Abstract
Colorectal cancer (CRC) is a prevalent malignancy, and its liver metastasis is a key factor leading to patient death. Research has shown that exosomal microRNA (miRNA) plays an important role as a mediator of communication between tumor cells and other cells in the growth, invasion, and metastasis of cancer cells. It also plays a critical role in the occurrence and development of CRC liver metastasis (CRC-LM). This article reviews the biological functions of exosomal miRNA and explores its significant involvement in the mechanism of CRC-LM. It also focuses on the latest progress of extracellular vesicle miRNA in the study of CRC-LM, providing new ideas and methods for future treatments.
Introduction
Colorectal cancer (CRC) is a widespread cancer affecting the digestive system, with both its incidence and mortality rates being notably high. According to the “2022 Global Cancer Statistical Report”, its incidence rate ranks third in malignant tumors and its mortality ranks second in malignant tumors (Figure 1). According to specific data, in 2022, the number of new cases of CRC worldwide reached 1,926,118, while the number of deaths from CRC reached 903,859 [1]. It is particularly noteworthy that men are more vulnerable in this battle against cancer, with a prevalence rate of 1.24 times that of women, and a mortality rate of 1.25 times that of women. The pathogenesis of CRC is primarily gradual, often manifesting initially from precursor lesions such as adenomatous polyps or serrated lesions, which typically lack a stalk. These precursor conditions can evolve over time if not monitored or managed appropriately [2]. Key demographic variables also contribute significantly to the development of CRC. The aging population, particularly in affluent nations, alongside dietary patterns, contributes significantly to increasing rates. Moreover, this disease correlates strongly with a series of modifiable risk factors, including obesity, sedentary lifestyles characterized by insufficient physical exercise, and tobacco use, highlighting the importance of lifestyle interventions and public health initiatives in combating this malignancy [3].
![Figure 1:
The incidence and mortality rates of the top 10 most frequently occurring cancers in 2022 (data from Bray et al. [1]).](/document/doi/10.1515/oncologie-2024-0571/asset/graphic/j_oncologie-2024-0571_fig_001.jpg)
The incidence and mortality rates of the top 10 most frequently occurring cancers in 2022 (data from Bray et al. [1]).
Cells in living organisms communicate crucial information through direct contact or by releasing substances, which is essential for normal cell growth and function. Disruptions in cellular communication can contribute to abnormal cell behavior, and in some cases, may be involved in the onset or advancement of cancer [4]. This is particularly relevant in CRC, where the dysregulation of cell-to-cell communication is crucial in the progression and spread of tumors. Research has found that extracellular vesicles (EVs) represent a novel form of cell-to-cell communication. EVs are different types of membrane structures produced by cells, such as exosomes and microvesicles, which fall from the interior or surface of cells. These vesicles are found everywhere in biological fluids and are important for life activities [5]. As a type of EVs, exosomes also participate in many life activities. They are important tools for cell signaling and are crucial for cellular research [6]. Especially, RNA in vesicles plays a significant role in CRC [7], 8]. Non-coding RNAs (ncRNAs) are functional RNA molecules categorized into housekeeping ncRNAs and regulatory ncRNAs. In recent years, regulatory ncRNAs, particularly miRNAs, lncRNAs, and circRNAs, have been extensively studied in exosomes [9]. miRNAs are particularly significant in cancer biology. Increasing evidence emphasizes that dysregulated miRNA expression plays functional roles in the metastasis of CRC [10]. Many studies have found that miRNAs with abnormal expression can help us study CRC [11], 12].
Furthermore, the liver is the primary site where CRC metastasizes, greatly affecting patient survival and recovery [13], 14]. About half of patients were discovered with lower TNM (Tumor Node Metastasis) stage ultimately develop metastases, predominantly in the liver [12], 15]. About 35–55 % of metastases in patients with metastatic CRC are limited to the liver [16], 17]. The primary reason for the failure of treatment for CRC liver metastasis (CRC-LM) is the occurrence of local recurrence or distant metastasis. In curative liver resection surgery, about 80 % of patients experience liver recurrence within 2 years [18]. Over the past decade, the emergence of new chemotherapy drugs and targeted therapies has improved the treatment efficacy for patients with metastatic CRC. Nowadays, there are many studies exploring how colon cancer metastasizes [19], [20], [21], [22], and for those patients who have already been transferred, their average survival time is approximately 30 months, a doubling from the past 20 years [23]. At present, there is a lack of clear biochemical and immunological indicators for early monitoring of CRC-LM; Finding efficient evaluation indicators for CRC-LM has important clinical value [24], 25]. This article aims to introduce the latest research advances on miRNAs in exosomes, reviewing the progress of exosomal miRNAs influencing CRC-LM, with the intention of providing insights for related research.
Origin and mechanism of exosomal miRNAs
Exosomes are intracellularly derived vesicles that are encapsulated by a double-layer lipid membrane structure, which provides them with the stability necessary to exist in the extracellular environment. This unique membrane encapsulation serves to safeguard biologically active molecules within these vesicles. The term “exosome” was first introduced by Trams et al. in 1981, who described the shedding of membrane vesicles that potentially possess physiological functions [26]. Exosomes generally have a diameter that ranges from 40 to 160 nm, with an average measuring approximately 100 nm, as depicted in Figure 2. They constitute a particular subset of EVs secreted by a majority of eukaryotic cells [27]. Although initially identified in the late 1980s and regarded merely as cellular debris, exosomes are now understood to play a crucial role in intercellular communication, contributing to a variety of biological activities in both healthy and diseased states, such as oncology [28]. Exosomes can originate from a wide range of cell types, including cancer cells, immune cells, and neurons, among others. The exosomes derived from distinct cell types may carry unique molecular compositions, which in turn influence their specific functions. The biological roles of exosomes are significantly dependent on their biologically active constituents, which include nucleic acids, proteins, lipids, and metabolites. The latest information from the exosome database (http://www.exocarta.org) indicates that there are many different molecular components in EVs, such as 9,769 proteins, 3,408 mRNAs, 2,838 miRNAs, and 1,116 lipids [29]. These molecules can be transferred between cells via exosomes, thereby affecting the functionality and behavior of target cells. Furthermore, exosomes are actively involved in cancer progression and metastasis, facilitating the transfer of biologically active molecules among various cells within the cancer microenvironment, whether locally or distally. They have the potential to serve as prognostic markers for cancer, represent therapeutic targets, and even act as carriers for anticancer drugs [30].
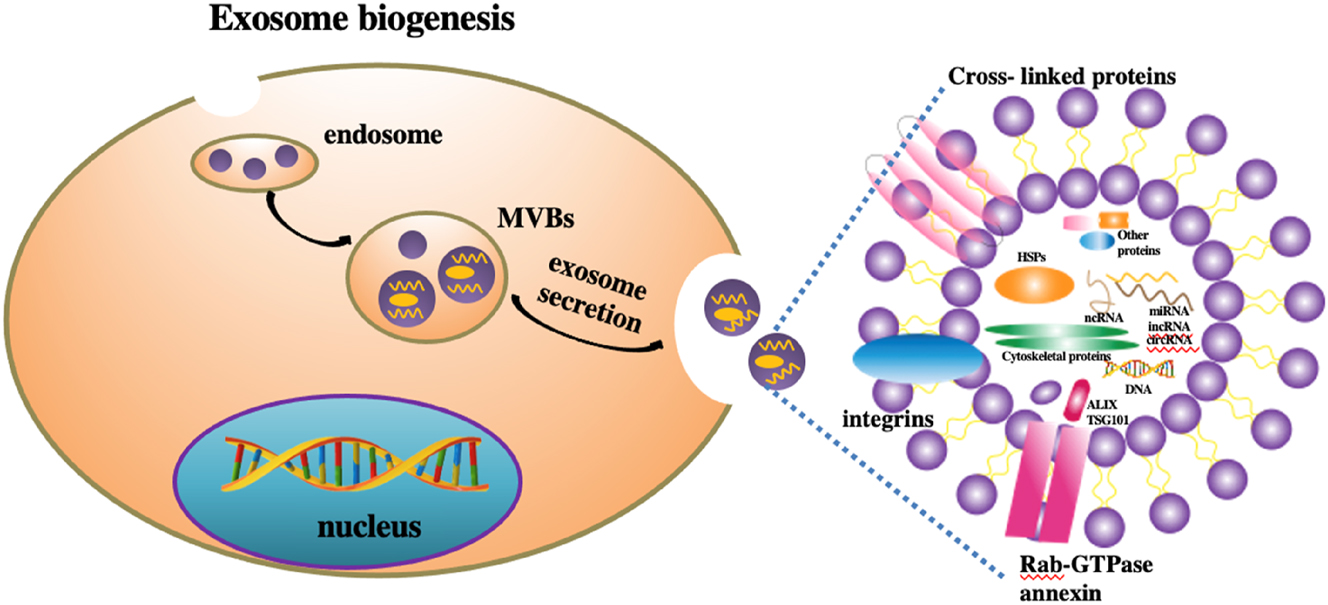
Biogenesis and contents of EVs.
Because EVs are highly stable, they can act as a mediator through various biological barriers and are abundant in our bodily fluids, positioning them as biomarkers for the early diagnosis of various diseases. For instance, prostate-specific membrane antigen (PSMA), secreted from exosomes of prostate cancer cells, is highly expressed in our prostate cancer cells, making it a key substance for prostate cancer diagnosis and monitoring [31]. Research has shown that circulating EVs obtained from the blood can also be used for the diagnosis of early CRC patients [32]. Exosomes miR-210 and miR-638 serve a crucial function in the early detection of hepatocellular carcinoma (HCC) [33]. In addition, the efficacy of many other EV miRNAs in early cancer detection has also been evaluated [34]. In endometrial cancer, the high expression of miR-15a-5p makes it a specific and promising diagnostic biomarker [35]. In the neurodegenerative disease Parkinson’s disease (PD), the EV miR-128 in blood samples can serve as a biomarker for early diagnosis [36]. Other diseases include ischemic stroke [37]. These studies demonstrate the important contribution of EVs to the early detection of various diseases, particularly cancer.
miRNA is a non-coding, single-stranded RNA molecule, about 22 nucleotides long, that is encoded by endogenous genes, usually ranging from 20 to 24 nucleotides, but there are also longer or shorter variants. miRNA is an important component of gene expression regulation, which can participate in multiple biological processes such as cell cycle regulation and biological developmental timing. miRNA can affect protein synthesis and cellular biological processes, mainly by regulating the translation and degradation of mRNA from target genes. When miRNA suppresses the translation of mRNA from target genes, it leads to a decrease in protein synthesis encoded by the target gene. In the case of degradation, the target gene is completely inhibited, affecting biological processes. Gene expression regulation has always been a subject of physiological and pathological research, as shown by Bajan et al. [38]. In the field of tumor research, miRNAs encapsulated in EVs are becoming promising biomarkers for detecting and evaluating cancer. It is noteworthy that miRNAs can reduce the stability and translation of target mRNAs that exhibit complete or partial sequence complementarity, demonstrating unique and diverse expression patterns that significantly impact numerous cellular processes and developmental pathways [39]. The production of miRNAs is driven by two crucial RNase III enzymes: Drosha and Dicer, as shown in Figure 3. This process involves two sequential steps for miRNA processing, taking place initially in the cell nucleus and subsequently in the cytoplasm. The first step occurs in the nucleus of the cell, where the primary miRNA (pri-miRNA) is transcribed from DNA, with a length of approximately 300–1,000 bases. It is then processed and cleaved by enzymes to become a precursor miRNA (pre-miRNA), which has a stem-loop structure and is about 70–90 bases long. Subsequently, pre-miRNA was transported to the cytoplasm by Exportin-5, where it undergoes a final processing stage mediated by Dicer, yielding mature miRNA duplexes. These duplexes then associate with Argonaute proteins, forming RNA-induced silencing complexes (RISC), where one strand is designated as the active, mature miRNA [38]. In the context of CRC, exosomal miRNAs not only function as biomarkers but also as signaling molecules, participating in tumor cell metastasis.
![Figure 3:
Biogenesis of miRNA (figure from Balacescu et al. [12], without any adaption, under the license of CC BY 4.0: http://creativecommons.org/licenses/by/4.0).](/document/doi/10.1515/oncologie-2024-0571/asset/graphic/j_oncologie-2024-0571_fig_003.jpg)
Biogenesis of miRNA (figure from Balacescu et al. [12], without any adaption, under the license of CC BY 4.0: http://creativecommons.org/licenses/by/4.0).
miRNAs exhibit significant stability in living organisms, such as research showing strong stability under freezing and extreme pH conditions, which makes them reliable in clinical applications [40]. In addition, when detecting miRNA, we can use less uncomfortable methods for patients, such as not having to undergo surgery or only making a small incision, so that patients will not feel so uncomfortable and their health status can be tracked for a long time [41]. More importantly, miRNA can serve as a super accurate biomarker to help doctors diagnose and predict disease progression more accurately in clinical practice. Especially for diseases like cancer and glaucoma, miRNA can detect them early because it can reflect subtle changes at the beginning of the disease [42]. Detecting miRNA is also very simple, and there are now many high-tech methods that can detect many samples at once, which are both convenient and fast to operate [43]. Moreover, by studying miRNA, scientists can uncover many complex biological processes, which helps us provide more personalized treatment plans for patients and make treatment more precise and effective [42].
Mechanisms of exosomal miRNA in CRC
Exosomal miRNAs are crucial in the development of CRC. They are like accelerators for tumors, not only allowing them to grow faster but also making it easier for them to spread to other places. These small RNA molecules are important “messengers” for communication between cells, especially between malignant cells, immune cells, and surrounding support cells, transmitting important information. Therefore, miRNA plays an irreplaceable role in constructing and maintaining the tumor microenvironment (TME), which is the critical setting for tumor growth [9], 44]. Moreover, miRNAs are essential for maintaining the normal functioning of our colon cells. Disruptions in the abundance of these miRNAs have been implicated in various aspects of CRC, including its initiation, progression, metastatic spread, development of drug resistance, and recurrence after treatment [45]. Research has established that miRNAs, which are secreted via exosomes or microvesicles, engage in direct communication processes between tumor cells and the surrounding TME [46]. Noteworthy examples of exosomal miRNAs identified as potential prognostic markers for CRC include miR-934, miR-25-3p, and miR-193a-5p, all of which correlate with the development of liver metastasis and more advanced disease stages [47]. Additionally, exosomal miRNAs such as miR-19b and miR-29a can respectively make cancer cells more resistant to radiation therapy and more prone to metastasis. So, if we can target these specific miRNAs, we may be able to find new methods for treating CRC [48]. In the serum of CRC patients, certain exosomal miRNAs, including miR-101-3p and miR-320d, have emerged as promising diagnostic biomarkers for metastatic CRC, providing a non-invasive method for early diagnosis and continuous tracking of the disease [49], 50]. Exosomal miRNAs have the capacity to modulate gene expression within recipient cells, affecting critical biological processes such as vascular permeability, immune response modulation, and cellular proliferation. For instance, miR-29a has been shown to diminish the expression of tight junction proteins, which consequently facilitates metastatic processes [51]. Bioinformatics analyses have illuminated the complexity of exosomal miRNA-mRNA regulatory networks in CRC, underscoring the intricate roles these miRNAs possess in cancer development and progression [52]. In terms of immune regulation, miRNAs in EVs can alter macrophages, making them more favorable for tumor growth. This creates an environment that makes tumors easier to grow and spread [44]. If we can fully understand how these EV miRNAs work in CRC, miRNA-based treatment methods may have greater development potential and help doctors better predict and diagnose this disease [47].
Exosomes are released by various types of cells, including malignant tumor cells, and are easily found in body fluids such as blood and cell culture media, etc. [53], 54]. People with CRC carry biologically active molecules from their EVs into liver cells, altering their characteristics [55], 56]. The tumor-derived exosomes (TEX) possess a unique ability to mediate organotropic metastasis, which is partly dictated by the expression of specific integrins on their surface. For example, integrin αvβ5 has been associated with liver metastasis, as it preferentially binds to Kupffer cells in the liver, promoting the formation of a pre-metastatic niche [57]. Similarly, integrin α6β4 and α6β1 have been linked to lung metastasis [58], 59]. These integrins bind to receptors on target cells, activate signals that cause inflammation in the body, and prepare “niches” in advance where cancer cells may go [60]. The process of cancer cells metastasizing to the liver is complex and undergoes many changes. This includes changes in the shape and function of circulating tumor cells (CTCs), such as transitioning from epithelial-like cells to mesenchymal-like cells (epithelial-mesenchymal transition, EMT) or vice versa (mesenchymal-epithelial transition, MET), as well as alterations in the liver’s surrounding environment and immune system suppression. A special type of EV released by CRC cells, called miRNA-TEX, plays a crucial role in this process. These EVs carry specific miRNAs, like miR-21, miR-181b, and miR-122, which affect both CTCs and the liver microenvironment. For example, miR-181b targets PTEN, resulting in enhanced activation of the PI3K/AKT pathway and promoting a pro-metastatic phenotype [61]. miR-122 has been shown to modulate glucose metabolism in the liver, creating a favorable energy environment for incoming cancer cells [62]. In addition, there are two receptors called CXCR4 and CCR6, which are essential in facilitating the transport of miRNA-TEX to the liver, as shown in Figure 4. And the liver itself also releases miRNAs, such as miR-203 and miR-155, which activate inflammatory pathways and promote tumor growth, further preparing the pre-metastatic niche for cancer cells [63], 64]. Furthermore, research investigating the metastatic patterns of CRC indicates that the liver is the primary site of metastasis, whereas other organs such as the lungs, bones, and brain are affected far less frequently [65].
![Figure 4:
CRC metastasis in liver via CTC (figure from Balacescu et al. [12], without any adaption, under the license of CC BY 4.0: http://creativecommons.org/licenses/by/4.0).](/document/doi/10.1515/oncologie-2024-0571/asset/graphic/j_oncologie-2024-0571_fig_004.jpg)
CRC metastasis in liver via CTC (figure from Balacescu et al. [12], without any adaption, under the license of CC BY 4.0: http://creativecommons.org/licenses/by/4.0).
Based on the extensive body of research available, miRNAs stand out as the most extensively studied encapsulated biomolecules within exosomes, particularly in their connection to the onset, advancement, and spread of cancer. A 2010 study demonstrated that non-coding regulatory miRNAs derived from cells and viruses can control gene expression by inhibiting the translation of mRNA into proteins, validating the role of miRNA-TEX in regulating gene expression within recipient cells [66], their notable contribution to cancer biomarkers has garnered significant recognition [67]. Wang et al. have demonstrated for the first time the crucial role of EVs in CRC-LM. In their study, using a nude mouse model, they found that EVs from a type of CRC cell (HT-29) that is particularly prone to liver metastasis can successfully induce strong liver metastasis in an environment composed of another type of cell (Caco-2) that is less prone to liver metastasis. Research has shown that the exosomes released by HT-29 cells can facilitate the spread of CRC to the liver because they can increase the production of a molecule called CXCR4 by various stromal cells. This molecule helps to alter the environment of the liver and provides a “foothold” for cancer cells before metastasis. In addition, when researchers added exosomes from HT-29 cells to Caco-2 cells, the mobility of Caco-2 cells was greatly enhanced, further demonstrating the important role of exosomes in CRC metastasis. Further research has also found that a small molecule called miRNA-TEX is an important “driver” in the process of CRC metastasis to the liver [68]. Among these miRNAs, miR-6803-5p and miR-548c-5p have shown significant influence on CRC progression. For example, miR-6803-5p enhances cancer cell proliferation and invasion via PTPRO/NF-κB Axis in CRC [69]. In contrast, in SW480 CRC cells overexpressing miR-548c-5p and stimulated by lipopolysaccharide (LPS), both cell proliferation and the production of inflammatory cytokines (TNF-α and IL-6) were notably reduced. Thus, miR-548c-5p may act as a tumor suppressor in CRC by targeting PGK1 [70].
Current research on exosomal miRNAs in liver metastasis of CRC
When diagnosed with CRC, 14–20 % of patients have already developed liver metastases. Among these patients who have already had liver metastases, one-third of them may experience worsening of their condition in the future, further affecting the liver [71]. Checking the level of carcinoembryonic antigen (CEA) in the blood is also important because it can tell us about the condition of the tumor and help us understand the effectiveness of treatment [72]. Now, doctors are also paying attention to inflammation indicators and CTCs before treatment, which may predict the risk of liver metastasis and help doctors decide on treatment plans [73]. Additionally, it is worth mentioning that non-alcoholic fatty liver disease (NAFLD) has been linked to an elevated risk for CRC-LM patients [74]. The presence of liver metastases considerably affects both the overall prognosis and the management strategies for CRC [71]. Surgical resection continues to be the main curative approach, and the incorporation of multimodal treatment strategies, including chemotherapy and targeted therapies, has been shown to enhance patient outcomes. Unfortunately, only 10–20 % of patients are eligible for radical surgical treatment at the time of diagnosis, highlighting the necessity for alternative therapeutic modalities for the larger proportion of patients.
Recently, numerous studies have demonstrated that EV miRNAs are crucial in regulating liver metastasis in CRC. Table 1 summarizes the roles of relevant miRNAs in liver metastasis. miR-21 is significantly enriched in exosomes sourced from CRC, and it plays an essential role in fostering a pro-inflammatory phenotype in the liver, as well as facilitating liver metastasis. Comprehensive microarray analysis has revealed that exosomes from CRC, which contain miR-21, induce an inflammatory microenvironment within murine livers, enhance the invasion of macrophages, and subsequently promote the localized liver metastasis of CRC cells [75]. Emerging data demonstrates that diminished levels of serum exosomal miR-638 correlate with an elevated risk of liver metastasis [76]. In an intriguing hypothesis put forth by Monzo et al., it is suggested that blood samples obtained from tumor drainage veins may yield more consistent and uniform insights compared to blood collected from peripheral veins (PV). Notably, in individuals diagnosed with liver metastasis, EV miR-328 levels in mesenteric vein (MV) plasma are notably higher compared to those in PV plasma. The finding of this study indicates that miR-328 may be involved in the mechanism of liver metastasis in colon cancer [77]. Additionally, research conducted by Teng et al. has demonstrated that exosomes extracted from the plasma of CRC patients with liver metastasis show increased levels of miR-193a, miR-126, and miR-148a, while exhibiting reduced levels of miR-196 in comparison to plasma obtained from patients devoid of metastasis [78].
miRNA worked on liver metastasis of CRC.
| Study | Findings | Key miRNA | Implications | Validation in human patients |
|---|---|---|---|---|
| Shao et al. (2018) [75] | miR-21-enriched CRC exosomes induce liver inflammation, enhance macrophage invasion, and promote metastasis. | miR-21 | miR-21 drives inflammatory niche formation and metastasis. | Yes |
| Yan et al. (2017) [76] | Low serum exosomal miR-638 correlates with increased liver metastasis risk. | miR-638 | miR-638 is a potential biomarker for metastasis risk. | Yes |
| Monzo et al. (2017) [77] | Higher miR-328 in mesenteric vein exosomes vs. peripheral veins in liver metastasis patients. | miR-328 | miR-328 may play a mechanistic role; mesenteric veins enhance biomarker analysis. | Yes |
| Teng et al. (2017) [78] | Elevated miR-193a, miR-126, miR-148a, and reduced miR-196 in CRC patient plasma with liver metastasis. | miR-193a, miR-126, miR-148a, miR-196 | These miRNAs are candidate biomarkers for metastasis monitoring. | No |
| Pei et al. (2023) [79] | Exosomal miR-203a-3p targets PTEN, inducing macrophage polarization and promoting liver metastasis. | miR-203a-3p | miR-203a-3p enhances metastasis via PTEN-mediated macrophage effects. | Yes |
| Zhao et al. (2020) [80] | Exosomal miR-934 induces M2 macrophage polarization, facilitating liver metastasis in CRC. | miR-934 | miR-934 promotes liver metastasis through macrophage polarization. | Yes |
| Shibamoto et al. (2023) [81] | Elevated plasma and tissue miR-4442 in CRC predicts liver metastasis and EMT, with levels reducing post-resection. | miR-4442 | miR-4442 is a potential marker for micrometastatic activity. | Yes |
| Liu et al. (2023) [51] | Exosomal miR-29a disrupts vascular endothelial barriers, promoting CRC-LM in murine models. | miR-29a | miR-29a is a target for metastasis prevention and therapeutic intervention. | No |
| Chen et al. (2023) [82] | hUC-MSC-derived exosomal miR-1827 inhibits M2 macrophage polarization and prevents liver metastasis. | miR-1827 | miR-1827 offers therapeutic potential via targeted macrophage modulation. | No |
| Zhao et al. (2022) [83] | EV miR-181a-5p stimulates hepatic stellate cells, enhancing CRC liver metastasis by TME remodeling. | miR-181a-5p | miR-181a-5p contributes to aggressive liver metastasis and TME modulation. | Yes |
| Sun et al. (2021) [84] | Hypoxia-induced exosomal miR-135a-5p activates Kupffer cells and immune suppression pathways in CRC-LM. | miR-135a-5p | miR-135a-5p promotes liver tropism via immune suppression signaling. | No |
| Tian et al. (2021) [85] | Exosomal miR-221/222 targets SPINT1, enhancing HGF secretion in the PMN and promoting CRC metastasis. | miR-221/222 | miR-221/222 drives PMN formation and metastasis through stromal cell activation. | Yes |
| Sur et al. (2021) [86] | miR-125b-5p, miR-17-5p, and miR-185-5p predict liver metastasis and chemotherapy response in late-stage CRC. | miR-125b-5p, miR-17-5p, miR-185-5p | Non-invasive biomarkers for metastasis risk and treatment outcomes. | Yes |
| Liu et al. (2021) [87] | miR-140-3p inhibits CRC-LM progression by targeting BCL9 and BCL2. Low miR-140-3p is linked to metastasis. | miR-140-3p | Potential therapeutic target and biomarker for CRC metastasis. | Yes |
| Wang et al. (2020) [88] | Exosomal miR-25-3p, miR-130b-3p, and miR-425-5p induce M2 polarization and enhance CRC metastasis via PTEN/PI3K/Akt signaling. | miR-25-3p, miR-130b-3p, miR-425-5p | Non-invasive biomarkers and therapeutic targets for metastasis. | No |
| Sun et al. (2020) [89] | Serum exosomal miR-122 is a diagnostic and prognostic marker for CRC-LM, linked to poor prognosis. | miR-122 | miR-122 is a specific biomarker for CRC with liver metastasis. | Yes |
| Matsumura et al. (2015) [90] | Identified 18 upregulated and 46 downregulated miRNAs in CRC patients with liver metastasis. miR-19a linked to poor prognosis. | miR-19a/b, miR-23a, miR-92a, miR-320a, miR-4437 | Exosomal miRNAs are markers for early detection and prognosis. | Yes |
Macrophages are a diverse group of immune cells that demonstrate remarkable adaptability in response to various stimuli by modifying their functional states within the TME. The intrinsic plasticity of macrophages plays a pivotal role in their functionality, enabling them to polarize into two distinct phenotypes, M1 macrophages, which are classically activated and characterized by their ability to secrete type I anti-inflammatory cytokines that exert anti-cancer effects [91]. and M2 macrophages, which display an opposing response characterized by anti-inflammatory properties and a pro-cancer profile [92]. Research indicates that interactions between tumor cells and tumor-associated macrophages (TAMs) occur through exosome-mediated communication. Specifically, miRNA derived from tumor cells can downregulate the expression of phosphatase and tensin homolog (PTEN), thereby activating the AKT signaling pathway and promoting polarization of M2 macrophages. In this interaction, TAMs may support or hinder tumor progression, which is influenced by the biological characteristics of the tumor cells they interact with. For instance, Pei et al. have suggested that exosomal miR-203a-3p released by CRC tumor cells may drive macrophage polarization by targeting PTEN, thereby enhancing the CRC-LM [79]; Additionally, Zhao et al. have investigated exosomal miR-934, which is also derived from CRC cells, and demonstrated its capability to promote M2 macrophage polarization, further facilitating CRC-LM (Figure 5) [80]. More precisely, CRC-derived EV miR-934 can trigger M2 macrophage polarization by reducing PTEN expression, thereby activating the PI3K/AKT signaling pathway. This polarization is driven by a positive feedback mechanism of the CXCL13/CXCR5/NFκB/p65/miR-934 axis, which facilitates liver metastasis of CRC. These findings can guide the development of effective CRC-LM prevention and treatment strategies. Notably, elevated levels of miR-934 detected in serum exosomes have been associated with CRC-LM, indicating its potential as a promising biomarker for liquid biopsy and an important predictor of future CRC-LM risk.
![Figure 5:
Exosomal miR-934 promotes liver metastasis of colorectal cancer (figure from Zhao et al. [80], without any adaption, under the license of CC BY 4.0: http://creativecommons.org/licenses/by/4.0).](/document/doi/10.1515/oncologie-2024-0571/asset/graphic/j_oncologie-2024-0571_fig_005.jpg)
Exosomal miR-934 promotes liver metastasis of colorectal cancer (figure from Zhao et al. [80], without any adaption, under the license of CC BY 4.0: http://creativecommons.org/licenses/by/4.0).
Shibamoto et al. discovered a varied overexpression of miR-4442 in CRC tissues, noting that certain CRC cells with elevated levels of miR-4442 were correlated with the progression and growth of liver metastases. Consequently, high concentrations of miR-4442 present in the preoperative plasma of CRC patients could serve as indicators of micrometastatic activity within lymph nodes and/or distant anatomical sites, particularly liver metastasis. In this investigation, miR-4442 exhibited significantly higher expression in tissues affected by metastatic liver cancer, yet its levels diminished post-surgical resection. Interestingly, the plasma concentrations of miR-4442 did not show a decrease following the successful surgical removal of primary CRC tissues [81]. Moreover, Liu et al. provided evidence through in vitro co-culture analyses that exosomal miR-29a facilitates liver metastasis in CRC-bearing murine models. These findings suggest that EMT CRC cells might transfer exosomal miR-29a to endothelial cells within the TME, thereby enhancing vascular permeability and advancing the progression and metastasis of CRC. The capability of exosomal miR-29a as a predictive biomarker for tumor metastasis and a promising target for CRC therapy has been highlighted [51]. A study conducted by Chen et al. indicates that human umbilical cord mesenchymal stem cell-derived exosomes (hUC-MSCs-Exos) play a role in inhibiting the polarizing effect of M2 macrophages and thereby impede liver metastasis in CRC by inducing miR-1827, which targets SUCNR1. This offers a theoretical framework for comprehending the mechanisms underlying exosome-mediated targeted therapies in CRC [82]. Moreover, Zhao et al. found the EV miR-181a-5p involved in high colorectal liver metastasis promotes liver metastasis by stimulating hepatic stellate cells and modifying TME. These highly aggressive CRC cells release an increased number of EVs enriched with miR-181a-5p, which consequently enhances CRC-LM [83]. Furthermore, Sun et al.’s study on the role of hypoxia-induced exosomes in promoting hepatic tropism of CRC revealed that Kupffer cells (KCs) can engulf exosomes that exhibit high levels of miR-135a-5p, which enter the liver from the bloodstream. Exosomal miR-135a-5p initiates the KSRP-YES-associated protein-7 axis to promote CRC-LM, and differentiation cluster 30-TNFR-associated factor 2-p65-mediated immune suppression signaling also contributes to this process [84]. Additionally, research indicates that CRC-secreted miR-221/222 can be transported via exosomes to hepatic stromal cells, inducing a favorable metastatic environment for incoming tumor cells, thus facilitating CRC metastasis. Specifically, miR-221/222-3p stimulates liver stromal cells to secrete hepatocyte growth factor (HGF) in the pre-metastatic niche (PMN) [85].
Sur et al.’s study results indicate that the studied miRNAs (namely miR-17-5p, miR-125b-5p, and miR-185-5p) are strongly linked to metastasis and therapeutic response in advanced CRC patients. These miRNAs may act as valuable non-invasive markers for identifying high-risk CRC patients for liver metastasis and assessing treatment outcomes in CRC trials [86]. Furthermore, Liu et al.’s in vivo experiments demonstrate that miR-140-3p inhibits CRC-LM, providing further evidence linking decreased miR-140-3p levels in plasma exosomes to CRC-LM, however, the role of exosomal miR-140-3p as a potential biomarker in CRC patients merits further exploration in forthcoming studies [87]. Meanwhile, the research carried out by Wang et al. indicates that CRC cells with overexpression of CXCR4 are capable of delivering a range of miRNAs, including miR-25-3p, miR-130b-3p, and miR-425-5p, to macrophages via exosomes, thereby triggering M2 polarization through the PTEN/PI3K/Akt signaling pathway. This pathway enhances CRC metastasis by promoting EMT and vascular endothelial growth factor (VEGF) secretion. Clinical findings suggest that these exosomal miRNAs have the potential to predict cancer progression and metastasis, making them promising non-invasive biomarkers for CRC [88]. Moreover, Sun et al. discovered that serum exosomal miR-122 serves as a unique diagnostic marker for CRC with liver metastasis, as its expression distinguishes CRC patients with liver metastasis from healthy individuals and those without liver metastasis. Furthermore, elevated expression of circulating exosomal miR-122 is linked to a poor prognosis in patients with CRC [89]. Notably, Matsumura et al. analyzed exosomal miRNA expression in 227 serum samples from CRC patients, finding increased expression of 18 exosomal miRNAs and decreased expression of 46 exosomal miRNAs compared to non-recurrent patients. Among these, 6 exosomal miRNAs, including miR-19a/b, miR-23a, miR-92a, miR-320a, and miR-4437, are linked to the development of liver metastasis, making them potential biomarkers for early detection of CRC-LM. Additional study shows that CRC patients with elevated exosomal miR-19a expression tend to have a worse prognosis than those with lower levels. This implies that exosomal miR-19a could serve as a potential prognostic indicator for CRC-LM [90].
Applications of exosomal miRNA in diagnosis, therapy, and prognosis of CRC-LM
Under the occurrence of CRC-LM, miRNAs in the EVs become important. It is not only being used more and more in diagnosis but has also become a focus of research in treatment and disease prediction. For example, research has found that in the blood of patients with CRC-LM, there is an EV called miR-122 that is particularly accurate, which can distinguish these patients from those without metastasis and healthy individuals [89]. In addition, studies have found that the EV miR-150 is downregulated in CRC patients with liver metastasis, and the low expression of this miRNA is associated with factors such as advanced TNM staging and elevated CA199 serum levels [93]. The plasma-derived EV miR-21 is positively correlated with liver metastasis in CRC patients, indicating that it can also be used for the diagnosis of CRC-LM [94]. MiR-17-5p and miR-92a-3p in EVs are linked to different stages of CRC, and they may be important markers for determining CRC and CRC-LM [25].
One significant advantage of exosomal miRNAs is their non-invasive detectability via blood, urine, or other bodily fluids, which positions them as potential candidates for diagnostic, prognostic, and predictive biomarkers specific to CRC-LM. Additionally, a distinct circulating exosomal miRNA signature has the potential to forecast prognosis and inform adjuvant chemotherapy strategies following liver metastasectomy in patients with CRC-LM [95]. However, the inherent heterogeneity of CRC and the variability observed in miRNA expression across tumor populations pose challenges in establishing universally applicable cut-off values for miRNA biomarkers [94], 96]. In addition to their diagnostic and prognostic capabilities, exosomal miRNAs are vital in affecting cancer metastasis by regulating the TME and orchestrating macrophage polarization, thus unveiling new targets for personalized therapeutic interventions. For example, exosomal miR-29a′s role in promoting liver metastases through increased vascular permeability marks it as a promising therapeutic target [51]. On a related note, exosomal miR-934 has been demonstrated to enhance liver metastasis by promoting the polarization of M2 macrophages, providing insights for targeted therapy [80]. In conclusion, exosomal miRNAs reveal extensive potential for application within the realms of diagnosis, treatment, and prognostic evaluation of CRC-LM. By utilizing them, we have the potential to improve early detection methods and optimize precise treatment plans for CRC.
Challenges of exosomal miRNA in CRC-LM research
Exosomal miRNA is a highly promising biomarker with great potential for diagnosing CRC and predicting disease progression. They provide valuable insights into various cell types, the progression of the disease, and potential therapeutic strategies, such as the utilization of anti-miRNA compounds [97]. Nevertheless, several challenges hinder their reliability and application in clinical settings. These include the absence of standardized procedures for exosome collection, issues related to contamination, and the limitations associated with existing analysis methods, including quantitative reverse transcription polymerase chain reaction (qRT-PCR), digital PCR, and next-generation sequencing (NGS) [98]. While the majority of research to date has concentrated primarily on serum and plasma as sources for exosomal miRNAs, there is a significant opportunity to broaden this focus to include other bodily fluids such as saliva, tears, and urine. This could potentially expand the diagnostic possibilities available for CRC. Moreover, detecting exosomal miRNAs in the context of colorectal liver metastasis presents additional complexities, specifically regarding the necessity for standardized methods of isolation and detection. Current techniques, such as size-exclusion chromatography, have proven reliable [99]; However, they face challenges regarding ease of implementation in clinical environments. Furthermore, the risk of contamination from plasma proteins coupled with the difficulties associated with the delivery of miRNA modulators highlight the need for the creation of novel treatment strategies and improved diagnostic methods [100]. Expanding research efforts into nontraditional sample types, along with leveraging advancements in bioinformatics for comprehensive miRNA profiling, could significantly improve early detection, prognostic accuracy, and the monitoring of therapeutic responses in cancer patients.
In summary, while exosomal miRNAs hold significant potential as diagnostic, prognostic, and therapeutic tools for CRC-LM, numerous challenges must be overcome to fully realize their clinical potential. Standardization of exosome isolation and analysis methods, overcoming miRNA heterogeneity, improving detection techniques, and addressing issues related to sample contamination and therapeutic delivery are essential steps in advancing the use of exosomal miRNAs in cancer management. Continued research and innovation in these areas will likely pave the way for the widespread application of exosomal miRNAs in precision oncology.
Conclusions
This study delves into the significant role of exosomal miRNAs in CRC-LM. Through a comprehensive analysis of existing research data, we found that exosomal miRNAs not only serve as key intermediaries in the communication between tumor cells and other cells, the initiation and progression of CRC-LM.
In particular, miRNAs like miR-21, miR-934, miR-29a, and miR-122 have been found to be strongly linked to CRC-LM, playing important roles in promoting inflammation, regulating macrophage polarization, affecting the liver microenvironment, and enhancing vascular permeability. Although progress has been made in revealing the potential mechanisms and clinical applications of exosomal miRNAs in CRC-LM, there are still many challenges and unknown areas. For example, detailed information on specific miRNA targets and their mechanisms of action remains to be further clarified, and more research is needed on how to effectively utilize these miRNAs as diagnostic, therapeutic, and prognostic biomarkers. In summary, exosomal miRNAs provide new perspectives and potential therapeutic targets for CRC-LM research. With the continuous advancement of technology and deeper research, we anticipate the development of more precise and effective diagnostic methods and treatment strategies to improve the prognosis and quality of life for CRC patients.
Funding source: Shaoguan Municipal Science and Technology Commission
Award Identifier / Grant number: 230330168035643
Acknowledgments
Dan Guo sincerely acknowledges the financial support provided by Shaoguan Municipal Science and Technology Commission (grant number 230330168035643) for carrying out this work.
-
Research ethics: Not applicable.
-
Informed consent: Not applicable.
-
Author contributions: Xuemei Zhong was involved in conceptualization and writing – review & editing of the manuscript. Dan Guo conceptualized and designed the writing – review & editing of the manuscript, and supervision. All authors have approved the final manuscript draft for publication.
-
Use of Large Language Models, AI and Machine Learning Tools: None declared.
-
Conflict of interest: Conflict of interests the authors have no relevant financial or non-financial interests to disclose.
-
Research funding: Shaoguan Municipal Science and Technology Commission (grant number 230330168035643).
-
Data availability: All data have been included.
References
1. Bray, F, Laversanne, M, Sung, H, Ferlay, J, Siegel, RL, Soerjomataram, I, et al.. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024;74:229–63. https://doi.org/10.3322/caac.21834.Suche in Google Scholar PubMed
2. Shaukat, A, Levin, TR. Current and future colorectal cancer screening strategies. Nat Rev Gastroenterol Hepatol 2022;19:521–31. https://doi.org/10.1038/s41575-022-00612-y.Suche in Google Scholar PubMed PubMed Central
3. Dekker, E, Tanis, PJ, Vleugels, JL, Kasi, PM, Wallace, MB. Colorectal cancer. Lancet 2019;394:1467–80. https://doi.org/10.1016/s0140-6736(19)32319-0.Suche in Google Scholar PubMed
4. Mittelbrunn, M, Sánchez-Madrid, F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol 2012;13:328–35. https://doi.org/10.1038/nrm3335.Suche in Google Scholar PubMed PubMed Central
5. Van Niel, G, d’Angelo, G, Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018;19:213–28. https://doi.org/10.1038/nrm.2017.125.Suche in Google Scholar PubMed
6. Hill, A, Shambrook, M. Exosomes and microvesicles. Methods Mol Biol 2017;1545:55–70. https://doi.org/10.1007/978-1-4939-6728-5_5.Suche in Google Scholar PubMed
7. Li, X, Wang, Q, Wang, R. Roles of exosome genomic DNA in colorectal cancer. Front Pharmacol 2022;13:923232. https://doi.org/10.3389/fphar.2022.923232.Suche in Google Scholar PubMed PubMed Central
8. Cheshomi, H, Matin, MM. Exosomes and their importance in metastasis, diagnosis, and therapy of colorectal cancer. J Cell Biochem 2019;120:2671–86. https://doi.org/10.1002/jcb.27582.Suche in Google Scholar PubMed
9. Vautrot, V, Chanteloup, G, Elmallah, M, Cordonnier, M, Aubin, F, Garrido, C, et al.. Exosomal miRNA: small molecules, big impact in colorectal cancer. J Oncol 2019;2019:8585276–18. https://doi.org/10.1155/2019/8585276.Suche in Google Scholar PubMed PubMed Central
10. Mitchell, PS, Parkin, RK, Kroh, EM, Fritz, BR, Wyman, SK, Pogosova-Agadjanyan, EL, et al.. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513–8. https://doi.org/10.1073/pnas.0804549105.Suche in Google Scholar PubMed PubMed Central
11. Hanahan, D. Hallmarks of cancer: new dimensions. Cancer Discov 2022;12:31–46. https://doi.org/10.1158/2159-8290.cd-21-1059.Suche in Google Scholar PubMed
12. Balacescu, O, Sur, D, Cainap, C, Visan, S, Cruceriu, D, Manzat-Saplacan, R, et al.. The impact of miRNA in colorectal cancer progression and its liver metastases. Int J Mol Sci 2018;19:3711. https://doi.org/10.3390/ijms19123711.Suche in Google Scholar PubMed PubMed Central
13. Jana, S, Krishna, M, Singhal, J, Horne, D, Awasthi, S, Salgia, R, et al.. Therapeutic targeting of miRNA-216b in cancer. Cancer Lett 2020;484:16–28. https://doi.org/10.1016/j.canlet.2020.04.020.Suche in Google Scholar PubMed
14. Bartel, DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. https://doi.org/10.1016/s0092-8674(04)00045-5.Suche in Google Scholar PubMed
15. Minagawa, M, Makuuchi, M. Surgical treatment of colorectal liver metastasis. Nihon Geka Gakkai Zasshi 2003;104:721–9.Suche in Google Scholar
16. Wang, D, Liu, J, Huo, T, Tian, Y, Zhao, L. The role of microRNAs in colorectal liver metastasis: important participants and potential clinical significances. Tumour Biol 2017;39:1010428317709640. https://doi.org/10.1177/1010428317709640.Suche in Google Scholar PubMed
17. Engstrand, J, Nilsson, H, Strömberg, C, Jonas, E, Freedman, J. Colorectal cancer liver metastases–a population-based study on incidence, management and survival. BMC Cancer 2018;18:1–11. https://doi.org/10.1186/s12885-017-3925-x.Suche in Google Scholar PubMed PubMed Central
18. Sorbye, H. Recurrence patterns after resection of liver metastases from colorectal cancer. In: Otto, F, Lutz, M, editors. Early gastrointestinal cancers II: rectal cancer. Cham: Springer; 2014, vol 203:243–52 pp.10.1007/978-3-319-08060-4_17Suche in Google Scholar PubMed
19. Tauriello, DV, Palomo-Ponce, S, Stork, D, Berenguer-Llergo, A, Badia-Ramentol, J, Iglesias, M, et al.. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018;554:538–43. https://doi.org/10.1038/nature25492.Suche in Google Scholar PubMed
20. Chai, W-X, Sun, L-G, Dai, F-H, Shao, H-S, Zheng, N-G, Cai, H-Y. Inhibition of PRRX2 suppressed colon cancer liver metastasis via inactivation of Wnt/β-catenin signaling pathway. Pathol Res Pract 2019;215:152593. https://doi.org/10.1016/j.prp.2019.152593.Suche in Google Scholar PubMed
21. Na, H, Liu, X, Li, X, Zhang, X, Wang, Y, Wang, Z, et al.. Novel roles of DC-SIGNR in colon cancer cell adhesion, migration, invasion, and liver metastasis. J Hematol Oncol 2017;10:28. https://doi.org/10.1186/s13045-016-0383-x.Suche in Google Scholar PubMed PubMed Central
22. Matsumura, H, Kondo, T, Ogawa, K, Tamura, T, Fukunaga, K, Murata, S, et al.. Kupffer cells decrease metastasis of colon cancer cells to the liver in the early stage. Int J Oncol 2014;45:2303–10. https://doi.org/10.3892/ijo.2014.2662.Suche in Google Scholar PubMed
23. Van Cutsem, E, Cervantes, A, Adam, R, Sobrero, A, Van Krieken, J, Aderka, D, et al.. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol 2016;27:1386–422. https://doi.org/10.1093/annonc/mdw235.Suche in Google Scholar PubMed
24. Zou, G, Wang, R, Wang, M. Clinical response and prognostic significance of serum miR-497 expression in colorectal cancer. Cancer Biomark 2019;25:11–8. https://doi.org/10.3233/cbm-181902.Suche in Google Scholar
25. Huang, S, Tan, X, Huang, Z, Chen, Z, Lin, P, Fu, SW. microRNA biomarkers in colorectal cancer liver metastasis. J Cancer 2018;9:3867–73. https://doi.org/10.7150/jca.28588.Suche in Google Scholar PubMed PubMed Central
26. Trams, EG, Lauter, CJ, Salem, JN, Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta 1981;645:63–70. https://doi.org/10.1016/0005-2736(81)90512-5.Suche in Google Scholar PubMed
27. Ruivo, CF, Adem, B, Silva, M, Melo, SA. The biology of cancer exosomes: insights and new perspectives. Cancer Res 2017;77:6480–8. https://doi.org/10.1158/0008-5472.can-17-0994.Suche in Google Scholar PubMed
28. Kalluri, R, LeBleu, VS. The biology, function, and biomedical applications of exosomes. Science 2020;367:eaau6977. https://doi.org/10.1126/science.aau6977.Suche in Google Scholar PubMed PubMed Central
29. Mathivanan, S, Fahner, CJ, Reid, GE, Simpson, RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res 2012;40:D1241–D4. https://doi.org/10.1093/nar/gkr828.Suche in Google Scholar PubMed PubMed Central
30. Tai, YL, Chen, KC, Hsieh, JT, Shen, TL. Exosomes in cancer development and clinical applications. Cancer Sci 2018;109:2364–74. https://doi.org/10.1111/cas.13697.Suche in Google Scholar PubMed PubMed Central
31. Sonbhadra, S, Mehak, PLM. Biogenesis, isolation, and detection of exosomes and their potential in therapeutics and Diagnostics. Biosensors 2023;13:802. https://doi.org/10.3390/bios13080802.Suche in Google Scholar PubMed PubMed Central
32. Wang, X, Tian, L, Lu, J, Ng, IO-L. Exosomes and cancer-Diagnostic and prognostic biomarkers and therapeutic vehicle. Oncogenesis 2022;11:54. https://doi.org/10.1038/s41389-022-00431-5.Suche in Google Scholar PubMed PubMed Central
33. Wang, C, Liu, J, Yan, Y, Tan, Y. Role of exosomes in chronic liver disease development and their potential clinical applications. J Immunol Res 2022;2022:1695802–15. https://doi.org/10.1155/2022/1695802.Suche in Google Scholar PubMed PubMed Central
34. Wong, C-H, Chen, Y-C. Clinical significance of exosomes as potential biomarkers in cancer. World J Clin Cases 2019;7:171–90. https://doi.org/10.12998/wjcc.v7.i2.171.Suche in Google Scholar PubMed PubMed Central
35. Yu, D, Li, Y, Wang, M, Gu, J, Xu, W, Cai, H, et al.. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer 2022;21:56. https://doi.org/10.1186/s12943-022-01509-9.Suche in Google Scholar PubMed PubMed Central
36. Gao, P, Li, X, Du, X, Liu, S, Xu, Y. Diagnostic and therapeutic potential of exosomes in neurodegenerative diseases. Front Aging Neurosci 2021;13:790863. https://doi.org/10.3389/fnagi.2021.790863.Suche in Google Scholar PubMed PubMed Central
37. Xu, Y, Hu, Y, Xu, S, Liu, F, Gao, Y. Exosomal microRNAs as potential biomarkers and therapeutic agents for acute ischemic stroke: new expectations. Front Neurol 2022;12:747380. https://doi.org/10.3389/fneur.2021.747380.Suche in Google Scholar PubMed PubMed Central
38. Bajan, S, Hutvagner, G. Regulation of miRNA processing and miRNA mediated gene repression in cancer. Microrna 2014;3:10–7. https://doi.org/10.2174/2211536602666140110234046.Suche in Google Scholar PubMed PubMed Central
39. Landgraf, P, Rusu, M, Sheridan, R, Sewer, A, Iovino, N, Aravin, A, et al.. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007;129:1401–14. https://doi.org/10.1016/j.cell.2007.04.040.Suche in Google Scholar PubMed PubMed Central
40. Marin, AM, Sanchuki, HBS, Namur, GN, Uno, M, Zanette, DL, Aoki, MN. Circulating cell-free nucleic acids as biomarkers for diagnosis and prognosis of pancreatic cancer. Biomedicines 2023;11:1069. https://doi.org/10.3390/biomedicines11041069.Suche in Google Scholar PubMed PubMed Central
41. Catuogno, S, Esposito, CL, Quintavalle, C, Cerchia, L, Condorelli, G, De Franciscis, V. Recent advance in biosensors for microRNAs detection in cancer. Cancers 2011;3:1877–98. https://doi.org/10.3390/cancers3021877.Suche in Google Scholar PubMed PubMed Central
42. Dobrzycka, M, Sulewska, A, Biecek, P, Charkiewicz, R, Karabowicz, P, Charkiewicz, A, et al.. miRNA studies in glaucoma: a comprehensive review of current knowledge and future perspectives. Int Int J Mol Sci 2023;24:14699. https://doi.org/10.3390/ijms241914699.Suche in Google Scholar PubMed PubMed Central
43. Martinez-Dominguez, MV, Zottel, A, Šamec, N, Jovčevska, I, Dincer, C, Kahlert, UD, et al.. Current technologies for RNA-directed liquid diagnostics. Cancers 2021;13:5060. https://doi.org/10.3390/cancers13205060.Suche in Google Scholar PubMed PubMed Central
44. Wadhonkar, K, Singh, N, Heralde, IIIFM, Parihar, SP, Hirani, N, Baig, MS. Exosome-derived miRNAs regulate macrophage-colorectal cancer cell cross-talk during aggressive tumor development. Colorectal Cancer 2023;12:CRC40. https://doi.org/10.2217/crc-2022-0012.Suche in Google Scholar
45. Shirafkan, N, Mansoori, B, Mohammadi, A, Shomali, N, Ghasbi, M, Baradaran, B. MicroRNAs as novel biomarkers for colorectal cancer: new outlooks. Biomed Pharmacother 2018;97:1319–30. https://doi.org/10.1016/j.biopha.2017.11.046.Suche in Google Scholar PubMed
46. Rupaimoole, R, Calin, GA, Lopez-Berestein, G, Sood, AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov 2016;6:235–46. https://doi.org/10.1158/2159-8290.cd-15-0893.Suche in Google Scholar PubMed PubMed Central
47. Mezher, M, Abdallah, S, Ashekyan, O, Shoukari, AA, Choubassy, H, Kurdi, A, et al.. Insights on the biomarker potential of exosomal non-coding RNAs in colorectal cancer: an in silico characterization of related exosomal lncRNA/circRNA–miRNA–target Axis. Cells 2023;12:1081. https://doi.org/10.3390/cells12071081.Suche in Google Scholar PubMed PubMed Central
48. Sun, T, Yin, YF, Jin, HG, Liu, HR, Tian, WC. Exosomal microRNA‐19b targets FBXW7 to promote colorectal cancer stem cell stemness and induce resistance to radiotherapy. Kaohsiung J Med Sci 2022;38:108–19. https://doi.org/10.1002/kjm2.12449.Suche in Google Scholar PubMed
49. Tao, L, Xu, C, Shen, W, Tan, J, Li, L, Fan, M, et al.. HIPK3 inhibition by exosomal hsa-miR-101-3p is related to metabolic reprogramming in colorectal cancer. Front Oncol 2022;11:758336. https://doi.org/10.3389/fonc.2021.758336.Suche in Google Scholar PubMed PubMed Central
50. Tang, Y, Zhao, Y, Song, X, Song, X, Niu, L, Xie, L. Tumor‐derived exosomal miRNA‐320d as a biomarker for metastatic colorectal cancer. J Clin Lab Anal 2019;33:e23004. https://doi.org/10.1002/jcla.23004.Suche in Google Scholar PubMed PubMed Central
51. Liu, K, Dou, R, Yang, C, Di, Z, Shi, D, Zhang, C, et al.. Exosome-transmitted miR-29a induces colorectal cancer metastasis by destroying the vascular endothelial barrier. Carcinogenesis 2023;44:356–67. https://doi.org/10.1093/carcin/bgad013.Suche in Google Scholar PubMed
52. Ma, J, Wang, P, Huang, L, Qiao, J, Li, J. Bioinformatic analysis reveals an exosomal miRNA-mRNA network in colorectal cancer. BMC Med Genom 2021;14:60. https://doi.org/10.1186/s12920-021-00905-2.Suche in Google Scholar PubMed PubMed Central
53. Squadrito, ML, Baer, C, Burdet, F, Maderna, C, Gilfillan, GD, Lyle, R, et al.. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep 2014;8:1432–46. https://doi.org/10.1016/j.celrep.2014.07.035.Suche in Google Scholar PubMed
54. Danac, JMC, Uy, AGG, Garcia, RL. Exosomal microRNAs in colorectal cancer: overcoming barriers of the metastatic cascade. Int J Mol Med 2021;47:112. https://doi.org/10.3892/ijmm.2021.4945.Suche in Google Scholar PubMed PubMed Central
55. Bebelman, MP, Smit, MJ, Pegtel, DM, Baglio, SR. Biogenesis and function of extracellular vesicles in cancer. Pharmacol Ther 2018;188:1–11. https://doi.org/10.1016/j.pharmthera.2018.02.013.Suche in Google Scholar PubMed
56. Zhang, X, Liu, D, Gao, Y, Lin, C, An, Q, Feng, Y, et al.. The biology and function of extracellular vesicles in cancer development. Front Cell Dev Biol 2021;9:777441. https://doi.org/10.3389/fcell.2021.777441.Suche in Google Scholar PubMed PubMed Central
57. Enns, A, Korb, T, Schlüter, K, Gassmann, P, Spiegel, H-U, Senninger, N, et al.. αvβ5-Integrins mediate early steps of metastasis formation. Eur J Cancer 2005;41:1065–72. https://doi.org/10.1016/j.ejca.2004.12.031.Suche in Google Scholar PubMed
58. Hsu, Y-L, Wu, C-Y, Hung, J-Y, Lin, Y-S, Huang, M-S, Kuo, P-L. Galectin-1 promotes lung cancer tumor metastasis by potentiating integrin α6β4 and Notch1/Jagged2 signaling pathway. Carcinogenesis 2013;34:1370–81. https://doi.org/10.1093/carcin/bgt040.Suche in Google Scholar PubMed
59. Mammadova-Bach, E, Zigrino, P, Brucker, C, Bourdon, C, Freund, M, De Arcangelis, A, et al.. Platelet integrin α6β1 controls lung metastasis through direct binding to cancer cell–derived ADAM9. JCI insight 2016;1:e88245. https://doi.org/10.1172/jci.insight.88245.Suche in Google Scholar PubMed PubMed Central
60. Hoshino, A, Costa-Silva, B, Shen, T-L, Rodrigues, G, Hashimoto, A, Tesic Mark, M, et al.. Tumour exosome integrins determine organotropic metastasis. Nature 2015;527:329–35. https://doi.org/10.1038/nature15756.Suche in Google Scholar PubMed PubMed Central
61. Jiang, Z-L, Zhang, F-X, Zhan, H-L, Yang, H-J, Zhang, S-Y, Liu, Z-H, et al.. miR-181b-5p promotes the progression of cholangiocarcinoma by targeting PARK2 via PTEN/PI3K/AKT signaling pathway. Biochem Genet 2022;60:223–40. https://doi.org/10.1007/s10528-021-10084-5.Suche in Google Scholar PubMed
62. Fong, MY, Zhou, W, Liu, L, Alontaga, AY, Chandra, M, Ashby, J, et al.. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol 2015;17:183–94. https://doi.org/10.1038/ncb3094.Suche in Google Scholar PubMed PubMed Central
63. Zhu, Y, Liu, Y, Xiao, B, Cai, H, Liu, M, Ma, L, et al.. The circular RNA PVT1/miR-203/HOXD3 pathway promotes the progression of human hepatocellular carcinoma. Biol Open 2019;8:bio043687.10.1242/bio.043687Suche in Google Scholar PubMed PubMed Central
64. Bala, S, Marcos, M, Kodys, K, Csak, T, Catalano, D, Mandrekar, P, et al.. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor α (TNFα) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem 2011;286:1436–44. https://doi.org/10.1074/jbc.m110.145870.Suche in Google Scholar PubMed PubMed Central
65. Qiu, M, Hu, J, Yang, D, Cosgrove, DP, Xu, R. Pattern of distant metastases in colorectal cancer: a SEER based study. Oncotarget 2015;6:38658–66. https://doi.org/10.18632/oncotarget.6130.Suche in Google Scholar PubMed PubMed Central
66. Pegtel, DM, Cosmopoulos, K, Thorley-Lawson, DA, van Eijndhoven, MA, Hopmans, ES, Lindenberg, JL, et al.. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 2010;107:6328–33. https://doi.org/10.1073/pnas.0914843107.Suche in Google Scholar PubMed PubMed Central
67. Pitzer, CR, Paez, HG, Alway, SE. The contribution of tumor derived exosomes to cancer cachexia. Cells 2023;12:292. https://doi.org/10.3390/cells12020292.Suche in Google Scholar PubMed PubMed Central
68. Wang, X, Ding, X, Nan, L, Wang, Y, Wang, J, Yan, Z, et al.. Investigation of the roles of exosomes in colorectal cancer liver metastasis. Oncol Rep 2015;33:2445–53. https://doi.org/10.3892/or.2015.3843.Suche in Google Scholar PubMed
69. Yan, S, Cheng, M, Duan, Q, Wang, Z, Gao, W, Ren, B, et al.. MiR‐6803‐5p promotes cancer cell proliferation and invasion via PTPRO/NF‐κB Axis in colorectal cancer. Mediat Inflamm 2019;2019:8128501–9. https://doi.org/10.1155/2019/8128501.Suche in Google Scholar PubMed PubMed Central
70. Ge, J, Li, J, Na, S, Wang, P, Zhao, G, Zhang, X. miR‐548c‐5p inhibits colorectal cancer cell proliferation by targeting PGK1. J Cell Physiol 2019;234:18872–8. https://doi.org/10.1002/jcp.28525.Suche in Google Scholar PubMed
71. Okholm, C, Mollerup, TK, Schultz, NA, Strandby, RB, Achiam, MP. Synchronous and metachronous liver metastases in patients with colorectal cancer. Dan Med J 2018;65:A5524.Suche in Google Scholar
72. Gupta, V, Chopde, A, Patkar, S, Deodhar, K, Goel, M. Oxaliplatin-induced sinusoidal obstruction syndrome masquerading as colorectal liver metastasis: a case Report. J Gastrointest Cancer 2023;54:682–6. https://doi.org/10.1007/s12029-022-00835-x.Suche in Google Scholar PubMed
73. Li, Q, Chen, L, Jin, H, Zhao, Y, Hao, Z, Ma, X. Pretreatment inflammatory markers predict outcomes and prognosis in colorectal cancer patients with synchronous liver metastasis. Clin Med Insights Oncol 2022;16:11795549221084851. https://doi.org/10.1177/11795549221084851.Suche in Google Scholar PubMed PubMed Central
74. Lv, Y, Patel, N, Zhang, H-J. The progress of non-alcoholic fatty liver disease as the risk of liver metastasis in colorectal cancer. Expert Rev Gastroenterol Hepatol 2019;13:1169–80. https://doi.org/10.1080/17474124.2019.1697231.Suche in Google Scholar PubMed
75. Shao, Y, Chen, T, Zheng, X, Yang, S, Xu, K, Chen, X, et al.. Colorectal cancer-derived small extracellular vesicles establish an inflammatory premetastatic niche in liver metastasis. Carcinogenesis 2018;39:1368–79. https://doi.org/10.1093/carcin/bgy115.Suche in Google Scholar PubMed
76. Yan, S, Han, B, Gao, S, Wang, X, Wang, Z, Wang, F, et al.. Exosome-encapsulated microRNAs as circulating biomarkers for colorectal cancer. Oncotarget 2017;8:60149–58. https://doi.org/10.18632/oncotarget.18557.Suche in Google Scholar PubMed PubMed Central
77. Monzo, M, Santasusagna, S, Moreno, I, Martinez, F, Hernáez, R, Muñoz, C, et al.. Exosomal microRNAs isolated from plasma of mesenteric veins linked to liver metastases in resected patients with colon cancer. Oncotarget 2017;8:30859–69. https://doi.org/10.18632/oncotarget.16103.Suche in Google Scholar PubMed PubMed Central
78. Teng, Y, Ren, Y, Hu, X, Mu, J, Samykutty, A, Zhuang, X, et al.. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun 2017;8:14448. https://doi.org/10.1038/ncomms14448.Suche in Google Scholar PubMed PubMed Central
79. Pei, W, Wei, K, Wu, Y, Qiu, Q, Zhu, H, Mao, L, et al.. Colorectal cancer tumor cell-derived exosomal miR-203a-3p promotes CRC metastasis by targeting PTEN-induced macrophage polarization. Gene 2023;885:147692. https://doi.org/10.1016/j.gene.2023.147692.Suche in Google Scholar PubMed
80. Zhao, S, Mi, Y, Guan, B, Zheng, B, Wei, P, Gu, Y, et al.. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol 2020;13:156. https://doi.org/10.1186/s13045-020-00991-2.Suche in Google Scholar PubMed PubMed Central
81. Shibamoto, J, Arita, T, Konishi, H, Kataoka, S, Furuke, H, Takaki, W, et al.. Roles of miR-4442 in colorectal cancer: predicting early recurrence and regulating epithelial-mesenchymal transition. Genes 2023;14:1414. https://doi.org/10.3390/genes14071414.Suche in Google Scholar PubMed PubMed Central
82. Chen, J, Li, Z, Yue, C, Ma, J, Cao, L, Lin, J, et al.. Human umbilical cord mesenchymal stem cell-derived exosomes carrying miR-1827 downregulate SUCNR1 to inhibit macrophage M2 polarization and prevent colorectal liver metastasis. Apoptosis 2023;28:549–65. https://doi.org/10.1007/s10495-022-01798-x.Suche in Google Scholar PubMed
83. Zhao, S, Mi, Y, Zheng, B, Wei, P, Gu, Y, Zhang, Z, et al.. Highly‐metastatic colorectal cancer cell released miR‐181a‐5p‐rich extracellular vesicles promote liver metastasis by activating hepatic stellate cells and remodelling the tumour microenvironment. J Extracell Vesicles 2022;11:e12186. https://doi.org/10.1002/jev2.12186.Suche in Google Scholar PubMed PubMed Central
84. Sun, H, Meng, Q, Shi, C, Yang, H, Li, X, Wu, S, et al.. Hypoxia‐inducible exosomes facilitate liver‐tropic premetastatic niche in colorectal cancer. Hepatology 2021;74:2633–51. https://doi.org/10.1002/hep.32009.Suche in Google Scholar PubMed
85. Tian, F, Wang, P, Lin, D, Dai, J, Liu, Q, Guan, Y, et al.. Exosome‐delivered miR‐221/222 exacerbates tumor liver metastasis by targeting SPINT1 in colorectal cancer. Cancer Sci 2021;112:3744–55. https://doi.org/10.1111/cas.15028.Suche in Google Scholar PubMed PubMed Central
86. Sur, D, Balacescu, L, Cainap, SS, Visan, S, Pop, L, Burz, C, et al.. Predictive efficacy of MiR-125b-5p, MiR-17-5p, and MiR-185-5p in liver metastasis and chemotherapy response among advanced stage colorectal cancer patients. Front Oncol 2021;11:651380. https://doi.org/10.3389/fonc.2021.651380.Suche in Google Scholar PubMed PubMed Central
87. Liu, D, Chen, C, Cui, M, Zhang, H. miR‐140‐3p inhibits colorectal cancer progression and its liver metastasis by targeting BCL9 and BCL2. Cancer Med 2021;10:3358–72. https://doi.org/10.1002/cam4.3840.Suche in Google Scholar PubMed PubMed Central
88. Wang, D, Wang, X, Si, M, Yang, J, Sun, S, Wu, H, et al.. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett 2020;474:36–52. https://doi.org/10.1016/j.canlet.2020.01.005.Suche in Google Scholar PubMed
89. Sun, L, Liu, X, Pan, B, Hu, X, Zhu, Y, Su, Y, et al.. Serum exosomal miR-122 as a potential diagnostic and prognostic biomarker of colorectal cancer with liver metastasis. J Cancer 2020;11:630–7. https://doi.org/10.7150/jca.33022.Suche in Google Scholar PubMed PubMed Central
90. Matsumura, T, Sugimachi, K, Iinuma, H, Takahashi, Y, Kurashige, J, Sawada, G, et al.. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer 2015;113:275–81. https://doi.org/10.1038/bjc.2015.201.Suche in Google Scholar PubMed PubMed Central
91. Gunassekaran, GR, Vadevoo, SMP, Baek, M-C, Lee, B. M1 macrophage exosomes engineered to foster M1 polarization and target the IL-4 receptor inhibit tumor growth by reprogramming tumor-associated macrophages into M1-like macrophages. Biomaterials 2021;278:121137. https://doi.org/10.1016/j.biomaterials.2021.121137.Suche in Google Scholar PubMed
92. Huang, C, Ou, R, Chen, X, Zhang, Y, Li, J, Liang, Y, et al.. Tumor cell-derived SPON2 promotes M2-polarized tumor-associated macrophage infiltration and cancer progression by activating PYK2 in CRC. J Exp Clin Cancer Res 2021;40:304. https://doi.org/10.1186/s13046-021-02108-0.Suche in Google Scholar PubMed PubMed Central
93. Zhang, Y, Liu, W-S, Zhang, X-Y, Tong, H-X, Yang, H, Liu, W-F, et al.. Low expression of exosomal miR-150 predicts poor prognosis in colorectal cancer patients after surgical resections. Carcinogenesis 2022;43:930–40. https://doi.org/10.1093/carcin/bgac059.Suche in Google Scholar PubMed
94. Nassar, FJ, Msheik, ZS, Itani, MM, Helou, RE, Hadla, R, Kreidieh, F, et al.. Circulating miRNA as biomarkers for colorectal cancer diagnosis and liver metastasis. Diagnostics 2021;11:341. https://doi.org/10.3390/diagnostics11020341.Suche in Google Scholar PubMed PubMed Central
95. Wang, Y, Chen, X, Lin, H, Sun, X, Fong, WP, Wu, X, et al.. A Prehepatectomy circulating Exosomal microRNA signature predicts the prognosis and adjuvant chemotherapeutic benefits in colorectal liver metastasis. Cancers 2021;13:4258. https://doi.org/10.3390/cancers13174258.Suche in Google Scholar PubMed PubMed Central
96. Jafari, A, Karimabadi, K, Rahimi, A, Rostaminasab, G, Khazaei, M, Rezakhani, L, et al.. The emerging role of exosomal miRNAs as biomarkers for early cancer detection: a comprehensive literature review. Technol Cancer Res Treat 2023;22:15330338231205999. https://doi.org/10.1177/15330338231205999.Suche in Google Scholar PubMed PubMed Central
97. Baumann, V, Winkler, J. miRNA-based therapies: strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med Chem 2014;6:1967–84. https://doi.org/10.4155/fmc.14.116.Suche in Google Scholar PubMed PubMed Central
98. Lin, B, Jiang, J, Jia, J, Zhou, X. Recent advances in exosomal miRNA biosensing for liquid biopsy. Molecules 2022;27:7145. https://doi.org/10.3390/molecules27217145.Suche in Google Scholar PubMed PubMed Central
99. Sempere, LF, Keto, J, Fabbri, M. Exosomal microRNAs in breast cancer towards diagnostic and therapeutic applications. Cancers 2017;9:71. https://doi.org/10.3390/cancers9070071.Suche in Google Scholar PubMed PubMed Central
100. Condrat, CE, Thompson, DC, Barbu, MG, Bugnar, OL, Boboc, A, Cretoiu, D, et al.. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells 2020;9:276. https://doi.org/10.3390/cells9020276.Suche in Google Scholar PubMed PubMed Central
© 2025 the author(s), published by De Gruyter on behalf of Tech Science Press (TSP)
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Frontmatter
- Review Articles
- Targeting HER2 in metastatic urothelial carcinoma: a contemporary review
- Advances in research on the impact of exosomal miRNAs on liver metastasis of colorectal cancer
- Sebaceous carcinoma of the intraoral origin: a literature review
- Role of emerging theranostic technologies in precision oncology: revolutionizing cancer diagnosis and treatment
- Research Articles
- Half-body irradiation with dose escalation in the era of advanced systemic therapies: unveiling new therapeutic opportunities
- Characteristics of MMR protein expression in colorectal cancer and MMR gene variations in Vietnamese patients with Lynch syndrome
- BMPR1A promotes the proliferation of colorectal cancer cells through the activation of Smad1
- Construction and validation of a diagnostic model for cholangiocarcinoma based on tumor-educated platelet RNA expression profiles
- Prognostic implications of PCSK9 expression in HER2-positive breast cancer
- Rapid Communication
- Potential impact of hormone replacement therapy on the risk of hepatocellular carcinoma in women of the PLCO cohort
- Article Commentary
- “Plant-based and ketogenic diets as diverging paths to address cancer”: a commentary concerning the supposed superiority of a plant-based diet
- Novel insights into molecular landscape of advanced renal cell carcinoma
- Corrigendum
- Corrigendum to “Y27632 induces tongue squamous cell carcinoma cell apoptosis through MAPK-ERK/JNK signal”
Artikel in diesem Heft
- Frontmatter
- Review Articles
- Targeting HER2 in metastatic urothelial carcinoma: a contemporary review
- Advances in research on the impact of exosomal miRNAs on liver metastasis of colorectal cancer
- Sebaceous carcinoma of the intraoral origin: a literature review
- Role of emerging theranostic technologies in precision oncology: revolutionizing cancer diagnosis and treatment
- Research Articles
- Half-body irradiation with dose escalation in the era of advanced systemic therapies: unveiling new therapeutic opportunities
- Characteristics of MMR protein expression in colorectal cancer and MMR gene variations in Vietnamese patients with Lynch syndrome
- BMPR1A promotes the proliferation of colorectal cancer cells through the activation of Smad1
- Construction and validation of a diagnostic model for cholangiocarcinoma based on tumor-educated platelet RNA expression profiles
- Prognostic implications of PCSK9 expression in HER2-positive breast cancer
- Rapid Communication
- Potential impact of hormone replacement therapy on the risk of hepatocellular carcinoma in women of the PLCO cohort
- Article Commentary
- “Plant-based and ketogenic diets as diverging paths to address cancer”: a commentary concerning the supposed superiority of a plant-based diet
- Novel insights into molecular landscape of advanced renal cell carcinoma
- Corrigendum
- Corrigendum to “Y27632 induces tongue squamous cell carcinoma cell apoptosis through MAPK-ERK/JNK signal”

