Abstract
Objective
The Glucose 6-phosphatase (G6Pase) catalytic subunit (G6PC) catalyzes glucose 6-phosphate (G6P) to inorganic phosphate and glucose, playing a critical role in endogenous energy supply. Here, the G6PC gene family was investigated and characterized in common carp (Cyprinus carpio).
Methods
Sequence alignment and phylogenetic analysis were performed using MEGA5. The HMM profiles, motif structure were analyzed using Pfam and MEME, respectively. Quantitative real-time PCR was used to test the expression profiles.
Results
Four assumptive members of G6PC family in common carp whole-genome sequence were identified as cg6pca.1, cg6pca.2a, cg6pca.2b and cg6pcb which were classified into g6pca and g6pcb subtypes, respectively. Evolutionary analysis revealed that cg6pca.2a and cg6pca.2b have a closer evolutionary relationship, and the same subtype members have higher homology among different species. A classical PAP2-glucose phosphates domain is found in four genes and were highly conserved. The expression patterns revealed that only cg6pca.2a elevated significantly after 12 and 24 h of both starvation and cold treatment (p < 0.05).
Conclusions
This study performed a comprehensive analysis of G6PC gene family in common carp. Moreover, cg6pca.2 may be the major functional gene in cold and fasting stress. And the transfactors, PLAG1 and Sox8, may be concerned with expression regulation of cg6pca.2.
Öz
Amaç
Glikoz 6-fosfatazın (G6Pase) katalitik alt birimi (G6PC), glikoz 6-fosfat (G6P)’ı inorganik fosfat ve glukoza katalize ederek endojen enerji kaynağında kritik bir rol oynar. Buradaki G6PC gen ailesi, sazanlarda (Cyprinus carpio) araştırılmış ve karakterize edilmiştir.
Yöntemler
Dizileme ve filogenetik analiz MEGA5 kullanılarak yapılmıştır. HMM profilleri, motif yapısı sırasıyla Pfam ve MEME kullanılarak analiz edilmiştir. Ekspresyon profillerini test etmek için kantitatif gerçek zamanlı PCR kullanılmıştır.
Bulgular
Sazanların tüm genom dizisinde G6PC ailesinin dört varsayım üyesi, sırasıyla g6pca ve g6pcb alt tipleri olarak sınıflandırılan cg6pca.1, cg6pca.2a, cg6pca.2b ve cg6pcb olarak tanımlanmaktadır. Evrimsel analizler, cg6pca.2a ve cg6pca.2b’nin daha evrimsel bir ilişkiye sahip olduğunu ve aynı alt tipteki üyelerin farklı türler arasında daha yüksek homolojiye sahip olduğunu ortaya çıkarmıştır. Dört gende klasik bir PAP2-glukoz fosfat bölgesi bulunur ve bu yüksek oranda korunmuştur. Ekspresyon paternleri, hem cg6pca.2a’nın hem açlık hem de soğuk tedavisinden 12 ve 24 saat sonra anlamlı derecede yükseldiğini ortaya koymuştur (p < 0.05).
Sonuçlar
Bu çalışma, sazanlarda G6PC gen ailesinin kapsamlı bir analizini gerçekleştirmiştir. Ayrıca, cg6pca.2, soğuk ve açlık stresinde temel fonksiyonel gen olabilir. PLAG1 ve Sox8 transfactors, cg6pca.2 ekspresyon regülasyonu ile ilgili olabilir.
Introduction
Glucose 6-phosphate (G6Pase, EC 3.1.3.9) is an important enzyme in the hepatic glucose production, catalysing glucose-6-phosphate (glu-6P) to glucose, which is then released into blood [1]. G6Pase catalysis (G6PC) carry out the last step in glycogenolysis and gluconeogenesis, and thus plays a crucial role in blood glucose homeostasis [2], [3]. At present three G6PC members were identified in Homo sapiens and Danio rerio [4], while only one member was identified in Branchiostoma japonicum [5].
In teleost, the regulation of glucose homeostasis is a complicated and species-specific process, opposite to mammals, in which blood glucose maintains at a stable level. The strictly carnivorous rainbow trout has a “glucose-intolerant” character testified by sustaining hyperglycemia when fed with a diet high in carbohydrate since the lack of molecular inhibition of gluconeogenic enzyme. Oppositely, common carp, an omnivorous fish, does not show postprandial hyperglycemia [6]. Marandel et al. [7] found that g6pcb2 may play a part in glucose intolerance of trout and may partly interpret its inefficiency of carbohydrate utilization. However, there are few studies about the phylogenetic relationships and functions of the G6PC genes in common carp.
Common carp (Cyprinus carpio) is not only the oldest, most domesticated commercial fishes in the world, but also is highly suitable for fundamental research for its taxonomic status and appropriate body size for obtaining sufficient tissue material for advanced studies [8]. While majority studies about G6Pase in common carp have been well documented when it comes to enzyme kinetics, this enzyme system has never been lucubrated from the respective of whole genome. Fortunately, the genome assembly of common carp published in 2014 [9] provides a prerequisite to studies of G6PC gene family in whole genome.
The purposes of this study, taking common carp hepatopancreas gland (the main gluconeogenic tissue) as object, were firstly to characterize the phylogenetic relationships, physicochemical properties, HMM profiles, motif structures in whole genome. Moreover, given the fact that abiotic stress, such as low temperature, can influence the synthesis of glucose in fish [10], expression profiles of cg6pca.1, cg6pca.2a, cg6pca.2b, and cg6pcb in common carp under cold or fasting conditions were analyzed using real-time quantitative PCR.
Materials and methods
Sequence information
The nucleotide and protein sequences of common carp genome assembly V1.0 and annotation file were extracted from the European Nucleotide Archive (ENA, PRJE7241) (https://www.ebi.ac.uk/ena/data/view/PRJEB7241) and the common carp database (http://www.carpbase.org/), respectively. The upstream sequences of the target genes were downloaded from the common carp genome via an in-house perl program. The amino acid sequences of zebrafish (Danio rerio) genome assembly (GRCz10, GCA_000002035.3) were extracted from Ensembl (ftp://ftp.ensembl.org/pub/release-90/fasta/danio_rerio/dna/). Other species’ protein sequences were obtained from the NCBI protein databases (http://www.ncbi.nlm.nih.gov/protein).
Sequence alignment and phylogenetic analysis
The multiple alignment of DNA or protein sequences was conducted with ClustalW (Belfield, Dublin, Ireland) [11], and gene phylogeny were analyzed by MEGA5 software (Hachioji, Tokyo, Japan) [12]. The phylogenetic trees, based on protein sequences, were constructed by the Neighbor-Joining method. The bootstrap method was used to test the reliability of the inferred trees with 1000 replications. Percent identity matrix of paralogous teleost sequences were calculated using the MUSCLE program.
Physicochemical properties and motifs analysis
The biophysical and biochemical parameters of the proteins belonging to common carp G6PC family were conducted using GPMAW program (Cambridge, MA, USA) [13]. Hidden Markov Model (HMM) profiles were analyzed using the Pfam program v27.0 (St. Louis, MO, USA) (http://pfam.xfam.org). The novel protein motif analysis was determined using MEME Suite v4.10 (Seattle, WA, USA) [14], and visual procedures were conducted by TBtools (Guangzhou, Guangdong, China) (https://github.com/CJ-Chen/TBtools).
Starvation or cold stress
For the starvation or cold stress, 270 juvenile common carps weighing 66.73±6.16 g that acclimated to 24°C (the temperature of control) were sampled and randomized to three groups equally (30 individuals per group): 24°C control group, 24°C starvation stress group and 4°C stress group. In the starvation test, none food was supplied during the test period while the temperature was kept to 24°C. While, in the cold stress, the reduction of water temperature from 24°C to 4°C for the experimental groups was finished in 1 h by a refrigerated cooler and ice block in an air-conditioning room (Gree, Hefei, Anhui, China) and the same food (30% protein, 8.0% total fat, 8.0% fiber, supplied by a commercial mixed feed, Yangzhou, Jiangsu, China) was supplied similar to control group. The control temperature was maintained at 24°C. During the course of the experiments, oxygen supplies was adequately given to the cold stress, starvation and control group. The sampling started when the water temperature of the cold-stress group reached at 4°C. The hepatic tissues of three fishes were collected in starvation or cold stress and the control groups were stored at liquid nitrogen until analyses of the transcript levels (Yangzhou, Jiangsu, China), with the time intervals of 0, 12, 24, 36 and 48 h. Every treatment was repeated three times.
Real-time quantitative PCR (qRT-PCR)
Total RNA was isolated from hepatopancreas gland using the RNAeasy Fibrous Tissue Mini-kit (Qiagen, Duesseldorf, Germany) following the instruction manual. Quality and integrity of mRNA were verified spectrophotometrically and by agarose gel electrophoresis. The cDNA was synthesized by PrimeScript®II 1st Strand cDNA Synthesis Kit (Takara, Dalian, Shandong, China) in a 20 μL reaction system with 5 μg total RNA using an iCycler (Bio-Rad, Hercules, CA, USA) programmed at 42°C for 60 min, and 95°C for 5 min. The real-time PCR reaction contained 1× Power SYBR Green Premix Ex Taq (Takara, Dalian, Shandong, China), 0.3 μM of each forward and reverse primer and 5 ng of cDNA template. Real-time qPCR was conducted in a Light Cycler 480® real-time qPCR system (Roche, Basel, Basel-Stadt, Switzerland) with the conditions: 3 min at 94°C, followed by 45 cycles of 10 s at 94°C, 20 s at 58°C. Melting curve assay was done to verify the primers specificity. Every sample was run in triplicate, and each assay was set three repetitions. When the procedure was completed, the threshold cycle (Ct) values were obtained from each reaction. The 2−ΔΔCT method was used to calculate the relative quantity [15]. According to previous researches [16], [17] and the preliminary experiment, actin-β was determined as the housekeeping control genes for the normalization of target genes. The specific primers showed in Table 1 were designed using Oligo v7.0 software (Vondelpark Colorado Springs, CO, USA) (Applied Biosystems) and Primer Premier 6.0 (PREMIER Biosoft, Palo Alto, CA, USA).
The sequences of primers used in this experiment.
| Target gene | Primer sequence(5′-3′) | Product length (bp) | Gene ID |
|---|---|---|---|
| actin-β | F:GAAGAGTTACGAGCTGCCTGA | 106 | M24113(GeneBank ID) |
| R:CATGGATACCGCAAGATTCC | |||
| cg6pca.1 | F:AGCCTGGCAGGCTGCCTTCA | 171 | cycg043353 |
| R:AGTCCAGGTACTCCGTCTGCA | |||
| cg6pca.2a | F:CATCCAACATTGACTGCTTAG | 125 | cycg044960 |
| R:GTAACCTGCTGCCACAAT | |||
| cg6pca.2b | F:ATCACTCTTGACCAGCT | 170 | cycg043354 |
| R:TCAGATCTGCCGCAA | |||
| cg6pcb | F:GACCTGCGCACAACA | 166 | cycg044064 |
| R:ACCAATAAGGACGCTCGC |
Statistical analyses
R statistical software (Version 3.1.3) (Auckland, Auckland Region, New Zealand) was used for statistical analysis. Before statistics and analyzing the information, the data were log transformed as to maintain assumptions of normality as determined by a Shapiro-Wilk’s test. Duncan’s multiple test was conducted to test the significance of differences in the time-course measurements at each sampling times (p<0.05). The data are presented as mean±SEM. The diagrams were generated by R and OriginPro 2015 (Origin Lab, Northampton, MA, USA).
Results and discussion
Phylogenetic relationships of common carp G6PC gene family
G6PC gene family has been identified in many species including both invertebrates and vertebrates [18], [19], [20]. By analyzing common carp genome, we identified four paralogs sharing high sequence homology with the zebrafish (Danio rerio) orthologs, namely cg6pca.1, cg6pca.2a, cg6pca.2b, and cg6pcb classed into g6pca and g6pcb subtypes, respectively (Figure 1A), encoding 354, 376, 252 and 353 amino acids, respectively (Table 2). In mammals, the G6PC gene family includes g6pc, g6pc2, and g6pc3 three members in all [19]. While, Marandel et al. [5] found rainbow trout (Oncorhynchus mykiss) has five members, which were grouped into g6pca.1, g6pca.2 and g6pcb orthologs, respectively. In this study, we identified in total four members in common carp. The identity of nucleotide and amino acid sequences showed that the four isoforms revealed a moderate sequence homology, except that cg6pca.2a and cg6pca.2b shared high sequence identity (Tables 3 and 4).
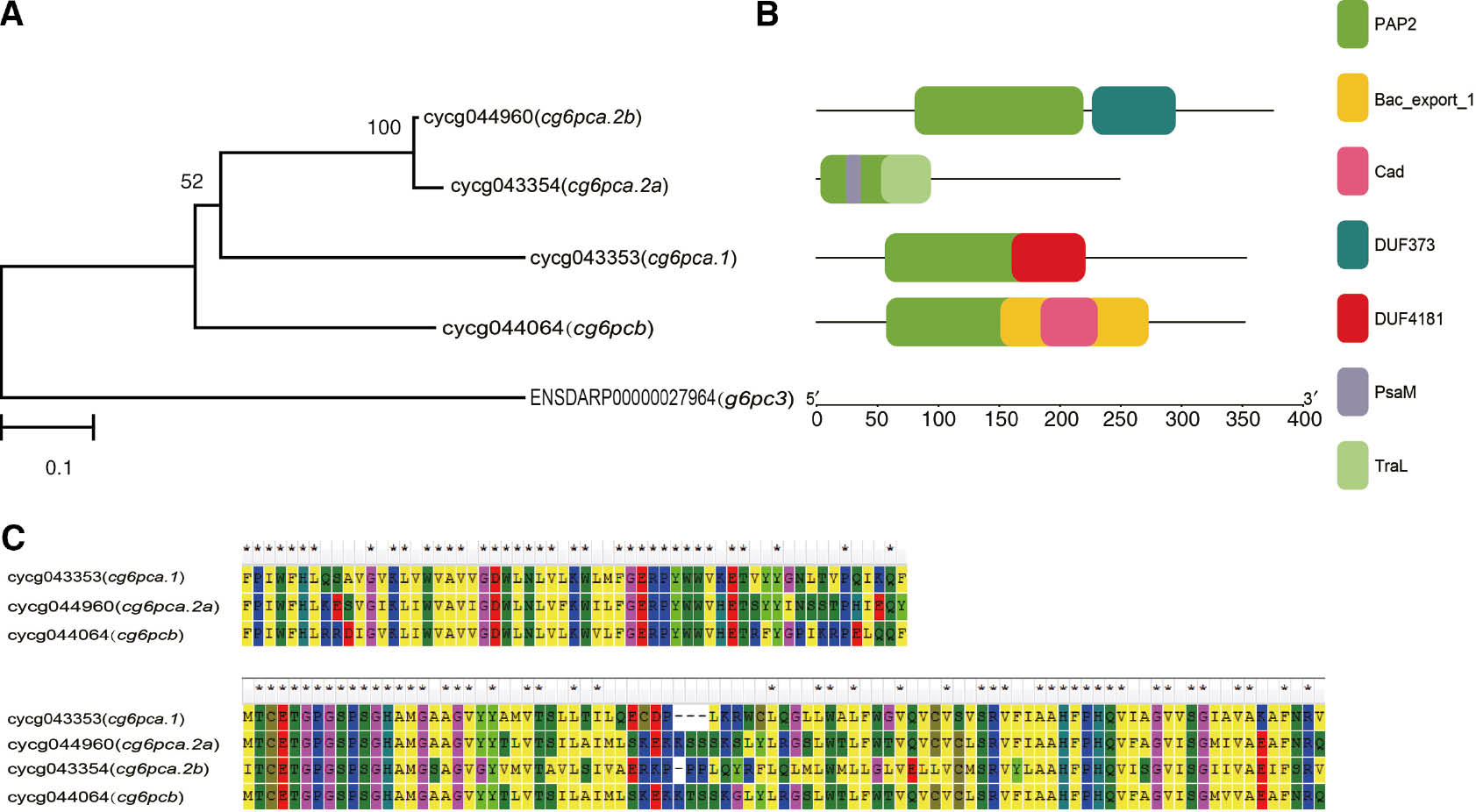
Phylogenetic relationship of common carp G6PC proteins (A), domain (B) and partial amino acid sequence of PAP2 (C).
(A) Shows the phylogenetic tree of G6PC proteins. (B) Indicates the protein motifs corresponding to (A). We used the Danio rerio g6pc3 amino acid sequence as outgroup. (C) Shows the partially conserved regions.
The G6PC gene family members in common carp V1.0 genome.
| Sub-family | Gene ID | Symbol | Locus tag | Localization | Gene length (CDS bp) | Protein length (aa) |
|---|---|---|---|---|---|---|
| g6pca | cycg043353 | cg6pca.1 | LN598274.1 | 44,873–48,794 | 1062 | 354 |
| g6pca | cycg044960 | cg6pca.2a | LN592114.1 | 13,565–15,903 | 1128 | 376 |
| g6pca | cycg043354 | cg6pca.2b | LN598274.1 | 52,465–54,957 | 756 | 252 |
| g6pcb | cycg044064 | cg6pcb | LN590703.1 | 8,925,238–8,927,076 | 1059 | 353 |
Matrix mRNA(CDS) identity of g6pc gene family among common carp.
| cycg044064(cg6pcb) | cycg043353(cg6pca.1) | cycg043354(cg6pca.2b) | cycg044960(cg6pca.2a) | |
|---|---|---|---|---|
| cycg044064 | – | 57.0 | 62.0 | 62.5 |
| cycg043353 | – | 60.5 | 61.3 | |
| cycg043354 | – | 92.5 | ||
| cycg044960 | – |
Phylogenetic tree analysis based on the full-length protein sequences showed that Teleost g6pc3 formed a clade itself, while the g6pca and g6pcb were clustered together, and the same subtype members have higher homology among different species. Common carp cg6pca.1, cg6pca.2a and cg6pca.2b have a closer evolutionary relationship than cg6pcb (Figures 1A and 2). This topology was similar with Oncorhynchus mykiss [6] and Amphioxus [5]. Yu et al. [5] constructed a genetic evolution model of G6PC family and considered that an original g6pc gene generated two paralogs in an early genome duplication event, and then one of the derived genes evolved into g6pc3, while the other one undergone a specific genetic multiplication on the vertebrate lineage when it had been split from the protochordate stem. The g6pc3 class is absent in common carp genome, contrary to relative species, Danio rerio. It could be that the gene was lost during the evolution. But for all this, we cannot rule out alignment errors caused by assembly of genome.
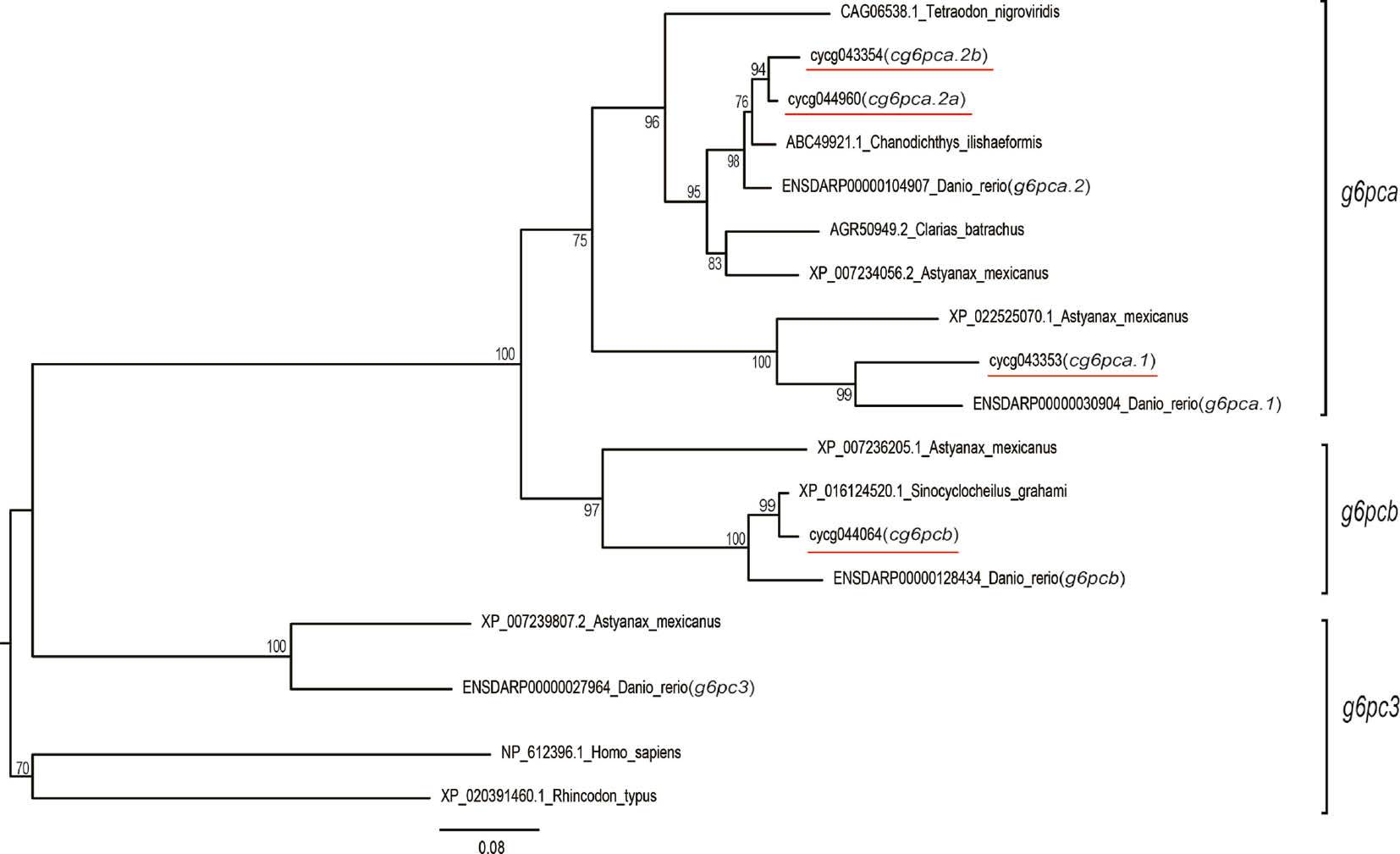
The phylogenetic relationships of common carp G6PC proteins among the fishes and other vertebrates.
The tree was constructed using the neighbor-joining method. The bootstrap values for 1000 replicates are represented at the nodes of clades.
Matrix protein identity of g6pc family among common carp.
| cycg044064(cg6pcb) | cycg043353(cg6pca.1) | cycg043354(cg6pca.2b) | cycg044960(cg6pca.2a) | |
|---|---|---|---|---|
| cycg044064 | – | 56.3 | 52.7 | 59.8 |
| cycg043353 | – | 53.1 | 55.2 | |
| cycg043354 | – | 96.4 | ||
| cycg044960 | – |
Structural properties of common carp G6PC gene members
The physicochemical characters of common carp G6PC family proteins indicated that the estimated molecular weight (MW) of cg6pca.1, cg6pca.2a, cg6pca.2b, and cg6pcb monomers are approximately 39.7, 42.2, 27.3, and 39.4 kDa, while isoelectric points (pI) are 9.75, 10.13, 10.27 and 9.91, respectively. The structural properties analysis revealed that common carp G6PC family genes all contain the classical PAP2-glucose-6-phosphatase (Figure 1) which is characterized of all G6Pase isoforms [5], and no signal peptide was identified by Signal P, which is also a typical property among all known G6PC members. Additionally, the conserved motifs were identified using the full-length protein sequences of common carp G6PC family members, and three motifs were screened out (Figure 3). Although the four isoforms showed a moderate amino acid sequence homology, their motif sequences were very similar (Figure 3C).
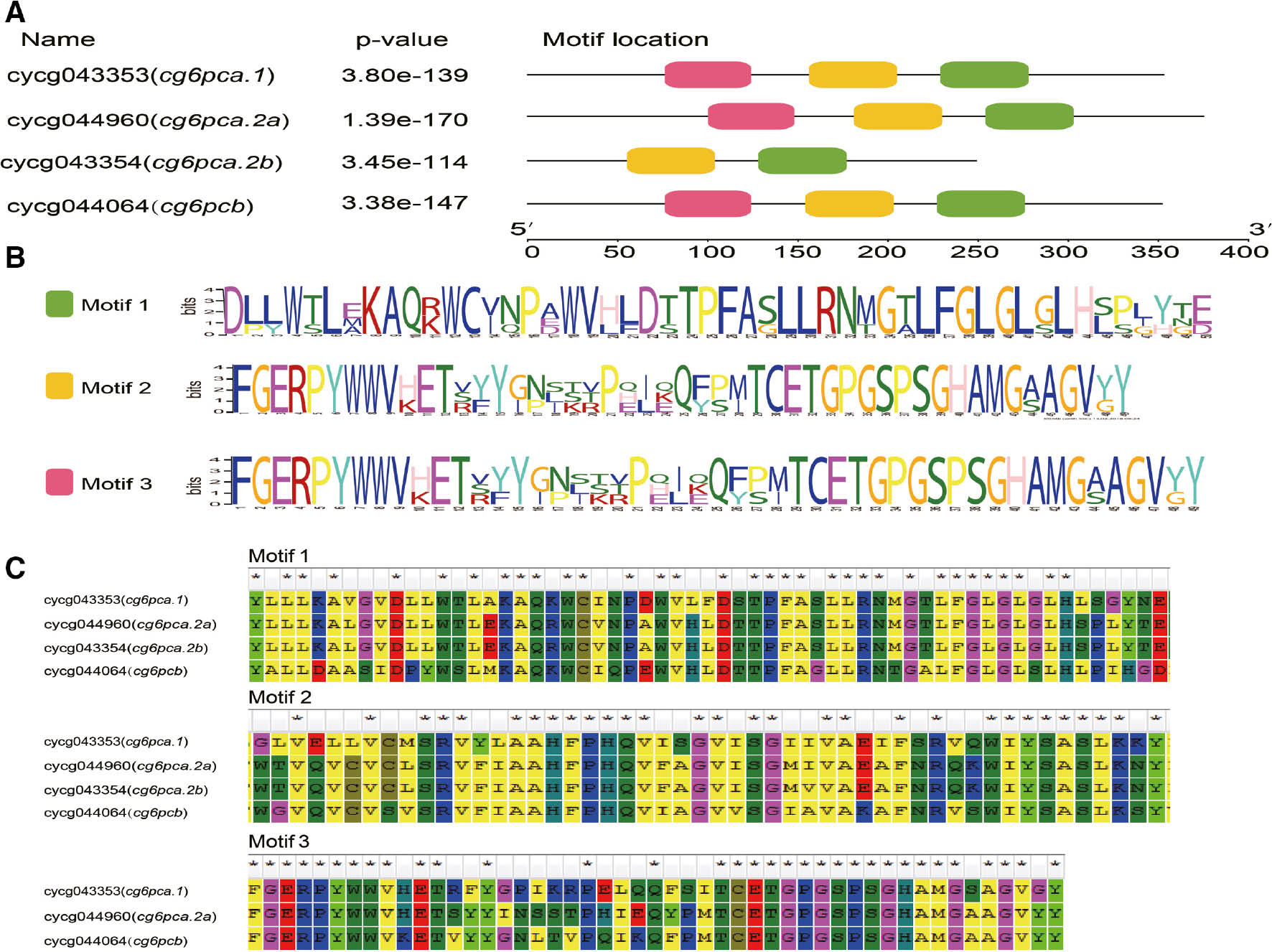
The motifs in common carp G6PC family (A), the motif logos (B) and partial amino acid sequence of the three motifs (C).
(A) The annotated amino acid sequence motifs and p-values. (B) The consensus motif sequences. (C) The specific motif sequences.
Gene expression variations under starvation or cold stress
In mammals, all the gluconeogenic enzymes are primarily determined by altering the content of the protein [20]. Moreover, G6Pase are also affected by short-term regulation via covalent or allosteric modification [21]. Previous studies mostly focused on the biological significances of G6PC in different nutritional status of fish, however few documents have presented the gene expression differential display under abiotic stress. Given the critical role of G6Pase in maintaining blood glucose homeostasis, in this study, the transcriptions of G6PC family were detected in hepatic tissues of common carp under two common abiotic stresses, starvation or cold condition. The results showed that cg6pca.1, cg6pca.2a and cg6pca.2b were transcribed under normal condition, except for that cg6pcb was not detected at control or any treatment condition. And contrary to expectations, only cgp6c.2a was significantly elevated after 12-h 4°C or after 24-h fasted treatment, respectively. Long et al. [22], Long et al. [23] and Hu et al. [24] also found Danio rerio g6pca2 were up-regulated by cold stress in the transcriptome analyses during cold stress. When the ambient temperature lower than the optimal temperature range, there will be a significant reduction in food intake and conversion rates [25]. Meanwhile, the response to cold stress itself is an energy-demanding process. The organisms have to mobilize more energy substance to cope with it. For the limited capacity to store abundant glycogen during cold stress, carps’ gluconeogenic pathway had been activated, in order to steadily support glucose. As the data represented in Figure 4, the levels of cg6pca.2a in cold stress was greater than starvation. This could be a sign that common carp need more glucose at low temperature than fast challenges.
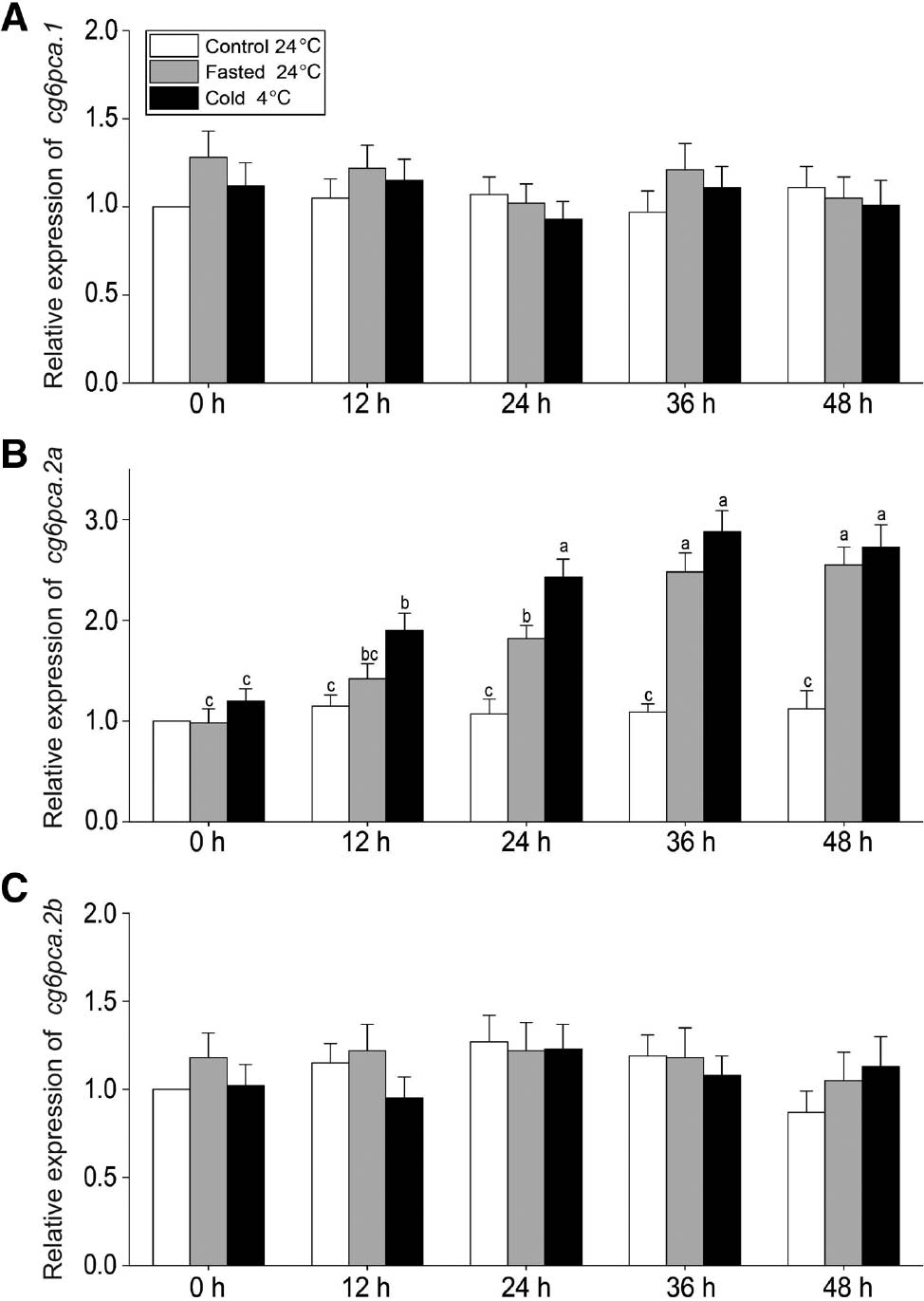
The relative expressions of common carp G6PC members in cold or starvation stress.
(A) The relative expression of cg6pca.1. (B) The relative expression of cg6pca.2a. (C) The relative expression of cg6pca.2b. Time-course changes in expression difference of cg6pca.1, cg6pca.2a, cg6pca.2a and cg6pcb under starvation (gray bars) or cold stress (black bars) were depicted (white bars for control group). Data were normalized to actin-β. Values represent mean±SEM (n=3). The letters indicate the differences are significant difference between any two means (p<0.05).
In consideration of the regulation of upstream control elements (UCE) to target gene, we identified the proximal common carp G6PC promoter binds multiple transcription factors as well. We scanned the 1 kb upstream regions of the 5′UTRs of common carp G6PC genes, which were the UCE-rich regions using the motif-finding algorithm MEME. Annotations of the detected motifs were conducted by TOMTOM program with JASPAR database. The workflow is shown in Figure 5A, copied from Hu et al. [26]. The proximal G6PC family promoter-binding transcription factors, including Alx1, CDX1, Foxa2, HNF4, PLAG1, RORA and SOX8 (Figure 5B). Among them, Foxa2 and HNF4 are the common factors in common carp and human [16], [19]. Except for cg6pcb, cg6pca.1, cg6cpa2.a and cg6pca2.b have the similar transfactor, PLAG1 and Sox8 are two unique transfactors in cg6cpa2.a among the three genes. Consequently, the different progress of cis-regulatory elements and trans-factors among common carp G6PC members may result in partially the diverse expression patterns, and PLAG1 and Sox8 may be concerned with abiotic stress in common carp.
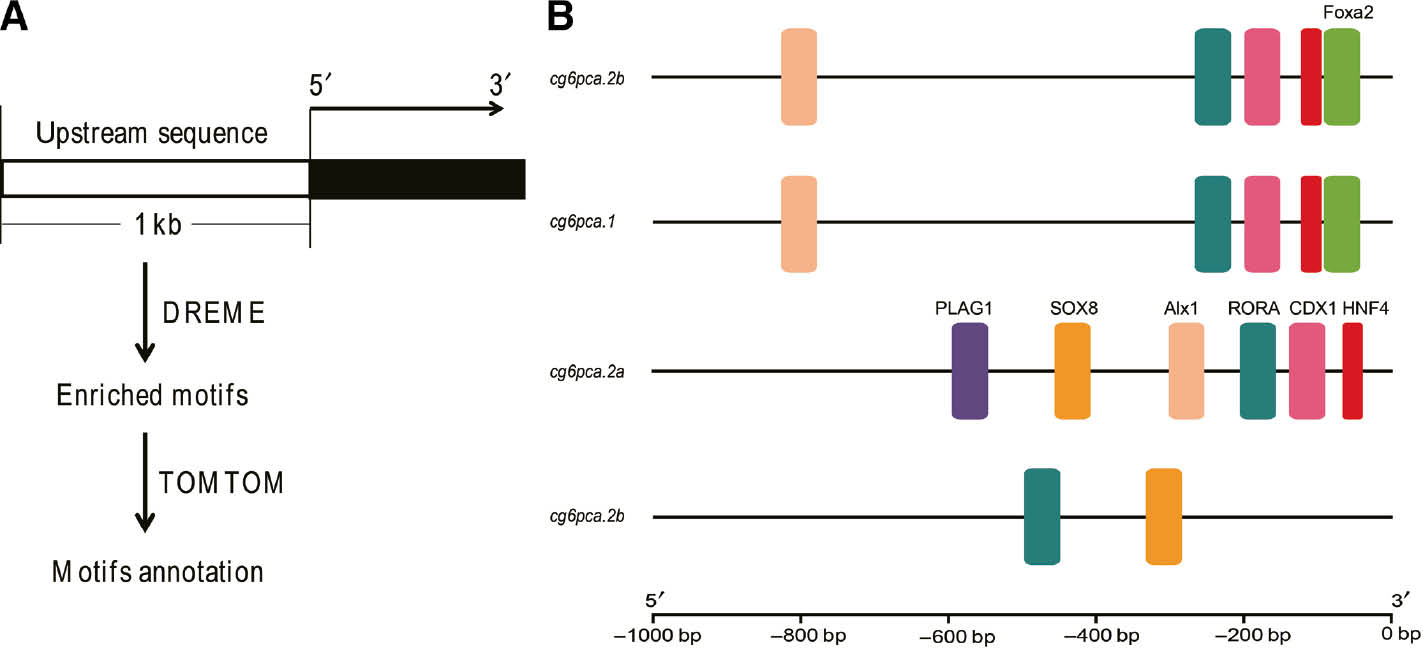
The identification of the cis-regulatory elements and transfactors.
(A) The screening process for enriched motifs in the proximal promoter regions of target genes. (B) Comparison of transcription factors binding the common carp G6PC members’ promoters.
In conclusion, this study firstly identified all G6PC members in common carp genome, revealed the phylogenetic relationships, physicochemical properties, HMM profiles, motif structures of common carp G6PC members. In addition, the expression profiles of common carp G6PC genes under different abiotic stresses figured out cg6pca.2a played an important role in resisting adverse situation.
Acknowledgements
The authors would like to thank the lab members for the Dr. Zongbin Cui and Xianghui Kong of the Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan and Henan Normal University, Xinxiang for technical assistance, respectively. This work was supported by grants from Strategic Pilot Science and Technology Projects of the Chinese Academy of Sciences (XDA08010208-3).
-
Conflict of interest statement: Authors have no conflict of interest.
References
1. Ghosh A, Shieh JJ, Pan CJ, Sun MS, Chou JY. The catalytic center of glucose-6-phosphatase. HIS176 is the nucleophile forming the phosphohistidine-enzyme intermediate during catalysis. J Biol Chem 2002;277:32837–42.10.1074/jbc.M201853200Search in Google Scholar
2. Enes P, Panserat S, Kaushik S, Oliva-Teles A. Nutritional regulation of hepatic glucose metabolism in fish. Fish Physiol Biochem 2009;35:519–39.10.1007/s10695-008-9259-5Search in Google Scholar
3. Moon TW, Foster GD. Chapter 4 tissue carbohydrate metabolism, gluconeogenesis and hormonal and environmental influences. Biochem Mol Bio Fish 1995;4:65–100.10.1016/S1873-0140(06)80007-XSearch in Google Scholar
4. Hutton JC, O’Brien RM. Glucose-6-phosphatase catalytic subunit gene family. J Biol Chem 2009;284:29241–5.10.1074/jbc.R109.025544Search in Google Scholar PubMed PubMed Central
5. Wang Y, Wang H, Li M, Gao Z, Zhang S. Identification, expression and regulation of amphioxus G6Pase gene with an emphasis on origin of liver. Gen Comp Endocrinol 2015;214:9–16.10.1016/j.ygcen.2014.12.021Search in Google Scholar PubMed
6. Panserat S, Médale F, Blin C, Brèque J, Vachot C, Plagnes-Juan E, et al. Hepatic glucokinase is induced by dietary carbohydrates in rainbow trout (Oncorhyncus mykiss), gilthead seabream (Sparus aurata) and common carp (Cyprinus carpio). Am J Physiol Regul Integr Comp Physiol 2000;278:R1164–70.10.1152/ajpregu.2000.278.5.R1164Search in Google Scholar PubMed
7. Marandel L, Seiliez I, Véron V, Skibacassy S, Panserat S. New insights into the nutritional regulation of gluconeogenesis in carnivorous rainbow trout (Oncorhynchus mykiss): a gene duplication trail. Physiol Genomics 2015;47:253–63.10.1152/physiolgenomics.00026.2015Search in Google Scholar PubMed
8. Kolder IC, van der Plas-Duivesteijn SJ, Tan G, Wiegertjes GF, Forlenza M, Guler AT, et al. A full-body transcriptome and proteome resource for the European common carp. BMC Genomics 2016;17:701.10.1186/s12864-016-3038-ySearch in Google Scholar PubMed PubMed Central
9. Xu P, Zhang X, Wang X, Li J, Liu G, Kuang Y, et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat Genetics 2014;46:1212–9.10.1038/ng.3098Search in Google Scholar PubMed
10. Magnoni LJ, Scarlato NA, Ojeda FP, Wöhler OC. Gluconeogenic pathway does not display metabolic cold adaptation in liver of Antarctic notothenioid fish. Polar Biol 2013;36:661–71.10.1007/s00300-013-1292-xSearch in Google Scholar
11. Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, Mcwilliam H, et al. Clustal w and clustal x version 2.0. Bioinformatics 2007;23:2947–8.10.1093/bioinformatics/btm404Search in Google Scholar
12. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. Mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2014;28:2731–9.10.1093/molbev/msr121Search in Google Scholar
13. Peri S, Steen H, Pandey A. Gpmaw – a software tool for analyzing proteins and peptides. Trends Biochem Sci 2001;26:687–9.10.1016/S0968-0004(01)01954-5Search in Google Scholar
14. Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. Meme suite: tools for motif discovery and searching. Nucleic Acids Res 2009;37(Web server issue):202–8.10.1093/nar/gkp335Search in Google Scholar PubMed PubMed Central
15. Huang HT, Kathrein KL, Barton A, Gitlin Z, Huang YH, Ward TP, et al. A network of epigenetic regulators guides developmental haematopoiesis in vivo. Nat Cell Biol 2013;15:1516–25.10.1038/ncb2870Search in Google Scholar PubMed PubMed Central
16. Xu LH, Chang YM, Liu CL, Liang LQ, Liu JL, Chi BJ, et al. Screening cold-acclimation differential expression candidate genes in the brain of common carp (Cyprinus carpio). Hereditas 2011;33:262.10.3724/SP.J.1005.2011.00262Search in Google Scholar
17. Liang L, Chang Y, He X, Tang R. Transcriptome analysis to Identify cold-responsive genes in amur carp (Cyprinus carpio haematopterus). PLoS One 2015;10:e0130526.10.1371/journal.pone.0130526Search in Google Scholar PubMed PubMed Central
18. Salgado MC, Metón I, Egea M, Baanante IV. Transcriptional regulation of glucose-6-phosphatase catalytic subunit promoter by insulin and glucose in the carnivorous fish, Sparus aurata. J Mol Endocrinol 2004;33:783.10.1677/jme.1.01552Search in Google Scholar PubMed
19. Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012;490:49–54.10.1038/nature11413Search in Google Scholar PubMed
20. And SJ, Granner DK. Molecular physiology of the regulation of hepatic gluconeogenesis and glycolysis. Ann Rev Physiol 1992;54:885.10.1146/annurev.ph.54.030192.004321Search in Google Scholar PubMed
21. Foster JD, Pederson BA, Nordlie RC. Glucose-6-phosphatase structure, regulation, and function: an update. Proc Soc Exp Biol Med Soc Exp Biol Med 1997;215:314–32.10.3181/00379727-215-44142Search in Google Scholar PubMed
22. Long Y, Song G, Yan J, He X, Li Q, Cui Z. Transcriptomic characterization of cold acclimation in larval zebrafish. BMC Genomics 2013;14:612.10.1186/1471-2164-14-612Search in Google Scholar PubMed PubMed Central
23. Long Y, Li L, Li Q, He X, Cui Z. Transcriptomic characterization of temperature stress responses in larval zebrafish. PLoS One 2012;7:e37209.10.1371/journal.pone.0037209Search in Google Scholar PubMed PubMed Central
24. Hu P, Liu M, Zhang D, Wang J, Niu H, Liu Y, et al. Global identification of the genetic networks and cis-regulatory elements of the cold response in zebrafish. Nucleic Acids Res 2015;43:9198–213.10.1093/nar/gkv780Search in Google Scholar PubMed PubMed Central
25. Abbink W, Blanco GA, Jac R, Partridge GJ, Kloet K, Schneider O. The effect of temperature and pH on the growth and physiological response of juvenile yellowtail kingfish Seriola lalandi in recirculating aquaculture systems. Aquaculture 2012;330–3:130–5.10.1016/j.aquaculture.2011.11.043Search in Google Scholar
26. Hu P, Liu M, Zhang D, Wang J, Niu H, Liu Y, et al. Global identification of the genetic networks andcis-regulatory elements of the cold response in zebrafish. Nucleic Acids Res 2015;43:9198–213.10.1093/nar/gkv780Search in Google Scholar
© 2019 Walter de Gruyter GmbH, Berlin/Boston
Articles in the same Issue
- Frontmatter
- Review Article
- Lipidomics and cognitive dysfunction – A Narrative review
- Research Articles
- Effects of topiramate on adipocyte differentiation and gene expression of certain carbonic anhydrase isoenzymes
- Heterologous expression of a plant WRKY protein confers multiple stress tolerance in E. coli Bir bitkinin heterolog ifadesi WRKY proteini çoklu stres yaratır E. coli’de tolerans
- Cytogenetic impact of sodium chloride stress on root cells of Vigna radiata L. seedlings
- Understanding the impacts of self-shuffling approach on structure and function of shuffled endoglucanase enzyme via MD simulations
- Evaluation ofTrichoderma atroviride and Trichoderma citrinoviride growth profiles and their potentials as biocontrol agent and biofertilizer
- Physio-biochemical analyses in seedlings of sorghum-sudangrass hybrids that are grown under salt stress under in vitro conditions
- The structural diversity of ginsenosides affects their cholinesterase inhibitory potential
- Extracellular acidity and oxygen availability conjointly control eukaryotic cell growth via modulation of cytoplasmic translation
- Genome-wide identification, phylogeny and expression analysis of G6PC gene family in common carp, Cyprinus carpio
- The effect of Diplotaenia turcica root extract in streptozotocin-induced diabetic rats
- Letter to the Editor
- Metabolomics analysis of medicinal insect Protaetia brevitarsis after Bacillus subtilis fermentation
- Opinion Paper
- Identifying and solving scientific problems in the medicine: key to become a competent scientist
Articles in the same Issue
- Frontmatter
- Review Article
- Lipidomics and cognitive dysfunction – A Narrative review
- Research Articles
- Effects of topiramate on adipocyte differentiation and gene expression of certain carbonic anhydrase isoenzymes
- Heterologous expression of a plant WRKY protein confers multiple stress tolerance in E. coli Bir bitkinin heterolog ifadesi WRKY proteini çoklu stres yaratır E. coli’de tolerans
- Cytogenetic impact of sodium chloride stress on root cells of Vigna radiata L. seedlings
- Understanding the impacts of self-shuffling approach on structure and function of shuffled endoglucanase enzyme via MD simulations
- Evaluation ofTrichoderma atroviride and Trichoderma citrinoviride growth profiles and their potentials as biocontrol agent and biofertilizer
- Physio-biochemical analyses in seedlings of sorghum-sudangrass hybrids that are grown under salt stress under in vitro conditions
- The structural diversity of ginsenosides affects their cholinesterase inhibitory potential
- Extracellular acidity and oxygen availability conjointly control eukaryotic cell growth via modulation of cytoplasmic translation
- Genome-wide identification, phylogeny and expression analysis of G6PC gene family in common carp, Cyprinus carpio
- The effect of Diplotaenia turcica root extract in streptozotocin-induced diabetic rats
- Letter to the Editor
- Metabolomics analysis of medicinal insect Protaetia brevitarsis after Bacillus subtilis fermentation
- Opinion Paper
- Identifying and solving scientific problems in the medicine: key to become a competent scientist

