Comparison of the Molecular International Prognostic Scoring System (IPSS-M) and Revised International Prognostic Scoring System (IPSS-R) in predicting the prognosis of patients with myelodysplastic neoplasms treated with decitabine
-
Quang Hao Nguyen
, Tuan Anh Tran
Abstract
Background
Molecular International Prognostic Scoring System (IPSS-M) is a newly developed prognostic model for myelodysplastic neoplasms (MDS), but has not yet been used widely. In this study, we aimed to compare the IPSS-M with the traditional Revised International Prognostic Scoring System (IPSS-R) in predicting the prognosis of decitabine treated-MDS patients.
Patients and methods
This retrospective cohort study was conducted on 19 newly diagnosed MDS patients who were examined for 51 gene mutations and received decitabine treatment. The survival analysis, including overall survival (OS), progression-free survival (PFS), and leukemia-free survival (LFS), was performed using the Kaplan–Meier method. Comparisons between the risk groups were carried out according to the IPSS-R and IPSS-M models.
Results
Among the 19 MDS patients, 12 (63.2 %) showed myeloid gene mutations, with the highest frequency of mutations in ASXL1, RUNX1, SRSF2, TET2, and TP53 (15.8 %). Survival analysis found that the OS was significantly different between the risk groups of both IPSS-R and IPSS-M models, but the PFS and LFS showed significant differences between the risk groups in only the IPSS-M model. The PFS of the moderate, high, and very high-risk groups were 34.66, 25.00, and 15.33 months (p=0.031); respectively. The LFS of the moderate, high, and very high-risk groups were 39.20, 25.00, and 18.37 months, (p=0.039); respectively.
Conclusions
Our results found that IPSS-M was better than IPSS-R in predicting the PFS and LFS of decitabine-treated MDS patients, IPSS-M may be superior to IPSS-R in predicting the prognosis of MDS patients.
Introduction
Myelodysplastic neoplasms (MDS), previously named myelodysplastic syndromes, are a heterogeneous group of hematopoietic clonal disorders characterized by ineffective hematopoiesis, cytopenia, bone marrow dysplasia, and an increased risk of leukemic transformation [1]. According to the 2022 World Health Organization (WHO) criteria, ‘myelodysplastic syndromes’ was renamed ‘myelodysplastic neoplasms’ (MDS) to emphasize the neoplastic nature, as well as the ability to transform into acute myeloid leukemia [2]. In addition, WHO 2022 proposed a new classification system, which classified MDS into two groups based on genetic and morphological abnormalities. This reclassification system aimed to emphasize the importance of genetic factors in hematopoietic clonal disorders [2]. These changes in terminology and the classification system have raised the question of whether a prognostic system like the Revised International Prognostic Scoring System (IPSS-R), which is based only on morphology and cytogenetics, is sufficient for predicting the prognosis of MDS patients.
Many studies have been conducted on the role of myeloid gene mutations in the prognosis of MDS [3], [4], [5], [6]. Based on the association between myeloid gene mutations and MDS prognosis, a new prognostic model Molecular International Prognostic Scoring System (IPSS-M) has been proposed, which combines the traditional IPSS-R with molecular data (31 genes) [7]. A few studies have investigated the superiority of the IPSS-M model compared to the IPSS-R model [8], [9], [10], [11], [12], [13], [14]; however, a complete consensus has not yet been reached. Baer et al. evaluated the importance of individual genes in the IPSS-M for risk prediction [12]. Zamanillo et al. and Aguirre et al. confirmed that the IPSS-M model can be used for therapeutic decision-making [8, 9]. Maurya et al. and Sabile et al. found that the IPSS-M model is superior in stratifying patients than the IPSS-R model [10, 11]. In contrast, Wu et al. suggested that the IPSS-M model is better than the IPSS-R model only in classifying patients aged ≥60 years [13]. Sauta et al. found that the IPSS-M model showed a significant improvement in predicting the overall survival (OS) and leukemia-free survival (LFS) of posttransplant patients, but not of hypomethylating agent (HMA)-treated patients [14].
However, the screening of 31 genes makes the application of the IPSS-M model more complicated, especially due to the lack of consensus over its superiority over the IPSS-R model. In this study, we aimed to compare the IPSS-M model with the traditional IPSS-R model in predicting the prognosis of MDS patients treated with decitabine (an HMA).
Materials and methods
Patients
The retrospective cohort study was conducted at the National Institute of Hematology and Blood Transfusion (NIHBT, Hanoi, Vietnam). All the newly diagnosed MDS patients who were treated with decitabine from January 2018 to June 2021 were enrolled in the study. The Review Board of the NIHBT approved the study (no. 939/QĐ-HHTM) and waived informed consent as it was a retrospective observational study. All the patient records were de-identified to protect patient privacy.
Cytogenetic and gene mutation analysis
We analyzed the bone marrow samples of the MDS patients obtained at the time of diagnosis. The karyotypes were determined based on the results of G-band staining. Fluorescence in situ hybridization was used to detect del(5q). Next-generation sequencing was performed to detect the 51 myeloid gene mutations, including mutations in the DNA methylation, chromatin modification, RNA splicing, cohesin complex, transcription, cytokine receptor/tyrosine kinase, RAS signaling, checkpoint/cell cycle, and DNA repair gene groups. Although there were 51 gene mutations were tested, 7 of the 31 genes listed by IPSS-M were lacking. They were BCORL1, ETKN1, GNB1, PHF6, PPM1D, PRPF8, and MLL PTD .
Treatment
The patients received a 3 h infusion of 15 mg/m2 decitabine every 8 h for 3 consecutive days. Patients received 4 to 9 treatment cycles, and each cycle was repeated every 6 weeks.
Definition
MDS diagnosis was conducted according to the newly proposed minimal diagnostic criteria for MDS [15], and MDS classification was based on the 2022 WHO criteria [2]. Risk stratification was conducted according to the IPSS-R and IPSS-M models [7, 16, 17]. The OS was calculated from the time of diagnosis to death or the last follow-up. The progression-free survival (PFS) was calculated from the start of treatment to relapse or death, whichever happened first. The LFS was calculated from the beginning of treatment to leukemic transformation.
Statistical analysis
The IPSS-R and IPSS-M were scored in all patients. Survival analyses, including the OS, PFS, and LFS, were performed using the Kaplan–Meier method. Comparisons between the IPSS-R and IPSS-M risk groups were carried out. The pLog-rank value<0.05 was considered statistically significant. Statistical analysis was performed using SPSS 25 software.
Bias was controlled because there was no missing data. Our study conforms to the STROBE guidelines [18].
Results
Patient characteristics
A total of 19 patients were retrospectively analyzed in this study. Among these, 12 were male (63.2 %) and 7 were female (36.8 %) with a median age of 64 years (28–81 years). According to the WHO 2022 criteria, the majority of patients (n=10; 52.6 %) were classified into MDS-IB1 type, while only one patient (5.3 %) was diagnosed with MDS-bi TP53, the rest were classified into MDS-IB2 (n=8; 42.1 %), (Table 1). The results of the cytogenetic analysis and the cell indices are presented in Table 1.
Patients’ characteristics.
| Classification | n, % | |
|---|---|---|
| MDS subtypes (n, %) | MDS-bi TP53 | 1 (5.3 %) |
| MDS-IB1 | 10 (52.6 %) | |
| MDS-IB2 | 8 (42.1 %) | |
| Cytogenetics category (n, %) | Very good | 0 (0 %) |
| Good | 15 (78.9 %) | |
| Intermediate | 0 (0 %) | |
| Poor | 2 (10.5 %) | |
| Very poor | 2 (10.5 %) | |
| IPSS-R (n, %) | Very low | 0 (0 %) |
| Low | 0 (0 %) | |
| Intermediate | 6 (31.6 %) | |
| High | 10 (52.6 %) | |
| Very high | 3 (15.8 %) | |
| IPSS-M (n, %) | Very low | 0 (0 %) |
| Low | 0 (0 %) | |
| Moderate low | 3 (15.8 %) | |
| Moderate high | 7 (36.8 %) | |
| High | 5 (26.3 %) | |
| Very high | 4 (21.1 %) | |
| Cell indices | Mean | Min–max |
|---|---|---|
| Hemoglobin, g/L | 82.58 | 47–105 |
| Total neutrophil count, ×109/L | 2.21 | 0.11–18.86 |
| Platelet count, ×109/L | 145.47 | 15–472 |
| Peripheral blood blast, % | 4.47 | 0–18 |
| Total bone marrow cell count, ×109/L | 74.25 | 5.8–448 |
| Bone marrow blast, % | 9.79 | 5–17 |
Gene mutation analysis
In this study, we detected a total of 14 myeloid gene mutations in MDS patients. Among the 19 MDS patients, 12 (63.2 %) showed myeloid gene mutations, with the number of mutations ranging between 1 and 7 per patient with a mean value of 2.5. The highest frequency of mutations was observed in the following genes: ASXL1, RUNX1, SRSF2, TET2, TP53 (15.8 %), (Table 2).
Frequency of gene mutations in MDS patients.
| Pathway/function | Gene | n, % |
|---|---|---|
| Chromatin modification | ASXL1 | 3 (15.8) |
| Cohesin complex | STAG2 | 2 (10.5) |
| Transcription | RUNX1 | 3 (15.8) |
| BCOR | 2 (10.5) | |
| RNA splicing | SF3B1 | 2 (10.5) |
| SRSF2 | 3 (15.8) | |
| U2AF1 | 1 (5.3) | |
| DNA methylation | TET2 | 3 (15.8) |
| IDH2 | 2 (10.5) | |
| DNMT3A | 2 (10.5) | |
| Cytokine receptor/tyrosine kinase | CSF3R | 1 (5.3) |
| KIT | 1 (5.3) | |
| RAS signaling | NRAS | 2 (10.5) |
| Checkpoint/cell cycle | TP53 | 3 (15.8) |
Risk stratification
The IPSS-R model classified 31.6 , 52.6, and 15.8 % of the MDS patients into intermediate, high, and very high-risk groups; respectively. In contrast, the IPSS-M model reclassified 15.8, 36.8, 26.3, and 21.1 % of the MDS patients into moderate low, moderate high, high, and very high-risk groups; respectively (Table 1).
Survival analysis
The OS analysis showed a significant difference between the risk groups in both the IPSS-R and IPSS-M categories. There was a significant difference between the intermediate, high, and very high-risk groups of the IPSS-R model and the moderate, high, and very high-risk groups of the IPSS-M model (p=0.005 and 0.043; respectively). This difference was more clearly observed between the very high-risk group and the remaining groups (Table 3). However, the PFS and LFS analysis found a significant difference between the risk groups in only the IPSS-M category. The PFS of the moderate, high, and very high-risk groups were 34.66, 25.00, and 15.33 months (p=0.031); respectively. The LFS of the moderate, high, and very high-risk groups were 39.20, 25.00, and 18.37 months, (p=0.039); respectively, (Table 4; Figures 1 and 2).
OS according to IPSS-R and IPSS-M.
| Risk | n | Months | p-Value | Risk | n | p-Value | ||
|---|---|---|---|---|---|---|---|---|
| IPSS-R | Intermediate + high | 16 | 36.50 | 0.002 | IPSS-R | Intermediate | 6 | 0.005 |
| Very high | 3 | 12.00 | High | 10 | ||||
| Very high | 3 | |||||||
| IPSS-M | Moderate + high | 15 | 38.19 | 0.016 | IPSS-M | Moderate (low and high) | 10 | 0.043 |
| Very high | 4 | 20.50 | High | 5 | ||||
| Very high | 4 | |||||||
-
Bold values represent values <0.05.
PFS and LFS according to IPSS-R and IPSS-M.
| Risk | n | PFS | LFS | |||
|---|---|---|---|---|---|---|
| Months | p-Value | Months | p-Value | |||
| IPSS-R | Intermediate | 6 | 25.50 | 0.368 | 27.83 | 0.211 |
| High | 10 | 32.74 | 39.30 | |||
| Very high | 3 | 12.00 | 13.00 | |||
| IPSS-M | Moderate (low and high) | 10 | 34.66 | 0.031 | 39.20 | 0.039 |
| High | 5 | 25.00 | 25.00 | |||
| Very high | 4 | 15.33 | 18.37 | |||
-
Bold values represent values <0.05.
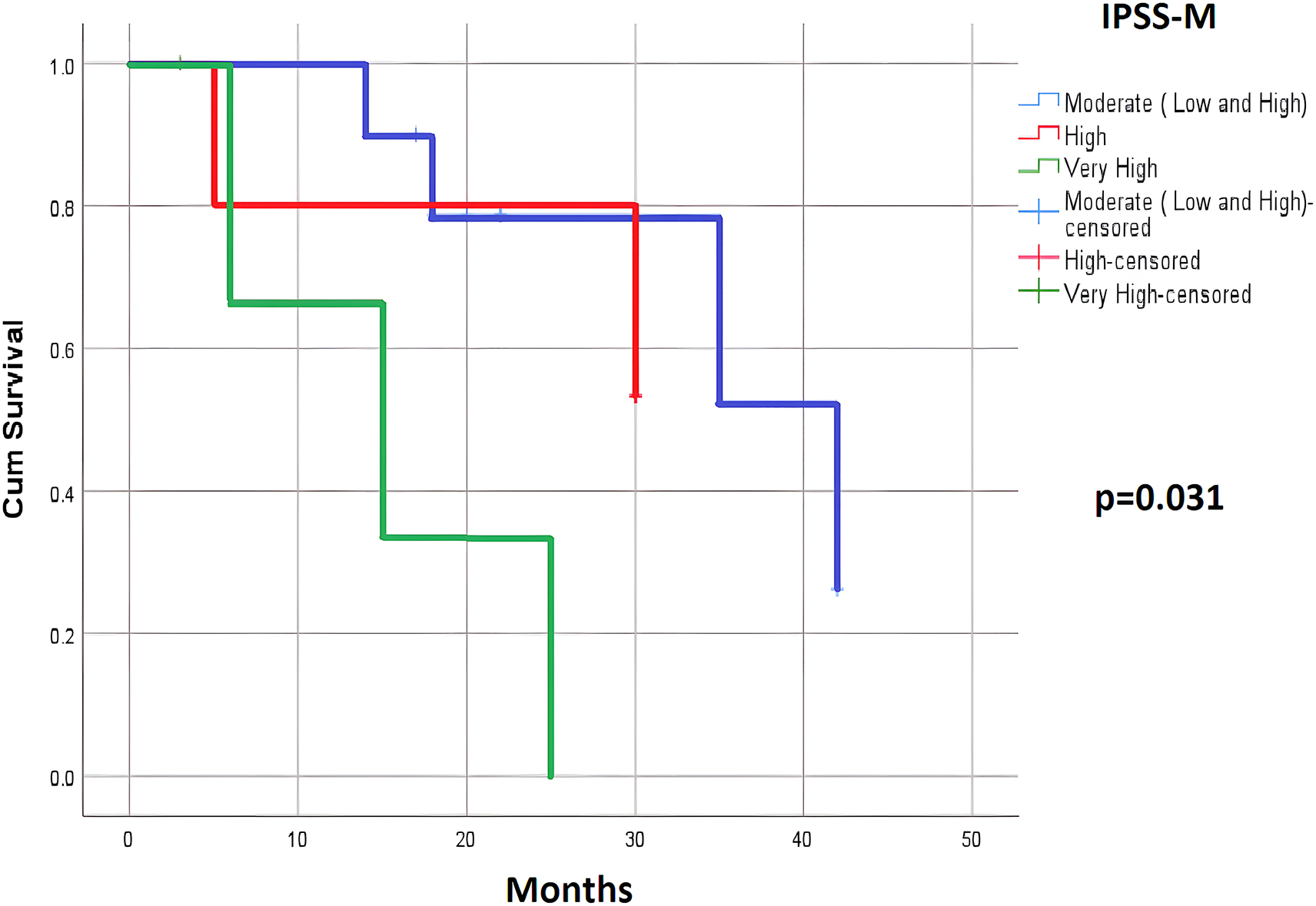
PFS according to the IPSS-M.
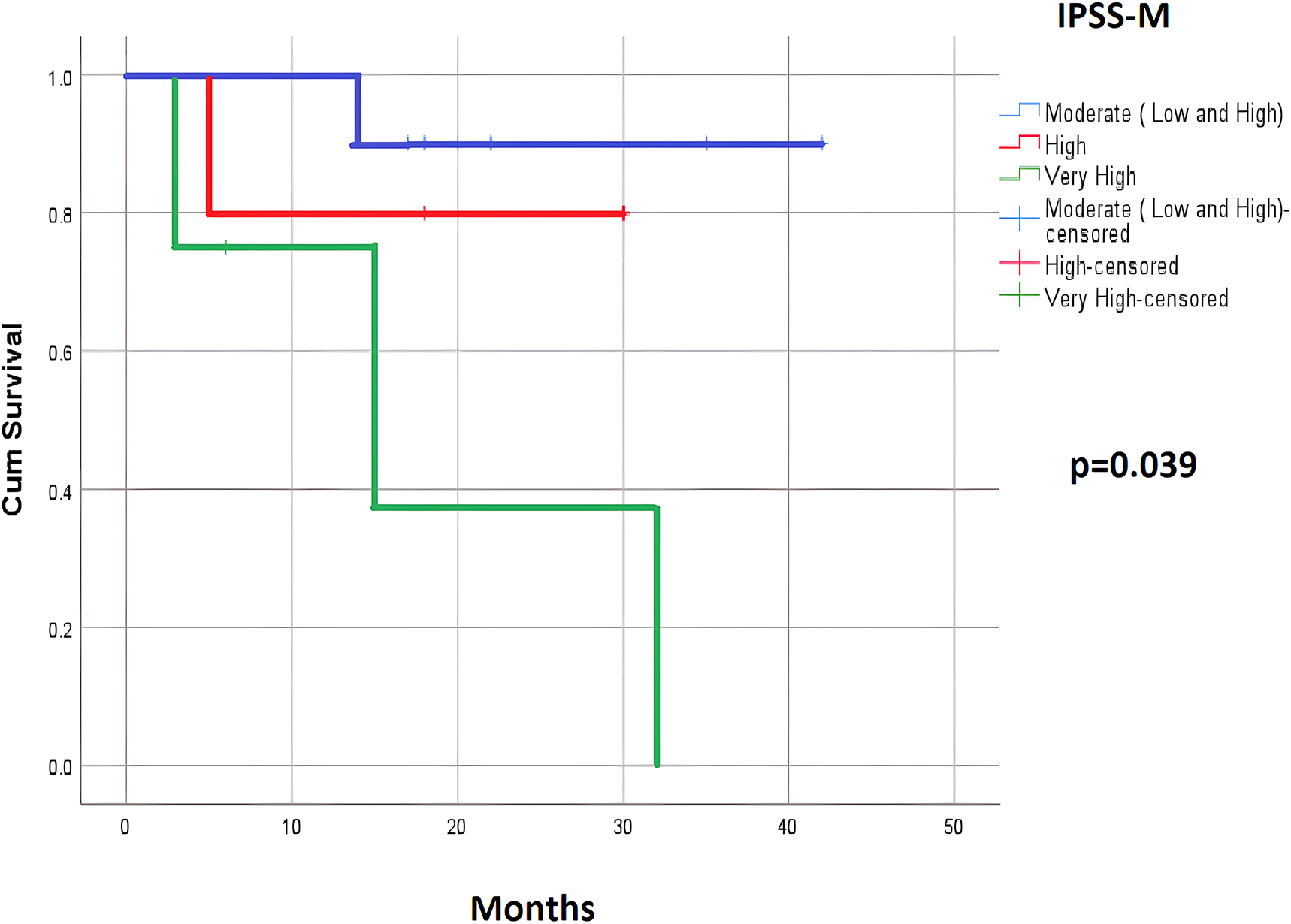
LFS according to the IPSS-M.
Discussion
Since its proposal in 2012, IPSS-R has become the primarily used prognostic system for MDS patients. It has also been used for therapeutic decision-making for MDS patients of different risk groups. For instance, the provision of supportive care and erythropoiesis-stimulating agents for low-risk patients (including very-low and low), HMA for high-risk patients (including high and very high), and varying treatments (erythropoiesis-stimulating agents or HMA) for intermediate-risk patients based on their status [16, 17, 19]. However, MDS pathogenesis is associated with genetic mutations [20], and with the development of genetic techniques, it is vital to add molecular data to the prognostic system. Some studies that supported this improvement [8], [9], [10], [11]. In addition, there are still some opinions that need further consideration. Since the IPSS-M online calculator can still assess risk even if data of some gene mutations are missing, it is not clear if it is essential to identify all 31 genes. Baer et al. suggested that TP53, FLT3 ITD , and KMT2A PTD (MLL PTD ) mutations are especially necessary for risk stratification in MDS patients [12]. However, in a study by Wu et al. KMT2A PTD (MLL PTD ) mutation was not detected in 852 MDS patients [13]. Therefore, in this study, we did not include KMT2A PTD (MLL PTD ), similar to the study by Zamanillo et al. [8]. Sauta et al. defined a minimum set of 15 essential genes for IPSS-M classification [14]. Therefore, the selection of genes to evaluate IPSS-M must be considered for their high efficiency and cost savings.
Wu et al. found that the IPSS-M was superior to the IPSS-R in the classification of MDS patients aged ≥60 years [13]. Wu et al. also suggested that the distinctive genetic factors of ethnic specificities affected the accuracy of IPSS-M risk categories [13]. Several studies showed that cytogenetic characteristics are associated with ethnicity [21], [22], [23], [24]. However, there are no studies on the frequency of myeloid gene mutations according to ethnic groups.
Sauta et al. suggested that IPSS-M improves MDS stratification and may be more effective in allotransplantation choice; however, IPSS-M could not effectively predict the prognosis of HMA-treated patients [14]. This may be attributed to the effect of HMA on some key genes in the DNA methylation group, such as DNMT3A and TET2. In contrast to the results reported by Sauta et al., our study found that compared to IPSS-R, IPSS-M could effectively predict the PFS and LFS of MDS patients treated with decitabine. In general, studies support the superiority of IPSS-M over IPSS-R; however, there are some inconsistencies between the target groups.
Some of the limitations of our study are that this was a small-scale study with a limited number of patients, and we presented an initial report of our analysis as a short communication. Therefore, further analysis is required on the IPSS-M based on the age, ethnicity, and treatment regimen of the patients, as well as the cost-effectiveness.
Conclusions
Our study found that compared to IPSS-R, IPSS-M could predict the PFS and LFS of decitabine-treated MDS patients more accurately, IPSS-M may be a better prognostic system than IPSS-R for predicting the prognosis of MDS patients.
Acknowledgments
The authors thank all technicians in Department of Molecular Cytogenetics, National Institute of Hematology and Blood Transfusion, for their efforts in supporting research.
-
Research ethics: The Review Board of the NIHBT approved the study (no. 939/QĐ-HHTM) and waived informed consent as it was a retrospective observational study.
-
Informed consent: The Review Board of the NIHBT waived informed consent as it was a retrospective observational study.
-
Author contributions: Minh Phuong Vu and Quang Hao Nguyen conceived the study. Minh Phuong Vu and Quang Hao Nguyen designed the study. Quang Hao Nguyen, Tuan Anh Tran, Quoc Chinh Duong and Duc Binh Vu participated in data collection and processing. Minh Phuong Vu, Quang Hao Nguyen, Tuan Anh Tran, Quoc Chinh Duong, Duc Binh Vu, Ha Thanh Nguyen and Quoc Khanh Bach participated in data analysis and interpretation, as well as in the literature search, wrote the manuscript. All authors have read and approved the final manuscript.
-
Competing interests: The authors declare that have no conflicts of interest.
-
Research funding: None declared.
-
Data availability: Data can be obtained from the corresponding author upon resonable request.
References
1. Zahid, MF, Malik, UA, Sohail, M, Hassan, IN, Ali, S, Shaukat, MHS. Cytogenetic abnormalities in myelodysplastic syndromes: an overview. Int J Hematol Oncol Stem Cell Res 2017;11:231–9.Search in Google Scholar
2. Khoury, JD, Solary, E, Abla, O, Akkari, Y, Alaggio, R, Apperley, JF, et al.. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia 2022;36:1703–19. https://doi.org/10.1038/s41375-022-01613-1.Search in Google Scholar PubMed PubMed Central
3. Zeng, X, Zhang, Y, Zhao, K, Zhou, L, Zhou, Y, Xuan, L, et al.. Somatic mutations predict prognosis in myelodysplastic syndrome patients with normal karyotypes. Signal Transduct Targeted Ther 2021;6:274. https://doi.org/10.1038/s41392-021-00606-3.Search in Google Scholar PubMed PubMed Central
4. Liu, M, Wang, F, Zhang, Y, Chen, X, Cao, P, Nie, D, et al.. Gene mutation spectrum of patients with myelodysplastic syndrome and progression to acute myeloid leukemia. Int J Hematol Oncol 2021;10:IJH34. https://doi.org/10.2217/ijh-2021-0002.Search in Google Scholar PubMed PubMed Central
5. Cook, MR, Karp, JE, Lai, C. The spectrum of genetic mutations in myelodysplastic syndrome: should we update prognostication? EJHaem 2021;3:301–13. https://doi.org/10.1002/jha2.317.Search in Google Scholar PubMed PubMed Central
6. Yu, J, Li, Y, Li, T, Li, Y, Xing, H, Sun, H, et al.. Gene mutational analysis by NGS and its clinical significance in patients with myelodysplastic syndrome and acute myeloid leukemia. Exp Hematol Oncol 2020;9:2. https://doi.org/10.1186/s40164-019-0158-5.Search in Google Scholar PubMed PubMed Central
7. Bernard, E, Tuechler, H, Greenberg Peter, L, Hasserjian Robert, P, Arango Ossa Juan, E, Nannya, Y, et al.. Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evid 2022;1:EVIDoa2200008. https://doi.org/10.1056/evidoa2200008.Search in Google Scholar PubMed
8. Zamanillo, I, Poza, M, Ayala, R, Rapado, I, Martinez-Lopez, J, Cedena, MT. Impact of IPSS-M implementation in real-life clinical practice. Front Oncol 2023;13:1199023. https://doi.org/10.3389/fonc.2023.1199023.Search in Google Scholar PubMed PubMed Central
9. Aguirre, LE, Al Ali, N, Sallman, DA, Ball, S, Jain, AG, Chan, O, et al.. Assessment and validation of the molecular international prognostic scoring system for myelodysplastic syndromes. Leukemia 2023;37:1530–9. https://doi.org/10.1038/s41375-023-01910-3.Search in Google Scholar PubMed
10. Maurya, N, Mohanty, P, Dhangar, S, Panchal, P, Jijina, F, Mathan, SLP, et al.. Comprehensive analysis of genetic factors predicting overall survival in Myelodysplastic syndromes. Sci Rep 2022;12:5925. https://doi.org/10.1038/s41598-022-09864-9.Search in Google Scholar PubMed PubMed Central
11. Sabile, JMG, Kaempf, A, Tomic, K, Manu, GP, Swords, R, Migdady, Y. A retrospective validation of the IPSS-M molecular score in primary and therapy-related myelodysplastic syndromes (MDS). Leuk Lymphoma 2023;64:1689–94. https://doi.org/10.1080/10428194.2023.2232491.Search in Google Scholar PubMed
12. Baer, C, Huber, S, Hutter, S, Meggendorfer, M, Nadarajah, N, Walter, W, et al.. Risk prediction in MDS: independent validation of the IPSS-M-ready for routine? Leukemia 2023;37:938–41. https://doi.org/10.1038/s41375-023-01831-1.Search in Google Scholar PubMed PubMed Central
13. Wu, J, Zhang, Y, Qin, T, Xu, Z, Qu, S, Pan, L, et al.. IPSS-M has greater survival predictive accuracy compared with IPSS-R in persons ≥60 years with myelodysplastic syndromes. Exp Hematol Oncol 2022;11:73. https://doi.org/10.1186/s40164-022-00328-4.Search in Google Scholar PubMed PubMed Central
14. Sauta, E, Robin, M, Bersanelli, M, Travaglino, E, Meggendorfer, M, Zhao, LP, et al.. Real-world validation of molecular international prognostic scoring system for myelodysplastic syndromes. J Clin Oncol 2023;41:2827–42. https://doi.org/10.1200/JCO.22.01784.Search in Google Scholar PubMed PubMed Central
15. Valent, P, Orazi, A, Steensma, DP, Ebert, BL, Haase, D, Malcovati, L, et al.. Proposed minimal diagnostic criteria for myelodysplastic syndromes (MDS) and potential pre-MDS conditions. Oncotarget 2017;8:73483–500. https://doi.org/10.18632/oncotarget.19008.Search in Google Scholar PubMed PubMed Central
16. Hong, M, He, G. The 2016 revision to the World Health Organization classification of myelodysplastic syndromes. J Transl Int Med 2017;5:139–43. https://doi.org/10.1515/jtim-2017-0002.Search in Google Scholar PubMed PubMed Central
17. Garcia-Manero, G. Myelodysplastic syndromes: 2023 update on diagnosis, risk-stratification, and management. Am J Hematol 2023;98:1307–25. https://doi.org/10.1002/ajh.26984.Search in Google Scholar PubMed
18. von Elm, E, Altman, DG, Egger, M, Pocock, SJ, Gøtzsche, PC, Vandenbroucke, JP, STROBE Initiative. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med 2007;147:573–7. https://doi.org/10.7326/0003-4819-147-8-200710160-00010.Search in Google Scholar PubMed
19. Chen-Liang, TH. Prognosis in myelodysplastic syndromes: the clinical challenge of genomic integration. J Clin Med 2021;10:2052. https://doi.org/10.3390/jcm10102052.Search in Google Scholar PubMed PubMed Central
20. Ogawa, S. Genetics of MDS. Blood 2019;133:1049–59. https://doi.org/10.1182/blood-2018-10-844621.Search in Google Scholar PubMed PubMed Central
21. Matsuda, A, Germing, U, Jinnai, I, Misumi, M, Kuendgen, A, Knipp, S, et al.. Difference in clinical features between Japanese and German patients with refractory anemia in myelodysplastic syndromes. Blood 2005;106:2633–40. https://doi.org/10.1182/blood-2005-01-0040.Search in Google Scholar PubMed
22. Elnahass, Y, Youssif, L. Cytogenetic features in primary myelodysplastic syndrome Egyptian patients. J Adv Res 2018;10:77–83. https://doi.org/10.1016/j.jare.2018.02.002.Search in Google Scholar PubMed PubMed Central
23. Li, L, Liu, XP, Nie, L, Yu, MH, Zhang, Y, Qin, TJ, et al.. Unique cytogenetic features of primary myelodysplastic syndromes in Chinese patients. Leuk Res 2009;33:1194–8. https://doi.org/10.1016/j.leukres.2008.11.021.Search in Google Scholar PubMed
24. Vu, MP, Ha, HQ, Nguyen, CN. Cytogenetic characteristics in Vietnamese patients diagnosed with primary myelodysplastic syndromes. Leuk Res Rep 2022;18:100343. https://doi.org/10.1016/j.lrr.2022.100343.Search in Google Scholar PubMed PubMed Central
© 2024 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Review Article
- Mesenchymal stem cell exosomes: a promising delivery system for glioma therapy
- Research Articles
- A multi-cancer analysis unveils ITGBL1 as a cancer prognostic molecule and a novel immunotherapy target
- MicroRNA-302a enhances 5-fluorouracil sensitivity in HepG2 cells by increasing AKT/ULK1-dependent autophagy-mediated apoptosis
- Integrated analysis of TCGA data identifies endoplasmic reticulum stress-related lncRNA signature in stomach adenocarcinoma
- Glioblastoma with PRMT5 gene upregulation is a key target for tumor cell regression
- BRCA mutation in Vietnamese prostate cancer patients: a mixed cross-sectional study and case series
- Microglia increase CEMIP expression and promote brain metastasis in breast cancer through the JAK2/STAT3 signaling pathway
- Integrative machine learning algorithms for developing a consensus RNA modification-based signature for guiding clinical decision-making in bladder cancer
- Transcriptome analysis of tertiary lymphoid structures (TLSs)-related genes reveals prognostic value and immunotherapeutic potential in cancer
- Prognostic value of a glycolysis and cholesterol synthesis related gene signature in osteosarcoma: implications for immune microenvironment and personalized treatment strategies
- Identification of potential biomarkers in follicular thyroid carcinoma: bioinformatics and immunohistochemical analyses
- Rapid Communication
- Comparison of the Molecular International Prognostic Scoring System (IPSS-M) and Revised International Prognostic Scoring System (IPSS-R) in predicting the prognosis of patients with myelodysplastic neoplasms treated with decitabine
- Case Reports
- Intrapericardial nonfunctional paraganglioma: a case report and literature review
- Treatment of central nervous system relapse in PLZF::RARA-positive acute promyelocytic leukemia by venetoclax combined with arubicin, cytarabine and intrathecal therapy: a case report
Articles in the same Issue
- Frontmatter
- Review Article
- Mesenchymal stem cell exosomes: a promising delivery system for glioma therapy
- Research Articles
- A multi-cancer analysis unveils ITGBL1 as a cancer prognostic molecule and a novel immunotherapy target
- MicroRNA-302a enhances 5-fluorouracil sensitivity in HepG2 cells by increasing AKT/ULK1-dependent autophagy-mediated apoptosis
- Integrated analysis of TCGA data identifies endoplasmic reticulum stress-related lncRNA signature in stomach adenocarcinoma
- Glioblastoma with PRMT5 gene upregulation is a key target for tumor cell regression
- BRCA mutation in Vietnamese prostate cancer patients: a mixed cross-sectional study and case series
- Microglia increase CEMIP expression and promote brain metastasis in breast cancer through the JAK2/STAT3 signaling pathway
- Integrative machine learning algorithms for developing a consensus RNA modification-based signature for guiding clinical decision-making in bladder cancer
- Transcriptome analysis of tertiary lymphoid structures (TLSs)-related genes reveals prognostic value and immunotherapeutic potential in cancer
- Prognostic value of a glycolysis and cholesterol synthesis related gene signature in osteosarcoma: implications for immune microenvironment and personalized treatment strategies
- Identification of potential biomarkers in follicular thyroid carcinoma: bioinformatics and immunohistochemical analyses
- Rapid Communication
- Comparison of the Molecular International Prognostic Scoring System (IPSS-M) and Revised International Prognostic Scoring System (IPSS-R) in predicting the prognosis of patients with myelodysplastic neoplasms treated with decitabine
- Case Reports
- Intrapericardial nonfunctional paraganglioma: a case report and literature review
- Treatment of central nervous system relapse in PLZF::RARA-positive acute promyelocytic leukemia by venetoclax combined with arubicin, cytarabine and intrathecal therapy: a case report

