Abstract
Objectives
Abnormal placentation may affect the maternal serum fraction of cell-free fetal DNA (fetal fraction) determined as part of non-invasive prenatal screening (NIPS). This study aimed to assess whether the fetal fraction can predict placenta accreta spectrum (PAS) with or without placenta previa (PP). We also investigated the impact of trophoblastic invasion depth on the fetal fraction.
Methods
This is a retrospective case-control study of pregnant women with and without abnormal placentation carrying a singleton and having undergone NIPS prior to 20 weeks of gestation. The eligible subjects were selected from a cohort managed at our institution for PAS suspected antenatally. We compared women with normal placentation (controls) to PAS, PP, or PAS + PP cases. Data were abstracted from electronic medical records, and PAS was confirmed histologically.
Results
Of the 146 patients in our cohort, 8 controls, 10 PP, 6 PAS, and 7 PAS + PP cases were eligible for the study. Among the groups, there were no significant differences in baseline demographic and clinical characteristics except the median number of prior uterine surgeries. Also, the groups did not significantly differ in their median fetal fraction. The fetal fraction did not discriminate any group when stratified according to the depth of placental invasion, i.e., no PAS, abnormally adherent, and abnormally invasive placenta.
Conclusions
The maternal serum fraction of cell-free fetal DNA measured before 20 weeks of gestation is not predictive of PAS with or without concurrent PP or the depth of trophoblastic invasion.
Introduction
Placenta accreta spectrum (PAS) refers to the range of abnormally adherent or invasive placenta, also known as placenta increta, placenta percreta, and placenta accreta [1]. The current concept of pathogenesis involves the preferential attachment of the blastocyst to an iatrogenic defect in the endometrium/decidua defect following uterine surgery, such as cesarean section. Early implantation to scar tissue facilitates abnormally deep trophoblastic invasion leading to interactions with the radial and arcuate arteries [2]. Even today, this age-old disease carries a heavy burden of complications with severe consequences, including maternal death. The most substantial risk factor for PAS is placenta previa in the index pregnancy after prior cesarean surgery [3, 4]. The incidence has markedly increased in recent years, parallel to the rising rate of cesarean deliveries [5].
Currently, women at risk for PAS are identified based on clinical risk factors mentioned above and imaging studies, ultrasound, and magnetic resonance imaging (MRI) [6], [7], [8]. However, their reported sensitivity and specificity of imaging vary substantially among studies, even when similar criteria are used, owing to interobserver variability [9]. Another challenge with imaging is its accuracy in predicting the depth of trophoblastic invasion. Of particular importance is to differentiate between abnormally adherent placenta (placenta accreta) and abnormally invasive placenta (placenta increta or percreta). The latter is associated with significantly higher mortality and morbidity [10, 11]. Determining the depth of villous invasiveness before delivery is essential to tailor individual care [12]. The current imaging modalities, both sonography [13] and MRI [14] fall short of accurate prediction of trophoblastic invasion.
The need for an accurate, rapid, and cost-effective diagnosis of PAS and its severity has led to a search for biomarkers (Reviewed in [15, 16]). One such biomarker is cell-free fetal DNA (cffDNA) which is widely used for non-invasive prenatal screening (NIPS) of common fetal aneuploidies. cffDNA derives from apoptotic trophoblastic cells, and the portion of the cff DNA of the total cell-free DNA in the maternal circulation is called the fetal fraction (FF) [17]. As the release of cffDNA is closely tied to placental morphogenesis, conditions that affect the placenta can directly impact the FF in maternal circulation [18]. An elevated FF is associated with placental disorders such as preeclampsia [19] and fetal growth restriction [20]. Dolin et al. [21] and Sekizawa et al. [22] reported increased levels of fetal fraction in cases of placenta previa (PP). Sekizawa et al. [22] also reported a high FF in two cases of PAS; however, they did not investigate the depth of invasion. Since concurrent placenta previa and PAS (placenta previa accreta) constitute two-thirds of the PAS cases, it is crucial to understand the interaction between placenta previa and PAS on the FF. In addition, we investigated the impact of the trophoblastic invasion depth on the FF.
Materials and methods
This is a retrospective case-control study approved by the Institutional Review Board (201,801,557). The eligible patients were selected from a cohort of 146 pregnant women managed at our institution for antenatally suspected PAS between January 2013 and December 2021. Only those women carrying a singleton and having NIPS before 20 weeks of gestation were included in the analyses. Those diagnosed with fetal anomalies and aneuploidies, delivered outside our institution, and lost to follow-up, and with missing FF results or histopathologic evaluation for PAS were excluded. The FF was determined by various commercial laboratories that perform NIPS. The patients’ source of reimbursement played a significant role in the choice of the commercial laboratory. Placental location was determined by sonography performed by maternal-fetal specialists. Transvaginal sonography was performed to evaluate the location of the placenta in relation to the internal cervical os. A patient with the leading edge of her placenta within 2 cm of the internal cervical os was labeled as having PP in our analyses. The diagnosis of PAS was established histologically according to the criteria proposed by the International Federation of Gynaecology and Obstetrics (FIGO). (1) Relevant demographic and clinical data were extracted from the patients’ medical records. The patients included in the study were divided into four groups: (1) normal placentation, (2) PAS), (3) PP, and (4) PAS + PP. Assuming a standard deviation of 3% for the population and using an α of 0.05 and β of 0.8., we estimated that we need at least six subjects in each group to detect a difference of 6% in means of cffDNA fraction. We used the Shapiro-Francia test to assess the distribution of our data for normality. One-way ANOVA and Kruskal-Wallis tests were used to compare parametric and non-parametric data.
Results
Figure 1 is the flowchart to depict selection of the patients for our analysis. Of the 146 patients in our cohort, 53 underwent NIPS and were delivered at our hospital. Of the remainder, 70 had no genetic screening or testing, 19 had either biochemical genetic screening or prenatal diagnosis, and 4 were delivered at another hospital and lost to follow-up. Of the 53 patients who underwent NIPS, we excluded 10 who lacked a documented FF and 12 due to other criteria. The final analyses were performed on 31 patients: 8 with normal placentation, 10 with PP, 6 with PAS, and 7 with PP + PAS. Among the groups, there were no significant differences in baseline demographic and clinical characteristics except the median number of prior uterine surgeries (Table 1). Also, the groups did not significantly differ in their median FF (Figure 2).
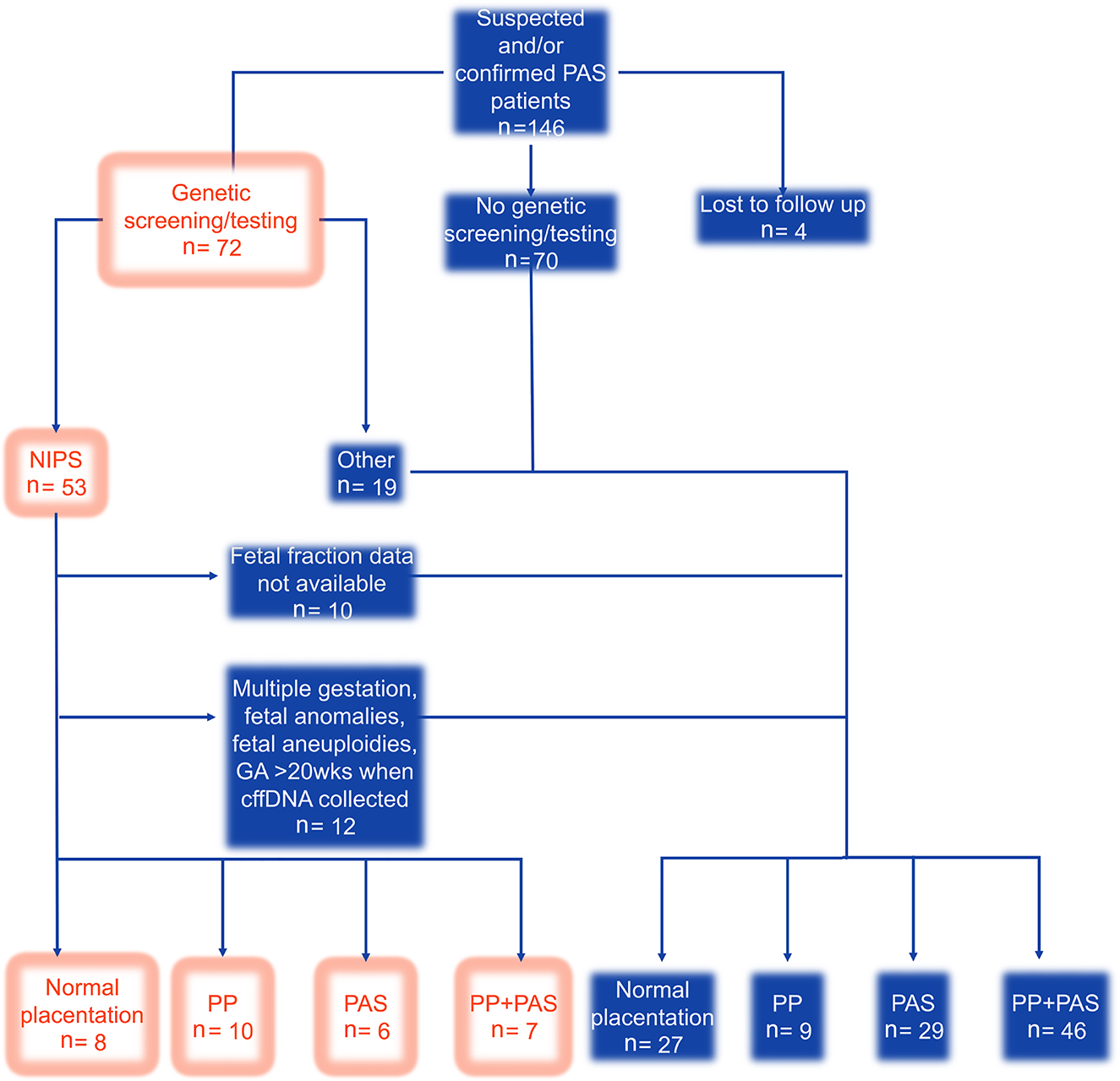
Flow diagram of patient selection. The unfilled boxes depict the selected patients for the study. n, number; wks, weeks; PP, placenta previa; PAS, placenta accreta spectrum.
Comparison of baseline demographic and clinical characteristics.
| Normal placentation | PP | PAS | PAS + PP | p-Value | |
|---|---|---|---|---|---|
| (n=8) | (n=10) | (n=6) | (n=7) | ||
| Mean age ± SD, year | 32.3 ± 4.92 | 33.3 ± 3.53 | 33.5 ± 4.55 | 32 ± 6.11 | 0.47 |
| Mean body mass index ± SD, kg/m2 | 36.7 ± 9.5 | 29.0 ± 8.2 | 30.9 ± 9.5 | 30.8 ± 4.8 | 0.19 |
| Race, n | 0.64 | ||||
| White | 6 | 6 | 3 | 3 | |
| Black | 2 | 3 | 2 | 2 | |
| Hispanic | 0 | 0 | 0 | 1 | |
| Other | 0 | 1 | 1 | 1 | |
| Median number of prior uterine surgeries (10–90% interquartile) | 3 (1–6) | 1 (0–3.9) | 1.0 (0–2) | 2 (1–4) | 0.03 |
| Median gestational age at blood sampling (10–90% interquartile), weeks | 11.5 (11–14) | 12 (9–17.8) | 12.5 (11–14) | 12 (11–12) | 0.76 |
| Pregnancy-induced hypertension, n | 1 | 2 | 4 | 1 | 0.08 |
| Tobacco use, n | 4 | 4 | 1 | 2 | 0.59 |
| In vitro fertilization, n | 0 | 3 | 1 | 0 | 0.18 |
| Female gender, n | 3 | 5 | 3 | 5 | 0.62 |
-
SD, standard deviation; kg/m2, kilogram/meter squared; n, number.

Fetal fraction and placentation. PAS, placenta accreta spectrum; PP, placenta previa.
Of the 13 patients with PAS with or without PP, 7 had abnormally adherent placenta, and 6 had abnormally invasive placenta. When stratified according to the depth of placental invasion, i.e., no PAS, abnormally adherent, and abnormally invasive placenta, the FF did not discriminate any group (Figure 3).
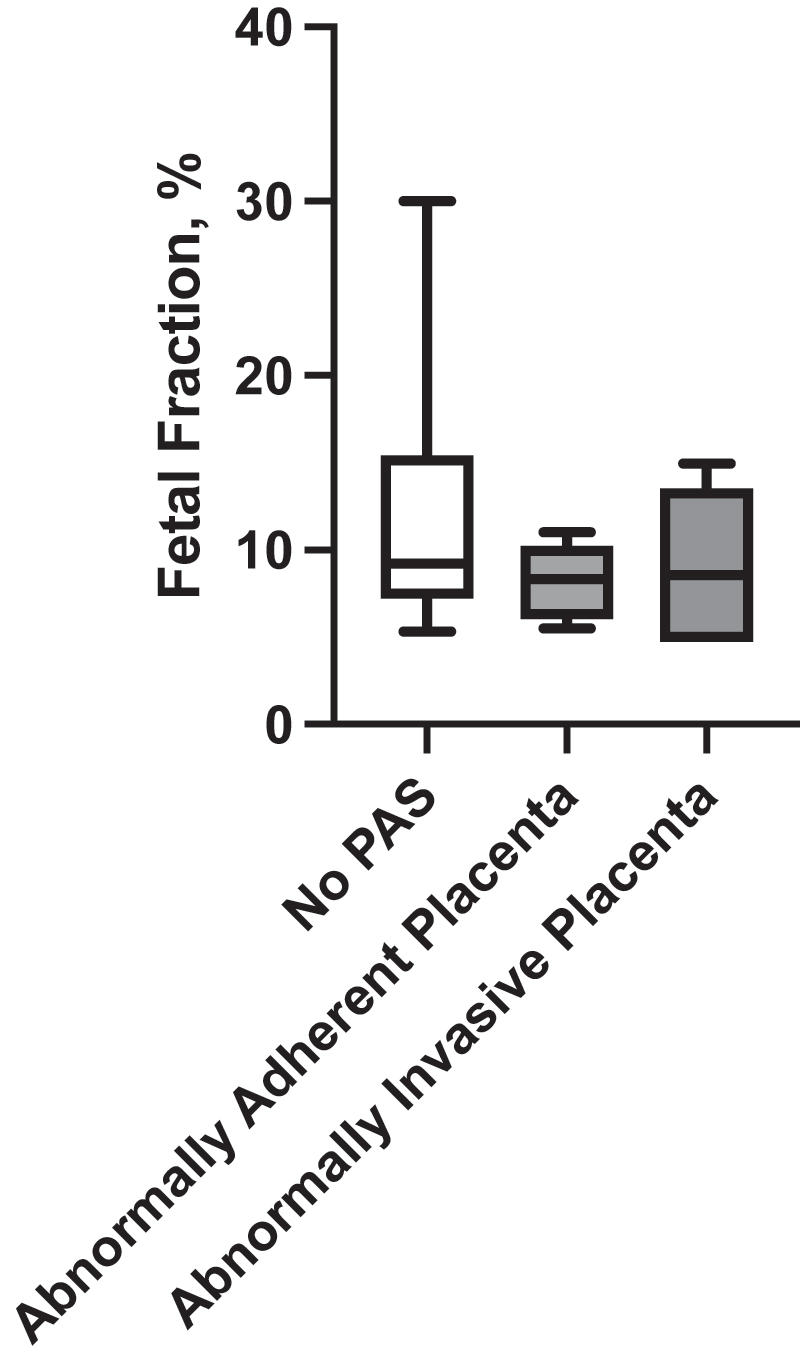
Fetal fraction and the depth of placental invasion. PAS, placenta accreta spectrum.
Discussion
FF determined before 20 weeks of gestation is not predictive of PAS in patients with or without PP. These findings are consistent two previous studies that did not find any significant relation between means or median FF and abnormal placentation, either PAS or PP. However, these studies did not control for the concurrent PAS and PP. We demonstrated that the fetal fraction does not differ between patients with placenta previa accreta or PAS alone.
To date, there has been no publications regarding the trophoblastic invasion depth on FF except a case report [22] of a patient with a high FF and the histologically confirmed invasive placenta. Here, we also demonstrated that FF does not correlate with the depth of trophoblast invasion among patients with PAS to be of clinical significance.
There was a significant difference between the median number of surgeries between the groups. This is potentially due to the fact that the patients were selected from a cohort of patients referred for PAS evaluation. Patients with greater than or equal to four prior cesareans were recommended to be referred for evaluation regardless of other ultrasound findings.
The major strength of our study was the number of PAS cases analyzed, which is almost twice as many as in the previous reports [23, 24]. Despite its relatively large size, our study may not be powered to detect small differences which are unlikely to be of clinical significance. Yet, a meta-analysis based on individual patient data from this and other studies [21], [22], [23], [24] would improve the power. Another strength is limiting our analysis to those patients who underwent NIPS before 20 weeks’ gestation, as often performed. This approach should minimize the effect of gestational age on the FF.
The major limitation is the retrospective design. The FF data were unavailable in 19% (10/53) of patients who had undergone NIPS at other institutions and then transferred to our care. Our population’s screening rate with cffDNA was only 35% (51/146). Failure to analyze the cffDNA of the entire cohort may have confounded our results. Nevertheless, this study represents the real-world clinical setting where patients chose other options over NIPS.
From a biological perspective, the lack of a relation between FF and PAS is not entirely surprising. Histologic studies by Kim et al. demonstrated no significant difference in apoptotic rates and proliferative index between abnormally adherent or invasive placentas compared to normally implanted placentas [25]. This is consistent with the current concept that abnormally deep trophoblastic invasion is the result of endometrium/decidua defect rather than excessive trophoblastic proliferation [2].
Although cffDNA is a valuable tool for screening common fetal aneuploidies, FF does not aid the clinical management of PAS. Imaging and clinical risk-based approaches remain the mainstay of identifying patients at risk for PAS, and the quest for biomarkers should continue.
-
Article note: A part of this study was presented as a poster at the 41st Virtual Annual Pregnancy Meeting of the Society of Maternal Fetal Medicine, January 25–30, 2021.
-
Research funding: None declared.
-
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
-
Competing interests: Authors state no conflict of interest.
-
Informed consent: Not applicable.
-
Ethical approval: The local Institutional Review Board deemed this study approved by the Institutional Review Board (201801557).
References
1. Jauniaux, E, Ayres-de-Campos, D, Langhoff-Roos, J, Fox, KA, Collins, S, Diagnosis, FPA, et al.. FIGO classification for the clinical diagnosis of placenta accreta spectrum disorders. Int J Gynaecol Obstet 2019;146:20–4. https://doi.org/10.1002/ijgo.12761.Search in Google Scholar PubMed
2. Jauniaux, E, Burton, GJ. Pathophysiology of placenta accreta spectrum disorders: a review of current findings. Clin Obstet Gynecol 2018;61:743–54. https://doi.org/10.1097/grf.0000000000000392.Search in Google Scholar PubMed
3. Bowman, ZS, Eller, AG, Bardsley, TR, Greene, T, Varner, MW, Silver, RM. Risk factors for placenta accreta: a large prospective cohort. Am J Perinatol 2014;31:799–804. https://doi.org/10.1055/s-0033-1361833.Search in Google Scholar PubMed
4. Clark, SL, Koonings, PP, Phelan, JP. Placenta previa/accreta and prior cesarean section. Obstet Gynecol 1985;66:89–92.Search in Google Scholar
5. Jauniaux, E, Gronbeck, L, Bunce, C, Langhoff-Roos, J, Collins, SL. Epidemiology of placenta previa accreta: a systematic review and meta-analysis. BMJ Open 2019;9:e031193. https://doi.org/10.1136/bmjopen-2019-031193.Search in Google Scholar PubMed PubMed Central
6. D’Antonio, F, Iacovella, C, Bhide, A. Prenatal identification of invasive placentation using ultrasound: systematic review and meta-analysis. Ultrasound Obstet Gynecol 2013;42:509–17. https://doi.org/10.1002/uog.13194.Search in Google Scholar PubMed
7. Familiari, A, Liberati, M, Lim, P, Pagani, G, Cali, G, Buca, D, et al.. Diagnostic accuracy of magnetic resonance imaging in detecting the severity of abnormal invasive placenta: a systematic review and meta-analysis. Acta Obstet Gynecol Scand 2018;97:507–20. https://doi.org/10.1111/aogs.13258.Search in Google Scholar PubMed
8. Reeder, CF, Sylvester-Armstrong, KR, Silva, LM, Wert, EM, Smulian, JC, Genc, MR. Outcomes of pregnancies at high-risk for placenta accreta spectrum following negative diagnostic imaging. J Perinat Med 2022;50:595–600. https://doi.org/10.1515/jpm-2021-0591.Search in Google Scholar PubMed
9. Bowman, ZS, Eller, AG, Kennedy, AM, Richards, DS, Winter, TC3rd, Woodward, PJ, et al.. Interobserver variability of sonography for prediction of placenta accreta. J Ultrasound Med 2014;33:2153–8. https://doi.org/10.7863/ultra.33.12.2153.Search in Google Scholar PubMed
10. Duzyj, CM, Cooper, A, Mhatre, M, Han, CS, Paidas, MJ, Illuzzi, JL, et al.. Placenta accreta: a spectrum of predictable risk, diagnosis, and morbidity. Am J Perinatol 2019;36:1031–8. https://doi.org/10.1055/s-0038-1676111.Search in Google Scholar PubMed
11. Yasin, N, Slade, L, Atkinson, E, Kennedy-Andrews, S, Scroggs, S, Grivell, R. The multidisciplinary management of placenta accreta spectrum (PAS) within a single tertiary centre: a ten-year experience. Aust N Z J Obstet Gynaecol 2019;59:550–4. https://doi.org/10.1111/ajo.12932.Search in Google Scholar PubMed
12. Sylvester-Armstrong, K, Reeder, C, Patrick, K, Genc, MR. Improved management of placenta accreta spectrum disorders: experience from a single institution. J Perinat Med 2022;50:286–93. https://doi.org/10.1515/jpm-2021-0263.Search in Google Scholar PubMed
13. Jauniaux, E, Collins, SL, Jurkovic, D, Burton, GJ. Accreta placentation: a systematic review of prenatal ultrasound imaging and grading of villous invasiveness. Am J Obstet Gynecol 2016;215:712–21. https://doi.org/10.1016/j.ajog.2016.07.044.Search in Google Scholar PubMed
14. Einerson, BD, Rodriguez, CE, Kennedy, AM, Woodward, PJ, Donnelly, MA, Silver, RM. Magnetic resonance imaging is often misleading when used as an adjunct to ultrasound in the management of placenta accreta spectrum disorders. Am J Obstet Gynecol 2018;218:618 e1–e7. https://doi.org/10.1016/j.ajog.2018.03.013.Search in Google Scholar PubMed
15. Bartels, HC, Postle, JD, Downey, P, Brennan, DJ. Placenta accreta spectrum: a review of pathology, molecular biology, and biomarkers. Dis Markers 2018;2018:1–11. https://doi.org/10.1155/2018/1507674.Search in Google Scholar PubMed PubMed Central
16. Zhang, T, Wang, S. Potential serum biomarkers in prenatal diagnosis of placenta accreta spectrum. Front Med 2022;9:860186. https://doi.org/10.3389/fmed.2022.860186.Search in Google Scholar PubMed PubMed Central
17. Hui, L, Bianchi, DW. Fetal fraction and noninvasive prenatal testing: what clinicians need to know. Prenat Diagn 2020;40:155–63. https://doi.org/10.1002/pd.5620.Search in Google Scholar PubMed PubMed Central
18. Taglauer, ES, Wilkins-Haug, L, Bianchi, DW. Review: cell-free fetal DNA in the maternal circulation as an indication of placental health and disease. Placenta 2014;35:S64–8. https://doi.org/10.1016/j.placenta.2013.11.014.Search in Google Scholar PubMed PubMed Central
19. Hahn, S, Rusterholz, C, Hosli, I, Lapaire, O. Cell-free nucleic acids as potential markers for preeclampsia. Placenta 2011;32:S17–20. https://doi.org/10.1016/j.placenta.2010.06.018.Search in Google Scholar PubMed
20. Al Nakib, M, Desbrière, R, Bonello, N, Bretelle, F, Boubli, L, Gabert, J, et al.. Total and fetal cell-free DNA analysis in maternal blood as markers of placental insufficiency in intrauterine growth restriction. Fetal Diagn Ther 2009;26:24–8. https://doi.org/10.1159/000236355.Search in Google Scholar PubMed
21. Dolin, C, Bennett, T-A, Pinson, K, Morgan, J, Madden, N, Yeager, S, et al.. 258: is there an association between placental location and cell-free DNA fetal fraction? Am J Obstet Gynecol 2017;216:S160. https://doi.org/10.1016/j.ajog.2016.11.164.Search in Google Scholar
22. Sekizawa, A, Jimbo, M, Saito, H, Iwasaki, M, Sugito, Y, Yukimoto, Y, et al.. Increased cell-free fetal DNA in plasma of two women with invasive placenta. Clin Chem 2002;48:353–4. https://doi.org/10.1093/clinchem/48.2.353.Search in Google Scholar
23. Adiyaman, D, Kuyucu, M, Atakul, BK, Can, D, Özeren, M, Koç, A, et al.. Can the cell-free DNA test predict placenta accreta spectrum or placenta previa totalis? Z Geburtshilfe Neonatol 2022;226:92–7. https://doi.org/10.1055/a-1579-1338.Search in Google Scholar PubMed
24. Samuel, A, Bonanno, C, Oliphant, A, Batey, A, Wright, JD. Fraction of cell-free fetal DNA in the maternal serum as a predictor of abnormal placental invasion-a pilot study. Prenat Diagn 2013;33:1050–3. https://doi.org/10.1002/pd.4195.Search in Google Scholar PubMed
25. Kim, K-R, Jun, S-Y, Kim, J-Y, Ro, JY. Implantation site intermediate trophoblasts in placenta cretas. Mod Pathol 2004;17:1483–90. https://doi.org/10.1038/modpathol.3800210.Search in Google Scholar PubMed
© 2022 Walter de Gruyter GmbH, Berlin/Boston
Articles in the same Issue
- Frontmatter
- Editorial
- Journal of Perinatal Medicine: Happy 50th anniversary
- Articles
- Fifty years of the Journal of Perinatal Medicine: an altmetric and bibliometric study
- Early origins of respiratory disease
- Oxygenation of the newborn. The impact of one molecule on newborn lives
- Emergency button cannula vs. umbilical catheter as neonatal emergency umbilical vein access – a randomized cross-over pilot study
- Covid and pregnancy in the United States – an update as of August 2022
- Facts and doubts on the beginning of human life – scientific, legal, philosophical and religious controversies
- Molecular subclasses of preeclampsia characterized by a longitudinal maternal proteomics study: distinct biomarkers, disease pathways and options for prevention
- Maternal telehealth: innovations and Hawaiʻi perspectives
- Prevention of risks of overweight and obesity in pregnant women
- Intraoperative ultrasound during repeat cesarean delivery facilitates sampling of uterine scar tissue
- The effect of abnormal placentation on maternal serum fetal fraction of cell-free DNA
- Prenatal predictors of adverse perinatal outcome in congenital cytomegalovirus infection: a retrospective multicenter study
- Diagnostic approach to fetal ventriculomegaly
- Clinical potential of human amniotic fluid stem cells
- Vaginal progesterone for the prevention of preterm birth: who can benefit and who cannot? Evidence-based recommendations for clinical use
- A second look at intrapartum fetal surveillance and future directions
- Computerized analysis of cardiotocograms in clinical practice and the SisPorto® system thirty-two years after: technological, physiopathological and clinical studies
- Acknowledgment
- Acknowledgment
Articles in the same Issue
- Frontmatter
- Editorial
- Journal of Perinatal Medicine: Happy 50th anniversary
- Articles
- Fifty years of the Journal of Perinatal Medicine: an altmetric and bibliometric study
- Early origins of respiratory disease
- Oxygenation of the newborn. The impact of one molecule on newborn lives
- Emergency button cannula vs. umbilical catheter as neonatal emergency umbilical vein access – a randomized cross-over pilot study
- Covid and pregnancy in the United States – an update as of August 2022
- Facts and doubts on the beginning of human life – scientific, legal, philosophical and religious controversies
- Molecular subclasses of preeclampsia characterized by a longitudinal maternal proteomics study: distinct biomarkers, disease pathways and options for prevention
- Maternal telehealth: innovations and Hawaiʻi perspectives
- Prevention of risks of overweight and obesity in pregnant women
- Intraoperative ultrasound during repeat cesarean delivery facilitates sampling of uterine scar tissue
- The effect of abnormal placentation on maternal serum fetal fraction of cell-free DNA
- Prenatal predictors of adverse perinatal outcome in congenital cytomegalovirus infection: a retrospective multicenter study
- Diagnostic approach to fetal ventriculomegaly
- Clinical potential of human amniotic fluid stem cells
- Vaginal progesterone for the prevention of preterm birth: who can benefit and who cannot? Evidence-based recommendations for clinical use
- A second look at intrapartum fetal surveillance and future directions
- Computerized analysis of cardiotocograms in clinical practice and the SisPorto® system thirty-two years after: technological, physiopathological and clinical studies
- Acknowledgment
- Acknowledgment

