Characteristics of MMR protein expression in colorectal cancer and MMR gene variations in Vietnamese patients with Lynch syndrome
-
Bich Mai Bui
, Cam Phuong Pham
, Van Thai Pham
, Thi Lan Anh Luong
, Van Tuyen Pham
, Thuan Loi Nguyen
, Quang Loc Bui
, Minh Khuy Doan
, Sy Tung Ha
, Thanh Huyen Nguyen
, Hong Phuc Dinh
and Thi Kim Phuong Doan
Abstract
Objectives
Detecting and diagnosing Lynch syndrome in colorectal cancer patients is essential for enhancing the quality of diagnosis, treatment, and management. This study aims to characterize mismatch repair (MMR) protein expression and MMR gene variants in Vietnamese patients.
Methods
A total of 218 patients diagnosed with colorectal cancer at the Nuclear Medicine and Oncology Center, Bach Mai Hospital (Hanoi, Vietnam) were included in this descriptive, retrospective study. Tumor tissue samples were evaluated for MMR protein expression. Cases with loss of MMR protein expression underwent genomic sequencing for MMR gene mutations.
Results
Among 218 colorectal cancer patients, 135 were men (61.9 %) and 83 were women (38.1 %), aged 16–86 years. The prevalence of MMR deficiency (dMMR) was 14.7 %. The expression loss rates of the MLH1, MSH2, MSH6, PMS2, MLH1/PMS2, and MSH2/MSH6 proteins were 9.4 %, 3.1 %, 3.1 %, 21.9 %, 46.9 %, and 15.6 %, respectively. Within the dMMR group, four variants were identified in the MLH1 gene, one in the MSH2 gene, two in the PMS2 gene, and one in the EPCAM gene, and all were classified as germline pathogenic variants associated with Lynch syndrome.
Conclusion
This is a study to apply next-generation sequencing (NGS) technology to diagnose Lynch syndrome from dMMR cases in Vietnamese colorectal cancer patients. Our research supports the screening and diagnosis of Lynch syndrome through testing using MMR protein expression and assessing pathogenic variants in MMR genes.
Introduction
Colorectal cancer (CRC) is among the most prevalent cancers globally. According to GLOBOCAN’s 2022 statistics, CRC ranks third in incidence at 9.6 % and second in cancer mortality rate at 9.3 % [1]. In Vietnam, CRC is among the 10 most common cancers, with an age-standardized incidence rate of 17.6/100,000 people in men and 11.6/100,000 people in women, ranking fifth among cancer. The pathogenesis of CRC has not been precisely determined, but there are at least three molecular pathways that cause colorectal tumors. These include chromosomal instability (CIN), observed in about 70–85 % of CRC; and microsatellite instability (MSI), observed in 15–20 % of cases, and CpG island methylator phenotype (CIMP) [2]. Each of these pathways is characterized by distinct pathological features, carcinogenic mechanisms, and tumor progression.
CRC is classified into acquired (sporadic), familial, and hereditary colorectal cancer. Although up to 25 % of colorectal cancer cases are related to familial factors, approximately 3–5% of cases are clearly described as hereditary cancer syndromes. The most prevalent hereditary colorectal cancer syndrome is Lynch syndrome, also known as hereditary nonpolyposis colorectal cancer (HNPCC). The other hereditary colorectal cancer syndromes are characterized by polyps such as familial polyposis syndrome, MUTYH-associated polyposis (MAP), hamartomatous polyposis syndrome, and serrated polyposis syndrome (SPS) [3]. Distinguishing colorectal cancer as acquired or hereditary is important for individualized and precise treatment and to help in predicting the progression of other cancers according to disease gene variations; to assist with genetic counseling regarding disease and cancer risk detection; and to screen, diagnose, and treat cancer early in genetically predisposed individuals. As the classic clinical guidelines for diagnosing Lynch syndrome, such as the Amsterdam criteria or the Bethesda guidelines, have several shortcomings including low sensitivity and the possibility of missing gene carriers, the development of combined diagnostic strategies is necessary. Molecular markers have been, and are being, developed to help improve the quality of screening and diagnosis of Lynch syndrome [4].
MSI results from the impaired function of a DNA mismatch to repair (MMR) genes, including MLH1, MSH2, MSH6, and PMS2. This impairment leads to changes in protein activity, inactivation of the DNA mismatch repair system, and the accumulation of mutations in the microsatellite regions. The most common MMR gene germline mutations are MLH1, MSH2, or less commonly, MSH6 and PMS2, which are responsible for the majority of cases of Lynch syndrome [3]. In addition, the germline mutation in the EPCAM gene is also detected at a rate of about 1–3% in Lynch cases [5] MSI can be identified in more than 90 % of colorectal cancers in people with Lynch syndrome, and in 15 % of cases in sporadic colorectal cancers, typically due to genetics. In such instances, MLH1 is inactivated due to epigenetic silencing, with the inactivation being caused by hypermethylation of the promoter region [6]. Tumors with MSI status typically have distinct characteristics (poorly differentiated morphology, lymphadenopathy and negative, mucin-rich, ring-shaped) and lymphocyte counts increase tumor infiltration (TIL) [7].
MSI testing plays an important role in deciding on adjuvant chemotherapy in stage II colorectal cancer and helps with selecting patients who probably respond well to immunotherapy and to identify cases of Lynch syndrome [7]. It is meaningful in genetic counseling with regard to assisting with the screening and early diagnosis of people carrying the disease gene. MSI may be assessed using PCR testing when a tumor has MSI high microsatellite instability (MSI-H). The test can identify the MSI-H cases. In addition, Microsatellite instability (MSI) cannot be directly detected by immunohistochemistry (IHC), but IHC can be used to identify deficiencies in MMR proteins, which are closely associated with MSI status. If one or more of these markers are missing, the tumor is considered to have mismatch repair deficiency. Loss of expression of MLH1 is often with PMS2; MSH2 expression is often lost with MSH6; and MSH6 and PMS2 expression can often be lost owing to the molecular combination of the above proteins. Tumors that exhibit loss of expression of an MMR protein may be collectively referred to as dMMR (Deficient Mismatch Repair) and are considered to be MSI-H; tumors with intact expression of all MMR proteins are classified as pMMR (Proficient Mismatch Repair), and are considered to have microsatellite (MSS) or MSI-L stability [8].
Currently, determining MSI status is a mandatory test according to numerous global colorectal cancer treatment guidelines, including the NCCN (National Comprehensive Cancer Network – US National Cancer Network) [9] and ESMO (European Society for Medical Oncology) [10]. This test is important for predicting disease prognosis and individualizing treatment. In Vietnam, there have been a number of studies published on the MSI status of colorectal cancer using IHC, but there is not much data on MMR protein expression status and corresponding gene variations. Therefore, the goal of our study is to describe the characteristics of the loss of MMR protein expression and MMR gene variants in patients with colorectal cancer.
Methods
Study design and population
This descriptive study was conducted among 218 patients diagnosed with colorectal carcinoma at the Center for Nuclear Medicine and Oncology, Bach Mai Hospital, Hanoi, Vietnam from January 2022 to June 2023. We conducted this study according to the STROBE guideline, please see Supplementary material for details.
This study was conducted in accordance with the Declaration of Helsinki (1964) and was approved by the Ethics Committee of Hanoi Medical University (Approval number 837/GCN-HĐĐĐNCYSH-ĐHYHN). After genetic counseling, all participants provided written consent for the anonymous reuse of their genomic data in this study. The genomic data were anonymized and aggregated for the cohort’s genetic analysis. Referring clinicians or participant interviews provided detailed information on personal and family cancer histories.
Data collection
The clinical and laboratory testing of patients with colorectal cancer were gathered. Immunohistochemical testing to evaluate the expression status of MMR proteins was conducted using Ventana Medical Systems (AZ, USA) and test kits VENTANA MMR RxDx panel (catalog number: MLH1: 760-5091, MSH2: 760-5093, MSH6: 760-5092, PMS2: 760-5094) on formalin-fixed paraffin-embedded (FFPE) colorectal cancer tissue samples after biopsy or surgery.
Cases with a loss of MMR protein expression continued to undergo genetic testing to identify gene variants related to Lynch Syndrome (gene panel includes MLH1, MSH2, MSH6, PMS2, EPCAM, APC, MUTYH) using next-generation sequencing (NGS). The genetic variations examined included point mutations, deletions and short insertions (less than 10 nucleotides) in the coding region and the region adjacent to the intron (−10/+10 nucleotides from the exon) of the investigated genes. Variant detection cases were further confirmed using Sanger sequencing.
IHC assessment process
IHC was performed using the BenchMark Ultra (Roche Diagnostics, Mannheim, Germany) automated immunostainer. The slides were counterstained with haematoxylin. The following antibodies were used: anti MLH1 (clone M1, ready to use concentration: 1 μg/mL; catalog number 760-5091, Roche/Ventana), anti PMS2 (clone A16-4, ready to use concentration: 1 μg/mL; catalog number 760-5094, Roche/Ventana), anti-MSH2 (clone G219-1129, ready to use concentration: 20 μg/mL; catalog number 760-5093, Roche/Ventana) and anti-MSH6 (clone SP93, ready to use concentration: 1 μg/mL; catalog number 760-5092, Roche/Ventana). Normal colonic crypt epithelium adjacent to the tumor, lymphoid cells, and stromal cells were used as internal positive controls. Additionally, slide-positive controls are routinely employed in our IHC laboratory.
Sample preparation and sequencing
Each participant provided either a 2 mL peripheral blood sample or a buccal swab sample. Genomic DNA was extracted from blood samples using the GeneJet Whole Blood Genomic DNA Purification MiniKit (catalog number: K0781, ThermoFisher, USA). DNA fragmentation and library preparation were performed using the NEBNext Ultra II FS DNA Library Prep Kit (catalog number: E7435S, New England Biolabs, USA) following the manufacturer’s instructions. The libraries were pooled and hybridized with pre-designed probes targeting seven specific genes (MLH1, MSH2, MSH6, PMS2, EPCAM, APC, MUTYH) (Integrated DNA Technologies, USA). Massive parallel sequencing was conducted using the NextSeq 500/550 High Output Kits v2 (150 cycles) on the Illumina NextSeq 550 system (Illumina, USA), with a minimum target coverage of 100×.
Variant calling and analysis
Quality control and data processing were carried out as previously described [11]. In brief, paired-end reads were aligned to the human reference genome (GRCh38) using the Burrows–Wheeler Aligner (BWA) version 0.7.12 [12]. The resulting alignment was used to compute the coverage depth of the targeted regions, and variant calling was performed using GATK 3.8.0.0 [13]. Variants were annotated using the dbSNP (https://www.ncbi.nlm.nih.gov/snp) [14], ClinVar (https://www.ncbi.nlm.nih.gov/clinvar) [15], and LOVD (https://www.lovd.nl/3.0/home) [16] databases and their molecular impacts were evaluated using the Ensemble Variant Effect Predictor 110 [17].
Variants were classified according to the guidelines of The American College of Medical Genetics and Genomics (ACMG) [18]. The ACMG employs a five-tier classification system: “pathogenic,” “likely pathogenic,” “variant of uncertain significance,” “likely benign,” and “benign,” based on existing literature, databases, and computational tools.
Sanger sequencing
Sanger sequencing was performed to confirm all pathogenic variants identified by NGS. Primers flanking the mutation location were designed using Primer3Plus (https://www.primer3plus.com) [19] and synthesized by Integrated DNA Technologies, USA. PCR amplification was performed using the same genomic DNA samples, with Q5 High-Fidelity 2X Mastermix (New England Biolabs, USA) according to the manufacturer’s protocol. The PCR products were purified and sequenced using the Genetic Analyzer 3500xl (Applied Biosystems, USA).
Study outcomes
The primary outcome of our study was the prevalence and distribution of MMR gene variants in colorectal cancer patients.
Statistical analysis
Characteristics of the study population are presented as means and standard deviations for continuous variables. All categorical factors are presented as percentages. Using Fisher’s exact test for categorical variables. We compare clinical and laboratory characteristics among gene variant groups. Statistical analyses were performed using Stata 17.0 (StataCorp, USA).
Results
Characteristics of the study population
Our study comprised 218 patients diagnosed with colorectal cancer, of whom 135 were men (61.9 %) and 83 were women (38.1 %) with ages ranging from 16 to 86 years old. The average age of the study participants was 59.9 and the age of diagnosis was typically over 50 years (accounting for 78.9 %). Of the 218 patients diagnosed with colorectal cancer, 32 had dMMR status. However, only 25 patients went on to undergo genetic testing. We found eight patients with pathogenic variants in the MMR gene and 17 patients with no known MMR gene variants (Figure 1).

Overview of research results.
The prevalence and distribution of the MMR protein expression
In the study population, the patients with a loss of MMR expression (dMMR) accounted for 14.7 % (Figure 2).

The prevalence of MMR protein expression in patients with colorectal cancer.
Among patients with dMMR, the proportion of patients with loss of MLH1 protein expression was 9.4 %; MSH2 was 3.1 %; MSH6 was 3.1 %; PMS2 was 21.9 %; MLH1/PMS2 was 46.9 %; and MSH2/MSH6 was 15.6 % (Table 1).
The distribution of lost MMR protein expression (n=32).
| Loss of MMR protein expression | Count, n | % |
|---|---|---|
| MLH1 | 3 | 9.4 |
| MSH2 | 1 | 3.1 |
| MSH6 | 1 | 3.1 |
| PMS2 | 7 | 21.9 |
| MLH1/PMS2 | 15 | 46.9 |
| MSH2/MSH6 | 5 | 15.6 |
| Total | 32 | 100.0 |
Here are three figures showing the results of MMR status assessment with different MMR protein loss patterns: loss of MLH1/PMS2 proteins (Figure 3), loss of MSH2/MSH6 proteins (Figure 4), and isolated loss of PMS2 (Figure 5).
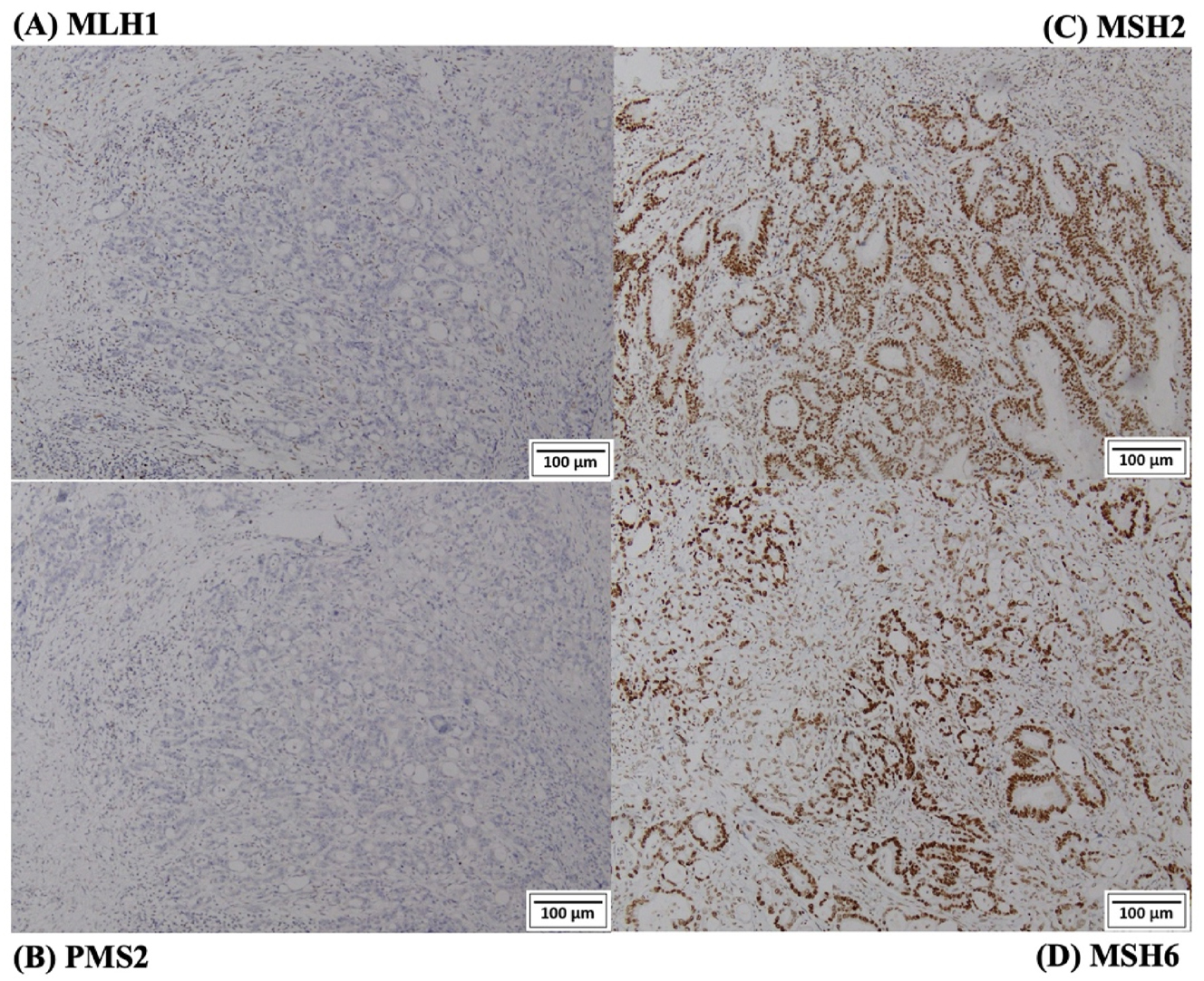
IHC staining images from a patient loss of expression of MLH1 and PMS2 proteins. (A) Loss of expression of MLH1 protein; (B) loss of expression of PMS2 protein; (C) normal expression of MSH2 protein; (D) normal expression of MSH6 protein.

IHC staining images from a patient loss of expression of MSH2 and MSH6 proteins. (A) Normal expression of MLH1 protein; (B) normal expression of PMS2; (C) loss of expression of MSH2 protein; (D) loss of expression of MSH6. These images were captured from the same region of the tumor across four different slides and appear to be similar in certain areas.

IHC staining images from a patient loss of expression of PMS2 protein alone. (A) Normal expression of MLH1 protein; (B) loss of expression of PMS2 protein; (C) normal expression of MSH2 protein; (D) normal expression of MSH6 protein.
The prevalence and distribution of MMR gene variants in dMMR patients
In 32 patients with dMMR, there was 25 patients agreed to have genetics testing. Of those, 32.0 % (8/25) patients carried pathogenic variants in MMR genes. In the group of patients with mutations in MMR genes, we detected 04 variations in the MLH1 gene, 01 variation in the MSH2 gene, 02 variations in the PMS2 gene, and 01 variation in the EPCAM gene – all classified as pathogenic variants, and causes of Lynch syndrome (Table 2). The most frequently affected gene is MLH1, with four cases, followed by PMS2 with three cases. EPCAM and MSH2 each have one case. This indicates a higher prevalence of MLH1 mutations in the studied population. The mutations include nonsense, frameshift, and missense variants. Frameshift mutations are the most common, appearing in four cases, which may suggest a significant impact on protein function leading to disease. The loss of MMR protein expression is consistent with the gene affected. For instance, MLH1 mutation result in the loss of MLH1 and PMS2 protein expression, while PMS2 mutation result in the loss of PMS2 expression. There is a strong familial component, with multiple cases reporting relatives who have had cancer, particularly colon cancer. This highlights the hereditary nature of these genetic variants and their association with cancer risk.
Characteristics of pathogenic variants detected by NGS sequencing and Sanger sequencing.
| Patient no. | Age of diag. | Gene | Variants | Results | Sanger sequencing | Loss of MMR protein expression | Family history | ACMG classification – phenotype |
|---|---|---|---|---|---|---|---|---|
| L1 | 29 | EPCAM | c.753T>G:p. Tyr251Ter | Nonsense | EPCAM | MSH6 | Grandmother passed away due to metastatic lung cancer at the age of 71 (cancer type not specified). | Pathogenic – Lynch syndrome |
| L2 | 58 | PMS2 | c.1738A>T:p. Lys580Ter | Nonsense | PMS2 | PMS2 | Mother passed away at the age of 40 due to illness (the exact illness is unknown). | |
| L3 | 31 | MLH1 | c.1740-1753del: p. Pro581fs | Frameshift | MLH1 | MLH1, PMS2 | Grandfather and paternal uncle both passed away around the age of 40 due to melena. | |
| L4 | 69 | MSH2 | c.2231-2235del:p. Thr744AsnfsTer12 | Frameshift | MSH2 | MSH2, MSH6 | Younger sister passed away at the age of 49 due to colon cancer, and mother passed away at the age of 51 due to illness (the exact illness is unknown). | |
| L5 | 29 | MLH1 | c.2102A>G:p. Gln701Arg | Missense | MLH1 | MLH1, PMS2 | Aunt was diagnosed with stomach cancer at the age of 46. | |
| L6 | 51 | MLH1 | c.2809del:p. Leu697fs | Frameshift | MLH1 | MLH1, PMS2 | Older brother was diagnosed with colon cancer at the age of 38, and mother passed away at the age of 37 due to a condition related to the colon (exact diagnosis unknown). | |
| L7 | 44 | MLH1 | c.109G>A:p. Glu37Lys | Missense | MLH1 | PMS2 | Father was diagnosed with rectal cancer at the age of 54, and two paternal uncles passed away from colon cancer before the age of 60 | |
| L8 | 68 | PMS2 | c.746-753del:p. Asp249fs | Frameshift | PMS2 | PMS2 | Mother passed away at the age of 60 due to illness (the exact illness is unknown). |
A 69-year-old female patient (L4) was diagnosed with colorectal cancer, histopathology showing an adenocarcinoma, immunohistochemistry staining revealing loss of MSH2-MSH6 protein expression. The patient’s medical history includes hypertension for 5 years, with no previous record of cancer in any organ. Family history includes mother’s death at 51 years old due to unknown illness, and the patient’s younger sister died at 49 years old due to colorectal cancer. Next generation sequencing test results detected a pathogenic variants on the MSH2 gene (c.2231_2235del) (p.Thr744AsnfsTer12) in a heterozygous state – Lynch syndrome – a hereditary colorectal cancer syndrome. Individuals with this gene variant are at risk for colorectal cancer, endometrial, gastric, and ovarian cancer. The patient received genetic counseling regarding the disease characteristics and cancer risks in other organs, and was advised to undergo genetic testing for family members to facilitate early screening and appropriate intervention (Figure 6).

Pedigree of a patient (L4).
Discussions
Characteristics of the study population
Among 218 patients diagnosed with CRC, men accounted for 61.9 % (135/218) and women for 38.1 % (83/218), so the male/female ratio was about 1.63. The incidence of colorectal cancer is higher in men than in women. The male/female ratio of studies in Vietnam ranges from 1.10 to 1.98 [20], 21]. The variation between studies may be owing to differences in sample size and inclusion criteria. Age is an important risk factor for colorectal cancer. Our study results indicated that the age of study participants ranged from 16 to 86 years; the average age was 59.9 and diagnosis typically occurred in those over 50 years, accounting for 78.9 %. Our study results are similar to other studies generated internationally and from Vietnam, revealing that the majority of colorectal cancers are detected after the age of 50 years [20], 22].
Screening methods to define high risk patients with Lynch syndrome
MSI-H/dMMR status has been linked to different features of CRC tumors, such as the primary tumor’s location, size, T stage, and spread to distant sites. Numerous retrospective studies have demonstrated a significant connection between dMMR/MSI-H status and early onset of the disease, larger tumor sizes, considerable tumor volume, primary tumor location, and advanced T stage in patients with stage I–III or I–IV CRC, including TNM stages [23]. MMR screening and MSI testing are crucial in diagnosing hereditary colorectal cancer syndrome; selecting treatment regimens for stage II/III colorectal cancer; and identifying indications for immunotherapy at the metastatic stage [24]. The proportion of dMMR patients in our study was 14.7 %, similar to the results of other studies in Vietnam (12.4 %) [25], China (13.8 %) [22], and the United States (15 %) [26].
In the typical dMMR case, immunostaining is completely absent for one of the two MMR protein heterodimers (MLH1/PMS2 or MSH2/MSH6), which represents by far the largest proportion, approximately 85 % of MSI-H/dMMR cases in the French dataset [27]. In our study, the percentage of patient with loss of MLH1 and PMS2 were 46.9 % and while the loss of MSH2 and MSH6 was 15.6 %. According to Hao et al., the most common finding was a simultaneous loss of expression of MLH1 and PMS2 (53.8 %), followed by a simultaneous loss of expression of MSH2 and MSH6 (30.8 %), loss of PMS2 expression accounts for the lowest proportion (15.4 %), and a single loss of MLH1, MSH2, MSH6 expression is not reported [25]. The majority of all non-typical findings consisted of isolated loss of PMS2 or MSH6 without loss of their respective heterodimerization partners MLH1 or MSH2 [27]. In our study, the proportion of patients with loss of MLH1 protein expression was 9.4 %; loss of MSH2 was 3.1 %; loss of MSH6 was 3.1 %; loss of PMS2 was 21.9 %. This difference may be owing to differences in study sample sizes and populations. Our study with a larger sample size observed patients with a single loss of expression of MLH1, MSH2, and MSH6. Isolated loss of PMS2 or MSH6 is typically due to germline mutations of the respective gene, and is therefore associated with Lynch syndrome [28].
The genetic testing to diagnose Lynch syndrome in dMMR patients
This is one of the studies in Vietnam using NGS techniques to analyze germline MMR gene variants and diagnose Lynch syndrome. The proportion of patients carrying mutations in our study was 32.0 % (8/25). This result is higher than in Li et al.’s study, in which the proportion was 23.5 % (154/656). This result may be because Li et al. tested both the dMMR and pMMR groups, whereas we conducted genetics testing only in the dMMR group [22].
In the eight patients diagnosed with Lynch syndrome among dMMR cases, the age of diagnosis ranged from 29 to 69 years, with two men and six women. All the patients had at least one relative (first-degree, second-degree, or third-degree) with a history of stomach cancer, colorectal cancer, rectal cancer, and premature death owing to illness (and not due to other causes such as accidents or war). This aligns with the established understanding of early onset of colorectal cancer and Lynch syndrome as described by Quach and Nguyen [29]. In a study by Sjursen et al. on 834 individuals with germline mutations in MMR genes, 221 different MMR variants were identified, including frame-shift mutations, nonsense mutations, insertions/deletions, missense mutations, and others [30]. Similarly, Pearlman et al. reported about 40 of 48 patients with MMR-deficient tumors had at least one mutation in a cancer susceptibility gene, and 37 patients had Lynch syndrome [31]. In Japan, Suzuki et al. conducted research on 119 young colorectal cancer patients (<50 years old), and eight patients were identified by the screening evaluations as candidates for germline MMR mutation analysis [32]. In Vietnam, according to research by Tran et al., which involved analyzing 17 genes in 1165 individuals across Vietnam who were either referred by physicians or self-enrolled in genetic testing, nine cases with genetic variants in the MLH1, MSH6, and PMS2 genes were identified. These mutations resulted in frameshift or nonsense mutations in the MMR genes [33]. This finding indicates that similar variants are present in the Vietnamese population as reported in the global literature. Apart from the study by Tran et al. [33], there is a notable absence of large-scale, comprehensive research in Vietnam addressing the prevalence of Lynch syndrome among colorectal cancer patients. Furthermore, Abu-Ghazaleh et al. highlighted that the interpretation of the prevalence data was hampered by poor reporting on key baseline characteristics of the patients, emphasizing the need for large, well-reported, epidemiological studies of Lynch syndrome prevalence [34]. According to Quach and Nguyen, the application of Bethesda and Amsterdam criteria for Lynch syndrome screening in 2012 was limited due to the unavailability of microsatellite instability testing methods and insufficient awareness of the disease in Vietnam [29]. While advancements have been made in testing methodologies, the general awareness of genetic diseases, particularly Lynch syndrome, is not fully established.
The association between MMR gene variants and loss protein expression
The pattern of nuclear staining depends on the specific gene involved, making it useful for predicting which gene carries a mutation. For instance, the absence of both MSH2 and MSH6 proteins in a tumor is associated with a germline mutation in MSH2 gene. In addition to subtle mutations impacting the MSH2 open reading frame, large deletions involving one or more exons are also a common cause of Lynch syndrome [35]. Our study results revealed that one patient had a mutation in the MSH2 gene, causing simultaneous loss of MSH2 and MSH6 expression; and two patients had a mutation in the PMS2 gene, causing loss of the corresponding protein PMS2 expression (Table 2). This result is consistent with previous studies reporting that loss of MSH2 expression is typically accompanied by loss of MSH6 expression and that expression and that the loss of PMS2 expression often occurs alone [31].
The patient who had lost MSH6 protein expression and carried EPCAM gene variant represents a case of a germline mutation in the EPCAM gene in Vietnamese individuals. On study in Mayo Clinic investigated the prevalence of EPCAM (TACSTD1) deletions in cases of Lynch Syndrome associated with MSH2. They found that 20–25 % of cases showed deletions suspected to affect MSH2, although no germline mutation was detected [21]. In dMMR patients, the most frequent constitutional epimutation involves hypermethylation of the MLH1 promoter on one allele, silencing gene expression across most somatic tissues, as well as MSH2 hypermethylation due to EPCAM gene mutation [36]. Families with Lynch syndrome who lack detectable sequence mutations in MMR genes should be evaluated for epigenetic mutations. Gene variants do not always correspond with loss of protein expression of the same gene and vice versa; therefore, genetic testing was highly recommended in the case of dMMR patients to confirm Lynch syndrome.
This research applies NGS technology for diagnosing Lynch syndrome in Vietnamese colorectal cancer patients with dMMR cases. We analyzed the association between MMR gene variants and loss of protein expression. Our study also has some limitations, first, selection bias may be present since only 25 out of 32 dMMR patients underwent genetic testing, potentially affecting the representativeness of our findings. Second, we did not assess the methylation status of genes, particularly the MLH1 gene. In addition, we only did genetics testing in dMMR group, so we could not examine the genetic variants in pMMR group. Future studies with larger prospective cohorts and genetic analysis of both dMMR and pMMR groups are need to further clarify the prevalence and genetic basis of Lynch syndrome in Vietnam.
Conclusions
This is a study to apply NGS technology to diagnose Lynch syndrome from dMMR cases in Vietnamese colorectal cancer patients. Our study results support the strategy of screening and diagnosing Lynch syndrome by assessing MMR protein expression status as well as pathogenic variants on MMR genes.
-
Research ethics: This study was conducted in accordance with the Declaration of Helsinki (1964) and was approved by the Ethics Committee of Hanoi Medical University (Approval number 837/GCN-HĐĐĐNCYSH-ĐHYHN). All research variables and indicators are collected honestly and scientifically. The patients consented to participate in the research and all personal information of the patient will be kept confidential and used only for research purposes.
-
Informed consent: Informed consent was obtained from all individuals included in this study, or their legal guardians or wards.
-
Author contributions: Conceptualization, Bich Mai Bui, Cam Phuong Pham, Thi Kim Phuong Doan; methodology, Bich Mai Bui, and Cam Phuong Pham; software, Van Thai Pham and Thuan Loi Nguyen; validation, Van Thai Pham, Quang Loc Bui and Van Thai Pham; formal analysis, Thi Lan Anh Luong and Quang Loc Bui; investigation, Bich Mai Bui and Sy Tung Ha; resources, Minh Khuy Doan and Sy Tung Ha; data curation, Sy Tung Ha and Thanh Huyen Nguyen; writing – original draft preparation, Bich Mai Bui and Hong Phuc Dinh; writing-review and editing, Bich Mai Bui and Thanh Huyen Nguyen; visualization, Thi Lan Anh Luong and Van Tuyen Pham; supervision, Cam Phuong Pham and Thi Kim Phuong Doan; project administration, Bich Mai Bui, Cam Phuong Pham and Thi Kim Phuong Doan. All authors have read and agreed to the published version of the manuscript. All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
-
Use of Large Language Models, AI and Machine Learning Tools: None declared.
-
Conflict of interest: The authors declare that the research was conducted without any commercial or financial relationships that could potentially create a conflict of interest.
-
Research funding: None declared.
-
Data availability: The data that support the findings of this study are available from the corresponding author, upon reasonable request.
References
1. Bray, F, Laversanne, M, Sung, H, Ferlay, J, Siegel, RL, Soerjomataram, I, et al.. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024;74:229–63. https://doi.org/10.3322/caac.21834.Search in Google Scholar PubMed
2. Mojarad, EN, Kuppen, PJ, Aghdaei, HA, Zali, MR. The CpG island methylator phenotype (CIMP) in colorectal cancer. Gastroenterol Hepatol Bed Bench 2013;6:120.Search in Google Scholar
3. Idos, G, Valle, L. Lynch syndrome. In: Adam, MP, Feldman, J, Mirzaa, GM, Pagon, RA, Wallace, SE, Amemiya, A, editors. Seattle, USA: University of Washington, Seattle; 1993.Search in Google Scholar
4. Tiwari, AK, Roy, HK, Lynch, HT. Lynch syndrome in the 21st century: clinical perspectives. QJM 2015;109:151–8. https://doi.org/10.1093/qjmed/hcv137.Search in Google Scholar PubMed
5. Pino, MS, Mino-Kenudson, M, Wildemore, BM, Ganguly, A, Batten, J, Sperduti, I, et al.. Deficient DNA mismatch repair is common in Lynch syndrome-associated colorectal adenomas. J Mol Diagn 2009;11:238–47. https://doi.org/10.2353/jmoldx.2009.080142.Search in Google Scholar PubMed PubMed Central
6. Leclerc, J, Vermaut, C, Buisine, M-P. Diagnosis of Lynch syndrome and strategies to distinguish Lynch-related tumors from sporadic MSI/dMMR tumors. Cancers 2021;13:467. https://doi.org/10.3390/cancers13030467.Search in Google Scholar PubMed PubMed Central
7. Boland, CR, Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010;138:2073–87.e3. https://doi.org/10.1053/j.gastro.2009.12.064.Search in Google Scholar PubMed PubMed Central
8. Kawakami, H, Zaanan, A, Sinicrope, FA. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol 2015;16:30. https://doi.org/10.1007/s11864-015-0348-2.Search in Google Scholar PubMed PubMed Central
9. National Comprehensive Cancer Network (NCCN). NCCN Clinical Practice Guidelines in Oncology: Colon Cancer. Version 6. Fort Washington, PA: National Comprehensive Cancer Network; 2024. [cited 2025 Jan].Search in Google Scholar
10. Cervantes, A, Adam, R, Roselló, S, Arnold, D, Normanno, N, Taïeb, J, et al.. Metastatic colorectal cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol 2023;34:10–32. https://doi.org/10.1016/j.annonc.2022.10.003.Search in Google Scholar PubMed
11. Tran, NH, Nguyen, TTH, Tang, HS, Hoang, LP, Nguyen, TL, Tran, NT, et al.. Genetic landscape of recessive diseases in the Vietnamese population from large-scale clinical exome sequencing. Hum Mutat 2021;42:1229–38. https://doi.org/10.1002/humu.24253.Search in Google Scholar PubMed
12. Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997; 2013. [Preprint].Search in Google Scholar
13. DePristo, MA, Banks, E, Poplin, R, Garimella, KV, Maguire, JR, Hartl, C, et al.. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–8. https://doi.org/10.1038/ng.806.Search in Google Scholar PubMed PubMed Central
14. Sherry, ST, Ward, M-H, Kholodov, M, Baker, J, Phan, L, Smigielski, EM, et al.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001;29:308–11. https://doi.org/10.1093/nar/29.1.308.Search in Google Scholar PubMed PubMed Central
15. Landrum, MJ, Lee, JM, Riley, GR, Jang, W, Rubinstein, WS, Church, DM, et al.. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 2014;42:D980–D5. https://doi.org/10.1093/nar/gkt1113.Search in Google Scholar PubMed PubMed Central
16. Fokkema, IF, Taschner, PE, Schaafsma, GC, Celli, J, Laros, JF, den Dunnen, JT. LOVD v. 2.0: the next generation in gene variant databases. Hum Mutat 2011;32:557–63. https://doi.org/10.1002/humu.21438.Search in Google Scholar PubMed
17. McLaren, W, Gil, L, Hunt, SE, Riat, HS, Ritchie, GR, Thormann, A, et al.. The ensembl variant effect predictor. Genome Biol 2016;17:122. https://doi.org/10.1186/s13059-016-0974-4.Search in Google Scholar PubMed PubMed Central
18. Richards, S, Aziz, N, Bale, S, Bick, D, Das, S, Gastier-Foster, J, et al.. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet Med 2015;17:405–24. https://doi.org/10.1038/gim.2015.30.Search in Google Scholar PubMed PubMed Central
19. Untergasser, A, Cutcutache, I, Koressaar, T, Ye, J, Faircloth, BC, Remm, M, et al.. Primer3—new capabilities and interfaces. Nucleic Acids Res 2012;40:e115–e. https://doi.org/10.1093/nar/gks596.Search in Google Scholar PubMed PubMed Central
20. Kempers, MJ, Kuiper, RP, Ockeloen, CW, Chappuis, PO, Hutter, P, Rahner, N, et al.. Risk of colorectal and endometrial cancers in EPCAM deletion-positive Lynch syndrome: a cohort study. Lancet Oncol 2011;12:49–55. https://doi.org/10.1016/s1470-2045(10)70265-5.Search in Google Scholar PubMed PubMed Central
21. Rumilla, K, Schowalter, KV, Lindor, NM, Thomas, BC, Mensink, KA, Gallinger, S, et al.. Frequency of deletions of EPCAM (TACSTD1) in MSH2-associated Lynch syndrome cases. J Mol Diagn 2011;13:93–9. https://doi.org/10.1016/j.jmoldx.2010.11.011.Search in Google Scholar PubMed PubMed Central
22. Li, Y, Fan, L, Zheng, J, Nie, X, Sun, Y, Feng, Q, et al.. Lynch syndrome pre-screening and comprehensive characterization in a multi-center large cohort of Chinese patients with colorectal cancer. Cancer Biol Med 2022;19:1235–48. https://doi.org/10.20892/j.issn.2095-3941.2021.0585.Search in Google Scholar PubMed PubMed Central
23. Mei, W-J, Mi, M, Qian, J, Xiao, N, Yuan, Y, Ding, P-R. Clinicopathological characteristics of high microsatellite instability/mismatch repair-deficient colorectal cancer: a narrative review. Front Immunol 2022;13:1019582. https://doi.org/10.3389/fimmu.2022.1019582.Search in Google Scholar PubMed PubMed Central
24. Lok, P, Dijk, S. Offer daily aspirin to cut risk of colorectal cancer in people with Lynch syndrome, says NICE. BMJ 2019;366:l5010. https://doi.org/10.1136/bmj.l5010.Search in Google Scholar PubMed
25. Hao, VTT, Tuyen, PV, Chuong, TV, Khuy, ĐM, Lan, TT, Trung, NV, et al.. Study on MMR protein expression status in patients with colorectal carcinoma at Bach Mai Hospital. Vietnam Med J 2023;533.Search in Google Scholar
26. Yurgelun, MB, Kulke, MH, Fuchs, CS, Allen, BA, Uno, H, Hornick, JL, et al.. Cancer susceptibility gene mutations in individuals with colorectal cancer. J Clin Oncol 2017;35:1086–95. https://doi.org/10.1200/jco.2016.71.0012.Search in Google Scholar PubMed PubMed Central
27. Rüschoff, J, Schildhaus, H-U, Rüschoff, JH, Jöhrens, K, Bocker Edmonston, T, Dietmaier, W, et al.. Testing for deficient mismatch repair and microsatellite instability: a focused update. Pathologie 2023;44:61–70. https://doi.org/10.1007/s00292-023-01208-2.Search in Google Scholar PubMed PubMed Central
28. National Comprehensive Cancer Network (NCCN). Genetic/Familial High-Risk Assessment: Colorectal, Endometrial, and Gastric. Version 3. Fort Washington, PA: National Comprehensive Cancer Network; 2024. [cited 2024 Oct].Search in Google Scholar
29. Quach, DT, Nguyen, OT. Clinical, endoscopic and pathological characteristics of early-onset colorectal cancer in Vietnamese. Asian Pac J Cancer Prev 2012;13:1767–70. https://doi.org/10.7314/apjcp.2012.13.5.1767.Search in Google Scholar PubMed
30. Sjursen, W, McPhillips, M, Scott, RJ, Talseth-Palmer, BA. Lynch syndrome mutation spectrum in New South Wales, Australia, including 55 novel mutations. Mol Genet Genomic Med 2016;4:223–31. https://doi.org/10.1002/mgg3.198.Search in Google Scholar PubMed PubMed Central
31. Pearlman, R, Frankel, WL, Swanson, B, Zhao, W, Yilmaz, A, Miller, K, et al.. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol 2017;3:464–71. https://doi.org/10.1001/jamaoncol.2016.5194.Search in Google Scholar PubMed PubMed Central
32. Suzuki, O, Eguchi, H, Chika, N, Sakimoto, T, Ishibashi, K, Kumamoto, K, et al.. Prevalence and clinicopathologic/molecular characteristics of mismatch repair-deficient colorectal cancer in the under-50-year-old Japanese population. Surg Today 2017;47:1135–46. https://doi.org/10.1007/s00595-017-1486-x.Search in Google Scholar PubMed
33. Tran, VT, Nguyen, ST, Pham, XD, Phan, TH, Nguyen, VC, Nguyen, HT, et al.. Pathogenic variant profile of hereditary cancer syndromes in a Vietnamese cohort. Front Oncol 2021;11:789659. https://doi.org/10.3389/fonc.2021.789659.Search in Google Scholar PubMed PubMed Central
34. Abu-Ghazaleh, N, Kaushik, V, Gorelik, A, Jenkins, M, Macrae, F. Worldwide prevalence of Lynch syndrome in patients with colorectal cancer: systematic review and meta-analysis. Genet Med 2022;24:971–85. https://doi.org/10.1016/j.gim.2022.01.014.Search in Google Scholar PubMed
35. Ligtenberg, MJ, Kuiper, RP, Geurts van Kessel, A, Hoogerbrugge, N. EPCAM deletion carriers constitute a unique subgroup of Lynch syndrome patients. Fam Can 2013;12:169–74. https://doi.org/10.1007/s10689-012-9591-x.Search in Google Scholar PubMed
36. Peltomäki, P. Epigenetic mechanisms in the pathogenesis of Lynch syndrome. Clin Genet 2014;85:403–12. https://doi.org/10.1111/cge.12349.Search in Google Scholar PubMed
Supplementary Material
This article contains supplementary material (https://doi.org/10.1515/oncologie-2024-0564).
© 2025 the author(s), published by De Gruyter on behalf of Tech Science Press (TSP)
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Review Articles
- Targeting HER2 in metastatic urothelial carcinoma: a contemporary review
- Advances in research on the impact of exosomal miRNAs on liver metastasis of colorectal cancer
- Sebaceous carcinoma of the intraoral origin: a literature review
- Role of emerging theranostic technologies in precision oncology: revolutionizing cancer diagnosis and treatment
- Research Articles
- Half-body irradiation with dose escalation in the era of advanced systemic therapies: unveiling new therapeutic opportunities
- Characteristics of MMR protein expression in colorectal cancer and MMR gene variations in Vietnamese patients with Lynch syndrome
- BMPR1A promotes the proliferation of colorectal cancer cells through the activation of Smad1
- Construction and validation of a diagnostic model for cholangiocarcinoma based on tumor-educated platelet RNA expression profiles
- Prognostic implications of PCSK9 expression in HER2-positive breast cancer
- Rapid Communication
- Potential impact of hormone replacement therapy on the risk of hepatocellular carcinoma in women of the PLCO cohort
- Article Commentary
- “Plant-based and ketogenic diets as diverging paths to address cancer”: a commentary concerning the supposed superiority of a plant-based diet
- Novel insights into molecular landscape of advanced renal cell carcinoma
- Corrigendum
- Corrigendum to “Y27632 induces tongue squamous cell carcinoma cell apoptosis through MAPK-ERK/JNK signal”
Articles in the same Issue
- Frontmatter
- Review Articles
- Targeting HER2 in metastatic urothelial carcinoma: a contemporary review
- Advances in research on the impact of exosomal miRNAs on liver metastasis of colorectal cancer
- Sebaceous carcinoma of the intraoral origin: a literature review
- Role of emerging theranostic technologies in precision oncology: revolutionizing cancer diagnosis and treatment
- Research Articles
- Half-body irradiation with dose escalation in the era of advanced systemic therapies: unveiling new therapeutic opportunities
- Characteristics of MMR protein expression in colorectal cancer and MMR gene variations in Vietnamese patients with Lynch syndrome
- BMPR1A promotes the proliferation of colorectal cancer cells through the activation of Smad1
- Construction and validation of a diagnostic model for cholangiocarcinoma based on tumor-educated platelet RNA expression profiles
- Prognostic implications of PCSK9 expression in HER2-positive breast cancer
- Rapid Communication
- Potential impact of hormone replacement therapy on the risk of hepatocellular carcinoma in women of the PLCO cohort
- Article Commentary
- “Plant-based and ketogenic diets as diverging paths to address cancer”: a commentary concerning the supposed superiority of a plant-based diet
- Novel insights into molecular landscape of advanced renal cell carcinoma
- Corrigendum
- Corrigendum to “Y27632 induces tongue squamous cell carcinoma cell apoptosis through MAPK-ERK/JNK signal”

