differentiation. Activation of TiPARP or overexpression of TiPARP upregulated BAT-related markers in primary BAT cells and WAT-related markers in 3T3-L1 cells, together with increased lipid accumulation. On the contrary, knockdown of TiPARP downregulated expression of specific markers in both BAT primary cells and 3T3-L1 cells, together with decreased lipid accumulation.
Conclusion
TiPARP regulates adipogenesis in both BAT primary cells and 3T3-L1 cells and therefore plays an important role in modulating maturity and lipid accumulation in brown and white adipocytes. These findings provide us with a new strategy for combating obesity.
Introduction
Adipose tissue is generally divided into white adipose tissue (WAT) and brown adipose tissue (BAT).[1] White adipocytes store energy in monolocular large lipid droplets,[2] while lipid droplets in brown adipocytes are usually small and scattered throughout the cytoplasm and account for >50% of the energy used in the synthesis of ATP released in the form of heat through mitochondria.[3] In both brown and white adipocytes, lipid droplets with triglyceride deposition are generated after adipogenic differentiation.[4]
Obesity is a metabolic disease with the increased volume of adipocytes resulting in body weight gain and local excessive fat deposition, leading to high blood pressure, hyperlipidemia, atherosclerosis, metabolic syndrome such as insulin resistance, and type 2 diabetes.[5] Therefore, looking for mechanism and effective treatment of obesity is increasingly important.
Poly(ADP-ribose) polymerase (PARP) is found in the nucleus. Its main role is to detect and signal single-strand DNA breaks (SSBs) to the enzymatic machinery involved in DNA repair.[6] PARP activation is an immediate cellular response to metabolic, chemical, or radiation-induced DNA SSBs.[7] Once PARP detects an SSB, it binds to the DNA, and, after a structural change, begins the synthesis of PAR as a signal for the other DNA-repair enzymes such as DNA ligase III, DNA polymerase b [polit binds to the DNA, and after a structural change, begins synthesis of PAR as a signal for poly (ADP-ribose) glycohydrolase].[8,9] Nicotinamide adenine dinucleotide (NAD+) is required as the substrate for generating ADP-ribose monomers.[10] Overactivation of PARP may deplete the stores of cellular NAD+ and induce progressive ATP depletion and necrotic cell death since glucose oxidation is inhibited.[11] It is suggested that inhibition of hexokinase activity by PARP1 leads to defects in glycolysis[12] and lipid metabolism.[13]
Large-scale genome-wide association study (GWAS) have identified that a genetic variant on chromosome 3, single nucleotide polymorphism (SNP) rs900399, is strongly associated with both HDL trait and triglyceride.[14] This variant lies in a noncoding region, in the vicinity of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-inducible poly-(ADP-ribose) polymerase (TiPARP; also known as PARP7 or ARTD14), which is widely expressed in many tissues and therefore might be closely related to lipid metabolism,[14] but the mechanisms remain unknown. The environmental toxin TCDD produces diverse toxic effects including a lethal wasting syndrome.[15] TiPARP is an aryl hydrocarbon receptor (AHR) target gene and is associated with hepatic steatohepatitis.[16] It has also been shown that TiPARP acts as a mediator of suppression of hepatic gluconeogenesis by TCDD.[17] The T allele of rs900400 is associated with higher expression of TiPARP in adipocytes,[18] although TiPARP affects liver metabolism, indicating a close relationship with energy and lipid metabolism, there is little research about TiPARP in adipose tissue. In the present study, we investigated the role of TiPARP in lipid metabolism in mouse primary brown adipocytes and 3T3-L1 cells.
Materials and methods
Materials
TiPARP rabbit polyclonal antibody was purchased from Signalway Antibody LLC (College Park, MA, USA). Mouse anti-β-actin was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Insulin, indomethacin, isobutylmethylxanthine, dexamethasone, and collagenase I were from Sigma Aldrich (St. Louis, MO, USA). IRDye-conjugated affinity purified anti-rabbit and anti-mouse IgGs were purchased from Rockland (Gilbertsville, PA, USA). Trizol reagent and the reverse transcription (RT) system were purchased from Invitrogen Inc. (Grand Island, NY, USA).
Animals and animal care
C57BL/6J mice (Vital River Laboratory Animal Technology, Beijing, China) were housed in a 12:12-h light/dark cycle. Regular chow and water were available ad libitum. The research related to animals use has been complied with all the relevant national regulations and institutional policies for the care and use of animals. All experimental protocols were approved by the Animal Care and Use Committee of Peking University Health Science Center (LA2016123).
Culture of primary brown adipocytes
BAT was harvested from decapitated suckling mice and brown preadipocytes were isolated by digesting the interscapular BAT of neonatal C57BL/6J mice with collagenase I and mechanistic dispersion, as described previously.[19] Isolated cells were randomly plated in tissue-culture dishes in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20% fetal bovine serum (FBS). After 24 h of culture at 37℃ (5% CO2), cells were rinsed twice with phosphate buffer saline (PBS), after which 70% of the initial cells were attached to the dish, forming a monolayer. Isolated brown preadipocytes were cultured in DMEM supplemented with 20% FBS for proliferation. To induce brown adipocyte differentiation, cells were cultured for 2 days in 10% FBS-DMEM supplemented with 20 nmol/L insulin, 1 nmol/L triiodothyronine (T3), 12.5 mmol/L indomethacin, 0.5 mmol/L isobutylmethylxanthine, and 2 mg/mL dexamethasone. On days 3–6, the induction medium was substituted by a maintenance medium consisting of DMEM supplemented with 20 nmol/L insulin and 1 nmol/L T3. For the undifferentiated controls, brown preadipocytes were cultured in 10% FBS-DMEM without other supplements.
Culture of 3T3-L1 cells
3T3-L1 cells (American Type Culture Collection, Manassas, VA, USA) were randomly plated in tissue-culture dishes in DMEM/F12 supplemented with 10% FBS and incubated at 37℃ (5% CO2) until confluence. Then cells were induced by adipogenesis as above.
Adenovirus infection
The TiPARP adenoviruses were expanded, titrated in 293 cells, and purified by cesium chloride as described previously.[20,21] For adenovirus-mediated gene transfer, primary brown preadipocytes or 3T3-L1 cells were infected with 106 titer adenovirus for 48 h. Infection efficiency was judged by green fluorescent protein (GFP) expression observed under the microscope (Leica Microsystems, GmbH, Wetzlar, Germany). Infected primary brown preadipocytes or 3T3-L1 adipocytes were differentiated for 6 days and then harvested for subsequent analysis.
shRNA plasmid and transfection
TiPARP pSH-U6-GFP vector cloning and pSH-U6-GFP negative control vector cloning were designed and purchased from Vigene Biosciences (Rockville, MD). The U6 target sequence was 5’-GGA CTA TCA TAT GCT TAC CG-3’. Full-length sequence of shTiPARP was 5’-GAT CCG CAG TTA TTT CGC AAA GAA TTC AAG AGA TTC TTT GCG AAA TAA CTG CTT TTT TA-3’. Primary brown adipocytes and 3T3-L1 cells were maintained in DMEM supplemented with 10% FBS and 50 U/mL penicillin and streptomycin and transfected with Neofect™ DNA transfection reagent from (Beijing, China). The original medium was removed from the cells and replaced with a fresh complete medium 2 h before transfection. Two micrograms of plasmid DNA was diluted with 100 μL DMEM, added with 2 μL Neofect™, blended gently, and incubated at 37℃ for 20 min to form a transfection complex. The transfected complex was added to the cell culture medium and cultured for 48 h.
RNA extraction and quantitative real-time PCR analysis
Total RNA was isolated using the TRIzol reagent. Reverse transcription was performed using the RT system according to the manufacturer’s instructions. PCR was conducted in a 25 μL volume containing 2.5 μL cDNA, 5 mmol/L MgCl2, 0.2 mmol/L dNTPs, 0.2 μmol/L of each primer, 1.25 U AmpliTaq polymerase, and 1 μL 800 × diluted SYBRGreen I stock using the Mx3000 multiple quantitative PCR system (Strata gene, La Jolla, CA, USA). PCR reactions were performed in duplicate, and each experiment was repeated four times. Primers used in this study are shown in Table 1. The mRNA expression was quantified using the comparative cross threshold (CT) method. The CT value of the housekeeping gene β-actin was subtracted from the CT value of the target gene to obtain ΔCT. The normalized fold changes of target gene mRNA expression were expressed as 2−ΔΔCT, where ΔΔCT equals to ΔCT (sample) ˗ ΔCT (control).
The required primer sequence
| Target genes | Upstream primer | Downstream primer |
|---|---|---|
| Mouse UCP1 | GGACGACCCCTAATCTAATG | CATTAGATTAGGGGTCGTCC |
| Mouse PGC1α | GATTGAAGTGGTGTAGCGAC | GTCGCTACACCACTTCAATC |
| Mouse PRDM16 | CCACCAGCGAGGACTTCAC | GGAGGACTCTCGTAGCTCGAA |
| Mouse Ednra | CGGAGATCAACTTTCTGG | TGGAGACGATTTCAATGG |
| Mouse resistin | TCCTTGTCCCTGAACTGC | ACGAATGTCCCACGAGC |
| Mouse adiponectin | TGTTGGAATGACAGGAGCTG | CGAATGGGTACATTGGGAAC |
| Mouse PAI-1 | AGGGCTTCATGCCCCACTTCTTCA | AGTAGAGGGCATTCACCAGCACCA |
| Human TiPARP | CGTCTGGGGAGTAGGCAAT | CCCGAGGGAGGATGTGAAAC |
| Mouse TiPARP | GACAAATGTTGAGGGCCAATTAC | TGCAAAAGGTCAGTTTGGAAGT |
| Mouse β-actin | ATCTGGCACCACACCTTC | AGCCAGGTCCAGACGCA |
Western blot analysis
Cultured cells were harvested and homogenized in an ice-cold fractionation buffer containing RIPA lysis buffer, phenylmethanesulfonyl fluoride (PMSF), and protein phosphatase inhibitor mixture. The cell lysate was treated with ultrasound for 3 s three times and then centrifuged at 15300 g for 10 min at 4℃. After centrifugation, the supernatant was used for Western blot analysis. Protein concentration was measured by Bradford’s method. A total of 40–60 μg protein from each sample was loaded. Proteins were transferred to polyvinylidene fluoride membranes. The membranes were incubated for 1 h at room temperature with 4% fat-free milk in Tris-buffered saline containing Tween-20, followed by incubation overnight at 4℃ with the individual primary antibodies. The specific reaction was detected using IRDye-conjugated second antibody and visualized using the Odyssey infrared imaging system (LICOR Biosciences, Lincoln, NE, USA). Quantification of image density in pixels was performed using the Odyssey infrared imaging system (LI-COR Biosciences).
Oil Red O staining
Brown adipocytes were rinsed in PBS three times and then fixed with 4% paraformaldehyde for 15 min. After washing, samples were incubated in 0.3% Oil Red staining solution for 45–60 min at room temperature. Samples were then counterstained with hematoxylin for 30 s, followed by washing in PBS for 5 min.
Statistical analysis
Data were expressed as mean ± SEM. Data analysis used GraphPad Prism software (Ver. 8.0.1). One-way ANOVA, Student–Newman–Keul test (comparisons between multiple groups), or unpaired Student’s t-test (between two groups) was used as appropriate. P < 0.05 denoted a statistical significance.
Results
TiPARP gene expression in different tissues
Eight-week-old C57BL/6J male mice were sacrificed, and TiPARP expression was detected by real-time PCR in BAT, subcutaneous (s)WAT, liver, aorta, heart, brain, lung, testis, kidney, small intestine, spleen, and skeletal muscle (Figure 1A). Protein levels of TiPARP in lipid metabolism-related tissue, BAT, sWAT, and liver were confirmed by Western blot (Figure 1B). TiPARP was expressed in a high percentage of sWAT and BAT, indicating the vital role of TiPARP in adipose tissue function.
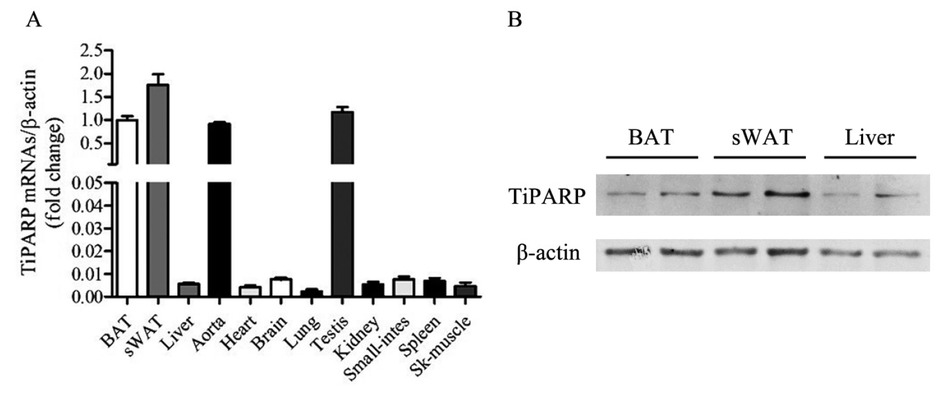
Expression of TiPARP in different organs and tissues. (A) TiPARP expression in BAT, sWAT, liver, aorta, heart, brain, lung, testis, kidney, small -intestine, spleen, and skeletal muscle in 8-week-old C57BL/6J male mice evaluated by real-time PCR. (B) TiPARP expression in BAT, sWAT, and liver in 8-week-old C57BL/6J male mice evaluated by Western blot. TiPARP: TCDD-inducible poly (ADP-ribose) polymerase; BAT: brown adipose tissue; sWAT: subcutaneous white adipose tissue.
Participation of TiPARP in brown adipocyte differentiation
In order to study the effect of TiPARP on brown adipocyte differentiation, we isolated primary brown preadipocytes and induced differentiation for 6 days. mRNA levels of brown adipocyte gene markers, such as UCP1, PGC1α, and PRDM16, were significantly increased after induction of differentiation, indicating the effective differentiation of brown adipocytes (Figure 2A). TiPARP mRNA level also increased along with differentiation (Figure 2B). The continuous activation of TiPARP gene during brown adipocyte differentiation suggests that TiPARP participates in brown adipocyte differentiation.

Effect of TiPARP activation during differentiation of brown adipocytes. Primary brown adipocytes were isolated from neonatal C57BL/6J mice and cultured for differentiation. Total RNA was extracted from primary brown adipocytes after 6 days of differentiation. Real-time PCR was performed to evaluate the expression of brown adipocyte marker genes (A) and TiPARP gene (B). β-Actin was used as a loading control. Results expressed as mean ± SEM (n = 4), *P < 0.05 versus nondifferentiation. Primary brown adipocytes were treated with 10-8 mol/L BaP for 8 h. Real-time PCR was performed to evaluate the expression of TiPARP (C) and genes related to brown adipocyte markers (D). Levels of mRNA expression were normalized to β-actin and expressed as mean ± SEM (n = 4), *P < 0.05 versus DMSO. TiPARP: TCDD-inducible poly(ADP-ribose) polymerase; DMSO: dimethyl sulfoxide.
The classic activator of TiPARP is TCDD. The structure of benzo[a]pyrene (BaP), another polyaromatic hydrocarbon, is similar to that of TCDD, and it is less toxic; therefore, we used BaP to activate TiPARP. Primary brown adipocytes were treated with 10-8 mol/L BaP. Real-time PCR was performed to evaluate the expression of TiPARP and genes related to brown adipocyte markers. TiPARP mRNA level was elevated distinctly by BaP (Figure 2C), together with brown adipocyte markers, such as UCP1, PGC1α, and PRDM16 (Figure 2D). Activation of TiPARP resulting in enhancement of brown adipocyte differentiation suggested that TiPARP plays an important role in brown adipocyte differentiation.
Overexpression of TiPARP accelerated adipogenic differentiation in brown adipocytes
To clarify the specific function of TiPARP in brown adipocyte differentiation, we used adenovirus to overexpress TiPARP in brown adipocytes and observe its lipid accumulation. Brown preadipocytes were isolated from suckling mice and fused for 4–6 days, transfected with 106 plaque forming unit (PFU) TiPARP adenoviruses for 48 h, and cultured for differentiation at the same time. Controls used GFP adenoviruses. GFP expression was observed in brown adipocytes under fluorescence microscopy (Figure 3A). Proteins were extracted after 6 days of differentiation, TiPARP protein level increased significantly (Figure 3B), along with TiPARP mRNA level (Figure 3C). The mRNA expression levels of brown adipocyte markers such as UCP1, PGC1α, and PRDM16 were also increased (Figure 3D). A higher fusion rate of brown adipocytes was observed under light microscopy (Figure 3E), and multilocular lipid droplets were detected by Oil Red O staining (Figure 3F) after adenovirus transfection compared with GFP controls. These results indicate that overexpression of TiPARP accelerates adipogenic differentiation and lipid accumulation in brown adipocytes.
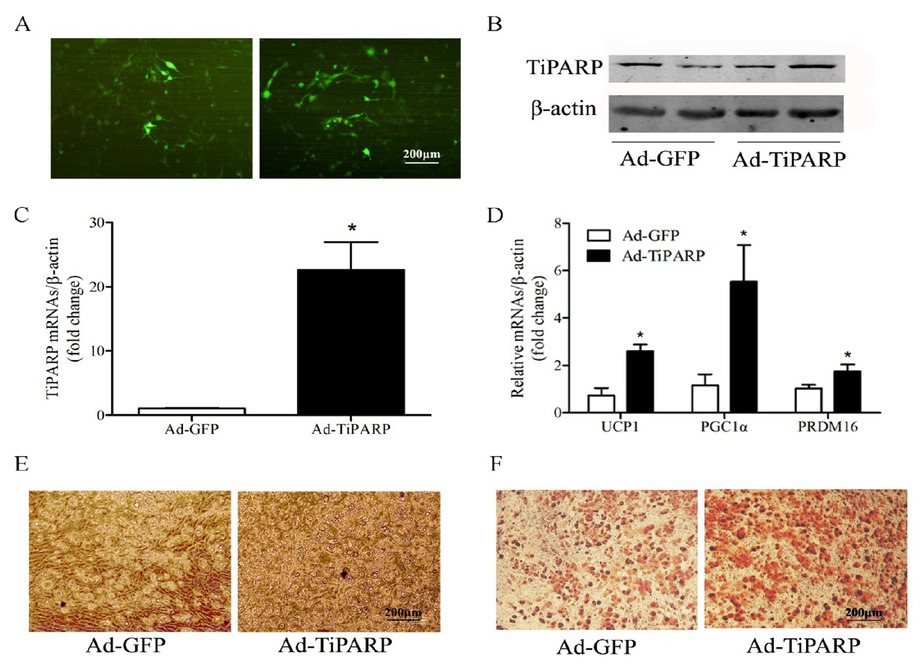
Overexpression of TiPARP accelerates adipogenic differentiation of brown preadipocytes. Primary brown adipocytes were isolated from neonatal C57BL/6J mice and transfected with 106 plaque forming unit (PFU) TiPARP adenoviruses for 48 h and cultured for differentiation at the same time, while controls used GFP adenoviruses. (A) GFP expression was observed in brown adipocytes under fluorescence microscopy. (B) Proteins were extracted after 6 days of differentiation, and Western blotting of TiPARP was performed. Total RNA was extracted after 6 days of differentiation. Real-time PCR was performed to evaluate the expression of TiPARP (C) and brown adipocyte marker genes (D). Levels of mRNA expression were normalized to β-actin and expressed as mean ± SEM (n = 4). (E) Brown adipocyte morphology was observed by light microscope after adenovirus transfection. (F) Oil red O staining was performed to demonstrate the lipid droplets after adenovirus transfection. *P < 0.05 relative to Ad-GFP control. TiPARP: TCDD-inducible poly (ADP-ribose) polymerase; GFP: green fluorescent protein; Ad-TiPARP: TiPARP-expressed adenovirus; Ad-GFP: GFP-expressed adenovirus.
Knockdown of TiPARP inhibits adipogenic differentiation in brown adipocytes
We used shRNA to knock down TiPARP in brown adipocytes and observe its lipid accumulation. Brown preadipocytes were isolated from suckling mice and fused for 4–6 days, transfected with 5 ng/mL shTiPARP for 48 h, and cultured for differentiation at the same time. Controls used shU6. GFP expression was observed in brown adipocytes under fluorescence microscopy (Figure 4A). Proteins were extracted after 6 days of differentiation, TiPARP protein level decreased significantly (Figure 4B), and TiPARP mRNA level also decreased (Figure 4C). mRNA expression of brown adipocyte markers such as UCP1, PGC1α, and PRDM16 was also clearly decreased (Figure 4D). There was a lower fusion rate of brown adipocytes observed under light microscopy (Figure 4E) and a lower positive rate of Oil Red O staining after shTiPARP transfection compared with shU6 (Figure 4F). These results suggest that the knockdown of TiPARP inhibits adipogenic differentiation and lipid accumulation in brown adipocytes.

Knockdown of TiPARP inhibits adipogenic differentiation of brown preadipocytes. Primary brown adipocytes were isolated from neonatal C57BL/6J mice and transfected with 5 ng/mL shTiPARP for 48 h and cultured for differentiation at the same time, while controls used shU6. (A) GFP expression was observed in brown adipocytes under fluorescence microscopy. (B) Proteins were extracted after 6 days of differentiation, and Western blotting of TiPARP was performed. Real-time polymerase chain reaction (PCR) was performed to evaluate the expression of TiPARP (C) and brown adipocyte marker genes (D). Levels of mRNA expression were normalized to β-actin and expressed as mean ± SEM (n = 4). (E) Brown adipocyte morphology was observed by light microscope after shRNA transfection. (F) Oil red O staining was performed to demonstrate lipid droplets after shRNA transfection. *P < 0.05 relative to shU6 controls. TiPARP: TCDD-inducible poly(ADP-ribose) polymerase; GFP: green fluorescent protein; shTiPARP: TiPARP-targeted short hairpin RNA; shU6: negative control.
TiPARP influences adipogenic differentiation of 3T3-L1 cells
TiPARP affected adipogenesis in 3T3-L1 cells. 3T3-L1 cells were fused for 4–6 days and transfected with 106 PFU TiPARP adenoviruses for 48 h. Controls used GFP adenoviruses. TiPARP mRNA expression increased significantly (Figure 5A), and white adipocyte markers such as Ednra, resistin, and adiponectin were also clearly increased (Figure 5B), indicating that overexpression of TiPARP accelerated adipogenic differentiation in 3T3-L1 cells.

Effect of TiPARP on adipogenic differentiation of 3T3-L1 cells. (A and B) 3T3-L1 cells were transfected with 106 plaque forming unit (PFU) TiPARP adenoviruses for 48 h, while controls used GFP adenoviruses. Total RNA was extracted after 48 h of transfection. Real-time PCR was performed to evaluate the expression of TiPARP (A) and white adipocyte marker genes (B). Levels of mRNA expression were normalized to β-actin and expressed as mean ± SEM (n = 4). *P < 0.05 relative to Ad-GFP control. (C and D) 3T3-L1 cells were transfected with 5 ng/mL shTiPARP for 48 h, while controls used shU6. Total RNA was extracted after 48 h of transfection. Real-time PCR was performed to evaluate the expression of TiPARP (C) and white adipocyte marker genes (D). Levels of mRNA expression were normalized to β-actin and expressed as mean ± SEM (n = 4). *P < 0.05 relative to shU6 controls.TiPARP: TCDD-inducible poly (ADP-ribose) polymerase; Ad-GFP: GFP-expressed adenovirus; shU6: negative control.
Furthermore, 3T3-L1 cells were fused for 4–6 days and transfected with 5 ng/mL shTiPARP for 48 h. Controls used shU6. TiPARP mRNA expression level decreased significantly (Figure 5C), and white adipocyte markers such as Ednra, resistin, PAI-1, and adiponectin were also decreased (Figure 5D), indicating that knockdown of TiPARP inhibited adipogenic differentiation in 3T3-L1 cells.
Discussion
In the present study, we reported that TiPARP regulates adipogenesis in both primary brown adipocytes and 3T3-L1 cells and therefore plays an important role in modulating maturity and lipid accumulation in brown and white adipocytes. This conclusion is supported by the following observations: (1) TiPARP gene level is increased during differentiation of primary brown adipocytes; (2) activation of TiPARP promotes differentiation of primary brown adipocytes; (3) overexpression of TiPARP promotes adipogenesis and lipid accumulation in primary brown adipocytes; (4) knockdown of TiPARP inhibits adipogenesis and lipid accumulation in primary brown adipocytes; (5) overexpression of TiPARP promotes adipogenesis in 3T3-L1 cells; and (6) knockdown of TiPARP inhibits adipogenesis in 3T3-L1 cells.
With the improvement of social and economic levels, the incidence of obesity and related metabolic diseases has become increasingly high. Obesity is not only related to high-sugar and high-fat diets but also closely related to environmental factors.[22] Current research shows that air pollution, especially PM2.5, has a significant role in aggravating obesity and type 2 diabetes in children and adults.[23, 24, 25] Air pollutants contain a variety of polycyclic aromatic hydrocarbons, which can easily activate AHRs in the human body[26] and induce a series of metabolic imbalances.[27,28]
The number of brown and white adipocytes in healthy adults remains in dynamic equilibrium,[29] so we are looking at the effect of TiPARP on adipogenic differentiation of adipocytes. Our study confirmed that TiPARP can induce lipid metabolism disorders under excess energy intake and suggested that TiPARP promotes adipogenic differentiation of brown and white adipocytes. TiPARP promoted differentiation of white and brown adipocytes, which revealed a new function of TiPARP, and suggested new potential activities of the PARP family. Other proteins in the PARP family play important roles in energy metabolism, and in mammalian cells and mouse tissues, NAD+ is the rate-limiting enzyme of the sirtuin family.[30] The reduced rate of NAD+ consumption of PARP-1 and CD38 can increase NAD+ bioavailability, leading to activation of sirtuin (Sirt)1 and Sirt3 to promote oxidative metabolism and prevent obesity induced by high-fat diets.[31] Another study of PARP2 showed that in skeletal muscle, miRNA-149 inhibited PARP2 by increasing cellular NAD+ levels and Sirt-1 gene activity, which in turn enhanced mitochondrial function through activation of PGC-1α.[32] PARP2 deletion in mice increased Sirt1 levels, promoted energy expenditure, and increased mitochondria.[33] In addition, PARP2-deficient mice are protected against diet-induced obesity.[34] Although insulin sensitivity was not affected, PARP2 mice exhibited significant pancreatic dysfunction and glucose intolerance.[33] During the process of weight loss in obese people, there was increased expression of NAD+/Sirt1 and decreased PARP activity,[35] suggesting that the PARP family occurs in obesity and is related to metabolic diseases. We speculate that the effect of TiPARP on adipose tissue may not be limited to its basic DNA repair function, but rather it regulates cell differentiation of adipocytes directly. Although a specific inhibitor of TiPARP is not available yet, it was reported that targeting AHR, the upstream molecule of TiPARP, prevents high-fat diet-induced insulin resistance and obesity.[36] Whether TiPARP can regulate the mutual conversion of WAT and BAT deserves in-depth study.
In conclusion, our study showed that TiPARP promotes adipogenic differentiation of adipocytes. These findings provide us with a new strategy for combating obesity. Inhibition of TiPARP expression may become a clinically relevant treatment for obesity-related metabolic disease, which needs further investigation.
-
Conflict of Interest
The authors declare no conflict of interest.
-
Source of Funding
This work was supported by the National Natural Science Foundation of China (81873568); the National Key Research and Development Program of China (2020YFA0803801); the Special project for central guidance of local science and technology development of Liaoning Province (2019JH6/10400006).
-
Ethics Approval
All experimental protocols were approved by the Animal Care and Use Committee of Peking University Health Science Center (LA2016123).
Acknowledgment
We appreciate Professor Eugene Y Chen of University of Michigan for kindly providing the TiPARP-expressing virus.
Reference
1 Kurylowicz A, Puzianowska-Kuznicka M. Induction of Adipose Tissue Browning as a Strategy to Combat Obesity. Int J Mol Sci 2020;21:6241.10.3390/ijms21176241Search in Google Scholar PubMed PubMed Central
2 Cinti S. The adipose organ. Prostaglandins Leukot Essent Fatty Acids 2005; 73: 9-15.10.1016/j.plefa.2005.04.010Search in Google Scholar PubMed
3 Porter C. Quantification of UCP1 function in human brown adipose tissue. Adipocyte 2017; 6: 167-74.10.1080/21623945.2017.1319535Search in Google Scholar PubMed PubMed Central
4 Zhang K, Yang X, Zhao Q, Li Z, Fu F, Zhang H, et al. Molecular Mechanism of Stem Cell Differentiation into Adipocytes and Adipocyte Differentiation of Malignant Tumor. Stem Cells Int 2020; 2020: 8892300.10.1155/2020/8892300Search in Google Scholar PubMed PubMed Central
5 Kopelman PG. Obesity as a medical problem. Nature 2000; 404: 635-43.10.1038/35007508Search in Google Scholar PubMed
6 El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res 2003; 31: 5526-33.10.1093/nar/gkg761Search in Google Scholar PubMed PubMed Central
7 Horton JK, Stefanick DF, Gassman NR, Williams JG, Gabel SA, Cuneo MJ, et al. Preventing oxidation of cellular XRCC1 affects PARP-mediated DNA damage responses. DNA Repair (Amst) 2013; 12: 774-85.10.1016/j.dnarep.2013.06.004Search in Google Scholar PubMed PubMed Central
8 Soni A, Siemann M, Grabos M, Murmann T, Pantelias GE, Iliakis G. Requirement for Parp-1 and DNA ligases 1 or 3 but not of Xrcc1 in chromosomal translocation formation by backup end joining. Nucleic Acids Res 2014; 42: 6380-92.10.1093/nar/gku298Search in Google Scholar PubMed PubMed Central
9 Fouquerel E, Sobol RW. ARTD1 (PARP1) activation and NAD(+) in DNA repair and cell death. DNA Repair (Amst) 2014; 23: 27-32.10.1016/j.dnarep.2014.09.004Search in Google Scholar PubMed PubMed Central
10 Alemasova EE, Lavrik OI. Poly(ADP-ribosyl)ation by PARP1: reaction mechanism and regulatory proteins. Nucleic Acids Res 2019; 47: 3811-27.10.1093/nar/gkz120Search in Google Scholar PubMed PubMed Central
11 Pieper AA, Verma A, Zhang J, Snyder SH. Poly (ADP-ribose) polymerase, nitric oxide and cell death. Trends Pharmacol Sci 1999; 20: 171-81.10.1016/S0165-6147(99)01292-4Search in Google Scholar
12 Fouquerel E, Goellner EM, Yu Z, Gagne JP, Barbi de Moura M, Feinstein T, et al. ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Rep 2014; 8: 1819-31.10.1016/j.celrep.2014.08.036Search in Google Scholar PubMed PubMed Central
13 Huang K, Du M, Tan X, Yang L, Li X, Jiang Y, et al. PARP1-mediated PPARalpha poly(ADP-ribosyl)ation suppresses fatty acid oxidation in non-alcoholic fatty liver disease. J Hepatol 2017; 66: 962-77.10.1016/j.jhep.2016.11.020Search in Google Scholar PubMed PubMed Central
14 Liu DJ, Peloso GM, Yu H, Butterworth AS, Wang X, Mahajan A, et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet 2017; 49: 1758-66.10.1038/ng.3977Search in Google Scholar PubMed PubMed Central
15 Hutin D, Tamblyn L, Gomez A, Grimaldi G, Soedling H, Cho T, et al. Hepatocyte-Specific Deletion of TIPARP, a Negative Regulator of the Aryl Hydrocarbon Receptor, Is Sufficient to Increase Sensitivity to DioxinInduced Wasting Syndrome. Toxicol Sci 2018; 165: 347-60.10.1093/toxsci/kfy136Search in Google Scholar PubMed PubMed Central
16 Ahmed S, Bott D, Gomez A, Tamblyn L, Rasheed A, Cho T, et al. Loss of the Mono-ADP-ribosyltransferase, Tiparp, Increases Sensitivity to Dioxin-induced Steatohepatitis and Lethality. J Biol Chem 2015; 290: 16824-40.10.1074/jbc.M115.660100Search in Google Scholar PubMed PubMed Central
17 Diani-Moore S, Ram P, Li X, Mondal P, Youn DY, Sauve AA, et al. Identification of the aryl hydrocarbon receptor target gene TiPARP as a mediator of suppression of hepatic gluconeogenesis by 2,3,7,8-tetrachlorodibenzo-p-dioxin and of nicotinamide as a corrective agent for this effect. J Biol Chem 2010; 285: 38801-10.10.1074/jbc.M110.131573Search in Google Scholar PubMed PubMed Central
18 Hivert MF, Scholtens DM, Allard C, Nodzenski M, Bouchard L, Brisson D, et al. Genetic determinants of adiponectin regulation revealed by pregnancy. Obesity (Silver Spring) 2017; 25: 935-44.10.1002/oby.21805Search in Google Scholar PubMed PubMed Central
19 Wu J, Boström P, Sparks LM, Ye L, Choi JH, Giang A-H, et al. Beige Adipocytes Are a Distinct Type of Thermogenic Fat Cell in Mouse and Human. Cell 2012; 150: 366-76.10.1016/j.cell.2012.05.016Search in Google Scholar PubMed PubMed Central
20 Wang N, Verna L, Chen NG, Chen J, Li H, Forman BM, et al. Constitutive Activation of qazFHL[Peroxisome Proliferator-activated Receptor-γ Suppresses Pro-inflammatory Adhesion Molecules in Human Vascular Endothelial Cells. Journal of Biological Chemistry 2002; 277: 34176-81.10.1074/jbc.M203436200Search in Google Scholar PubMed
21 Li Z, Xu G, Qin Y, Zhang C, Tang H, Yin Y, et al. Ghrelin promotes hepatic lipogenesis by activation of mTOR-PPARgamma signaling pathway. Proc Natl Acad Sci U S A 2014; 111: 13163-8.10.1073/pnas.1411571111Search in Google Scholar PubMed PubMed Central
22 Zou MH. Is NAD(P)H oxidase a missing link for air pollution-enhanced obesity? Arterioscler Thromb Vasc Biol 2010; 30: 2323-4.10.1161/ATVBAHA.110.216648Search in Google Scholar PubMed PubMed Central
23 Zhai L, Zhao J, Xu B, Deng Y, Xu Z. Influence of indoor formaldehyde pollution on respiratory system health in the urban area of Shenyang, China. Afr Health Sci 2013; 13: 137-43.10.4314/ahs.v13i1.19Search in Google Scholar PubMed PubMed Central
24 Jung DY, Leem JH, Kim HC, Kim JH, Hwang SS, Lee JY, et al. Effect of Traffic-Related Air Pollution on Allergic Disease: Results of the Children’s Health and Environmental Research. Allergy Asthma Immunol Res 2015; 7: 359-66.10.4168/aair.2015.7.4.359Search in Google Scholar PubMed PubMed Central
25 Vrijheid M, Fossati S, Maitre L, Marquez S, Roumeliotaki T, Agier L, et al. Early-Life Environmental Exposures and Childhood Obesity: An Exposome-Wide Approach. Environ Health Perspect 2020; 128: 67009.10.1289/EHP5975Search in Google Scholar PubMed PubMed Central
26 Suzuki T, Hidaka T, Kumagai Y, Yamamoto M. Environmental pollutants and the immune response. Nat Immunol 2020; 21: 1486-95.10.1038/s41590-020-0802-6Search in Google Scholar PubMed
27 Dixon AE, Poynter ME. Mechanisms of Asthma in Obesity. Pleiotropic Aspects of Obesity Produce Distinct Asthma Phenotypes. Am J Respir Cell Mol Biol 2016; 54: 601-8.10.1165/rcmb.2016-0017PSSearch in Google Scholar PubMed PubMed Central
28 Umetsu DT. Mechanisms by which obesity impacts upon asthma. Thorax 2017; 72: 174-7.10.1136/thoraxjnl-2016-209130Search in Google Scholar PubMed
29 Moser C, Straub LG, Rachamin Y, Dapito DH, Kulenkampff E, Ding L, et al. Quantification of adipocyte numbers following adipose tissue remodeling. Cell Rep 2021; 35: 109023.10.1016/j.celrep.2021.109023Search in Google Scholar PubMed
30 Covarrubias AJ, Perrone R, Grozio A, Verdin E. NAD(+) metabolism and its roles in cellular processes during ageing. Nat Rev Mol Cell Biol 2021; 22: 119-41.10.1038/s41580-020-00313-xSearch in Google Scholar PubMed PubMed Central
31 Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab 2012; 15: 838-47.10.1016/j.cmet.2012.04.022Search in Google Scholar PubMed PubMed Central
32 Mohamed JS, Hajira A, Pardo PS, Boriek AM. MicroRNA-149 inhibits PARP-2 and promotes mitochondrial biogenesis via SIRT-1/PGC-1alpha network in skeletal muscle. Diabetes 2014; 63: 1546-59.10.2337/db13-1364Search in Google Scholar PubMed
33 Bai P, Canto C, Brunyanszki A, Huber A, Szanto M, Cen Y, et al. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab 2011; 13: 450-60.10.1016/j.cmet.2011.03.013Search in Google Scholar PubMed PubMed Central
34 Szanto M, Bai P. The role of ADP-ribose metabolism in metabolic regulation, adipose tissue differentiation, and metabolism. Genes Dev 2020; 34: 321-40.10.1101/gad.334284.119Search in Google Scholar PubMed PubMed Central
35 Rappou E, Jukarainen S, Rinnankoski-Tuikka R, Kaye S, Heinonen S, Hakkarainen A, et al. Weight Loss Is Associated With Increased NAD(+)/ SIRT1 Expression But Reduced PARP Activity in White Adipose Tissue. J Clin Endocrinol Metab 2016; 101: 1263-73.10.1210/jc.2015-3054Search in Google Scholar PubMed
36 Kumar A, Ren Y, Sundaram K, Mu J, Sriwastva MK, Dryden GW, et al. miR-375 prevents high-fat diet-induced insulin resistance and obesity by targeting the aryl hydrocarbon receptor and bacterial tryptophanase (tnaA) gene. Theranostics 2021; 11: 4061-77.10.7150/thno.52558Search in Google Scholar PubMed PubMed Central
© 2022 Lingchao Liu, Bahetiyaer Huwatibieke, Xiaoqing Lu, Dexin Li, Haiyan Dong, Jingyan Han, Weizhen Zhang, Yi Zhang, Yin Li, published by Sciendo
This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 3.0 License.
Articles in the same Issue
- Editorial
- Inhaled antibiotics and airway bacterial decolonization for patients with chronic obstructive pulmonary disease: The rationale and future
- Perspective
- Retinal examination modalities in the early detection of Alzheimer's disease: Seeing brain through the eye
- Commentary
- Hydrogel: A promising new technique for treating Alzheimer’s disease
- Metabolic adaptation in lactation: Insulin-dependent and -independent glycemic control
- Review Article
- Aberrant energy metabolism in Alzheimer’s disease
- Obesity and coronavirus disease 2019
- Crosstalk between adipose tissue and the heart: An update
- Delta variant: Partially sensitive to vaccination, but still worth global attention
- Benefits of physical activity on cardiometabolic diseases in obese children and adolescents
- Original Article
- TCDD-inducible poly (ADP-ribose) polymerase promotes adipogenesis of both brown and white preadipocytes
- Functional evaluation of intermediate coronary lesions with integrated computed tomography angiography and invasive angiography in patients with stable coronary artery disease
- A novel wide-band dielectric imaging system for electro-anatomic mapping and monitoring in radiofrequency ablation and cryoablation
- Brief Report
- Impact of COVID-19 pandemic control measures on infection of other respiratory pathogens: A real-world data research in Guangzhou, China
Articles in the same Issue
- Editorial
- Inhaled antibiotics and airway bacterial decolonization for patients with chronic obstructive pulmonary disease: The rationale and future
- Perspective
- Retinal examination modalities in the early detection of Alzheimer's disease: Seeing brain through the eye
- Commentary
- Hydrogel: A promising new technique for treating Alzheimer’s disease
- Metabolic adaptation in lactation: Insulin-dependent and -independent glycemic control
- Review Article
- Aberrant energy metabolism in Alzheimer’s disease
- Obesity and coronavirus disease 2019
- Crosstalk between adipose tissue and the heart: An update
- Delta variant: Partially sensitive to vaccination, but still worth global attention
- Benefits of physical activity on cardiometabolic diseases in obese children and adolescents
- Original Article
- TCDD-inducible poly (ADP-ribose) polymerase promotes adipogenesis of both brown and white preadipocytes
- Functional evaluation of intermediate coronary lesions with integrated computed tomography angiography and invasive angiography in patients with stable coronary artery disease
- A novel wide-band dielectric imaging system for electro-anatomic mapping and monitoring in radiofrequency ablation and cryoablation
- Brief Report
- Impact of COVID-19 pandemic control measures on infection of other respiratory pathogens: A real-world data research in Guangzhou, China

