Abstract
The number of shrew species in Israel has been and still is the subject of debate. In this work we used for the first time a molecular marker, the cytochrome b gene, to investigate the number and identity of shrew species in Israel. Our molecular results confirmed the presence of four species: Crocidura leucodon, Crocidura suaveolens gueldenstaedtii, Crocidura ramona, and Suncus etruscus. The C. ramona sequences were found to differ from all other Crocidura species sequenced to date, supporting its status as a distinct species. Whether it is conspecific with Crocidura portali (described in 1920 from Israel and usually synonymized with C. suaveolens), will require additional study. The sequences of Israeli C. suaveolens were found to be very similar to those of Iran, Turkey, and Georgia (i.e., C. suaveolens gueldenstaedtii), in agreement with previous studies. The Israeli C. leucodon sequences, however, formed a distinct clade among C. leucodon. Finally, the S. etruscus sequences clustered with sequences from France, Italy, and Iran.
1 Introduction
Shrews (family: Soricidae) are among the most diverse and abundant mammals (Burgin et al. 2018b; Wilson and Reeder 2005). Among them, the Old-World genus Crocidura, contains 198 species, more than any other mammalian genus (Burgin et al. 2018a). However, being mainly small, nocturnal, and rather unremarkable-looking, their diversity tends to have escaped the notice of the general public, environmental decision-makers, conservation agencies, and even scientists. The number of shrew species thought to exist in Israel is under debate (Table 1). Tristram (1884) recognized five species in the Levant and assigned them to the genus Sorex: Sorex araneus, Sorex tetragonurus (currently known as S. araneus), Sorex pygmaeus (currently known as Suncus etruscus), Sorex crassicaudus (currently known as Suncus murinus), and Sorex fodiens (currently known as Neomys fodiens). N. fodiens is a water-shrew and, together with S. araneus and S. murinus, it has not been reported again in Israel since Tristram’s original work (Bodenheimer 1935). Thomas (1919, 1920) assigned two Israeli species to the genus Crocidura: Crocidura russula judaica (Thomas 1919) and a new species that he described: Crocidura portali (Thomas 1920). Bodenheimer (1935) contended that three species of Crocidura occur in Israel: C. russula, Crocidura judaica, and C. portali. Later, he considered C. portali as a subspecies of Crocidura suaveolens (Bodenheimer 1958); however, no explanation was given for this decision. Regarding Suncus,Bodenheimer (1935) indicated that the presence of S. crassicaudus is a consequence of misidentification and Bodenheimer (1958) reassigned the shrew described by Tristram (1884) as S. pygmaeus, to S. etruscus. Harrison (1963), later contended that Thomas’s C. russula judaica (1919) is in fact a local synonym of Crocidura leucodon, based on morphological traits. He further identified C. russula and C. suaveolens portali as inhabiting Israel.
Number and identities of Israeli shrew species.
| Source | Shrew species inhabiting Israel |
|---|---|
| Tristram (1884) | Sorex araneus, S. tetragonurusa, S. pygmaeusb, S. crassicaudusc, S. fodiensd |
| Thomas (1919, 1920)h | C. russula judaica, C. portalie |
| Bodenheimer (1935; 1937) | Sorex araneus, Sorex minutus (=pygmaeus), Neomys fodiens, C. russula, C. r. judaica, C. portalie, S. tristramif |
| Bodenheimer (1958) | C. russula, C. judaica, C. suaveolens portali, S. etruscus |
| Harrison (1963)i | C. russula, C. leucodon judaica, C. suaveolens portali |
| Mendelssohn and Yom-Tov (1987) | C. russula, C. leucodon, C. suaveolens, S. etruscus |
| Mendelssohn and Yom-Tov (1999); Harrison and Bates (1991) | C. leucodon judaica, C. suaveolens monacha, S. etruscus etruscus |
| Dolev and Perevolotsky (2004); Meiri et al. (2019), this work | C. ramona, C. leucodon, C. suaveolens, S. etruscus |
| Wilson and Reeder (2005) | C. gmelinig, C. katinka, C. ramona, C. leucodon, C. suaveolens, S. etruscus |
| IUCN red list (2020) | C. katinka, C. ramona, C. leucodon, C. suaveolens, S. etruscus, perhaps C. gmelini |
| Burgin et al. (2018b) | C. ramona, C. leucodon judaica, C. gueldenstaedtii gueldenstaedtii, S. etruscus etruscus |
aSorex tetragonurus has been synonymized with Sorex araneus (Wilson and Reeder 2005).
bSorex pygmaeus has been synonymized with Suncus etruscus (Wilson and Reeder 2005).
cSorex crassicaudus has been synonymized with Suncus murinus (Wilson and Reeder 2005). Its presence in Israel was considered to be the result of a misidentification by Bodenheimer (1958) and later sources.
dSorex fodiens has been synonymized with Neomys fodiens (Wilson and Reeder 2005).
eCrocidura portali has been synonymized either with C. suaveolens (Bodenheimer 1958) or under C. gmelinig (Wilson and Reeder 2005), or kept at the species rank (Kryštufek and Vohralík 2001).
fSuncus tristrami has been synonymized with C. suaveolens (Wilson and Reeder 2005).
gCrocidura gmelini has been synonymized with C. suaveolens (Bannikova et al. 2006; Burgin et al. 2018b; Saeedzadeh et al. 2017).
hDescription of specific specimens.
iHarrison (1963) focused on the genus Crocidura.
Molecular studies of shrews started in the 1980s. Based on karyotype and allozyme comparisons, Catzeflis et al. (1985) demonstrated that the individuals called C. russula in the Middle East were in fact members of C. suaveolens, and further placed the subspecies gueldenstaedtii and monacha within suaveolens. Harrison and Bates (1991) treated C. suaveolens specimens from Israel as C. suaveolens monacha. They also noted that a smaller form is present in southern Israel, Sinai, and Arabia, and that, consequently, a second subspecies may be present “If this proves to be the case the name portali is available”. Relationships among members of the suaveolens species complex have been found to be intricate and obscured by introgression events (Bannikova et al. 2006; Castiglia et al. 2017; Dubey et al. 2006, 2007a; Ohdachi et al. 2004). C. suaveolens specimens from the Caucasus, the Balkans, and the Levant have been either treated as a distinct species within the suaveolens group, Crocidura gueldenstaedtii (Bannikova et al. 2006; Burgin et al. 2018b), or as a subspecies: C. suaveolens gueldenstaedtii (Burgin et al. 2018a; Dubey et al. 2008; Palomo et al. 2016). Of note, the subspecies assignment of Israeli specimens is further complicated by the use of the subspecies monacha (Harrison and Bates 1991; Mendelssohn and Yom-Tov 1999). In the absence of a formal revision of the C. suaveolens group, we refer throughout the text to such specimens as C. suaveolens gueldenstaedtii.Ivanitskaya et al. (1996) described a new species of Israeli endemic shrew: Crocidura ramona, which presents different morphological characteristics and karyotype to those of C. leucodon and C. suaveolens.
It is thus currently considered that there are at least four shrew species in Israel: C. leucodon, C. suaveolens/C. gueldenstaedtii, C. ramona, and S. etruscus (Dolev and Perevolotsky 2004; Meiri et al. 2019; while there is debate regarding two additional species, C. portali and Crocidura katinkaTable 1, Gerrie and Kennerley 2017; Wilson and Reeder 2005). C. katinka was originally described in 1937 from a Pleistocene skull discovered in the Tabun Cave near Mount Carmel (Bate 1937). The species was thought to be extinct until the recent discovery of skulls similar to Bate’s C. katinka in owl pellets from Syria (Hutterer and Kock 2002). While there has not been recent evidence of C. katinka’s presence in Israel, it is often listed as a member of the Israeli fauna (Gerrie and Kennerley 2017; Wilson and Reeder 2005). Similarly, the exact status of C. portaliThomas 1920 remains unresolved. It has been treated variously as a distinct species (Kryštufek and Vohralík 2001) or suggested as a senior synonym of C. ramona (Burgin et al. 2018b; Kryštufek and Vohralík 2001), synonymized with C. suaveolens (Bodenheimer 1958) or with Crocidura gmelini (Hoffmann 1996; Wilson and Reeder 2005). C. gmelini is found in arid environments from Iran to Kazakhstan (Hutterer 2017) and has sometimes been synonymized with C. suaveolens (Bannikova et al. 2006; Burgin et al. 2018b; Saeedzadeh et al. 2017). It is thus unclear as to whether or not another member of the C. suaveolens species complex (Bannikova et al. 2006; Dubey et al. 2006, 2007a) is present in Israel.
Neither C. leucodon nor S. etruscus from Israel have been the subject of any molecular phylogenetic study to date. Based on the sequences of two specimens, Dubey et al. (2007a) suggested that “C. suaveolens” from Israel is part of the gueldenstaedtii clade (their ‘C. suaveolens clade V’), which includes individuals from the southern Balkans, the Caucasus, Transcaucasia, Turkey, the Near East, Arabia, and several Mediterranean islands. In their molecular study of the Crocidura radiation, Dubey et al. (2008) sequenced part of the BRCA1 (254 bp) and part of the Apolipoprotein B gene (840 bp) of a C. ramona specimen from Israel. A careful look at their published sequence revealed the ramona BRCA1 sequence (accession number: EF525159) to be identical to the C. suaveolens sequences, while the Apolipoprotein B gene (accession number: EF525041) differs, suggesting that at least one of the sequences could be a contamination. This casts doubt on Dubey et al.’s (2008) inference concerning the phylogenetic position of this species.
According to the International Union for Conservation of Nature’s Red List of Threatened Species (IUCN Red List; last accessed April 2020) all the Israeli shrews are listed as ‘least-concern’. Nonetheless, only C. suaveolens. is assessed as having stable populations (Palomo et al. 2016), while the three other species’ population trends are considered unknown (Aulagnier et al. 2017; Hutterer and Shenbrot 2017; Shenbrot et al. 2016). Consequently, there is sparse knowledge regarding the biodiversity patterns of shrews in Israel, and an inadequate understanding of their conservation needs. This is especially true for C. ramona, which has been described as endemic to Israel and Palestine. It is known from the Negev, the Arava, Samaria and the Judean Desert.
In this work we performed a barcoding analysis using cytochrome b (cyt b) sequences of representatives of the Israeli shrew diversity in order to determine the number of shrew species present in Israel.
2 Materials and methods
2.1 Sample origin
Thirty-one samples were used in this work. We received eight C. leucodon, 12 C. suaveolens, six S. etruscus, and three C. ramona samples (all from Israel) from the Steinhardt National Collection of Natural History, Zoological Museum at Tel Aviv University (Israel). The location of the samples is indicated in Figure 1A. In addition, the Field Museum of Natural History (Chicago, USA) kindly provided two samples of Crocidura nana from Tanzania, which were also considered in order to examine the possible relationship of this species to C. ramona. Both ramona and nana share a small size and a greyish tinge to their fur (Dobson 1890; Ivanitskaya et al. 1996). Voucher number, collection location, and date of all samples are provided in Supplementary Table S1.
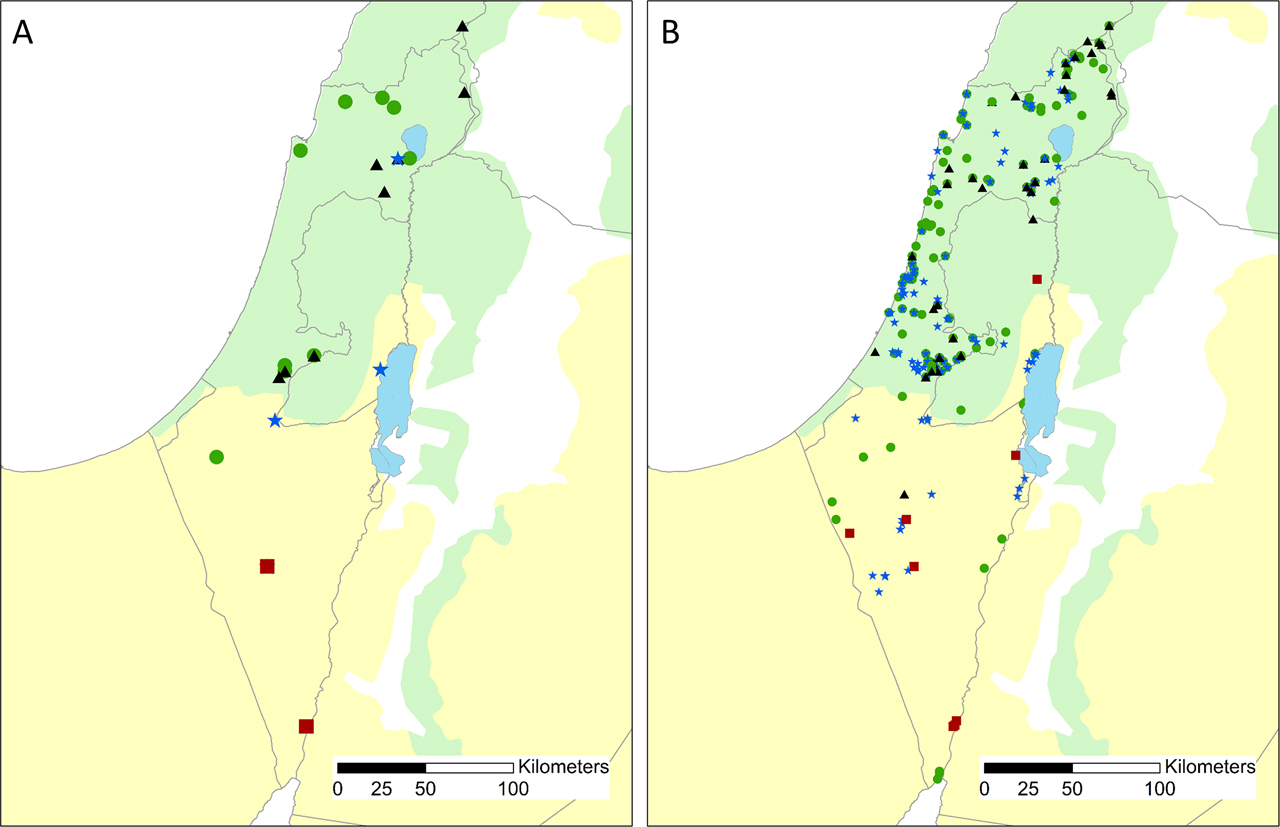
Shrew sample locations. A. samples sequenced in this work. B. shrew specimen present in the Steinhardt National Collection of Natural History, Zoological Museum at Tel Aviv University (Israel). The two major biomes of Israel: the Mediterranean biome, and the desert, are indicated in green and yellow, respectively. Red squares, blues stars, green circles and black triangles indicate C. ramona, S. etruscus, C. suaveolens gueldenstaedtii and C. leucodon specimen respectively. The museum records encompass 79 C. leucodon, 594 C. suaveolens gueldenstaedtii, 13 C. ramona and 443 S. etruscus.
2.2 DNA isolation
Two types of museum samples were used. The first was that of tissue samples from specimens captured relatively recently (2002–2015) and preserved in 70–100% ethanol. The second type of sample was that of skin pieces of older specimens (collected 1969–1995) preserved as study skins (Supplementary Table S1). Each type of sample was extracted following a different protocol.
For the specimens preserved in alcohol, a sample of the leg muscle (about 0.5 gr), ear (about 0.5 gr/sample), or kidney/heart (about the same weight) was cut into small parts and digested using 0.5 mL of lysis buffer (1% SDS, 10 mMTris-HCl pH8, 125mMNaCl, 5 mM EDTA, and 0.5 mg/mL Proteinase K) at 55 °C. Following homogenization, the DNA was extracted using a standard phenol-chloroform protocol followed by ethanol/sodium-acetate precipitation (Sanbrook et al. 1989).
For the four specimens preserved as skins, we used the QiagenDNeasy Blood and Tissue Kit following the protocol of Iudica et al. (2001) for small bones from dried mammalian museum specimens. This protocol starts with washing the tissue 3 times in 250 µL of 1% PBS for 10 min at 55 °C. This step allows the removal of any inhibitors and residual fixatives present in the skin specimens. Iudica et al.’s (2001) protocol includes a long digestion time (in our case digestion was conducted for 15–36 h, depending on the sample) with repeated additions of proteinase K (specifically 10 μL every 6 h, overnight 20 μL). The long digestion was necessary since the skin tissues used were hard and difficult to digest.
2.3 Amplification and sequencing of the cytochrome b
Amplification of the complete cyt b gene was conducted differently for the ethanol-preserved samples and the skin samples. For the ethanol-preserved samples the DNA quality following extraction allowed us to amplify large fragments. Consequently, amplification of the cyt b gene was conducted with the primers D1 and R1, which are located in the tRNA-glu and tRNA-thr, respectively (primer sequences are presented in Supplementary Table S2). The PCR amplifications were conducted using the BIOTAQ™ DNA polymerase (Bioline, London). Following a denaturation at 94 °C for 2 min, the PCR was set for 40 cycles of denaturation at 94 °C for 40 s, annealing at 59 °C for 40 s, and elongation at 72 °C for 2.5 min. The 40 cycles were followed by a final elongation step at 72 °C for 10 min.
Because the yield of the amplification was insufficient to directly sequence the PCR fragments obtained, these products were re-amplified. The re-amplifications were conducted using the nested primers D2 and R2, which are located in the tRNA(Glu) and tRNA(Pro) genes, respectively. The PCR conditions were as described above.
For the four samples preserved as skins, the DNA was too degraded to enable the amplification of fragments longer than 400 bp. The sequencing of the complete cyt b gene was conducted by amplifying small overlapping fragments of ∼200–350 bp. Different primer sets were used for each species (all primer sequences are given in Supplementary Table S2). Following denaturation at 94 °C for 3 min, the PCR was set for 34 cycles of denaturation at 94 °C for 45 s, annealing at 51 °C for 45 s, and elongation at 72 °C for 45 s. The 34 cycles were followed by a final elongation step at 72 °C for 10 min.
PCR products were directly sequenced using Big Dye Terminator v1.1 (Applied Biosystems) on an ABI 310 sequencer by the DNA Sequencing Unit at the G.S. Wise Faculty of Life Sciences, Tel Aviv University.
2.4 Phylogenetic reconstructions
Four datasets were used to reconstruct the phylogenetic position of the obtained Israeli sequences. The first dataset comprised representatives of the Crocidura diversity. Specifically, sequence searches were conducted on the National Center for Biotechnology Information (NCBI) nucleotide collection database, with the queries ‘Crocidura cyt b’ and ‘Crocidura cytochrome b’, and all sequences longer than 500 bp were downloaded. We also downloaded all sequences from Dubey et al.’s (2008) dataset, and sequences from the same individuals were concatenated. From the list of sequences obtained from the NCBI searches and the Dubey dataset, we selected the longest sequence for each Crocidura species. In addition, two C. leucodon, two C. suaveolens, three C. ramona, two S. etruscus (all from Israel), and two C. nana (from Tanzania), obtained as described above, were added to the dataset. We also added S. etruscus (LC126597 and DQ630397), S. dayi (DQ630389 and DQ630432), two S. montanus (GQ290374 and DQ630388), and three S. murinus (LC126460, LC126565, and LC126577) sequences from different countries as outgroups (Dubey et al. 2008) (Supplementary Table S3).
The C. leucodon dataset was created by downloading C. leucodon cyt b sequences from NCBI using the keywords “C. leucodon cyt b” and selecting only nucleotide results. A total of 56 sequences longer than 500 bp were downloaded. Eight newly-obtained C. leucodon sequences from Israel were added to this dataset. The tree was rooted with cyt b sequences of Crocidura musseri (FJ813927 and FJ813929) and Crocidura obscurior (KC684154, KC684155, and KC684158) downloaded from NCBI. Again, the outgroup choice was based on Dubey et al. (2008) (Supplementary Table S4).
The C. suaveolens complex dataset was created by downloading C. suaveolens cyt b sequences from NCBI using the keywords “C. suaveolens cyt b” and selecting only nucleotide results. A total of 203 sequences longer than 500 bp were downloaded. Four Crocidura shantungensis cyt b sequences (KF144163, AB077278, KF144159, and AY843447) and one Crocidura zarudnyi sequence (AY925211) were also added since they are known to be closely related to C. suaveolens (Dubey et al. 2008). A total of 12 C. suaveolens sequences from Israel were added to this dataset. The tree was rooted with cyt b sequences from Crocidura brunnea (DQ059025), Crocidura lasiura (AY843503), and Crocidura nigripes (DQ059024), since these species are closely related to C. suaveolens (Dubey et al. 2008) (Supplementary Table S5).
The S. etruscus dataset was prepared by first downloading Suncus cyt b sequences from NCBI using the search “Suncus cyt b” and selecting only nucleotide results. A total of 250 sequences was downloaded and sequences shorter than 500 bp were removed. From this dataset we selected all representatives of S. etruscus, Suncus fellowesgordoni, S. madagascariensis, S. malaynus, and S. dayi. In addition, we selected three S. montanus, five S. murinus, and two S. stoliczkanus sequences as outgroups. This choice was based on previous phylogenetic works (Dubey et al. 2008; Meegaskumbura et al. 2012a, b; Omar et al. 2011). Finally, we added six S. etruscus sequences from Israel to this dataset (Supplementary Table S6).
The nucleotide sequences were aligned using a codon-based alignment as implemented in Geneious 7.1.9, with MAFFT version 7.0.1.7 under the L-ins-i algorithm (Katoh and Standley 2013). Following alignment, columns with at least 50% gaps were excluded. The alignment files are provided in Nexus format in Supplementary Datasets S1–S4. Phylogenetic trees were reconstructed for each dataset separately under the maximum likelihood (ML) and Bayesian criteria. ML analyses were performed with the program RaxML version 8.1.2 (Stamatakis 2014) as implemented in RAxMLGUI 1.5b2 beta (Silvestro and Michalak 2012). The analyses were run with the options “ML + thorough bootstrap”, “20 runs”, “1000 bootstrap repetitions”, and “GTRGAMMA”. Bayesian reconstructions were performed with MrBayes version 3.2.6 (Ronquist et al. 2012) under the GTR + Gamma model. The parameters of the analyses were: two runs with four chains each, sampling every 100 generations, and Burninfrac set to 0.25. The analyses were run for 15,000,000 generations for the C. leucodon and S. etruscus datasets and for 30,000,000 generations for the C. suaveolens complex and the entire Crocidura dataset. For each dataset, we verified that the average standard deviation of split frequencies was below 0.01 before the burnin threshold was reached. We also verified that the Potential Scale Reduction Factor (PSRF) parameters were always close to 1.0 at the end of the run.
2.5 Network analyses
Median-joining networks (Bandelt et al. 1999) were inferred using cyt b haplotypes and the program NETWORK v 10.0.0.0 under default parameters (available at http://www.fluxes-engineering com/sharenet.htm). The datasets used in the network analyses were constructed from the matrices used in the phylogenetic analyses by removing outgroups and shorter sequences as well as excluding all positions containing ambiguous or missing data. The C. leucodon dataset comprised 51 taxa and 1041 positions, of which 155 were variable. The C. suaveolens complex dataset comprised 75 taxa and 901 positions, of which 76 were variable. The S. etruscus dataset comprised 23 taxa and 516 positions, of which 98 were variable.
3 Results
3.1 Phylogenetic relationships among Crocidura species
The phylogenetic tree reconstructed based on the Crocidura dataset, and which encompassed representatives of the Crocidura species diversity, agreed overall with the findings of Dubey et al. (2008). Because our analyses were based on a single gene (1140 bp), whereas Dubey et al. (2008) had used four genes (3306 bp), branch support is much lower in our case. It should be noted that the topological differences between Dubey’s trees and the trees in Figure 2 and Supplementary Figure S1 only relate to the lowly supported nodes. Specifically, our results recover the monophyly of the Afrotropical clade and the Asian clade described by Dubey et al., but with no support (ML Bootstrap Percentage, BP < 50%; Bayesian Posterior Probabilities, PP = 0.65 for the Asian clade and PP = 0.98 for the Afrotropical clade). The monophyly of the Old-World clade could, however, not be recovered.
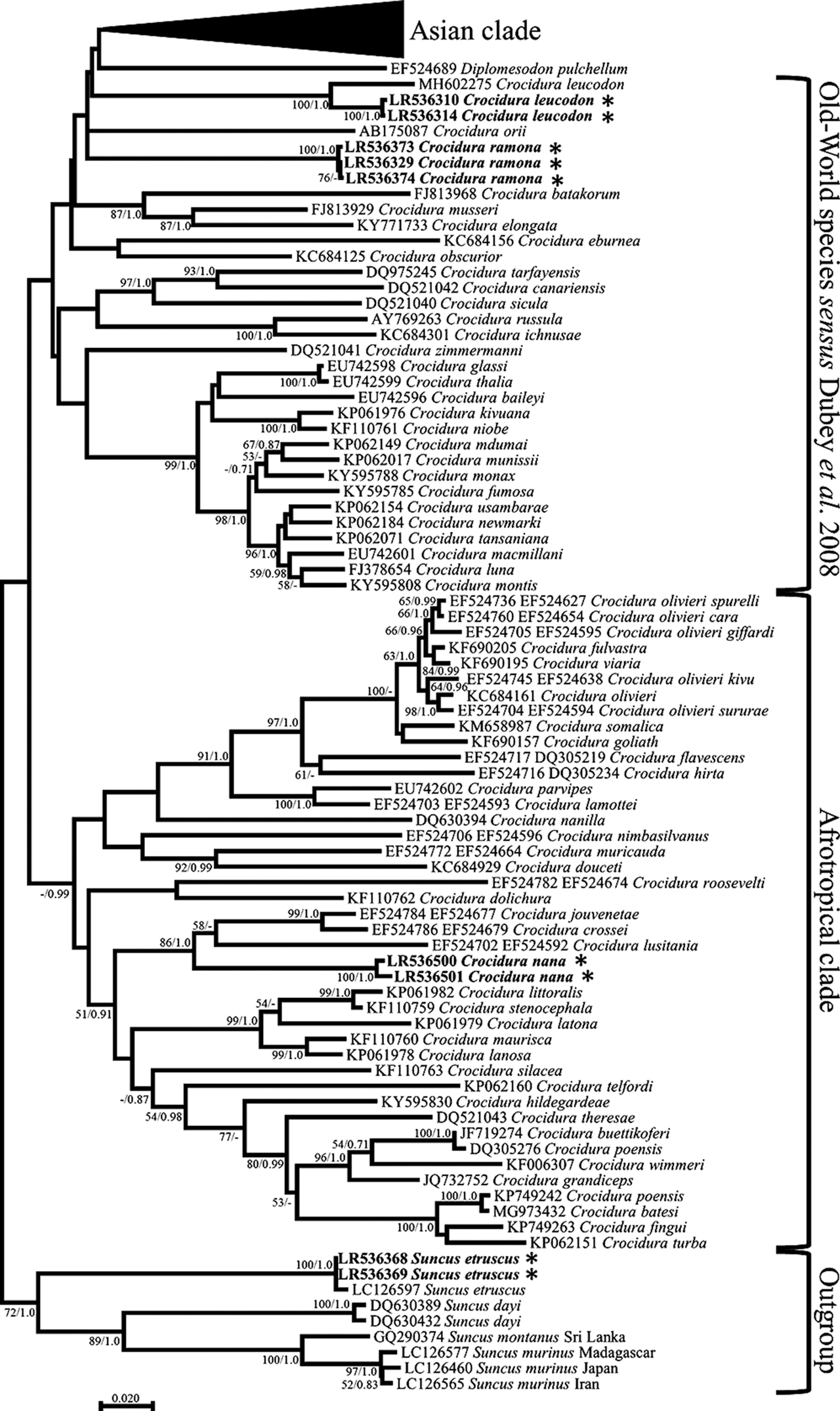
Maximum likelihood tree of Crocidura cytb sequences with emphasis on the Asian and Old World species. Phylogenetic relationships inferred from a matrix of 1,128 nucleotide positions for 131 individuals. Maximum likelihood bootstrap supports above 50% and Bayesian posterior probabilities above 0.70 are indicated near the corresponding node separated with a slash. Sequences obtained in this work are indicated in bold. Authors of sequence data in Supplementary Table S3.
Regarding the Israeli samples, the phylogenetic results confirmed the presence of four distinct species: S. etruscus, C. leucodon, C. suaveolens gueldenstaedtii, and C. ramona (Figure 2 and Supplementary Figure S1). The first three species strongly clustered (BP = 100, PP = 1) with sequences of the same species. In agreement with Dubey et al. (2008), C. ramona sequences formed a distinct lineage whose phylogenetic position could not be determined since the monophyly of the Old-World clade was not supported in our analyses (Figure 2). The C. ramona sequences are clearly unrelated to the C. nana sequences that we obtained from Tanzania, which are nested within an Afrotropical clade with Crocidura jouvenetae, Crocidura crossei, and Crocidura lusitania (BP = 86, PP = 1.0; Supplementary Figure S1).
3.2 Phylogenetic relationships within the C. leucodon clade
The C. leucodon tree recovered the monophyly of the main known C. leucodon clades (Dubey et al. 2007b; Mahmoudi et al. 2019). We found a western clade (with samples from western Europe and western Turkey; BP = 71; PP = 0.99), an eastern clade (with samples from Bulgaria, Romania, Georgia, and eastern Turkey; BP = 99%; PP = 1.0), and an Iranian clade (with samples from the Hyrcanian region of Iran; BP = 85%; PP = 1.0) (Figure 3; Dubey et al. 2007b; Mahmoudi et al. 2019). Within this tree, the Israeli C. leucodon cyt b sequences formed an isolated and strongly supported clade (BP = 91; PP = 1.0) (Figure 3). This Israeli clade is thus placed as a sister clade to the eastern clade, with (BP = 79; PP = 0.99). The average p-distance between sequences from Israel and sequences of the eastern clade was 0.023 (minimum = 0.019; maximum = 0.030). The results of the network analyses support the division into the four distinct C. leucodon lineages observed in the phylogenetic analysis (Supplementary Figure S2).
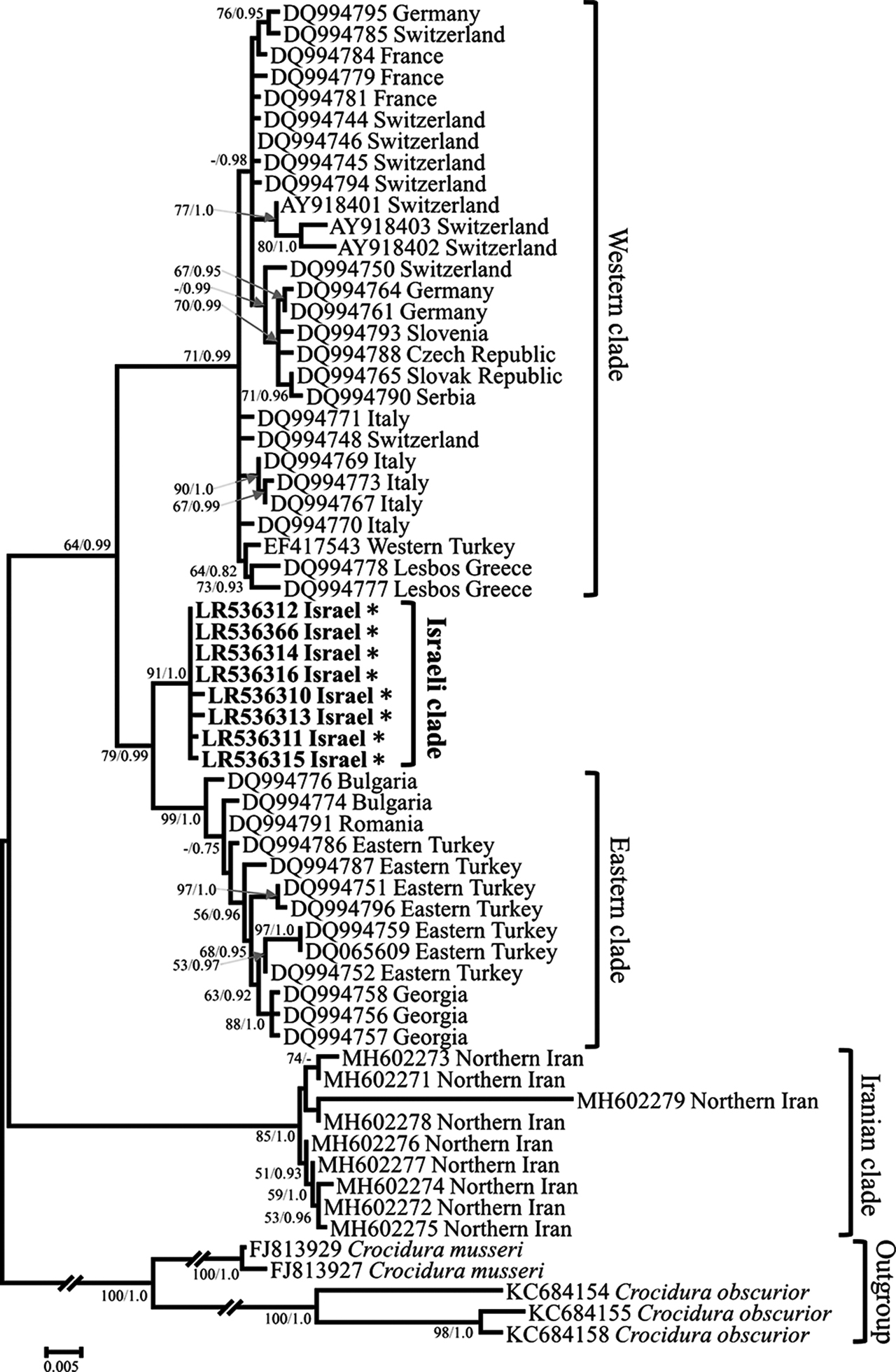
Maximum likelihood tree of Crocidura leucodon cytb sequences. Phylogenetic relationships inferred from a matrix of 1,077 nucleotide positions for 63 individuals. Maximum likelihood bootstrap supports above 50% and Bayesian posterior probabilities above 0.70 are indicated near the corresponding node separated with a slash. Sequences obtained in this work are indicated in bold. Authors of sequence data in Supplementary Table S4.
3.3 Phylogenetic relationships within the C. suaveolens complex
The C. suaveolens complex is a highly diverse lineage. Dubey et al. (2007a) divided this complex, and its closest outgroup, C. shantungensis, into ten different clades, all of which were recovered in our phylogenetic reconstruction (Figure 4). All Israeli “C. suaveolens” sequences were found to be nested within clade V, ‘gueldenstaedtii’, of Dubey et al. (2007a) (BP = 100; PP = 1.0; Figure 4). This clade includes samples from western Asia, Corsica, Minorca, and Crete. The Israeli sequences did not form a distinct clade within clade V. The average p-distance between sequences from Israel and other sequences of the ‘gueldenstaedtii’ clade was 0.005 (minimum = 0; maximum = 0.018). Similarly the network analysis of the ‘gueldenstaedtii clade’ sequences did not separate Israeli haplotypes from Iranian, Georgian, Turkish, or Corsican haplotypes (Supplementary Figure S3).

Maximum likelihood tree of Crocidura suaveolens complex cytb sequences. Phylogenetic relationships inferred from a matrix of 996 nucleotide positions for 215 individuals. Maximum likelihood bootstrap supports above 50% and Bayesian posterior probabilities above 0.70 are indicated near the corresponding node separated with a slash. Sequences obtained in this work are indicated in bold. Authors of sequence data in Supplementary Table S5.
3.4 Phylogenetic relationships within the S. etruscus clade
S. etruscus is a widespread Eurasian species whose distribution extends from France to Vietnam. Previous phylogeographic studies have shown that this species is probably paraphyletic and may be polyphyletic (Meegaskumbura et al. 2012a). Our phylogenetic tree (Figure 5) separated S. etruscus into two major clades: an eastern one and a western one. In agreement with Meegaskumbura et al. (2012a), the eastern clade included S. etruscus sequences from South Asia and S. madagascariensis sequences (BP = 77; PP = 0.99), supporting the view that S. madagascariensis is a junior synonym of S. etruscus. However unlike the findings of Meegaskumbura et al. (2012a), sequences from western European and the Near-East clustered together to form the Western clade (BP = 67; PP = 0.86). The two S. etruscus clades weakly cluster together (BP = 59; PP = 0.76) and are closely related to S. malayanus and S. fellowesgordoni sequences (BP = 100; PP = 1.0). One S. etruscus sequence from Vietnam (KF110756) formed a distant lineage, suggesting it could be a different species. Within our tree, the Israeli sequences did not form a distinct clade but, rather, clustered within the western clade (BP 67/0.86) together with specimens from France and Iran (Figure 5). The average p-distance between sequences from Israel and other sequences of the western clade was 0.003 (minimum = 0; maximum = 0.013). The results of the network analyses support the phylogenetic results (Supplementary Figure S4).
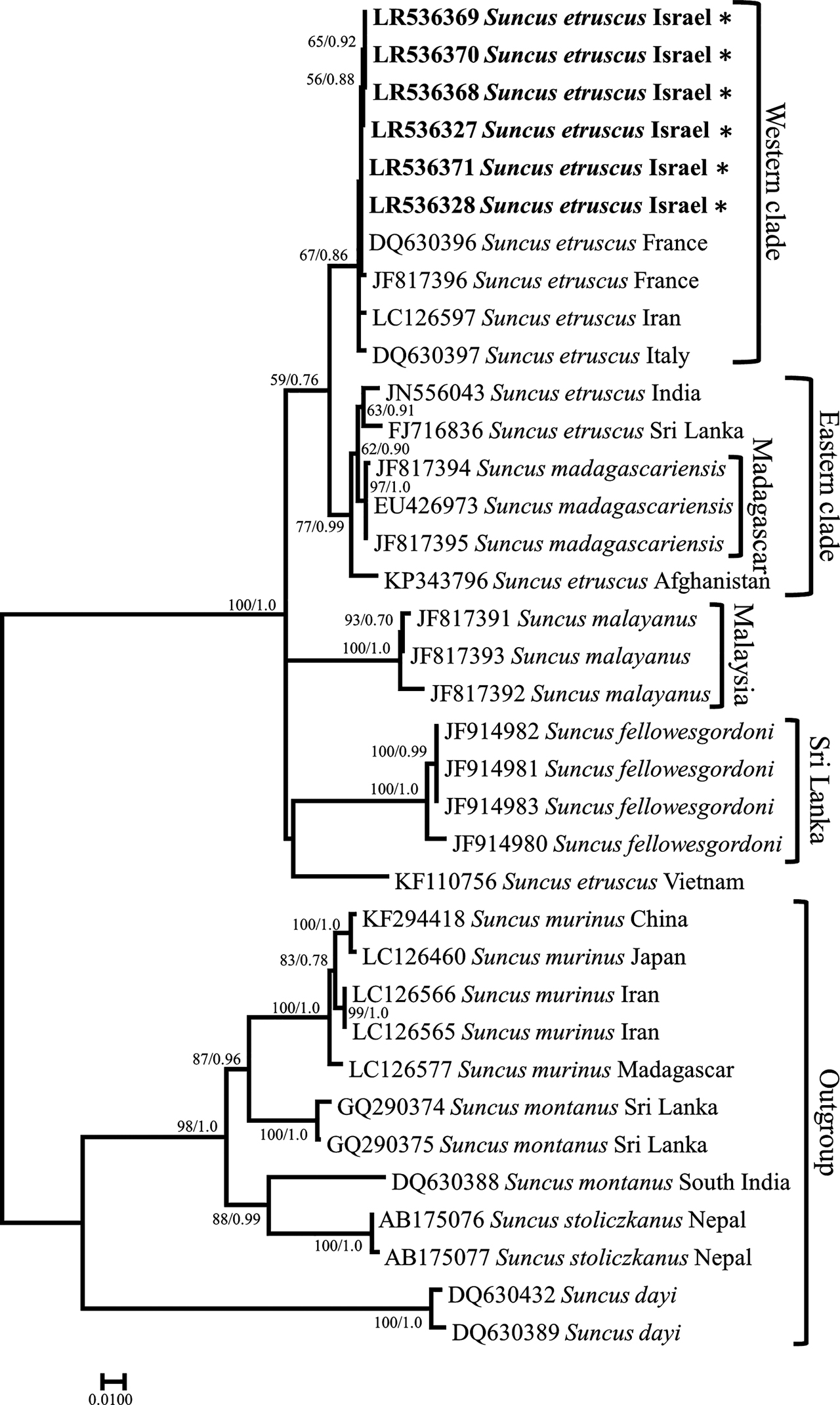
Maximum likelihood tree of Suncus cytb sequences. Phylogenetic relationships inferred from a matrix of 1,140 nucleotide positions for 36 individuals. Maximum likelihood bootstrap supports above 50% and Bayesian posterior probabilities above 0.70 are indicated near the corresponding node separated with a slash. Sequences obtained in this work are indicated in bold. Authors of sequence data in Supplementary Table S6.
4 Discussion
In agreement with Burgin et al. (2018b), Dolev and Perevolotsky (2004), and Meiri et al. (2019), our current findings confirm the presence of four different species of shrew in Israel: C. leucodon, C. suaveolens gueldenstaedtii, C. ramona, and S. etruscus. Although we attempted to sequence animals with atypical external characteristics, such as a black shrew (voucher number TAUM12603) or specimens with debated assignment (e.g., TAUM12207 was only identified as Crocidura sp.), we did not detect any additional species.
4.1 Crocidura ramona
Our work, based on sequences from type specimens, corroborates the view that C. ramona is a distinct species and probably endemic to Israel and the West Bank (Dubey et al. 2008; Ivanitskaya et al. 1996). C. ramona has been suggested to be related to the Palearctic “flat-headed rock-shrews” (i.e., Crocidura pergrisea, C.arispa, Crocidura armenica, Crocidura serezkyensis and C. zarudnyi) (Burgin et al. 2018b; Kryštufek and Vohralík 2001). Although the DNA of most of the latter has not yet been sequenced, our analysis indicates that C. ramona and C. zarudnyi are distantly related (Figure 2). Similarly, we have shown here that C. ramona is not closely related to another silvery-gray shrew – C. nana, as the two formed two distinct and distant clades in our phylogenetic analyses (Supplementary Figure S1). We note that there is some doubt regarding C. nana’s distribution. It is usually considered to be restricted to Somalia and Ethiopia (Cassola 2019; Hutterer 2005), whereas the samples sequenced in this work originated from Tanzania. We cannot be certain therefore that C. nana is the correct species assignment for the Tanzanian samples that we sequenced.
Because no morphological comparisons have been carried out between the skulls of C. katinka and C. ramona, the exact relationship between these two species remains to be determined. Although C. katinka has been suggested to be related to certain African species with cranial similarities (e.g., Crocidura bottegi, C. obscurior, Crocidura bottegoides) (Burgin et al. 2018b; Hutterer and Kock 2002), our analyses have demonstrated that C. ramona is unrelated to C. obscurior.
It is also possible that C. ramona is conspecific with C. portali. Thomas (1920) described C. portali as a small shrew (“though not excessively so”), with “pale drab-grey” pelage; and indicated that it has “clearly nothing to do with the C. russula group”. The gross morphology of the holotype (BMNH #19.4.11.9) and its size agree with C. ramona – though a detailed examination is still needed in order to confirm or refute this. Kryštufek and Vohralík (2001) suggested that C. portali may be a valid species, related to Crocidura arispa (and other members of the pergrisea group), and a senior synonym of C. ramona. Hutterer and Kock (2002) indicated that C. ramona and C. portali are similar in skull dimensions as well as in pelage, but note that they are also similar to C. gmelini, a species that they note requires “a better definition”. C. gmelini (ranging from Iran to Mongolia) has been synonymized with C. suaveolens, a species distantly related to C. ramona (Figure 2). C. arispa, C. ramona, and C. katinka are currently all considered valid species, while C. portali is not (Burgin et al. 2018b; IUCN 2020). Clearly, a taxonomic revision of these taxa is warranted. Neither the DNA of the C. portali type, nor that of any shrews identified as C. arispa, C. pergrisea, or C. katinka, has been sequenced and, unfortunately, we failed to amplify any cyt b fragment from a tissue of a specimen identified as C. portali (BMNH ZD 1971.817). We thus tentatively ascribe the sequence we obtained to C. ramona, pending a taxonomic revision.
4.2 Crocidura leucodon
The phylogenetic reconstruction indicates that the Israeli C. leucodon forms a distinct clade related to the eastern clade (Figure 4, Dubey et al. 2007b). The different C. leucodon clades have been suggested to have diverged during the Pleistocene glaciations (Dubey et al. 2007b; Mahmoudi et al. 2019). It is unlikely however that the Israeli clade corresponds to a fourth refugium from the Ice Age. Rather, the observed mitochondrial genetic divergence is possibly the result of the edge position of the Israeli population at the southernmost part of the C. leucodon range. In agreement, Mendelssohn and Yom-Tov (1999) noted that C. leucodon is less abundant than C. s. gueldenstaedtii, basing their conclusion on the number of shrews deposited in museum collections. The current data available at the Steinhardt Museum of Natural History support the view that the range of C. leucodon is more limited than that of C. suaveolens in Israel, since C. leucodon samples are rarer in the collection (79 leucodon vs. 594 suaveolens), and with one exception, restricted to the Mediterranean biome (Figure 1B). Edge populations have been suggested to harbor adaptive traits and genetic variability that may be important when considering future conservation needs (Hampe and Petit 2005; Mátýas et al. 2009). We thus also recommend that further population and genomic studies be carried out on this species, since its population status is currently unknown (Shenbrot et al. 2016), and assessment from museum collections alone may provide a biased representation of the population status.
4.3 Crocidura suaveolens gueldenstaedtii
The phylogenetic analysis indicated that all Israeli “C. suaveolens” sequences are part of clade V – the ‘gueldenstaedtii’ clade (Dubey et al. 2007a), and close to the Turkish and Georgian sequences (Figure 5). While the Israeli populations represent the edge of this species complex range, they do not present a mitochondrial genetic difference from the rest of its clade, unlike the situation in C. leucodon. This may be due to their larger population size and wider range in Israel compared to that of C. leucodon (Figure 1B). However, it is also possible that genetic differences exist in the nuclear genome. Based on the high similarity between the Israeli and Balkan sequences we agree with the status of “least concern” listed by the IUCN (Palomo et al. 2016). Our molecular cyt b data unequivocally place Israeli ‘suaveolens’ specimens within the ‘gueldenstaedtii’ clade. The specific status of the form awaits a thorough taxonomic revision, including an examination of animals from the type localities, preferably alongside specimens from the type localities of other members of the complex (e.g., C. ‘gmelini’, C. suaveolens monacha, C. portali) as well as phylogenetic information from nuclear markers. Until such a study is carried out, we tentatively refer to Israeli specimens as members of C. suaveolens gueldenstaedtii.
4.4 Suncus etruscus
The Israeli S. etruscus sequences appear to be closely related to the European ones and one Iranian sequence. The differences between the Israeli sequences and those of other western clade members are very small and comparable to the distances observed within the C. suaveolens gueldenstaedtii clade. This suggests that the western clade encompasses individuals from France in the west to Iran in the east and Israel in the south. However, this species has been poorly sampled in molecular studies to date. As a case in point, only a few S. etruscus specimens have been sequenced from Iran where members of both the western and eastern clade are present (Darvish et al. 2017; Ohdachi et al. 2016). More data are needed in order to decipher the population structure of this species.
5 Conclusions
Our molecular analyses have confirmed here the distribution in Israel of four shrew species: C. suaveolens gueldenstaedtii, C. leucodon, C. ramona, and S. etruscus (Table 1) (Dolev and Perevolotsky 2004; Meiri et al. 2019; Mendelssohn and Yom-Tov 1999). With the exception of C. suaveolens gueldenstaedtii, this is the first time that the cyt b of Israeli samples has been sequenced. Our results also confirm that C. ramona is a distinct species, though what name should be assigned to it remains to be decided. The Israeli C. leucodon belong to a phylogenetically distinct clade within the C. leucodon tree. Our results emphasize the need for deeper phylogeographic analyses in order to complement our barcoding identifications based on sequence similarity. Because mitochondrial genetic information does not always reflect the nuclear information, studies should be performed with nuclear markers in order to corroborate these results.
Acknowledgments
We would like to thank the Steinhardt Museum of Natural History, the Harrison Institute, the Hungarian National Museum, the Natural History Museum (London, UK), and the Field Museum of Natural History (Chicago, USA) for lending us the samples for this study. We also address special thanks to Arieh Landsman (Steinhardt Museum of Natural History) and the sequencing Unit of Tel Aviv University. Finally, we wish to thank Naomi Paz for English editing of the text.
Author contribution: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
Research funding: None declared.
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
Aulagnier, S., Hutterer, R., Jenkins, P., Bukhnikashvili, A., Kryštufek, B., and Kock, D. (2017). Suncus etruscus. The IUCN Red List of Threatened Species 2017. e.T90389138A22288134.Suche in Google Scholar
Bandelt, H.J., Forster, P., and Rohl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16: 37–48, https://doi.org/10.1093/oxfordjournals.molbev.a026036.Suche in Google Scholar
Bannikova, A.A., Lebedev, V.S., Kramerov, D.A., and Zaitsev, M.V. (2006). Phylogeny and systematics of the Crocidura suaveolens species group: corroboration and controversy between nuclear and mitochondrial DNA markers. Mammalia 70: 106–119, https://doi.org/10.1515/mamm.70.1-2.106.Suche in Google Scholar
Bate, D.M.A. (1937). XLIII — New Pleistocene mammals from Palestine. Ann. Mag. Nat. Hist. 20: 397–400, https://doi.org/10.1080/00222933708655355.Suche in Google Scholar
Bodenheimer, F. (1958). The present taxonomic status of the terrestrial mammals of Palestine. Bull. Res. Counc. Isr 7: 165–190.Suche in Google Scholar
Bodenheimer, F.S. (1935). Animal life in Palestine. Jerusalem, Israel: L. Mayer, pp. 506.Suche in Google Scholar
Bodenheimer, F.S. (1937). Prodromus Faunae Palestinae. Essai sur les éléments zoogéographiques et historiques du sud-ouest du sous-règne paléarctique. Mémoires présentés à l’Institut d’Égypte 33: 1–61.Suche in Google Scholar
Burgin, C.J., Colella, J.P., Kahn, P.L., and Upham, N.S. (2018a). How many species of mammals are there? J. Mammal. 99: 1–14, https://doi.org/10.1093/jmammal/gyx147.Suche in Google Scholar
Burgin, C.J., He, K., Haslauer, R., Sheftel, B.I., Jenkins, P.D., Ruedi, M., Hintsche, S., Motokawa, M., Hinckley, A., and Hutterer, R. (2018b). Family Soricidae (shrews). In: Wilson, D.E., and Mittermeier, R.A. (Eds.), Handbook of the Mammals of the World. Vol. 8. Insectivores, Sloths and Colugos. Barcelona: Lynx Edicions, pp. 332–551.Suche in Google Scholar
Cassola, F. (2019). Crocidura nana. The IUCN Red List of Threatened Species 2019. e.T41341A22306927.Suche in Google Scholar
Castiglia, R., Annesi, F., Amori, G., Solano, E., and Aloise, G. (2017). The phylogeography of Crocidura suaveolens from southern Italy reveals the absence of an endemic lineage and supports a Trans-Adriatic connection with the Balkanic refugium. Hystrix 28: 104–106.Suche in Google Scholar
Catzeflis, F., Maddalena, T., Hellwing, S., and Vogel, P. (1985). Unexpected findings on the taxonomic status of east mediterranean Crocidura russula Auct (Mammalia, Insectivora). Z. Säugetierkd. 50: 185–201.Suche in Google Scholar
Darvish, J., Mahmoudi, A., Pehpuri, A., and Saeidzadeh, S. (2017). New data on distribution and taxonomy of the genus Suncus (Mammalia: Soricidae) in Iran; molecular evidence. Iran. J. Anim. Biosyst. 13: 229–235.Suche in Google Scholar
Dobson, G.E. (1890). Description of new species of Crocidura from Africa. Ann. Mag. Nat. Hist. ser. 6: 225–227, https://doi.org/10.1080/00222939009460817.Suche in Google Scholar
Dolev, A., and Perevolotsky, A. (2004). The red book: vertebrates in Israel. Jerusalem: Israel Nature and Parks Authority and the Society for the Protection of Nature in Israel, p. 318.Suche in Google Scholar
Dubey, S., Cosson, J.F., Magnanou, E., Vohralík, V., Benda, P., Frynta, D., Hutterer, R., Vogel, V., and Vogel, P. (2007a). Mediterranean populations of the lesser white-toothed shrew (Crocidura suaveolens group): an unexpected puzzle of Pleistocene survivors and prehistoric introductions. Mol. Ecol. 16: 3438–3452, https://doi.org/10.1111/j.1365-294x.2007.03396.x.Suche in Google Scholar
Dubey, S., Cosson, J.F., Vohralík, V., Kryštufek, B., Diker, E., and Vogel, P. (2007b). Molecular evidence of Pleistocene bidirectional faunal exchange between Europe and the Near East: the case of the bicoloured shrew (Crocidura leucodon, Soricidae). J. Evol. Biol. 20: 1799–1808, https://doi.org/10.1111/j.1420-9101.2007.01382.x.Suche in Google Scholar
Dubey, S., Salamin, N., Ruedi, M., Barriere, P., Colyn, M., and Vogel, P. (2008). Biogeographic origin and radiation of the Old World crocidurine shrews (Mammalia: Soricidae) inferred from mitochondrial and nuclear genes. Mol. Phylogenet. Evol. 48: 953–963, https://doi.org/10.1016/j.ympev.2008.07.002.Suche in Google Scholar
Dubey, S., Zaitsev, M., Cosson, J.F., Abdukadier, A., and Vogel, P. (2006). Pliocene and Pleistocene diversification and multiple refugia in a Eurasian shrew (Crocidura suaveolens group). Mol. Phylogenet. Evol. 38: 635–647, https://doi.org/10.1016/j.ympev.2005.11.005.Suche in Google Scholar
Gerrie, R., and Kennerley, R. (2017). Crocidura katinka. The IUCN Red List of Threatened Species 2017. e.T136634A22303188.Suche in Google Scholar
Hampe, A., and Petit, R.J. (2005). Conserving biodiversity under climate change: the rear edge matters. Ecol. Lett. 8: 461–467, https://doi.org/10.1111/j.1461-0248.2005.00739.x.Suche in Google Scholar
Harrison, D. L. (1963). Some observation on the white-toothed shrews (genus Crocidura Wagler 1832) of Israel. Bull. Res. Counc. Isr. Sect. B Zool. 11: 177–182.Suche in Google Scholar
Harrison, D.L., and Bates, P.J.J. (1991). The mammals of Arabia. Sevenoaks, Kent: Harrison Zoological Museum Publ., p. 354.Suche in Google Scholar
Hoffmann, R.S. (1996). A research information system for mammals with Palaearctic examples. Bonn. Zool. Beitr. 46: 15–32.10.1002/latj.201800019Suche in Google Scholar
Hutterer, R. (2005). Order Soricomorpha. In Wilson, DE., Reeder, DM. (ed.), Mammal species of the world: a taxonomic and geographic reference. 3rd ed. Vol. 1, Baltimore: The John Hopkins University Press, pp. 230-311.Suche in Google Scholar
Hutterer, R. (2017). Crocidura gmelini. The IUCN Red List of Threatened Species 2017. e.T41319A22307461.Suche in Google Scholar
Hutterer, R., and Kock, D. (2002). Recent and ancient records of shrews from Syria, with notes on Crocidura katinka Bate 1937 (Mammalia: Soricidae). Bonn. Zool. Beitr. 50: 249–258.Suche in Google Scholar
Hutterer, R., and Shenbrot, G. (2017). Crocidura ramona. The IUCN Red List of Threatened Species 2017. e.T136722A89475013.Suche in Google Scholar
IUCN. 2020. The IUCN red list of threatened species. Gland: Switzerland. Version 2020-1. Available at: https://www.iucnredlist.org. Downloaded on April 2020.Suche in Google Scholar
Iudica, C.A., Whitten, W.M., and Williams, N.H. (2001). Small bones from dried mammal museum specimens as a reliable source of DNA. Biotechniques 30: 732–736, https://doi.org/10.2144/01304bm04.Suche in Google Scholar
Ivanitskaya, E., Shenbrot, G., and Nevo, E. (1996). Crocidura ramona sp. nov. (Insectivora, Soricidae): a new species of shrew from the central Negev desert, Israel. Z. Säugetierkd. 61: 93–103.Suche in Google Scholar
Katoh, K., and Standley, D.M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780, https://doi.org/10.1093/molbev/mst010.Suche in Google Scholar
Kryštufek, B., and Vohralík, V. (2001). Mammals of Turkey and Cyprus: introduction, checklist, Insectivora. Koper, Republic of Slovenia: Knjinica Annales Majora, p. 140.Suche in Google Scholar
Mahmoudi, A., Darvish, J., Siahsarvie, R., Dubey, S., and Kryštufek, B. (2019). Mitochondrial sequences retrieve an ancient lineage of bicolored shrew in the Hyrcanian refugium. Mamm. Biol. 95: 160–163, https://doi.org/10.1016/j.mambio.2018.06.006.Suche in Google Scholar
Mátýas, C., Vendramin, G.G., and Fady, B. (2009). Forests at the limit: evolutionary — genetic consequences of environmental changes at the receding (xeric) edge of distribution. Report from a research workshop. Ann. For. Sci. 66, https://doi.org/10.1051/forest/2009081.Suche in Google Scholar
Meegaskumbura, S., Meegaskumbura, M., and Schneider, C. (2012a). Phylogenetic position of Suncus fellowesgordoni with pigmy shrews from Madagascar and Southeast Asia inferred from cytochrome-b. Ceylon J. Sci. 41: 83–87, https://doi.org/10.4038/cjsbs.v41i1.4541.Suche in Google Scholar
Meegaskumbura, S., Meegaskumbura, M., and Schneider, C.J. (2012b). Re-evaluation of the taxonomy of the Sri Lankan pigmy shrew Suncus fellowesgordoni (Soricidae: Crocidurinae) and its phylogenetic relationship with S. etruscus. Zootaxa 3187: 57–68, https://doi.org/10.11646/zootaxa.3187.1.5.Suche in Google Scholar
Meiri, S., Belmaker, A., Berkowic, D., Kazes, K., Maza, E., Bar-Oz, G., and Dor, R. (2019). A checklist of Israeli land vertebrates. Isr. J. Ecol. Evol. 65: 43–50, https://doi.org/10.1163/22244662-20191047.Suche in Google Scholar
Mendelssohn, H., and Yom-Tov, Y. (1987). Mammals. In: Alon, A. (Ed.), Plants and animals of the land of Israel (in Hebrew), Vol. 7. Tel Aviv: Ministry of Defense, The Publishing House Society for the Protection of Nature in Israel, p. 295.Suche in Google Scholar
Mendelssohn, H., and Yom-Tov, Y. (1999). Fauna Palaestina: Mammalia of Israel. Jerusalem: The Israel Academy of Sciences and Humanities, pp. 439.Suche in Google Scholar
Ohdachi, S.D., Iwasa, M.A., Nesterenko, V.A., Abe, H., Masuda, R., and Haberl, W. (2004). Molecular phylogenetics of Crocidura shrews (Insectivora) in east and central Asia. J. Mammal. 85: 396–403, https://doi.org/10.1644/1545-1542(2004)085<0396:mpocsi>2.0.co;2.10.1644/1545-1542(2004)085<0396:MPOCSI>2.0.CO;2Suche in Google Scholar
Ohdachi, S.D., Kinoshita, G., Oda, S.-i., Motokawa, M., Jogahara, T., Arai, S., Nguyen, S.T., Suzuki, H., Katakura, K., Bawm, S., et al. (2016). Intraspecific phylogeny of the house shrews, Suncus murinus-S. montanus species complex, based on the mitochondrial cytochrome b gene. Mamm. Stud. 41: 229–238, https://doi.org/10.3106/041.041.0408.Suche in Google Scholar
Omar, H., Adamson, E., Bhassu, S., Goodman, S., Soarimalala, V., Hashim, R., and Ruedi, M. (2011). Phylogenetic relationships of Malayan and Malagasy pygmy shrews of the genus Suncus (Soricomorpha: Soricidae) inferred from mitochondrial Cytochrome b gene sequences. Raffles Bull. Zool. 59: 237–243.Suche in Google Scholar
Palomo, L., Kryštufek, B., Amori, G., and Hutterer, R. (2016). Crocidura suaveolens. The IUCN Red List of Threatened Species 2016.Suche in Google Scholar
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M. A., and Huelsenbeck, J. P. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61: 539–542, https://doi.org/10.1093/sysbio/sys029.Suche in Google Scholar
Saeedzadeh, S., Mehdipour, A., Darvish, J., Aliabadian, M., and Mahmoudi, A. (2017). New look to the con-specificity of the two shrews, Crocidura gmelini and C. suaveolens from Iran; geometric morphometrics approach. Iran. J. Anim. Biosyst. 13: 237–246.Suche in Google Scholar
Sanbrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular cloning: a laboratory manual, 2nd edn. New York: Cold Spring Harbor Laboratory Press, p. 1546.Suche in Google Scholar
Shenbrot, G., Hutterer, R., Amori, G., Kryštufek, B., Yigit, N., Mitsain, G., and Palomo, L. J. (2016). Crocidura leucodon. The IUCN Red List of Threatened Species 2016. e.T29651A115169304.Suche in Google Scholar
Silvestro, D., and Michalak, I. (2012). raxmlGUI: a graphical front-end for RAxML. Org. Divers. Evol. 12: 335–337, https://doi.org/10.1007/s13127-011-0056-0.Suche in Google Scholar
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313, https://doi.org/10.1093/bioinformatics/btu033.Suche in Google Scholar
Thomas, O. (1919). The white-toothed shrew of Palestine. Ann. Mag. Nat. Hist. 3: 32, https://doi.org/10.1080/00222931908673797.Suche in Google Scholar
Thomas, O. (1920). A new shrew and two new foxes from Asia Minor and Palestine. Ann. Mag. Nat. Hist. 5: 199–122, https://doi.org/10.1080/00222932008632347.Suche in Google Scholar
Tristram, H.B. (1884). The survey of western Palestine. The fauna and flora of Palestine. London: The Committee of the Palestine Exploration Fund, p. 538.10.5962/bhl.title.7594Suche in Google Scholar
Wilson, D.E., and Reeder, D.M. (2005). Mammal species of the world: a taxonomic and geographic reference, 3rd edn. Baltimore: Johns Hopkins University Press, p. 2142.10.56021/9780801882210Suche in Google Scholar
Supplementary material
The online version of this article offers supplementary material (https://doi.org/10.1515/mammalia-2019-0143).
© 2020 Erez Shpirer et al., published by De Gruyter.
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Frontmatter
- Evolutionary biology
- Covid-19: natural or anthropic origin?
- Ecology
- Relative abundance and activity patterns of terrestrial carnivorous mammals in Península Valdés, Patagonia, Argentina
- Understanding population baselines: status of mountain ungulate populations in the Central Tien Shan Mountains, Kyrgyzstan
- Do prairie voles (Microtus ochrogaster) change their activity and space use in response to domestic cat (Felis catus) excreta?
- An expandable radio collar for monitoring young terrestrial mammals
- Conservation
- Stranding cases of endangered Ganges river dolphins in the Ghaghara–Sharada irrigation canals, Ganges river basin, India: conservation implications
- Relative rarity of small wild cats in the Brazilian Pantanal
- Biogeography
- New records of bats (Chiroptera) in the Atlantic Forest of Espírito Santo, southeastern Brazil
- Leucism and updated geographic distribution of Molossus nigricans Miller, 1902 (Chiroptera: Molossidae) in Honduras
- Northernmost finding and further information on water deer Hydropotes inermis in Primorskiy Krai, Russia
- First record of a Nathusius’ pipistrelle (Pipistrellus nathusii) overwintering at a latitude above 60°N
- Taxonomy/phylogeny
- Molecular relationships of the Israeli shrews (Eulipotyphla: Soricidae) based on cytochrome b sequences
- Validating the relationships: which species of Myotis “nattereri” group (Chiroptera: Vespertilionidae) actually inhabits the Caucasus
- Annual reviewer acknowledgement
- Reviewer acknowledgement Mammalia volume 84 (2020)
Artikel in diesem Heft
- Frontmatter
- Evolutionary biology
- Covid-19: natural or anthropic origin?
- Ecology
- Relative abundance and activity patterns of terrestrial carnivorous mammals in Península Valdés, Patagonia, Argentina
- Understanding population baselines: status of mountain ungulate populations in the Central Tien Shan Mountains, Kyrgyzstan
- Do prairie voles (Microtus ochrogaster) change their activity and space use in response to domestic cat (Felis catus) excreta?
- An expandable radio collar for monitoring young terrestrial mammals
- Conservation
- Stranding cases of endangered Ganges river dolphins in the Ghaghara–Sharada irrigation canals, Ganges river basin, India: conservation implications
- Relative rarity of small wild cats in the Brazilian Pantanal
- Biogeography
- New records of bats (Chiroptera) in the Atlantic Forest of Espírito Santo, southeastern Brazil
- Leucism and updated geographic distribution of Molossus nigricans Miller, 1902 (Chiroptera: Molossidae) in Honduras
- Northernmost finding and further information on water deer Hydropotes inermis in Primorskiy Krai, Russia
- First record of a Nathusius’ pipistrelle (Pipistrellus nathusii) overwintering at a latitude above 60°N
- Taxonomy/phylogeny
- Molecular relationships of the Israeli shrews (Eulipotyphla: Soricidae) based on cytochrome b sequences
- Validating the relationships: which species of Myotis “nattereri” group (Chiroptera: Vespertilionidae) actually inhabits the Caucasus
- Annual reviewer acknowledgement
- Reviewer acknowledgement Mammalia volume 84 (2020)

