Legionella pneumophila is a Gram-negative intracellular pathogen that can replicate within freshwater amoeba and mammalian alveolar macrophages. In order to establish an intracellular niche permissive for its replication, Legionella translocates over 300 different effector proteins into the host cytosol via its Dot/Icm secretion system.[1] Among these, members of the SidE effector family (SidE, SdeA, SdeB, and SdeC) regulate multiple cellular processes by a unique phosphoribosyl (PR) ubiquitination mechanism that bypasses the canonical ubiquitination machinery,[1] and two phosphoglycosyl ubiquitin (PR-Ub)-specific deubiquitinases of Legionella (DupA and DupB) could cleave the PR-Ub induced by SidEs from PR-ubiquitinated substrates. The activity of the SidE family is regulated by a calmodulin (CaM)-dependent glutamylase effector of Legionella, named SidJ. Activated SidJ inhibits the adenosine diphosphate (ADP)-ribotransferase activity of SidEs by covalently attaching one or more glutamate moieties on the first glutamate residue of the ExE (where “x” represents any amino acid) element of the mono-ADP-ribosyl transferase (mART) domain that is essential for ubiquitin activation.[2,3] Interestingly, SdjA, a member of SidJ family in some Legionella strains, shares high level of sequence and structural identities to SidJ.[4] However, the function of SdjA is still unknown.
In a recent paper in mBio, Song et al. reveal that SdjA is a bifunctional enzyme that distinctly regulates PR-ubiquitination of members of the SidE family.[5] Firstly, Song et al. noted that SdjA was unable to alleviate the toxicity of SdeA, but effectively suppressed the yeast toxicity caused by SdeB or SdeC. Then, they examined the ubiquitination of Rab33b induced by the SidE family in a series of L. pneumophila-mutant strains and found that SdjA selectively inhibited the ubiquitin ligase activity of SdeB and SdeC, but not SidE and SdeA. Following subsequent biochemical experiments, the authors demonstrated that SdjA was another CaM-dependent glutamylase against SdeB and SdeC by catalyzing covalent attachment of a glutamate moiety to the first glutamate residue of the ExE element in the mART domains of SdeB and SdeC. Moreover, the authors unexpectedly found that SdjA removed SidJ-mediated glutamylation from Glu-SdeA and restored the Ub ligase activity of SdeA, and CaM was not required for the deglutamylase activity. The results reveal that SdjA is a bifunctional enzyme which inhibits the ubiquitin ligase activity of SdeB and SdeC by glutamylation and functions as a deglutamylase that reverses SidJ-induced glutamylation on SdeA (Figure 1).
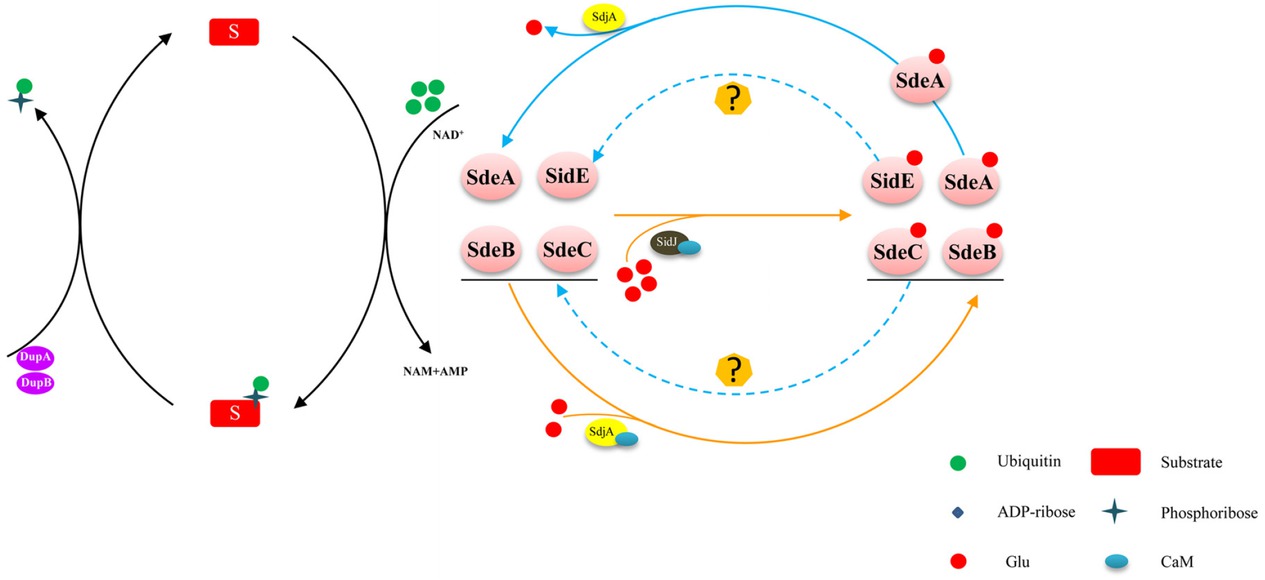
The PR-ubiquitination pathway and regulation of the SidE effector family. The SidE effector family (SidE, SdeA, SdeB, and SdeC) catalyzes a NAD-dependent PR ubiquitination. Two PR-Ub–specific deubiquitinases (DupA and DupB) cleave the PR-Ub from PR-ubiquitinated substrates. SidJ activated by CaM inhibits the ADP-ribosyltransferase activity of SidEs. In addition, SdjA inhibits the ubiquitin ligase activity of SdeB and SdeC by glutamylation and functions as a deglutamylase that reverses SidJ-induced glutamylation on SdeA. ADP: adenosine diphosphate; CaM: calmodulin; NAD: nicotinamide adenine dinucleotide; PR: phosphoribosyl; PR-Ub: phosphoribosyl ubiquitin.
The discovery of SdjA as a bifunctional enzyme raises several intriguing questions. First, how does SdjA selectively recognize and differently impact the activity of members of the SidE family? A recent study showed that the N-terminal domain (NTD) is involved in recognizing members of the SidE family.[6] Song et al. also noted that the NTD of SdjA was essential for its glutamylation activity and a mutant of SdjA lacking the helix-turnhelix motif (HTH) and the NTD showed an indiscriminate deglutamylation activity toward all members of the SidE family. However, how SdjA recognizes members of the SidE family in the absence of NTD and the key motif or residues in SdjA for its deglutamylase activity are still under investigation. Second, CaM is essential for the glutamylation activity of SdjA, while it is dispensable for the deglutamylation activity, indicating their different catalytic mechanisms. Future structure-based analysis will likely provide insights into the exact catalytic mechanism for the peptidase activity of SdjA. Third, Song et al. noted that SdjA interferes with Rab33b ubiquitination induced only by SdeB or SdeC, but not by SidE. The result is not entirely consistent with Osinski et al.’s report, which revealed that SdjA glutamylated the active site Glu in SdeB, SdeC, and SidE and completely abolished Ub-ligase activity of SdeB, SdeC, and SidE in vitro.[6] Also, how the induced glutamylation on SdeB, SdeC, and SidE was revered remained to be investigated.
Remarkably, Song et al. revealed that an enzyme can catalyze two completely opposite biochemical reactions in regulating PR ubiquitination, which is a valuable contribution expanding our understanding of enzyme functions. The activity of SdjA and SidJ is reminiscent of MavC and MvcA, effectors of L. pneumophila which share 62% similarity in their primary sequences and function in regulation of the activity of the E2 enzyme UBE2N by opposite biochemical activities.[7, 8, 9] Due to these examples, it would not be surprising if other effectors of this pathogen co-opt the host ubiquitin network by opposite biochemical activities.
Funding statement: This work is supported by the National Natural Science Foundation of China (No. 31970178 and 32000139).
-
Conflict of Interest
None declared.
Reference
1 Qiu J, Luo ZQ. Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol 2017;15:591–605.10.1038/nrmicro.2017.67Search in Google Scholar PubMed
2 Gan N, Zhen X, Liu Y, Xu X, He C, Qiu J, et al. Regulation of phosphoribosyl ubiquitination by a calmodulin-dependent glutamylase. Nature 2019;572:387–91.10.1038/s41586-019-1439-1Search in Google Scholar PubMed PubMed Central
3 Black MH, Osinski A, Gradowski M, Servage KA, Pawlowski K, Tomchick DR, et al. Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases. Science 2019;364:787–92.10.1126/science.aaw7446Search in Google Scholar PubMed PubMed Central
4 Liu Y, Luo ZQ. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun 2007;75:592–603.10.1128/IAI.01278-06Search in Google Scholar PubMed PubMed Central
5 Song L, Xie Y, Li C, Wang L, He C, Zhang Y, et al. The Legionella Effector SdjA Is a Bifunctional Enzyme That Distinctly Regulates Phosphoribosyl Ubiquitination. mBio 2021;12:e0231621.10.1128/mBio.02316-21Search in Google Scholar PubMed PubMed Central
6 Osinski A, Black MH, Pawlowski K, Chen Z, Li Y, Tagliabracci VS. Structural and mechanistic basis for protein glutamylation by the kinase fold. Mol Cell 2021;81:4527–39.10.1016/j.molcel.2021.08.007Search in Google Scholar PubMed PubMed Central
7 Gan N, Nakayasu ES, Hollenbeck PJ, Luo ZQ. Legionella pneumophila inhibits immune signalling via MavC-mediated transglutaminase-induced ubiquitination of UBE2N. Nat Microbiol 2019;4:134–43.10.1038/s41564-018-0282-8Search in Google Scholar PubMed PubMed Central
8 Gan N, Guan H, Huang Y, Yu T, Fu J, Nakayasu ES, et al. Legionella pneumophila regulates the activity of UBE2N by deamidase-mediated deubiquitination. EMBO J 2020;39:e102806.10.15252/embj.2019102806Search in Google Scholar PubMed PubMed Central
9 Guan H, Fu J, Yu T, Wang ZX, Gan N, Huang Y, et al. Molecular Basis of Ubiquitination Catalyzed by the Bacterial Transglutaminase MavC. Adv Sci (Weinh) 2020;7:2000871.10.1002/advs.202000871Search in Google Scholar PubMed PubMed Central
© 2022 Jun Jiao, Xuan Ouyang, You Xu, Xiaolu Xiong, published by Sciendo
This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 3.0 License.
Articles in the same Issue
- Editorial
- Research progress on the role of probiotics in acute liver failure
- Commentary
- A bifunctional enzyme of Legionella that distinctly regulates phosphoribosyl ubiquitination of the SidE family effectors
- The potential role of the brain–gut axis in the development and progression of Alzheimer's disease
- Review Article
- Novel insights into alcoholic liver disease: Iron overload, iron sensing and hemolysis
- Right ventricle remodeling in chronic thromboembolic pulmonary hypertension
- Original Article
- Prevalence and risk factors of hyperuricemia and gout: a cross-sectional survey from 31 provinces in mainland China
- Binding domain characterization of growth hormone secretagogue receptor
- Pan-cancer landscape of the RUNX protein family reveals their potential as carcinogenic biomarkers and the mechanisms underlying their action
- Letter to Editor
- Extracorporeal membrane oxygenation using a modified cardiopulmonary bypass system
- A Case of abdominal pain and diarrhea post immunotherapy: Hypophysitis associated with immune checkpoint inhibitors
Articles in the same Issue
- Editorial
- Research progress on the role of probiotics in acute liver failure
- Commentary
- A bifunctional enzyme of Legionella that distinctly regulates phosphoribosyl ubiquitination of the SidE family effectors
- The potential role of the brain–gut axis in the development and progression of Alzheimer's disease
- Review Article
- Novel insights into alcoholic liver disease: Iron overload, iron sensing and hemolysis
- Right ventricle remodeling in chronic thromboembolic pulmonary hypertension
- Original Article
- Prevalence and risk factors of hyperuricemia and gout: a cross-sectional survey from 31 provinces in mainland China
- Binding domain characterization of growth hormone secretagogue receptor
- Pan-cancer landscape of the RUNX protein family reveals their potential as carcinogenic biomarkers and the mechanisms underlying their action
- Letter to Editor
- Extracorporeal membrane oxygenation using a modified cardiopulmonary bypass system
- A Case of abdominal pain and diarrhea post immunotherapy: Hypophysitis associated with immune checkpoint inhibitors

