Abstract
This study explores the efficacy and reliability of high-pressure freezing (HPF) as a sample preparation technique for electron microscopy (EM) analysis across a diverse range of biological samples. Utilizing the HPM Live µ technology, based on the historical hydraulic HPM010 from BalTec, we demonstrate the reliability of our industrial equipment to achieve the critical parameters necessary for vitrification (2076 bars, cooling rate above 2000 K/s). By directly measuring physical values within the HPF chamber, we evaluate the proper functioning of the equipment, contributing to the technique’s reliability. A meticulous approach was adopted for each sample type, acknowledging the uniqueness of each specimen, and associating the final sample analysis with its HPF curve, aiding in protocol optimization. Samples including human cell pellets, cell monolayer, mouse brain and liver biopsies, and Arabidopsis thaliana root and seedlings were processed for EM analysis following HPF. The ultrastructure of each sample type was rigorously examined, revealing homogeneous preservation and minimal ice nucleation artefacts. Overall, our work underscores the robustness and versatility of our HPM Live µ in preserving biological ultrastructure, offering valuable insights for researchers employing EM techniques in diverse biological studies.
1 Introduction
Vitrification through high pressure freezing (HPF) represents a pivotal technique for immobilizing biological samples, facilitating their analysis via electron microscopy (EM). By rapidly freezing specimens under high pressure, HPF capitalizes on the water phase diagram to prevent ice crystal formation, instead inducing the formation of amorphous ice – a glass-like structure [1–4]. Achieving optimal vitrification conditions necessitates reaching 2,076 bars, where water solidifies only at −22 °C [5], followed by rapid temperature decrease below −100 °C [6] at rates exceeding 2,000 K/s [7]. Under these conditions, water transitions into a metastable amorphous state [6], characterized by density comparable to its liquid form, thereby preserving the ultrastructure of surrounding biological material.
To attain these extreme physical conditions, we employ a liquid nitrogen flow pressurized at 2,100 bars, released within a mere 10 ms onto the sample. To safeguard delicate biological material during this process, the sample is positioned between two small metal carriers, typically composed of aluminium or gold-coated copper. These carriers transmit pressure to the hydrated sample while aiding heat extraction through their thermal conductive properties [4], [7], [8].
Following vitrification, samples can be prepared for subsequent analysis using electron microscopy techniques, notably freeze substitution (FS) and plastic-embedded electron microscopy or cryo-electron microscopy (cryo-EM). These techniques yield high-resolution images of the sample’s ultrastructure, which is vital for elucidating the function and behaviour of biological molecules and complexes [9].
HPF/FS offers several advantages over conventional chemical fixation and room temperature (RT) dehydration methods. Firstly, it preserves samples in a state closely resembling their native condition, enhancing the accuracy of structural and functional analyses. Secondly, it mitigates distortions that may arise during dehydration and embedding, common pitfalls in RT chemical fixation processes, that is particularly beneficial when studying wide sample areas. Nonetheless, HPF is acknowledged as a complex and expert technique, necessitating substantial optimization.
This article showcases the versatility of our newly developed high-pressure freezing machine, the HPM Live µ [4], across a diverse array of biological specimens commonly studied in research laboratories. On each sample type, we tested diverse cryo-protection and diverse freeze-substitution protocols to provide a common ground for researchers looking to test novel biological material in their own lab.
All samples in our study were vitrified on the HPM Live µ using standard preset parameters, aiming to reach 2,076 bars prior to decreasing the temperature below the vitrification point of −100 °C at cooling rates exceeding 2,000 K/s [2], [7], [10]. Samples were loaded either in aluminum carrier sandwiches to achieve a specimen cavity depth of 100–200 µm [7] with a radius of 2 or 5 mm, or cultured in CryoCapsules® [11], [12] (Table 1). We also explored the use of various cryoprotectants to minimize variability from the culture medium [13–15] (Table 1). Cryoprotection is a recurrent question associated with high pressure freezing protocols. Cryoprotectants are usually sugars, acting as water cages. They may be categorized in two forms: penetrating (short) and non-penetrating (long-chain-sugars). While it significantly protects the biological sample from ice damage during vitrification – by increasing the ice nucleation critical point barrier – it acts on the sample’s osmolarity. Their use must therefore be considered wisely and may require preliminary tests by the experimentalist to evaluate osmolarity impact on the sample’s physiology. Alternatively, cryo-filler like, hexadecene, may be used. These compounds aim at reducing the water content without affecting the sample’s osmolarity. We chose to focus only on non-penetrating long-chain-sugar cryo-protectants in this work.
HPF conditions. HPF and freeze substitution conditions for 20 independent experiments.
| Sample | Carrier | Depth | Cryoprotectant | Freeze substitution | |
|---|---|---|---|---|---|
| Cell pellet | A | 3 mm | 200 µm | Polyvinylpyrrolidone | 95 % Acetone, 5 % H2O, 1 % OsO4, 0.5 % glutaraldehyde, 0.2 % uranyl acetate |
| B | 3 mm | 200 µm | Polyvinylpyrrolidone | 95 % Acetone, 5 % H2O, 2.5 % OsO4, 0.5 % glutaraldehyde, 0.2 % uranyl acetate | |
| C | 3 mm | 100 µm | 12.5 % Dextran | 95 % Acetone, 5 % H2O, 1 % OsO4, 0.5 % glutaraldehyde, 0.2 % uranyl acetate | |
| Cell monolayer | D | CC | 100 µm | 8 % Dextran | 95 % Acetone, 5 % H2O, 1 % OsO4, 0.5 % glutaraldehyde, 0.2 % uranyl acetate |
| E | CC | 100 µm | 8 % Dextran | 95 % Acetone, 5 % H2O, 1 % OsO4, 0.5 % glutaraldehyde, 0.2 % uranyl acetate | |
| F | CC | 100 µm | 8 % Dextran | 95 % Acetone, 5 % H2O, 1 % OsO4, 0.5 % glutaraldehyde, 0.2 % uranyl acetate | |
| Cell monolayer (CLEM) | G | CC | 100 µm | Without cryoprotectant | 95 % Acetone, 5 % H2O, 0.5 % glutaraldehyde, 0.2 % uranyl acetate |
| H | CC | 100 µm | Without cryoprotectant | 95 % Acetone, 5 % H2O, 0.5 % glutaraldehyde, 0.2 % uranyl acetate | |
| I | CC | 100 µm | Without cryoprotectant | 95 % Acetone, 5 % H2O, 0.5 % glutaraldehyde, 0.2 % uranyl acetate | |
| Mouse brain | J | 6 mm | 100 µm | 20 % Dextran | 95 % Acetone, 5 % H2O, 1 % OsO4, 0.5 % glutaraldehyde, 0.2 % uranyl acetate |
| K | 6 mm | 100 µm | 20 % Dextran | 95 % acetone, 5 % H2O, 1 % OsO4, 0.5 % glutaraldehyde, 0.2 % uranyl acetate | |
| L | 6 mm | 100 µm | 20 % Dextran | 95 % acetone, 5 % H2O, 1 % OsO4, 0.5 % glutaraldehyde, 0.2 % uranyl acetate | |
| M | 6 mm | 100 µm | 20 % Dextran | 95 % Acetone, 5 % H2O, 1 % OsO4, 0.5 % glutaraldehyde, 0.2 % uranyl acetate | |
| Mouse liver | N | 6 mm | 100 µm | 20 % Dextran | 95 % Acetone, 5 % H2O, 0.5 % glutaraldehyde, 0.1 % uranyl acetate |
| O | 6 mm | 100 µm | 20 % Dextran | 95 % Acetone, 5 % H2O, 0.5 % glutaraldehyde, 0.1 % uranyl acetate | |
| P | 6 mm | 100 µm | 20 % Dextran | 95 % acetone, 5 % H2O, 1 % OsO4, 0.5 % glutaraldehyde, 0.1 % uranyl acetate | |
| A. thaliana root | Q | 3 mm | 200 µm | 7 mM Tris pH 6.6, 140 mM Sucrose, 7 mM Trehalose | 95 % Acetone, 5 % H2O, 2.5 % OsO4, 0.5 % glutaraldehyde, 0.2 % uranyl acetate |
| R | 3 mm | 200 µm | 7 mM Tris pH 6.6, 140 mM Sucrose, 7 mM Trehalose | 95 % Acetone, 5 % H2O, 2.5 % OsO4, 0.5 % glutaraldehyde, 0.2 % uranyl acetate | |
| A. thaliana seedling | S | 6 mm | 200 µm | 7 mM Tris pH 6.6, 140 mM Sucrose, 7 mM Trehalose | 95 % Acetone, 5 % H2O, 1 % OsO4, 0.5 % glutaraldehyde, 0.2 % uranyl acetate |
| T | 6 mm | 200 µm | 7 mM Tris pH 6.6, 140 mM Sucrose, 7 mM Trehalose | 95 % Acetone, 5 % H2O, 0.5 % glutaraldehyde, 0.2 % uranyl acetate |
Freeze substitution was conducted using standard cocktails, often used in labs as primary trial cocktail when exploring a novel sample using osmium tetroxide or uranyl acetate as a sole contrasting agent. Embedding was performed in EPON resin, widely adopted in the field [16–19]. Unstained sections of 70 nm were subsequently imaged by TEM.
We present this manuscript as a foundational reference for biological exploration, allowing scientists to utilize our protocol with confidence, knowing it can yield satisfactory preliminary results. Our images may serve as a reference point for project-specific optimization, advancing the collective understanding and application of HPF techniques in biological research.
2 Materials and methods
High pressure freezing is a reliable method, long established in electron microscopy biology laboratories. However, to ensure the highest reliability in sample preparation, a couple of precautions must be taken [4], [20]. Our routine workflow is presented in Figure 1. We generally prepare the experiment the day before, we ensure that all material is available and ready to use (Day 0, Figure 1). We recapitulate a list of points we apply when testing a novel sample:
A rule of thumb is that a sample that keeps a natural living state vitrifies better and is worth the effort (avoid starvation if not the topic of research, feed the cells with fresh medium in the morning of the experiment if possible).
All samples were vitrified as fresh as possible to limit degradation prior to HPF (limit necrosis and fixation artefacts).
Work quickly. All samples were prepared and vitrified in the shortest possible time, from their culture media to the aluminium carrier. Less than a minute is ideal.
Avoid mechanical damages by over-filling your carrier assembly.
Never dry the sample out.
No air bubbles should be left in the sample to avoid pressure plateau reduction during vitrification.
Use the shallowest combination of carriers to optimize your vitrification success rate.
No carrier coating was applied to limit variability among experiments.
The carrier sandwich is composed of an A carrier, with a selected cavity of 100 or 200 µm depth. Once the sample is deposited into it, it is enclosed using the flat side of a cover B carrier. The B carrier may be prewetted on a filter paper impregnated with the appropriate cryoprotectant. This prewetting step reduces the formation of air bubbles during cavity closure.
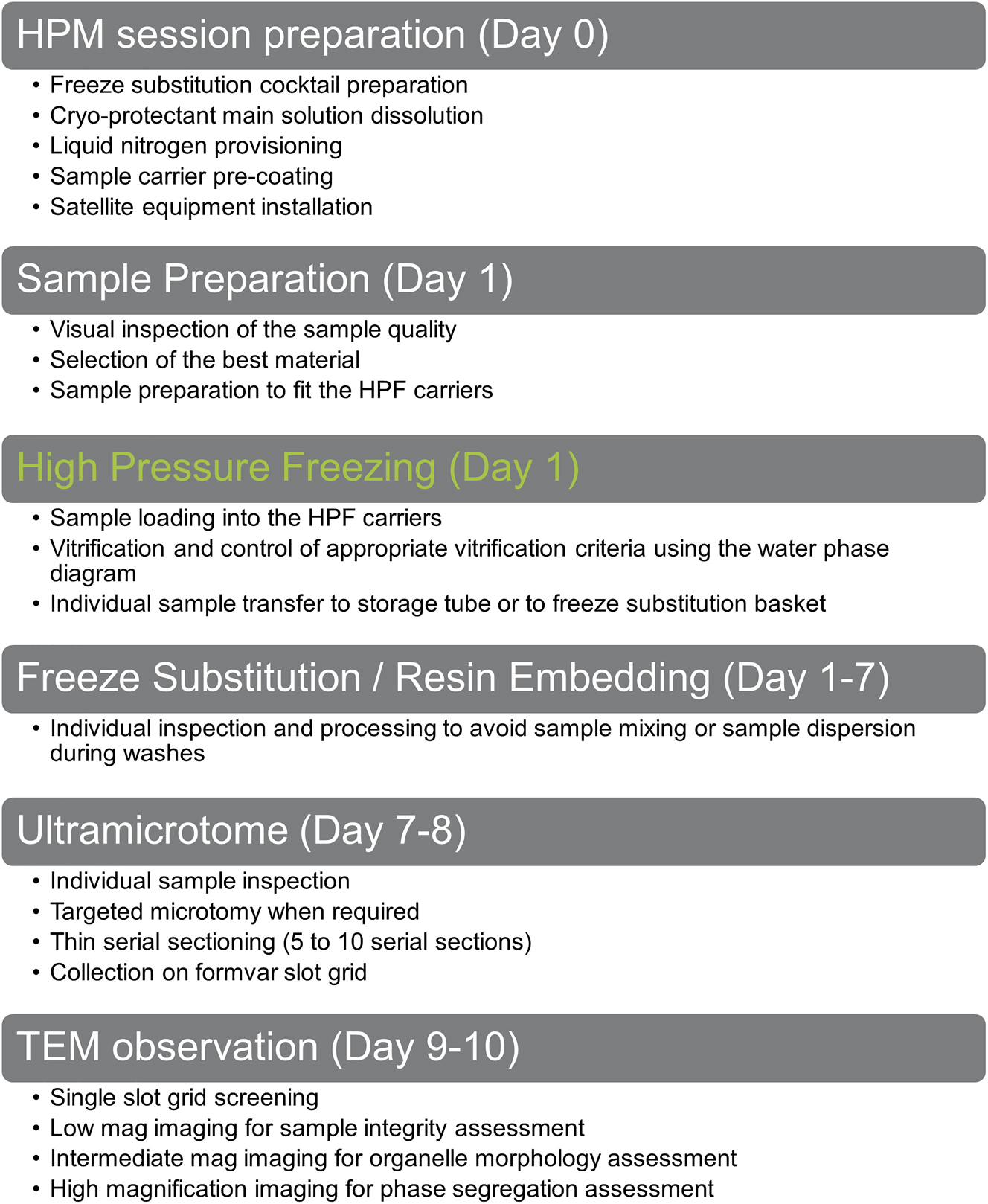
Global workflow to assess sample vitrification versatility of our HPM Live µ over 6 different samples.
More extensive and graphical illustrations of various sample preparations are presented in detail in a dedicated book chapter [4].
2.1 More specifically regarding the sample presented in this work
2.1.1 Monolayer cell into a pellet
Neuroblastoma cells, type SHSY-5Y, were cultured in complete medium (1x DMEM + GlutaMAX, 20 % fetal bovine serum, G418) at 37° up to 70 % confluence. Treated by Trypsin, resuspended in complete medium and pelleted by centrifugation. The cell pellets were resuspended in cryoprotectant (Table 1) and then pipetted in an A carrier (3 mm) of 100 µm or 200 µm depth (Table 1). The carrier was closed by a B carrier flat side to create a 100 µm or 200 µm deep cavity prior to loading into the HPM clamp for vitrification.
2.1.2 Monolayer cell cultured onto a cryocapsule
Neuroblastoma cells, type SHSY-5Y, were cultured in complete medium (1× DMEM + GlutaMAX, 20 % fetal bovine serum, G418) at 37° up to 40 % confluence into a CryoCapsule [11]. On the day of vitrification, the CryoCapsule was closed using a covering sapphire disc, and loaded into the specific CryoCapsule clamp [12]. The cells involved in a correlative light and electron microscopy (CLEM) project were imaged at 5 or 20× prior to vitrification in the absence of cryo-protectant (Figure 4, line 0). Cells not involved in a CLEM project were exposed to 8 % dextran, loaded as previously described and vitrified.
2.1.3 Mouse biopsy (brain, and liver)
F8-deficient mice obtained from Kazazian et al. [21] were backcrossed (>10 times) on a C57Bl/6 background. Housing and experiments were performed following French regulations and the experimental guidelines of the European Community. Mice were deeply anaesthetized and subjected to intracardiac perfusion with 4 % paraformaldehyde in Phosphate Buffered Saline (PBS) (Gibco™ PBS, pH 7.4, Fisher Scientific, Hampton, New Hampshire, USA), prior to biopsy to ensure proper fixation of the tissues. Prefixation offers several advantages: it slows down sample degradation and enhances tissue rigidity, facilitating subsequent slicing steps using an oscillating knife (vibratome VTS 2000, Leica Microsystems). Tissues or organs were maintained at 4 °C between biopsy and thinning. All manipulations, from thinning to vitrification, were conducted at room temperature.
Large organs were reduced to blocks no more than 5 mm in height and affixed onto the vibratome tray from their larger face using cyanoacrylate adhesive (super glue (XGLU super forte, Roldan, Toulouse, France). The excess adhesive was carefully removed using filter paper, and the sample was submerged in PBS. It is notable that super glue exhibits strong reactivity to PBS, and any excess may result in the generation of contaminating fibres in the bath, although this observation is not directly presented in our data.
The amplitude and frequency of the razor blade in the vibratome were empirically adjusted until a smooth, gently floating 100 µm slice of tissue was obtained (approximately 1.5 mm/s at 50 Hz, namely speed 7 and frequency 5 on our Vibratome VTS 2000). These parameters were adjusted by eye until a satisfying slice was obtained. We recommend the user to test on a dummy sample and find out the most appropriate parameters according to his own sample. The floating slice was carefully moved to a clean area of the PBS bath using a fine paintbrush. A biopsy puncher (Miltex® Biopsy Punch w/Plunger, 4.0 mm, Blue, Electron Microscopy Sciences, Hatfield, Pennsylvania, USA) was then used to extract a 4 mm disc from the slice, which was lifted out of the PBS bath with the paintbrush and deposited into the A carrier. A drop of cryoprotectant was added (Table 1, following comments about cryoprotectant refer to Table 1), and excess solution was removed with filter paper until a tiny concave meniscus remained. Subsequently, the sample was encapsulated by a flat side of the B carrier, which had been pre-wetted with the cryoprotectant solution to minimize air bubble encapsulation. The assembly was then loaded into the HPM clamp and subjected to vitrification. A short video is available on our website: https://www.cryocapcell.com/tips-and-tricks-for-em and on YouTube https://www.youtube.com/shorts/xz17Wy_l_64.
2.1.4 Arabidopsis thaliana (seedlings and roots)
Three to five-day-old Arabidopsis thaliana (Col-0) seedlings cultured on Murashige and Skoog medium [22] plates under room temperature with continuous light were utilized in this study. All manipulations were conducted at room temperature. The seedlings were gently removed from the culture plate using tweezers and carefully deposited onto a clean glass slide. To prevent desiccation, several drops of cryoprotectant solution (7 mM Tris pH 6.6, 140 mM Sucrose, 7 mM Trehalose) were added to cover the plants.
Subsequently, the seedling, or the root tip excised using a microscalpel (Microscalpel plastic P-715, Feather safety razor, Osaka, Japan) was transferred to an A carrier (refer to Table 1). Another drop of cryoprotectant was added to the carrier to ensure proper protection of the sample. The carrier was then sealed using a flat side of the B carrier, which had been pre-wetted with cryoprotectant, and subjected to vitrification.
2.1.5 High-pressure freezing
Following the aforementioned preparations, each sample was subjected to vitrification using our HPM Live µ® apparatus (CryoCapCell, Paris, France). Prior to the initiation of the high-pressure liquid nitrogen flow, a brief pre-injection of ethanol lasting 75 (Sample J, K, L, and M) or 150 ms (all other samples) was administered. This pre-injection serves to delay cooling while the system reaches the desired pressure. Throughout the process, temperature and pressure were directly measured inside the HPF chamber using sensors positioned above and below the specimen (Temperature sensor: TS, Pressure sensor: PS, in Supplementary Figure 1). Notably, no data manipulation or calculation was applied to the collected data, which is presented in its raw form within this manuscript. This unprocessed data serves to illustrate the repeatability of our apparatus and the precise trajectory of each sample across the water phase diagram.
Consistency and standardization were upheld by systematically applying the same HPF parameters to all samples, thereby minimizing variability in our protocol. This approach ensures robust and reproducible results across the experimental cohort.
2.1.6 Freeze substitution and embedding
Following vitrification, the samples underwent freeze substitution using an FS-8500 RMC (Boeckeler Instruments, Tucson, Arizona) in a solution mix detailed in Table 1. The freeze substitution process followed a temperature ramp program as outlined below:
For the standard freeze substitution protocol:
18 h at −90 °C
15 h from −90° to −60 °C (with a temperature increase of 2 °C per hour)
8 h at −60 °C
15 h from −60° to −30 °C (with a temperature increase of 2 °C per hour).
Upon completion of the program, the samples were kept on ice for 1 h, followed by three washes in acetone. Subsequently, the samples were embedded in EPON 812 resin (EmBed-812 Embedding Kit, Electron Microscopy Sciences, Hatfield, Pennsylvania, USA) through a series of steps below.
Preparation of EPON 812: Mix1 (2 mL Embed-812, 3.1 mL Dodecenyl Succinic Anhydride) and Mix2 (2 mL Embed-812, 1.7 mL methyl-5-norbornene-2, 3-dicarboxylic anhydride) were mixed just before the use and added 170 µL DMP-30. We used this solution as 100 % resin.
Dehydration and embedding step: 2 h each at 30, 60, and 100 % resin concentration in acetone. At 100 % was repeated twice. The embedding process concluded with 48 h of polymerization at 45 °C. 60 °C is recommended for normal polymerisation. In our case, an oven fault limited the temperature to 45 °C. For this reason, we extended the polymerisation time, after which no impact of the lower temperature on sample preservation was observed.
2.1.7 Ultra microtome
70 nm–80 nm thin sections were made on an Ultracut S (Leica) with either an ultra 45° or an oscillating diamond knife 35° (ultra-sonic, Diatome, Switzerland) then collected on homemade formvar coated mesh grids, or formvar coated slot grids. The use of either diamond knife was dictated by availability on the sectioning day, and in this project, does not affect the observations.
2.1.8 Transmission electron microscope imaging
TEM imaging was performed with a Tecnai 12 transmission electron microscope, 120 kV (FEI) equipped with a CCD camera (OneView 4Kx4K Gatan) controlled by GMS software.
2.1.9 Individual sample datasheet
All presented material is individually registered into F°Low [23], our project management software, and their unique sample preparation protocol is synthesized into a 2-page PDF for easy reproduction of the protocol presented in this article. All 20 files are presented as supplementary material (Supplementary Material).
3 Results
3.1 Twenty independent HPFs with optimised conditions
High pressure freezing (HPF) is often perceived as a sophisticated technique, subject to variability influenced by factors such as sample type, equipment, and operator proficiency. However, we assert that HPF is inherently reliable, with the initial mastery of biological sample preparation (culture, biopsy, loading) being the pivotal aspect achievable within a few trials. Leveraging our HPM technology, derived from the established hydraulic HPM010 technology from BalTec, we consistently applied critical parameters to achieve vitrification. Furthermore, our direct measurement of physical values inside the HPF chamber serves as a valuable tool for assessing equipment functionality.
Recognizing the labour-intensive nature of obtaining electron microscopy (EM) images following vitrification, we refrained from pooling samples randomly and instead treated each sample as unique. Associating TEM analysis with its corresponding HPF curve proves invaluable when establishing protocols for novel sample types. When observing a degraded sample while a good HPF curve is associated gives a clear insight towards the need to optimize: either the sample preparation (see first paragraph of ‘material and method’ section’), or the post-vitrification handling (avoid warming during all transfers, to avoid re-crystalization).
To validate this reliability, we rigorously tested a diverse array of samples, all prepared on the same apparatus by the same team of experimentalists. Our sample selection encompassed what we identified as established model systems in the field, aiming to propose reference protocols for their analysis: human cell cultures, mouse biopsies (including brain, and liver tissues) and A. thaliana (both root and seedling specimens). Our comprehensive procedure is delineated in Figure 1, with vitrified samples undergoing freeze substitution, resin embedding in EPON, thin sectioning, and subsequent TEM observation.
During vitrification sessions, meticulous attention was given to each sample’s journey across the water phase diagram, serving as a reliable indicator of HPF efficacy. Any samples showing signs of inadequate vitrification were promptly discarded, and the vitrification was repeated, given the importance of this initial step in the process. We favour the water phase diagram display over conventional temperature and pressure crossing points, as it provides more informative insights and avoids potential inaccuracies due to uncalibrated scales. The various criteria we base our evaluation on are: pressure rise time (over 200 bar/ms), pressure when crossing the Liquid to Solid phase transition line (STL, above 1800 bars), the cooling rate between the crossing of STL and the first order transition line (TLL (above 2000 K/s), the critical pressure maintenance time (time spent below the TLL line (above 250 ms) (Supplementary Figure 2). These criteria are extensively described in the book chapter on the HPM Live µ [4] and we would refer the reader to this manuscript for a deeper understanding of our analysis.
Following this rationale, we conducted at least two independent HPF sessions with valid HPF curves for each sample type. In total, we present data from 20 independent experiments covering six distinct sample types (including cell pellets, tissues, and plants), showcasing the capability of our HPM Live µ to effectively vitrify a wide range of biological materials within the physical depth limitation of 200 µm [7].
The criteria we used to evaluate the vitrification quality were based on the ice nucleation pattern. Ice nucleation is a random phenomenon, occurring at the cost of energy to pass the phase transition energy barrier [24], [25]. Once the barrier is breached, it is more energy efficient to continue growing the ice crystal by adding surrounding water molecules than nucleating a novel ice crystal [26]. A great summary of this phenomenon is described here [27]. Following this logic, free floating molecules in the culture medium will be the most prone to nucleate (least energy to escape the liquid phase) and aggregate to grow a larger crystal. Hence, ice crystals have the tendency to grow until they meet with neighbouring ice crystals and connect. To grow, an ice crystal captures the surrounding water molecule. These molecules escape from the surrounding medium from the least bound to the most [26]. The presence of cryo-protectants (sugars) or biological material (proteins, sugars, lipids) increases the energy barrier and therefore reduces ice nucleation and slows down ice growth [28], [29]. Since the ice grows by capturing the surrounding molecules, the most obvious ice damage to observe in electron microscopy already at low magnification is an empty region surrounded by dehydrated material [26]. The same pattern may be observed at a finer scale while increasing magnification. We used this understanding of ice behaviour to define our analysis strategy:
Low magnification observation. The sample should look homogeneous, with even contrast and smooth gradients across the whole tissue. The culture medium contains less proteins than the sample, and stains less with the chemicals used during freeze-substitution. A darker region indicates the presence of biological material.
Intermediate magnification: membranes should appear regular and smooth; the nucleus looks smooth with an even gradient of the chromatin.
High magnification: microtubules or membranes are continuous, without brutal interruption (caused by an ice needle interrupting the continuity), and chromatin looks homogenous.
3.2 Vitrification of human cell pellet
Three SHSY-5Y cell pellets (Table 1, Sample A, B, and C) were prepared and subjected to high pressure freezing (HPF). The resulting HPF curves met the criteria for further processing for electron microscopy (Figure 2, line 1). To analyse our sample, we observed the biological material and the surroundings. Observation of the surrounding culture medium mixed with PVP (sample A and B) displays globally poor preservation and segregation patterns are observed (Figure 2, A-2, and 3, B-2, and 3, asterisks), while the culture medium mixed with dextran (sample C) shows no sign of ice nucleation (Figure 2, C-2, and 3 arrowheads). In all three samples, spaces between the cells and the surrounding material are observed, suggesting a separation due to shrinkage. Notably, examination of the cell’s ultrastructure revealed well-preserved samples in all three conditions, without major ice nucleation causing large segregation (Figure 2, line 2). Sample B presents higher contrast at the mitochondria (Figure 2, A-3, B-3) and at membranes (Figure 2, A-4, B-4) compared to sample A. This is likely related to the increased Osmium tetroxide concentration (Table 1). Interestingly, a comparison of sample A and C shows a higher contrast of mitochondria for C, while a lower staining of the membranes in general. At intermediate magnification, organelles such as the Golgi apparatus, endosomes and mitochondria display regular morphology in all 3 samples (Figure 2, line 3). At higher magnification, continuous and sharp membranes were observed across all samples and no segregation pattern appeared in the cytoplasm (Figure 2, line 4).
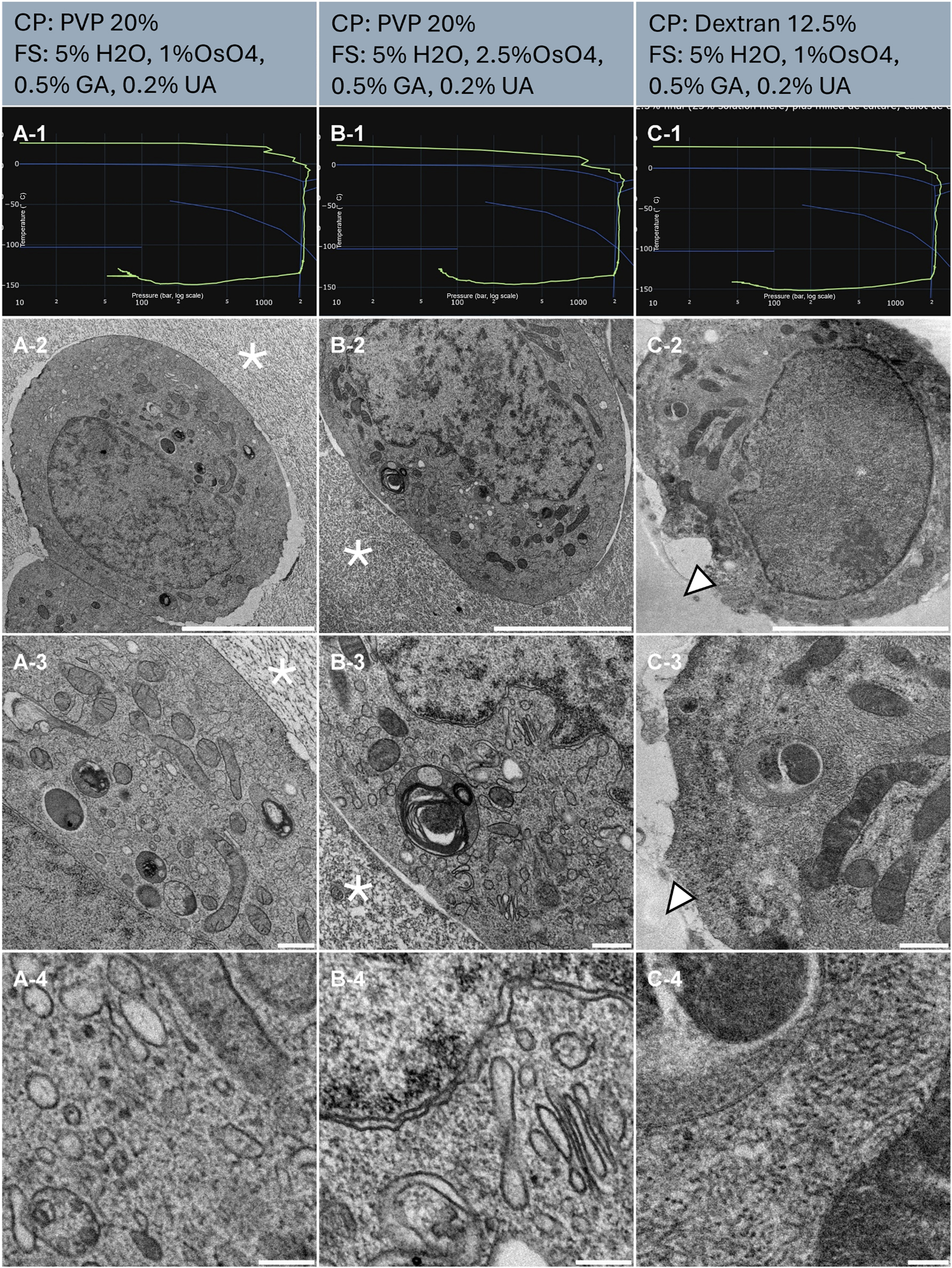
Morphological observation of 3 samples (column A, B and C) of human cell pellet, concentrated by centrifugation and resuspended in cryo-protectants. The top of the lines presents a cryo-protectant (CP) and a freeze substitution (FS) cocktail for each sample. Line 1 presents each individual HPF curve. X axis: pressure (bar, log scale), Y axis: temperature (°C), green line: experimental curve (pressure/temperature). Blue lines draw a water phase diagram in the region of interest. Line 2 presents an overview of the samples, scale bar 5 µm. Line 3 presents general organelle preservation and distribution, scale bar 1 µm. Line 4 presents a higher magnification of a detailed view, scale bar 0.2 µm. Asterisks represent ice crystals. Arrowheads point to the dextran surrounding the cell without damage.
3.3 Vitrification of cell monolayer on CryoCapsule
SHSY-5Y cells were cultured onto CryoCapsule (Table 1, Samples D, E and F), exposed to 8 % dextran as a cryo-protectant and high pressure frozen. The resulting HPF curves of sample E and F met the criteria for further processing for electron microscopy (Figure 3, E-1, F-1). An overview examination of the cell culture revealed E and F to be homogenously well-preserved samples (Figure 3, E-2, F-2, arrowheads).
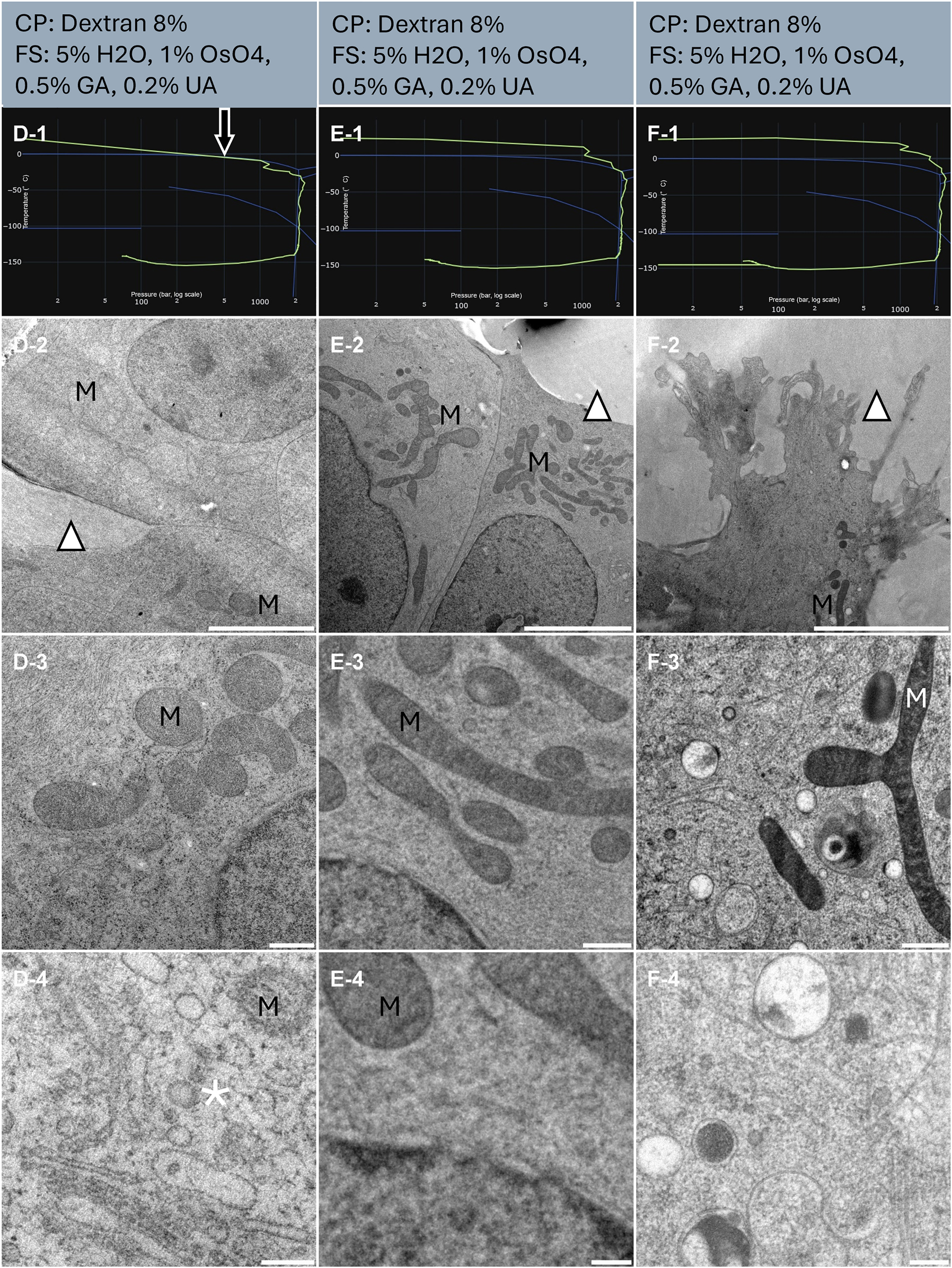
Morphological observation of 3 samples (column D, E and F) of human cell monolayer cultured onto CryoCapsule. The top of the lines presents a cryo-protectant (CP) and a freeze substitution (FS) cocktail for each sample. Line 1 presents each individual HPF curve. X axis: pressure (bar, log scale), Y axis: temperature (°C), green line: experimental curve (pressure/temperature). Blue lines draw a water phase diagram in the region of interest. Line 2 presents an overview of the samples, scale bar 5 µm. Line 3 presents general organelle preservation and distribution, scale bar 0.5 µm. Line 4 presents a higher magnification of a detailed view, scale bar 0.2 µm. The arrow on the HPF curves points to the early entry into the solidification phase. Asterisks represent ice crystals. Arrowheads point towards the dextran that displays smooth preservation. M localize to the mitochondria.
Sample D presents an HPF curve entering the solidification area 7 ms before the critical 2,000 bars are achieved (Figure 3, D-1, arrow). In experimental conditions, this sample would be discarded, but to explore the impact of this HPF curve, it was further processed by freeze-substitution. All freeze substitutions were comparable, containing osmium tetroxide, and the contrast at the membranes is clearly delineated in all conditions.
At low magnification, in all three samples, the culture medium, mixed with dextran, displays no evidence of ice nucleation. The cells are homogenous, without large void areas or acute contrast of large ice segregation pattern (Figure 3, line 2) and all samples look adequately preserved. Of note, the nucleus and the mitochondria (M) appear lighter in sample D, compared to E and F (Figure 3, D-2 to 4, E-2 to 4, F-2, and 3).
At intermediate magnification, it is noticeable that mitochondria appear light (Figure 3, D-3) or dark (Figure 3, E-3, F-3).
At higher magnification, microcrystalline ice damages are clearly visible in sample D, while sample E and F display good morphological preservation (Figure 3, D-4, E-4, and F-4).
Sample E and F display a difference in contrast despite the same freeze substitution cocktail (Figure 3, column middle and right). These samples were freeze-substituted in separate experiments and our apparatus for freeze substitution being manual, we suspect a time difference between reaching 4 °C and the first acetone rinses.
3.4 Vitrification of cell monolayer on CryoCapsule for CLEM analysis
SHSY-5Y cells were cultured onto CryoCapsule (Table 1, Sample G, H and I) to conduct live-CLEM experiments. To preserve sample preparation as close as possible to native, we did not add cryo-protectants to the sample prior to vitrification. The resulting HPF curves met the criteria for further processing for electron microscopy (Figure 4, G-1, H-1, and I-1). In this experiment, we avoided osmium tetroxide in the freeze-substitution cocktail to avoid turning the whole sample black and facilitate sample retrieval at the ultramicrotome. As a consequence, membranes are not visible and only proteins inserted into them reveal their presence through the uranyl acetate stain (Figure 4, G-2 to 4, H-2 to 4, and I-2 to 4). Cytoskeleton and proteins however are clearly observable. No discontinuity in the cytoskeleton nor protein alignment along the membrane is observed, highlighting proper ultrastructure preservation of the sample (Figure 4, G-3, and 4, H-3, and 4, and I-3, and 4). Interestingly, this lack of contrast at the membranes is specific to Embed812 epoxy resin and is occurring systematically in our hands. A similar freeze-substitution cocktail followed by methacrylate resin embedding produces nicely stained membranes. This is illustrated as a supplementary figure in the other article published in this journal edition: “Universal FS Basket: a simple device to increase freeze-substitution and electron microscopy embedding reliability”.
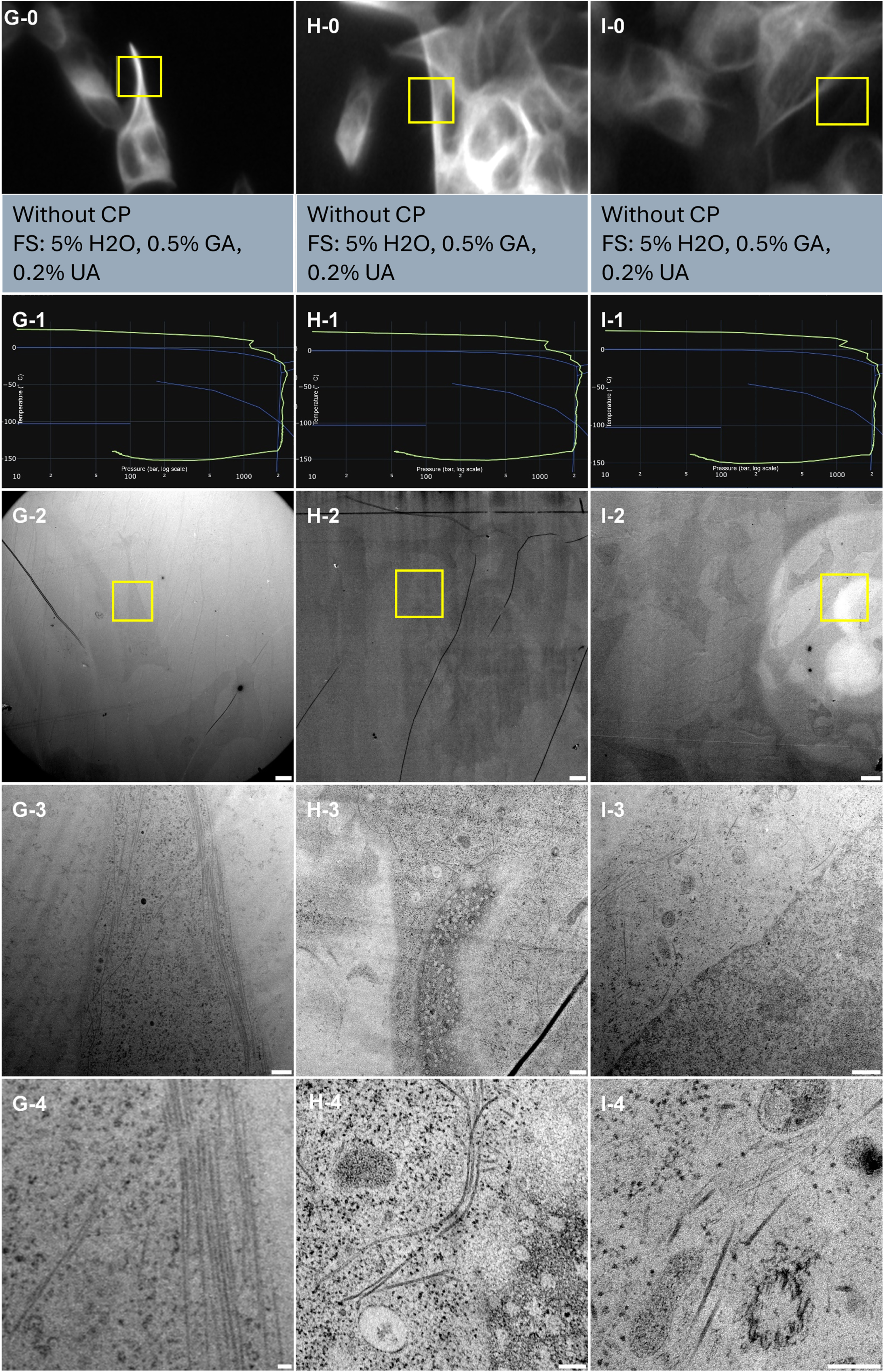
Correlative light and electron microscopy analysis of 3 samples (column G, H and I) of cell monolayer cultured onto CryoCapsule. Line 0 presents each ROI observed live on the HPM Live µ with a 20× magnification prior to HPF, scale bars 20 µm. Yellow boxes in Line 0 and Line 2 indicate the same ROI. The information between Line 0 and Line 1 presents a cryo-protectant (CP) and a freeze substitution (FS) cocktail for each sample. Line 1 presents each individual HPF curve. X axis: pressure (bar, log scale), Y axis: temperature (°C), green line: experimental curve (pressure/temperature). Blue lines draw a water phase diagram in the region of interest. Line 2 presents an overview of the samples, scale bar 20 µm. Line 3 presents general organelle preservation and distribution, scale bar 2 µm. Line 4 presents a higher magnification of microtubules, endosomes (G, H, I), mitochondria (H, I), nuclear pore complexes top view (H) and microtubule organizing centre (I), scale bar 0.5 µm.
From the CLEM perspective, we could locate all the cells observed live, back in the electron microscope (Figure 4, yellow squares), demonstrating the high reliability and reproducibility of the CryoCapsule for live-CLEM approached on cell monolayers.
3.5 Vitrification of mouse brain biopsy
Four punches from perfused brain slice 100 µm thick (Table 1, Samples J, K, L and M) were randomly selected for further vitrification. The resulting HPF curves of sample J met the criteria for further processing to electron microscopy (Figure 5, J-1). Sample K crosses the solidification phase above 1200 bars and reaches 2000 bars 4 ms later (Figure 5, K-1). The overview examination of the ultrastructure revealed homogeneous preservation across both tissues with regular spacing between the myelinated neurons (Figure 5, J-2, K-2). At intermediate magnification, organelles display a smooth and regular morphology (Figure 5, J-3, K-3). It is interesting to note that the myelinated neurons do not appear densely packed as usually observed in the volume EM dataset prepared by the OTO protocol [30]. This results from the gentler dehydration process obtained using freeze-substitution [31]. At higher magnification, individual myelin sheaths, dense vesicular regions, and mitochondrial cristae with double leaflet membranes are clearly discernible (Figure 5, J-4, K-4).
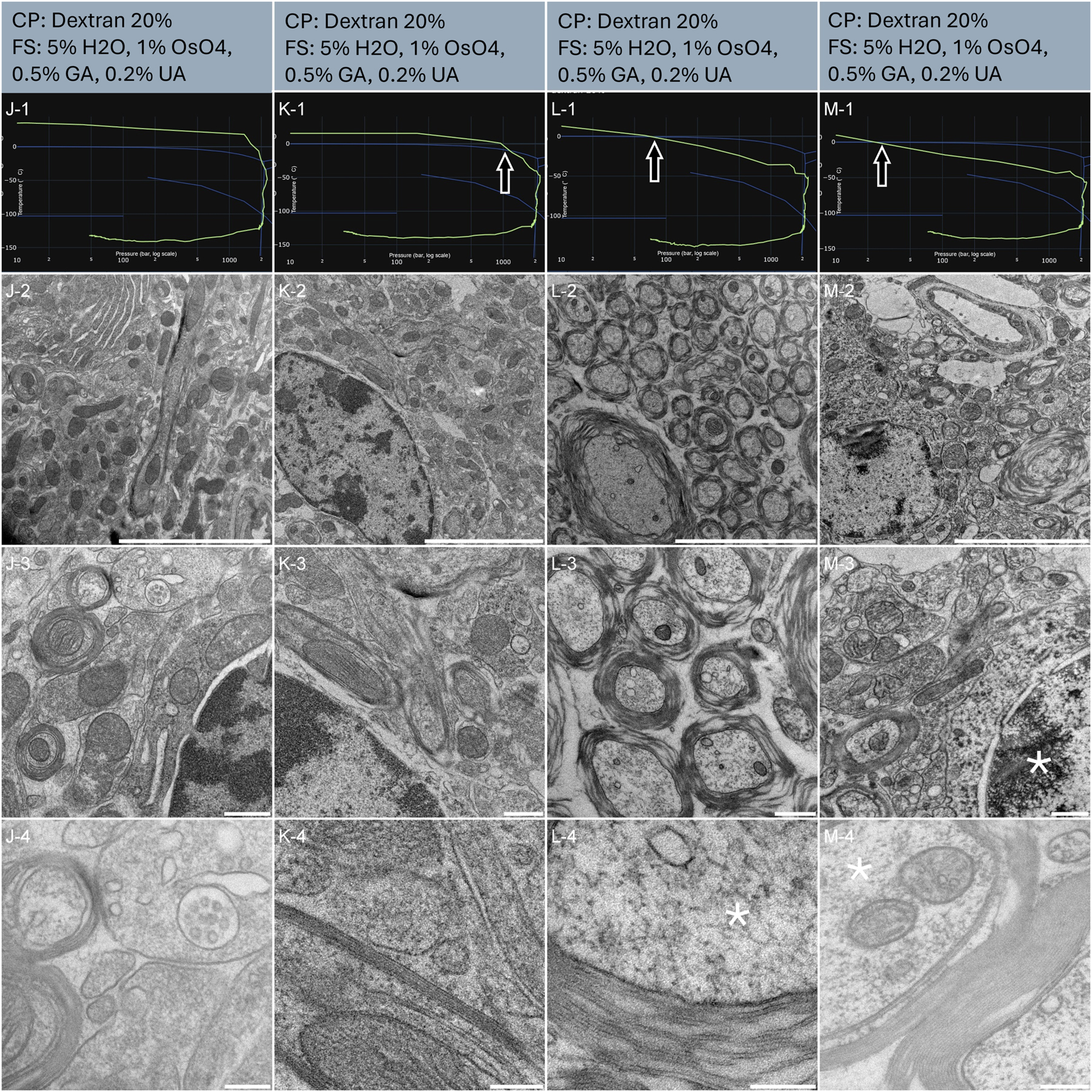
Morphological observation of 4 samples (column J, K, L and M) of mouse brain biopsies, sliced using vibratome. The top of the lines presents a cryo-protectant (CP) and a freeze substitution (FS) cocktail for each sample. Line 1 presents each individual HPF curve. X axis: pressure (bar, log scale), Y axis: temperature (°C), green line: experimental curve (pressure/temperature). Blue lines draw a water phase diagram in the region of interest. Line 2 presents an overview of the samples, scale bar 5 µm. Line 3 presents general organelle preservation and distribution, scale bar 0.5 µm. Line 4 presents a higher magnification of a detailed view with membranes, myelin sheath, neuronal vesicles and cytoplasm, scale bar 0.2 µm. Arrows on the HPF curves present problematic points of the curves. Asterisks represent ice crystals.
Sample L and M had bad HPF curves (Figure 5, L-1, M-1): the samples entered the solidification area 10 and 8 ms respectively before the critical 2000 bars were reached. We further processed these samples out of curiosity. Both samples display a degraded ultrastructure, already visible at low magnification (Figure 5, L-2, M-2). Acute contrast gradients are visible in the myelin sheath (Figure 5, L-2) and the nucleus displays chromatin segregation (Figure 5, M-2). At intermediate magnification, the myelin sheath appears discontinuous (Figure 5, L-3) and the crystalline structure of the chromatin appears strongly defined (Figure 5, M-3). At high magnification, segregation artefacts are visible in the cytoplasm of the myelinated neurons (Figure 5, L-4, and M-4). Of note, no large ice crystal damages are observed at low magnification. Importantly, the preservation of tissue L and M presents a very high heterogeneity. Some regions initially observed displayed some good preservation (data not shown). Further sectioning and observation revealed damaged structures. Proximity to the carrier, or better local infiltration of dextran might explain this discrepancy.
3.6 Vitrification of mouse liver biopsy
Liver slices of 100 µm thick were randomly selected and punched into three samples (Table 1 sample N, O and P). The resulting HPF curves met the criteria for further processing to electron microscopy (Figure 6, line 1). An overview assessment of the ultrastructure revealed homogenous preservation across all tissues (Figure 6, line 2). It is worth noting that the cytoplasm of the cells presenting in the tissue has a fairly different organization from other more commonly studied tissues. Large areas of the cytoplasm seem void at low magnification (Figure 6, line 3, white circles). This morphology is coherent with other work, describing these areas as ‘amorphous’ [32].
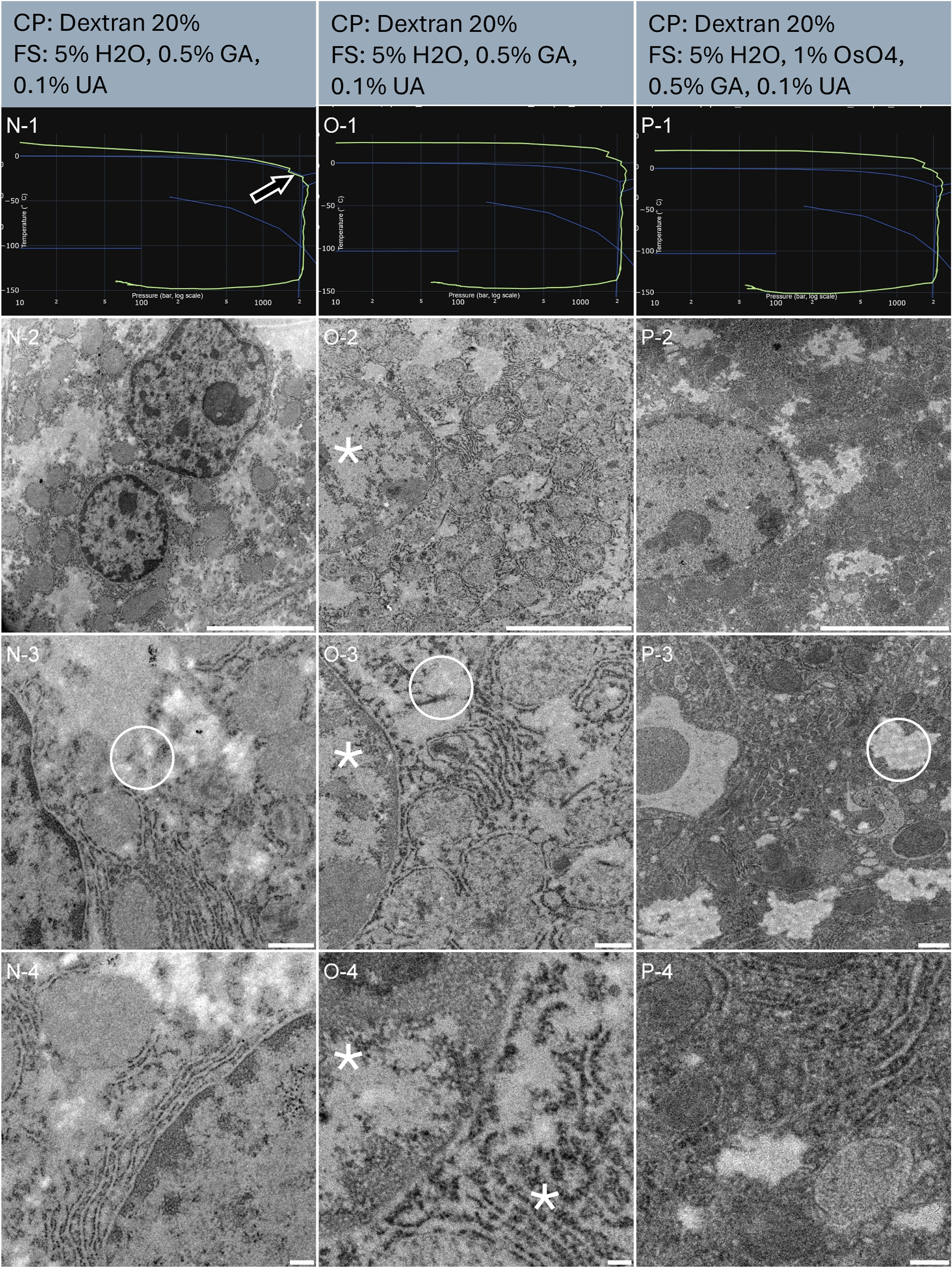
Morphological observation of 3 samples (column N, O, and P) of mouse liver biopsies, sliced using an oscillating knife ultramicrotome. The top of the lines presents a cryo-protectant (CP) and a freeze substitution (FS) cocktail for each sample. Line 1 presents each individual HPF curve. X axis: pressure (bar, log scale), Y axis: temperature (°C), green line: experimental curve (pressure/temperature). Blue lines draw a water phase diagram in the region of interest. Line 2 presents an overview of the samples, scale bar 5 µm. Line 3 presents general organelle preservation and distribution, scale bar 0.5 µm. Line 4 presents a higher magnification of a detailed view, scale bar 0.2 µm. The arrow on the HPF curve points to the entry into the solidification phase, here at the expected pressure. Asterisks represent ice crystals. Circles are the cytoplasm void areas, containing electron-lucent vesicles, as seen in chemical fixation protocols.
Sample N’s HPF curve was at the edge of the crystallization line, but remained in the liquid phase until proper pressure was attained (Figure 6, N-1). At all magnifications, sample N displays comparable morphological preservation to sample P (Figure 6, N-2 to 4). Thereby confirming that the HPF parameters were met to achieve good ultrastructure preservation without ice nucleation.
Sample N and O were freeze substituted without osmium tetroxide. As a result, the membranes are less prominently marked or appear lighter at higher magnification (Figure 6, N-4, and O-4). Sample P was exposed to osmium tetroxide during freeze-substitution, resulting in darkly stained membranes (Figure 6, P-4).
At low magnification, no obvious ice damages are visible on any of the three samples. The void areas of the cytoplasm have been identified also in chemically fixed material. Yet, we notice in a side by side comparison of all three samples that the nucleus in sample O is less prominently visible and displays a similar morphology to the surrounding material, in opposition to sample N and P (Figure 6, line 2).
At intermediate magnification the void cytoplasm displays circular void regions, that would be at first interpreted as ice damage, but in fact, are identical in chemically fixed material. The nucleus in O shows some ice damage (Figure 6, line 3, asterisk in O-3).
At high magnification, sample O clearly displays ice damage (Figure 6, asterisks in O-4). N and P have nicely preserved ultrastructure (Figure 6, line 4).
This comparison illustrates the benefit of associating each sample with its HPF curve. Out of 3 samples, sample O has a great HPF curve and yet displays degraded ultrastructure preservation at high magnification. Considering that the other two samples are well preserved, we can argue that an air bubble was trapped in the carrier prior to HPF, or that some devitrification occurred post HPF.
3.7 Vitrification of A. thaliana root
In plant science, the root tip is one structure largely studied (Table 1, sample Q and R). The resulting HPF curves met the criteria for further processing to electron microscopy (Figure 7, line 1). Sample Q displays the typical plant root tip structure with organized cell arrangement (Figure 7, Q-2). For sample R, the orientation of the tissue prevented us from getting oriented with certainty. However, the imaged structure is also expected to be near the root tip (Figure 7, R-2).
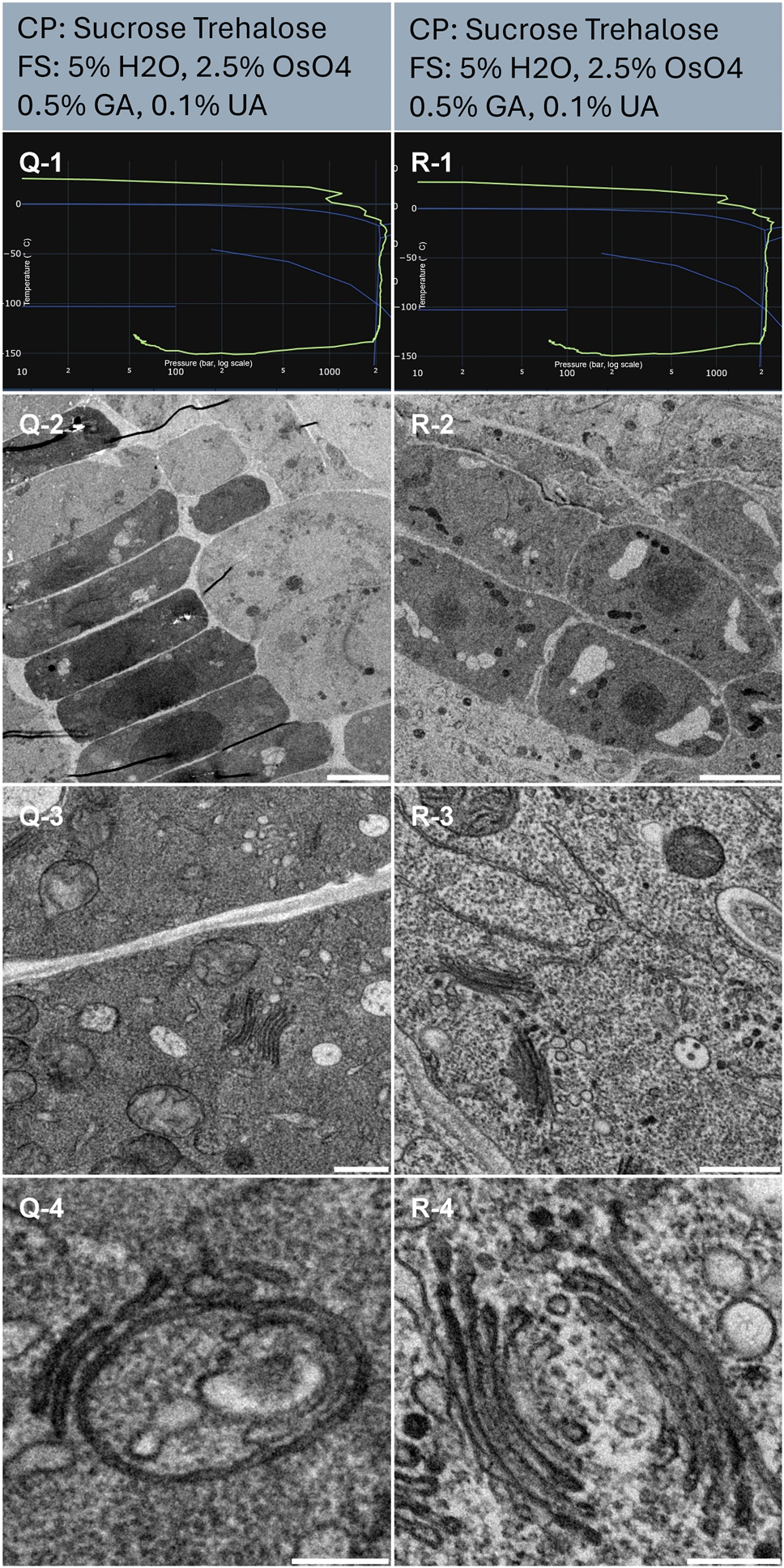
Morphological observation of 2 samples (column Q and R) of Arabidopsis thaliana root. The top of the lines presents a cryo-protectant (CP) and a freeze substitution (FS) cocktail for each sample. Line 1 presents each individual HPF curve. X axis: pressure (bar, log scale), Y axis: temperature (°C), green line: experimental curve (pressure/temperature). Blue lines draw a water phase diagram in the region of interest. Line 2 presents an overview of the samples, scale bar 10 µm. Line 3 presents general organelle preservation and distribution, scale bar 1 µm. Line 4 presents a higher magnification of a detailed view of membranes from Golgi apparatus and endosomes, scale bar 0.2 µm.
From the global view, the ultrastructure of both samples (Figure 7, Q-2, and R-2) is heterogenous with very nicely preserved structures in smaller cells near the tip (Figure 7), and more degraded structures in elongated cells with large vacuoles (data not shown). Focusing on the nicely preserved tip, the ultrastructure is well preserved, and no ice nucleation artifacts are visible (Figure 7, Q-2, and R-2). At intermediate magnification, the organelles appear well preserved (Figure 7, Q-3, and R-3).
At high magnification, no ice damage is visible (Figure 7, Q-4, and R-4).
3.8 Vitrification of A. thaliana seedling
Plant leaves are notably challenging to vitrify [33]. The presence of large vacuoles and air spaces prevents the proper building of the pressure up to 2,100 bars which is necessary to achieve fast glass transition and limit ice nucleation. In this figure, we present two samples (Table 1, sample S and T) that were well preserved. The resulting HPF curves met the criteria for further processing to electron microscopy (Figure 8, line 1). We observe from the large perspective (Figure 8, line 2), some membrane ruptures and sample fractures. This is likely caused by mechanical rupture around the vacuoles during vitrification. However, closer inspection (Figure 8, line 3) displays nicely preserved organelles typical of this plant tissue, the chloroplast (Figure 8, T-3). At higher magnifications, trafficking vesicles and other membrane structures were very smooth and distinguishable (Figure 8, S-3, and 4).
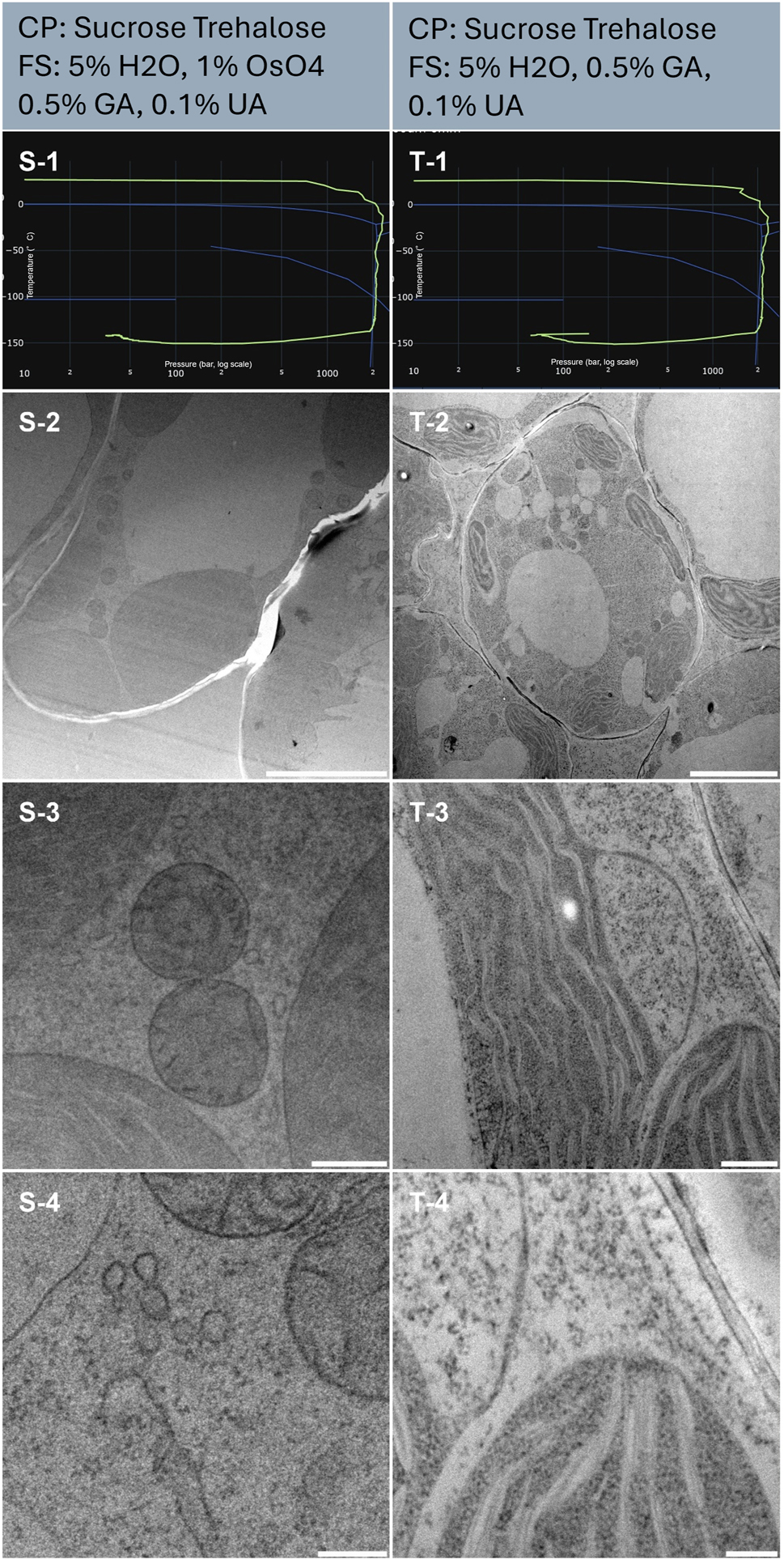
Morphological observation of 2 samples (column S and T) of Arabidopsis thaliana seedling. The top of the lines presents a cryo-protectant (CP) and a freeze substitution (FS) cocktail for each sample. Line 1 presents each individual HPF curve. X axis: pressure (bar, log scale), Y axis: temperature (°C), green line: experimental curve (pressure/temperature). Blue lines draw a water phase diagram in the region of interest. Line 2 presents an overview of the samples, scale bar 5 µm. Line 3 presents general organelle preservation and distribution, scale bar 0.5 µm. Line 4 presents a higher magnification of a detailed view of mitochondria, endosome (S-4) and thylakoid, cytoplasm (T-4), scale bar 0.2 µm.
Sample T was freeze-substituted in the absence of osmium tetroxide, however, the general contrast of the sample is rather strong (Figure 8, column right). At higher magnification, we note that the membranes are not clearly marked but rather the protein dense structure of the chloroplast stands out (Figure 8, T-3, and 4). Comparatively, the membranes of sample S, freeze-substituted in the presence of osmium display clear membranes (Figure 8, S-3, and 4).
4 Discussion
In the realm of biological specimen preservation, high pressure freezing (HPF) stands out as the exclusive method capable of vitrifying specimens ranging from 5 to 200 µm in thickness. Since its introduction in laboratories in the late 1980s [34–36], HPF has recently garnered renewed interest from the scientific community, particularly with the emergence of cryo-electron microscopy spurred by the resolution revolution [37] and the quiet revolution of volume electron microscopy [38]. Both fields will inevitably need to shift towards good frozen hydrated material to preserve and observe ‘life-like state’ of biological structures, and High-Pressure Freezing monitoring will play a key role in stablishing efficient workflow.
The HPM Live µ discussed in this article represents a significant advancement from the established HPM010 by BalTec, renowned as the pioneering and most dependable apparatus in the market. To validate the reliability of this newer iteration, we conducted extensive testing on a diverse array of biological specimens, including isolated cells, tissues, and plants. Our evaluation encompassed sample preservation quality at varying scales, from multiple cells to individual organelles, employing freeze substitution and room temperature imaging techniques.
By leveraging freeze substitution and room temperature imaging, we were able to comprehensively assess sample preservation homogeneity across different locations, up to millimetres apart on a formvar slot grid. High-magnification observations enabled the identification of microcrystalline ice-induced damages such as membrane rupture or protein segregation.
This sample preparation and imaging strategy facilitated the exploration of a wide range of samples, irrespective of prior expertise. By establishing comparable assessment criteria and employing widely used protocols, we aim to provide facility laboratories with a foundational framework for handling novel samples. Our detailed protocol serves as a reference point, enabling researchers to gauge the adequacy of their own sample preparations. Nonetheless, we acknowledge that sample preparation may require further refinement, with post-processing techniques like freeze-substitution tailored to specific research objectives, such as membrane analysis or cytoskeletal studies, drawing upon existing literature knowledge [16], [18], [39–42].
Plant material is notably challenging to cryo-preserve due to the presence of large vacuoles and air gaps where gas exchange occurs. These characteristics necessitate the use of cryo-protectants or fillers prior to high-pressure freezing (HPF). In this study, we opted to vitrify samples submerged in cryo-protectant in the simplest manner possible. However, it is advisable to further prepare plant samples by degassing in a vacuum bell for a few minutes to empty the airspaces before loading them into carriers. This step optimizes the pressure build-up on the sample during vitrification, enhancing preservation quality [33].
While cryo-protection is ideally avoided, it can be beneficial for novel or challenging samples. In pre-fixed samples, cryo-protection causes minimal osmotic shock and helps preserve sample ultrastructure homogeneity. The method presented here might be sufficient depending on the scientific question and if the experiment can accommodate selective observation.
Our selection of EPON embedding resin for all samples was primarily based on its widespread use and established reputation for embedding various biological specimens with consistent quality [43–45]. However, it is important to note that EPON resin tends to reduce the contrast generated by the heavy metal salts present in the freeze substitution cocktail, often necessitating post-staining procedures for enhanced visualization. In our study, we deliberately avoided post-staining to minimize variability and showcase the true effectiveness of each sample preparation protocol.
An evident consequence of this choice was observed in samples that did not undergo osmium tetroxide exposure during freeze substitution, resulting in notably weak contrast, especially in membrane structures, in which uranyl acetate alone at low concentration (0.2 % or less) could not adequately compensate for. Osmium tetroxide, while effective for contrast enhancement, poses significant challenges in terms of health and safety regulations, as well as disposal requirements. Additionally, it can cause the entire sample pellet to turn black, hindering the identification of specific samples for targeted microtomy.
Addressing this technical challenge presents complexities, as alternatives such as soft X-ray apparatus, while capable of mitigating osmium tetroxide-related issues, are not universally available and entail substantial costs, making them impractical for routine screening protocols like the one presented in our study. To circumvent these challenges, we propose the consideration of more electron-transparent resins, such as HM20 Lowicryl® [46] (EMS, USA), or contrast-enhancing electro-conductive resins like R221® [47] (CryoCapCell, France), which could offer improved visibility of cellular structures without compromising sample integrity. A comparative example of this is presented as Supplementary Material in a second article presented in this journal edition: “Universal FS Basket: a simple device to increase freeze-substitution and electron microscopy embedding reliability”.
During the two-year duration of this work, we thoroughly analysed various aspects of the high-pressure freezing (HPF) protocol, including pre-fixation, cryoprotection, HPF curve evaluation, freeze substitution cocktail impact, and the influence of resin on contrast (though not presented in this manuscript). Meeting the expected vitrification criteria, namely reaching 2000 bars before cooling and achieving a temperature decrease rate of over 2000 K/s, is crucial. We deliberately examined samples that did not meet these criteria out of scientific curiosity, as exemplified by samples D, L and M. As expected, these samples turned out to display globally a bad ultrastructure preservation.
5 Conclusion and outlook
In summary, our study demonstrates the preservation of high-quality ultrastructure across a broad range of samples, meeting the requirements of most facilities. The sample preparation parameters proposed here are likely applicable to all high-pressure freezing apparatuses. By ensuring that samples undergo the appropriate journey across the water phase diagram, researchers can use this manuscript as a guide to establish internal HPF protocols.
Funding source: CryoCapCell
Acknowledgments
We thank Yvette Akwa (INSERM UMR 1195) and Kevin Guillemeau (INSERM UMR 1195) for neuroblastoma cells, Christian Specht (INSERM UMR 1195) for mouse brain tissue, Thibaud Sefiane (INSERM UMR 1176), Marie Clavel (INSERM UMR 1176), and Peter Lenting (INSERM UMR 1176) for mouse liver tissue. Cindy Degerny (INSERM UMR 1195) and Marcel Tawk (INSERM UMR 1195) for Zebrafish, Claire Boulogne (Plateforme Imagerie-Gif Institut de Biologie Intégrative de la Cellule, UMR 9198) for A. thaliana, RMC Boeckeler Instrument, and the Francis Crick institute for the kind lending of the RMC FS8500 freeze substitution apparatus. We acknowledge the ImagoSeine core facility of the Institut Jacques Monod, member of the France BioImaging infrastructure (ANR-10-INBS-04) and GIS-IBiSA” for the use of TEM.
-
Research ethics: F8-deficient mice obtained from Kazazian et al. were backcrossed (>10 times) on a C57Bl/6 background. Housing and experiments were performed following French regulations and the experimental guidelines of the European Community. All animal experiments were conducted with approved protocols at Inserm by DDPP Val de Marne, France, under license number F 94-043-013.
-
Author contributions: C. K., M. B. and X. H. have designed the research. C. K. and X. H. performed the experiments and wrote the manuscript. Y. B., F. E., J. H., M. B., and X. H. contributed to the development and regulation of the HPM Live µ. All the authors read and approved the manuscript.
-
Competing interests: The authors are the manufacturers and distributors of the HPM Live µ, used in this manuscript.
-
Research funding: This research program was internally supported by CryoCapCell.
-
Data availability: All data presented in this manuscript are stored and registered following the FAIR principles. Each sample presented in the manuscript figure is related to its own sample datasheet generated with the F°Low database (CryoCapCell property). The datasheet is available to facilitate protocol implementation in readers laboratories.
References
[1] A. Kaech and U. Ziegler, “High-pressure freezing: current state and future prospects,” Methods Mol. Biol., vol. 1117, pp. 151–171, 2014, https://doi.org/10.1007/978-1-62703-776-1_8.Search in Google Scholar PubMed
[2] D. Studer, M. Michel, M. Wohlwend, E. B. Hunziker, and M. D. Buschmann, “Vitrification of articular cartilage by high-pressure freezing,” J. Microsc., vol. 179, no. Pt 3, pp. 321–332, 1995. https://doi.org/10.1111/j.1365-2818.1995.tb03648.x.Search in Google Scholar PubMed
[3] D. Studer, M. Michel, and M. Müller, “High pressure freezing comes of age,” Scanning Microsc. Suppl., vol. 3, pp. 253–268, 1989, discussion 268–9. http://www.ncbi.nlm.nih.gov/pubmed/2694271 Search in Google Scholar
[4] X Heiligenstein, et al.., “HPM Live μ for a Full CLEM Workflow,” Method. Cell Biol., vol. 162, pp. 115–149, 2021. https://doi.org/10.1016/bs.mcb.2020.10.022.Search in Google Scholar PubMed
[5] M. Chaplin, “Water structure and science.” Available: https://water.lsbu.ac.uk/water/water_phase_diagram.html.Search in Google Scholar
[6] C. A. Tulk, J. J. Molaison, A. R. Makhluf, C. E. Manning, and D. D. Klug, “Absence of amorphous forms when ice is compressed at low temperature,” Nature, vol. 569, no. 7757, pp. 542–545, 2019, https://doi.org/10.1038/s41586-019-1204-5.Search in Google Scholar PubMed
[7] E. Shimoni and M. Müller, “On optimizing high-pressure freezing: from heat transfer theory to a new microbiopsy device,” J. Microsc., vol. 192, no. Pt 3, pp. 236–247, 1998. https://doi.org/10.1046/j.1365-2818.1998.00389.x.Search in Google Scholar PubMed
[8] E. Hunziker, W. Herrmann, R. Schenk, M. Mueller, and H. Moor, “Cartilage ultrastructure after high pressure freezing, freeze substitution, and low temperature embedding. I. Chondrocyte ultrastructure – implications for the theories of mineralization and vascular invasion,” J. Cell Biol., vol. 98, no. 1, pp. 267–276, 1984, https://doi.org/10.1083/JCB.98.1.267.Search in Google Scholar PubMed PubMed Central
[9] S. S. Biel, K. Kawaschinski, K.-P. Wittern, U. Hintze, and R. Wepf, “From tissue to cellular ultrastructure: closing the gap between micro- and nanostructural imaging,” J. Microsc., vol. 212, no. Pt 1, pp. 91–99, 2003, https://doi.org/10.1046/j.1365-2818.2003.01227.x.Search in Google Scholar PubMed
[10] P. Walther, C. Buser, M. Hagedorn, and M. Wohlwend, “A new compact high pressure freezing device,” Micron, vol. 33, no. 1994, 2003.Search in Google Scholar
[11] X. Heiligenstein, et al.., “The CryoCapsule: simplifying correlative light to electron microscopy,” Traffic, vol. 15, no. 6, pp. 700–716, 2014, https://doi.org/10.1111/tra.12164.Search in Google Scholar PubMed PubMed Central
[12] X. Heiligenstein, I. Hurbain, C. Delevoye, J. Salamero, C. Antony, and G. Raposo, “Step by step manipulation of the cryocapsule with HPM high pressure freezers,” Methods Cell Biol., vol. 124, no. 5478, pp. 259–274, 2014, https://doi.org/10.1016/B978-0-12-801075-4.00012-4.Search in Google Scholar PubMed
[13] F. Franks, M. H. Asquith, C. C. Hammond, H. B. Skaer, and P. Echlin, “Polymer cryoprotectants in the preservation of biological ultrastructure. I. Low temperature states of aqueous solutions of hydrophilic polymers,” J. Microsc., vol. 110, no. 3, pp. 223–228, 1977.10.1111/j.1365-2818.1977.tb00034.xSearch in Google Scholar PubMed
[14] P. Echlin, H. B. Skaer, B. O. Gardiner, F. Franks, and M. H. Asquith, “Polymeric cryoprotectants in the preservation of biological ultrastructure. II. Physiological effects,” J. Microsc., vol. 110, no. 3, pp. 239–255, 1977. https://doi.org/10.1111/j.1365-2818.1977.tb00035.x.Search in Google Scholar PubMed
[15] H. B. Skaer, F. Franks, M. H. Asquith, and P. Echlin. “Polymeric cryoprotectants in the preservation of biological ultrastructure. III. Morphological aspects.” J. Microsc., vol. 110, no. 3, pp.257–270, 1977. https://doi.org/10.1111/j.1365-2818.1977.tb00036.x.Search in Google Scholar PubMed
[16] C. Buser and P. Walther, “Freeze-substitution: the addition of water to polar solvents enhances the retention of structure and acts at temperatures around −60 degrees C,” J. Microsc., vol. 230, no. Pt 2, pp. 268–277, 2008, https://doi.org/10.1111/j.1365-2818.2008.01984.x.Search in Google Scholar PubMed
[17] R. Steinbrecht and M. Müller, “Freeze-substitution and freeze-drying,” in Cryotechniques in Biological Electron Microscopy, Heidelberg, Springer, 1987.10.1007/978-3-642-72815-0Search in Google Scholar
[18] P. Hawes, C. L. Netherton, M. Mueller, T. Wileman, P. Monaghan, and A. Road, “Rapid freeze-substitution preserves membranes in high-pressure frozen tissue culture cells,” J. Microsc., vol. 226, no. Pt 2, pp. 182–189, 2007, https://doi.org/10.1111/j.1365-2818.2007.01767.x.Search in Google Scholar PubMed
[19] E. Kellenberger, “The potential of cryofixation and freeze substitution: observations and theoretical considerations,” J. Microsc., vol. 161, no. Pt 2, pp. 183–203, 1991. https://doi.org/10.1111/j.1365-2818.1991.tb03083.x.Search in Google Scholar PubMed
[20] K. L. McDonald, H. Schwarz, M. Thomas, R. Webb, C. Buser, and M. Morphew, “Author’s personal copy ‘tips and tricks’ for high-pressure freezing of model systems,” Methods Cell Biol., vol. 96, 2010, https://doi.org/10.1016/S0091-679X(10)96028-7.Search in Google Scholar PubMed
[21] L. Bi, A. M. Lawler, S. E. Antonarakis, K. A. High, J. D. Gearhart, and H. H. Kazazian, “Targeted disruption of the mouse factor VIII gene produces a model of Haemophilia A,” Nat. Genet., vol. 10, no. 1, pp. 119–121, 1995, https://doi.org/10.1038/NG0595-119.Search in Google Scholar PubMed
[22] T. Murashige and F. Skoog, “A revised medium for rapid growth and bio assays with tobacco tissue cultures,” Physiol. Plant., vol. 15, no. 3, pp. 473–497, 1962.10.1111/j.1399-3054.1962.tb08052.xSearch in Google Scholar
[23] X. Heiligenstein and M. Belle, “F°Low: EM goes FAIR. A novel database software to track with high accuracy the journey of a biological sample towards electron microscopy analysis,” Res. Sq., 2024. https://doi.org/10.21203/RS.3.RS-4133153/V1.Search in Google Scholar
[24] M. A. Holden, J. M. Campbell, F. C. Meldrum, B. J. Murray, and H. K. Christenson, “Active sites for ice nucleation differ depending on nucleation mode,” Proc. Natl. Acad. Sci. USA, vol. 118, no. 18, 2021, https://doi.org/10.1073/PNAS.2022859118/-/DCSUPPLEMENTAL.Search in Google Scholar
[25] B. J. Murray, “Cracking the problem of ice nucleation,” Science (1979), vol. 355, no. 6323, 346–347, 2017, https://doi.org/10.1126/SCIENCE.AAM5320.Search in Google Scholar PubMed
[26] J. Liu, C. Zhu, K. Liu, Y. Jiang, Y. Song, J. S. Francisco, X. C. Zeng, and J. Wang, “Distinct ice patterns on solid surfaces with various wettabilities,” Proc. Natl. Acad. Sci. USA, vol. 114, no. 43, pp. 11285–11290, 2017, https://doi.org/10.1073/PNAS.1712829114.Search in Google Scholar
[27] “7.3: Nucleation of ice crystals – geosciences LibreTexts.” [Online]. Available: https://geo.libretexts.org/Bookshelves/Meteorology_and_Climate_Science/Practical_Meteorology_(Stull)/07:_Precipitation_Processes/7.03:_Nucleation_of_Ice_Crystals [accessed: June 06, 2024].Search in Google Scholar
[28] K. Matsumura, F. Hayashi, T. Nagashima, R. Rajan, and S. H. Hyon, “Molecular mechanisms of cell cryopreservation with polyampholytes studied by solid-state NMR,” Commun. Mater., vol. 2, no. 1, pp. 1–12 2021, https://doi.org/10.1038/s43246-021-00118-1.Search in Google Scholar
[29] “(2) (PDF) Positron annihilation study of eutectic crystallization of water-DMSO mixture and its relevance in cryobiology.” [Online]. Available: https://www.researchgate.net/publication/362722402_Positron_annihilation_study_of_eutectic_crystallization_of_water-DMSO_mixture_and_its_relevance_in_cryobiology?channel=doi&linkId=62fc0b25eb7b135a0e3f28bd&showFulltext=true [accessed: June 06, 2024].Search in Google Scholar
[30] T. J. Deerinck, E. Bushong, and A. Thor, “A new protocol for preparation of biological specimens for serial block face scanning electron microscopy Microscopy,” NCMIR Methods 3D EM, vols. 6–8, 2010.Search in Google Scholar
[31] R. M. Rajani, N. Dupré, V. Domenga‐Denier, G. Van Niel, X. Heiligenstein, and A. Joutel, “Characterization of early ultrastructural changes in the cerebral white matter of CADASIL small vessel disease using high pressure freezing/freeze‐substitution,” Neuropathol. Appl. Neurobiol., pp. 1–11, 2021, https://doi.org/10.1111/nan.12697.Search in Google Scholar PubMed
[32] V. B. Vays, I. M. Vangeli, O. A. Averina, M. L. Lovat, and L. E. Bakeeva, “Ultrastructure of hepatocytes from laboratory mice fed a standard dry laboratory animal diet,” Biochemistry (Moscow), vol. 85, no. 9, pp. 1082–1092, 2020, https://doi.org/10.1134/S0006297920090084/METRICS.Search in Google Scholar
[33] M. W. Hess, “Cryopreparation methodology for plant cell biology,” Methods Cell Biol., vol. 79, no. 79, pp. 57–100, 2007, https://doi.org/10.1016/S0091-679X(06)79003-3.Search in Google Scholar PubMed
[34] H. Moor, ”Theory and practice of high pressure freezing,” Cryotechniques in Biological Electron Microscopy, R. A. Steinbrecht and K. Zierold, Eds., Berlin, Heidelberg, Springer, 1987, 175–191. https://doi.org/10.1007/978-3-642-72815-0_8.Search in Google Scholar
[35] H. Moor, G. Bellin, C. Sandri, and K. Akert, “The influence of high pressure freezing on mammalian nerve tissue,” Cell Tissue Res, vol. 209, no. 2, pp. 201–216, 1980, https://doi.org/10.1007/BF00237626.Search in Google Scholar PubMed
[36] E. B. Hunziker, W. Herrmann, R. K. Schenk, M. Mueller, and H. Moor, “Cartilage ultrastructure after high pressure freezing, freeze substitution, and low temperature embedding. I. Chondrocyte ultrastructure--implications for the theories of mineralization and vascular invasion,” J. Cell Biol., vol. 98, no. 1, pp. 267–276, 1984. Available at: https://doi.org/10.1083/jcb.98.1.267.Search in Google Scholar PubMed PubMed Central
[37] W. Kühlbrandt, “The resolution revolution,” Science (1979), vol. 343, no. 6178, pp. 1443–1444, 2014, https://doi.org/10.1126/SCIENCE.1251652.Search in Google Scholar PubMed
[38] M. Eisenstein, “Seven technologies to watch in 2023,” Nature, vol. 613, no. 7945, pp. 794–797, 2023, https://doi.org/10.1038/D41586-023-00178-Y.Search in Google Scholar PubMed
[39] N. Feder and R. L. Sidman, “Methods and principles of fixation by freeze-substitution,” J. Biophys. Biochem. Cytol., vol. 4, no. 5, pp. 593–600, 1958, https://doi.org/10.1083/jcb.4.5.593.Search in Google Scholar PubMed PubMed Central
[40] P. Wild, E. M. Schraner, H. Adler, and B. M. Humbel, “Enhanced resolution of membranes in cultured cells by cryoimmobilization and freeze-substitution,” Microsc. Res. Tech., vol. 53, no. 4, pp. 313–321, 2001, https://doi.org/10.1002/jemt.1098.Search in Google Scholar PubMed
[41] R. A. Steinbrecht, “Freeze-substitution for morphological and immunocytochemical studies in insects,” Microsc. Res. Tech., vol. 24, no. 6, pp. 488–504, 1993, https://doi.org/10.1002/jemt.1070240605.Search in Google Scholar PubMed
[42] I. Anatomie, H. Embryologie, and A. Histologie, “Of plants and other pets: practical aspects of freeze-substitution,” J. Microsc., vol. 212, no. October, pp. 44–52, 2003.10.1046/j.1365-2818.2003.01237.xSearch in Google Scholar PubMed
[43] H. H. Mollenhauer, “Artifacts caused by dehydration and epoxy embedding in transmission electron microscopy,” Microsc. Res. Techn., vol. 26, no. 6, pp. 496–512, 1993, https://doi.org/10.1002/jemt.1070260604.Search in Google Scholar PubMed
[44] K. L. McDonald, “Rapid embedding methods into epoxy and LR white resins for morphological and immunological analysis of cryofixed biological specimens,” Microsc. Microanal., vol. 20, no. 01, pp. 152–163, 2014, https://doi.org/10.1017/S1431927613013846.Search in Google Scholar PubMed
[45] N. Matsko and M. Mueller, “Epoxy resin as fixative during freeze-substitution,” J. Struct. Biol., vol. 152, no. 2, pp. 92–103, 2005, https://doi.org/10.1016/j.jsb.2005.07.005.Search in Google Scholar PubMed
[46] E. Carlemalm, W. Villiger, J. A. Hobot, J. D. Acetarin, and E. Kellenberger, “Low temperature embedding with Lowicryl resins: two new formulations and some applications,” J. Microsc., vol. 140, no. 1, pp. 55–63, 1985, https://doi.org/10.1111/j.1365-2818.1985.tb02660.x.Search in Google Scholar PubMed
[47] N. Nešić, et al.., “Automated segmentation of cell organelles in volume electron microscopy using deep learning,” Microsc. Res. Tech., vol. 87, no. 8, pp. 1718–1732, 2024. https://doi.org/10.1002/jemt.24548.Search in Google Scholar PubMed
Supplementary Material
This article contains supplementary material (https://doi.org/10.1515/mim-2024-0002).
© 2024 the author(s), published by De Gruyter on behalf of Thoss Media
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Editorials
- Empowering the Microscopy Community: Welcome to the Inaugural Issue of Methods in Microscopy
- Embracing innovation in microscopy
- News
- Microscopy Community News
- Special Issue: Volume Microscopy Across Scales
- View
- Array tomography: trails to discovery
- Tutorial
- Quantification of collective signalling in time-lapse microscopy images
- Research Articles
- A systematic approach of vitrification by high pressure freezing
- FAST-EM array tomography: a workflow for multibeam volume electron microscopy
- Converting a symmetrical Gaussian beam into a thin tunable light sheet
- Serial block-face scanning electron microscopy of adherent cells on thin plastic substrate
- How to increase freeze-substitution and electron microscopy embedding reliability
Articles in the same Issue
- Frontmatter
- Editorials
- Empowering the Microscopy Community: Welcome to the Inaugural Issue of Methods in Microscopy
- Embracing innovation in microscopy
- News
- Microscopy Community News
- Special Issue: Volume Microscopy Across Scales
- View
- Array tomography: trails to discovery
- Tutorial
- Quantification of collective signalling in time-lapse microscopy images
- Research Articles
- A systematic approach of vitrification by high pressure freezing
- FAST-EM array tomography: a workflow for multibeam volume electron microscopy
- Converting a symmetrical Gaussian beam into a thin tunable light sheet
- Serial block-face scanning electron microscopy of adherent cells on thin plastic substrate
- How to increase freeze-substitution and electron microscopy embedding reliability

