Lnc5q21.2, a novel long intergenic RNA, sensitizes colorectal cancer cells to ATR inhibitor by activating Wnt pathway
-
Meiying Zhang
Abstract
Background and objectives
Colorectal cancer (CRC) is still the leading cause of cancer-related death. With the recognizing the importance of long non-coding RNA (LncRNA) in development and cancer, it is urgently to identify new LncRNA and understand the mechanism to develop novel therapeutic strategies.
Methods
Nine CRC cell lines, 52,146 and 285 cases of normal colorectal mucosa, adenoma and CRC samples were utilized. Northern blot, rapid amplification of cloned cDNA ends, RNA pulldown, Mass spectrum, RNA immunoprecipitation, CRISPR/Cas9, fluorescence in situ hybridization assays and xenograft mice model were employed.
Results
Lnc5q21.2 is identified to be a novel long intergenic non-coding RNA and its full length is 668 nt. Lnc5q21.2 is mainly located in cell nucleus and its expression is regulated by N6-methyladenine (6mA) modification. Compared to adjacent tissue, the levels of Lnc5q21.2 were increased significantly in CRC samples (P < 0.001), with a progression tendency from noncancerous colorectal mucosa, adjacent tissue, adenoma to CRC samples (P < 0.001). Lnc5q21.2 highly expression is associated with alcohol consumption and poor prognosis (both P < 0.05). Lnc5q21.2 promotes cell proliferation, migration, invasion and cell cycle progression. Lnc5q21.2 activates Wnt signaling by interacting with homeobox A10 (HOXA10) and down regulating empty spiracles homeobox 2 expression. Further study demonstrates that Lnc5q21.2 promotes DNA damage repair by enhancing ATR signaling. Lnc5q21.2 sensitizes CRC cells to ATR inhibitor both in vitro and in vivo.
Conclusions
Lnc5q21.2 is a novel lncRNA. Lnc5q21.2 promotes ATR pathway by activating Wnt signaling via interacting with HOXA10. Lnc5q21.2 sensitizes CRC cells to ATR inhibitor both in vitro and in vivo.
Introduction
Colorectal cancer (CRC) is one of the major causes of cancer-related death.[1] The 5-year survival rate is 91% for localized CRC patients and 14% for metastatic disease.[2] The discovery of regulatory elements is becoming the primary part of cancer biology research in the postgenomic era.[3] Long noncoding RNAs (LncRNA) have emerged as important modulators in cancer initiation and development.[4] Recently, lncRNAs were recognized to play a determining role in cellular phenotype during development.[5] LncRNAs have been linked to chromatin remodeling, DNA damage repair, necroptosis and drug resistance in cancer and other diseases.[6, 7, 8, 9] More than 70% of the human genome may yield noncoding RNA transcripts.[4,10] With the development of new techniques, more important lncRNA fragments related to diseases, tissue structure or cell type specificity will be discovered. Using spatial transcriptomics, a batch of lncRNA fragments were found linked to CRC metastatic tissues.[11,12] However, very limited numbers of lncRNAs were identified with full-length sequences. Deep understanding the function of lncRNAs and their regulation network remains a challenge. Some of lncRNAs were exhibited cancer type specificity and related to malignant transformation.[13, 14, 15] Many genomic mutations were discovered not in protein encoding region, while they were inside lncRNAs transcribed location.[15,16] By interacting with DNA, RNA and proteins, lncRNAs take part in gene regulatory networks, which have important implications for cancer therapeutic strategy. However, we are far from incorporating lncRNAs into the clinic, which is impeded by the limitation of fully understanding their mechanisms.[4] It is a big challenge to identify the full-length sequences and structural elements to develop novel, rational, tailored therapeutic strategies.
Here, we identified a novel lncRNA, Lnc5q21.2, and dug its role in CRC. Lnc5q21.2 promotes CRC growth by activating Wnt signaling via interacting with homeobox A10 (HOXA10) and its highly expression increased the sensitivity of ATR inhibitor.
Materials and methods
Cancer cell lines and tissue samples
All CRC cells were established from primary CRC. Normal colorectal mucosa from noncancerous patients (52), adenomas (146) and matched CRC and adjacent tissue samples (285) were obtained from the Chinese PLA General Hospital, under the guidelines approved by the institutional review board at the Chinese PLA General Hospital (NO. 20090701-015). Informed consent was obtained from all patients.
Identification of Lnc5q21.2 with northern blot and obtaining its full-length sequence by RACE assay
Biotin RNA labeling mix (Roche, Switzerland, Cat#11685597910) and T7 RNA polymerase (Life, USA, Cat#AM2718) were utilized to synthesize Lnc5q21.2 probes for northern blot. The full-length sequence was obtained by 5’ and 3’ rapid amplification of cloned cDNA ends (RACE) assay according to the instruction of the FirstChoice RLM-RACE Kit (Ambion, USA, AM1700). The primer sequences are listed in Supplementary Table S1.
RT-PCR and Lnc5q21.2 stably expressed and deleted cells establishment
RNA preparation and semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) were followed previously.[17] SYBR Green was applied for Real-time RT-PCR. GAPDH was used as an internal reference to calculate 2−ΔCt, representing the relative expression level of lncRNAs.[18] Primer sequences for Lnc5q21.2 and GAPDH are listed in Supplementary Table S1.
The human full-length cDNA of Lnc5q21.2 was cloned into the pLenti6-GFP vector for stably expressed cells. The primer sequences were listed in Supplementary Table S1. The single guide RNA (sgRNA) sequences for targeting Lnc5q21.2 were designed by zlab (https://www.zlab.bio/resources). Monoclonal Lnc5q21.2 deleted cells were selected by limited dilution and verified by PCR. The primer sequences were listed in Supplementary Table S2.
Lnc5q21.2 was cloned into pcDNA3.1+ vectors for transient expression. The full-length and three possible distinct coding sequences were constructed. The possible starting sites (ATG) are as follow 5’-ATGATATTGG…Flag-3’, 5’-ATGTATTGG… Flag-3’ and 5’-ATGATTGG… Flag-3’. TMEM176A-pcDNA3.1-3×Flag plasmid was used as control.[19]
Fluorescence in situ hybridization
To clarify the location of Lnc5q21.2 in cells, fluorescence in situ hybridization assay (FISH) was employed. Probes for Lnc5q21.2 detection were designed according to LNA probe software (https://sg.idtdna.com/pages), which were labeled with DIG following the manufacturer’s instruction. Lnc5q21.2 highly expressed HCT116 cells were plated on sterile glass coverslips and incubated for 24 h. Cells were washed with PBS and fixed with 4% formaldehyde for 15 min at room temperature. Then, the cells were permeabilized in PBS containing 0.5% Triton X-100 on ice for 5 min, and washed for 3 times using PBS. Labeled Lnc5q21.2 probes were applied for FISH following the protocols of TSA ™ plus fluorescence systems (PerkinElmer, USA, Cat# NEL741). A Leica TCS SP2 confocal laser microscopy was utilized to observe the location of labeled Lnc5q21.2 probes in cells. The labeled probe sequences for Lnc5q21.2 were listed in Supplementary Table S3.
RNA pull down
To gain the interacting proteins of Lnc5q21.2, six distinct parts of Lnc5q21.2 were labeled by biotin to serve as detection probes. The labeled lacZ mRNA probes were served as negative control (BGI, China). The sequences of these probes are listed in Supplementary Table S3. Cell lysate prepared from HCT116 cells was applied for RNA pull down.[20]
By using RNA pull down, the interacting sequence of Lnc5q21.2 to related protein was obtained by truncating Lnc5q21.2. Different parts of Lnc5q21.2 fragments were labeled by biotin using in vitro transcription assay, including Lnc5q21.2F1-200, Lnc5q21.2F1-400, Lnc5q21.2F1-668, Lnc5q21.2F200-668 and Lnc5q21.2F400-668.
RNA immunoprecipitation and chromatin immunoprecipitation
To validate the interaction of proteins to Lnc5q21.2, RNA immunoprecipitation (RIP) approach was employed by using antibodies against proteins gained from RNA pull down.[20] The antibodies were listed in Supplementary Table S4.
Chromatin immunoprecipitation (ChIP) was performed according to the instruction of EpiTect ChIP One Day Kit (Qiagen, Germany, Cat# 334471). The primers sequences for promoter region detection were listed in Supplementary Table S1.
Chromatin isolation by RNA purification assay
To find more clues for Lnc5q21.2 interacting with empty spiracles homeobox 2 (EMX2), Chromatin isolation by RNA purification (ChIRP) assay was utilized.[21] Using labeled Lnc5q21.2 probes, the promoter region sequence of EMX2 gene was detected from DNA extracted from cross-linked HCT116 cells lysates.
Additional materials & methods
Part of material & methods were described in the supplementary data, including MTT, colony formation, flow cytometry, transwell assays and xenograft mouse model, nucleus and cytoplasm isolation, luciferase reporter assay and siRNA knockdown.
Statistical analysis
SPSS 23.0 software (IBM, USA) for χ2, Fisher’s exact and Student’s t-test were used for statistical analysis. Kaplan-Meier plots and the log-rank test were used to estimate overall survival (OS). The five-year OS was assessed by univariate and multivariate Cox proportional hazards regression models. P < 0.05 was determined as significance difference.
Results
Lnc5q21.2 was highly expressed in CRC
One of the lncRNA fragments was isolated differentially expressed from a small number of cancers and matched adjacent tissue samples by high-throughput assay (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70880). The original length of this fragment is 273 nt. It is located on human Chromosome 5q21.2 and named as Lnc5q21.2 according to the genomic location. To deep understand Lnc5q21.2 in CRC, the expression was detected in 9 CRC cell lines first. Lnc5q21.2 was highly expressed in HCT116, SW480, SW620, DLD1, HT29 and DKO cells, and unexpressed in RKO, LOVO and LS180 cells (Figure 1A). Then, the expression was examined in tissue samples. The expression level of Lnc5q21.2 was increased significantly in CRC tissue compared to adjacent tissue samples (P < 0.001, Figure 1B), with a progression tendency from noncancerous colorectal mucosa, adjacent tissue, adenoma to CRC samples (P < 0.001, Figure 1C). Increased level of Lnc5q21.2 was associated with alcohol (P < 0.05, Table 1), while no association with age, gender, tumor size, TNM stage, lymph node metastasis, smoking or family tumor history (all P > 0.05, Table 1).
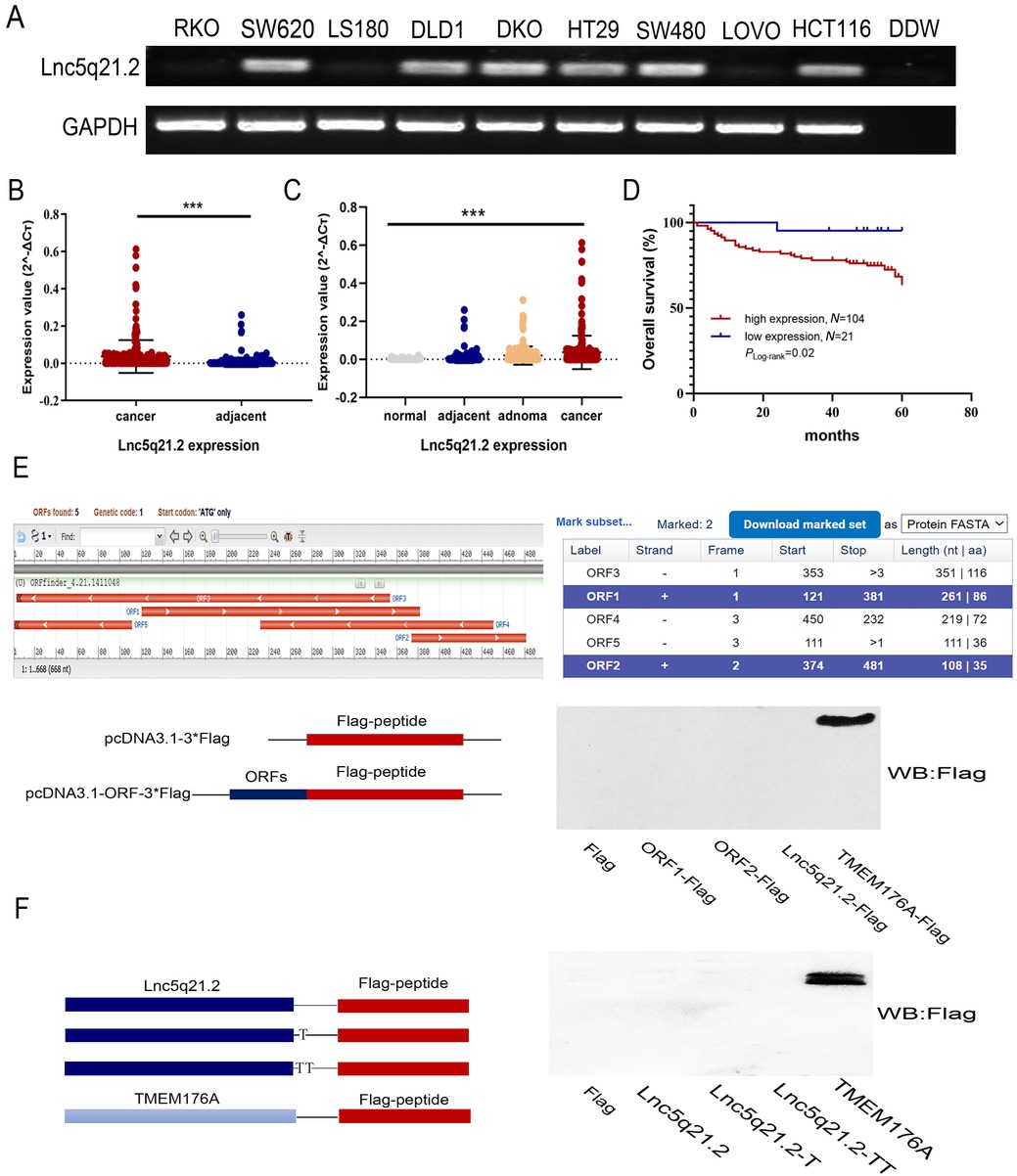
The expression of Lnc5q21.2 in CRC. A: Semi-quantitative RT-PCR shows the expression of Lnc5q21.2 in CRC cells. DDW: double distilled water; GAPDH, internal control. B: The expression of Lnc5q21.2 in normal colorectal mucosa, adenoma, CRC and adjacent tissue samples. normal: normal colorectal mucosa; adjacent: adjacent tissue samples; adenoma: colonic adenoma samples; cancer: CRC samples. C: The expression of Lnc5q21.2 in CRC and adjacent tissue samples.D: The association of Lnc5q21.2 and 5-year OS of CRC. E: The potential predicted ORFs was presented by ORF Finder. ORF1, 121-381bp; ORF2, 374-481bp, ORF3, 353-3bp, ORF4, 450-232bp, ORF5, 111-1bp; +, forward; -, reverse. TMEM176A with Flag tag severs as a positive control. F: Full-length Lnc5q21.2 was cloned into the eukaryotic expression vector pcDNA3.1 with three possible coding patterns. TMEM176A with Flag tag severs as a positive control. ***P < 0.001. CRC, colorectal cancer; RT-PCT, reverse transcription polymerase chain reaction; OS, overall survival.
The association of Lnc5q21.2 expression and clinical factors in human CRC
| Clinical parameter | NO. 285 | High expression n = 236 | Reduced/Loss expression n = 49 | P value* | |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 179 | 154 | 25 | 0.061 | |
| Female | 106 | 82 | 24 | ||
| Age (years) | |||||
| ≤50 | 48 | 41 | 7 | 0.599 | |
| >50 | 237 | 195 | 42 | ||
| Differentiation | |||||
| Moderately/Well | 214 | 182 | 32 | 0.082 | |
| Poorly | 71 | 54 | 17 | ||
| Tumor stage | |||||
| I/II | 170 | 143 | 27 | 0.476 | |
| III/IV | 115 | 93 | 22 | ||
| Metastasis | |||||
| Positive | 107 | 83 | 24 | 0.069 | |
| Negative | 178 | 153 | 25 | ||
| Tumor size | |||||
| ≤4 cm | 119 | 100 | 19 | 0.642 | |
| >4 cm | 166 | 136 | 30 | ||
| Smoking | |||||
| No | 197 | 159 | 38 | 0.161 | |
| Yes | 88 | 77 | 11 | ||
| Drinking | |||||
| No | 234 | 188 | 46 | 0.018* | |
| Yes | 51 | 48 | 3 | ||
| Inherited | |||||
| No | 275 | 228 | 47 | 0.811 | |
| Yes | 10 | 8 | 2 |
*P values are obtained from χ2 test, significant difference, P < 0.05. CRC, colorectal cancer.
For 125 cases of available CRC with survival data, a Cox multivariable proportional hazards and Kaplan-Meier model were applied to analyze the association of Lnc5q21.2 expression and survival time. Lnc5q21.2 expression is significantly associated with poor 5-year OS (P < 0.05, Figure 1D) and is an independent prognostic factor (P < 0.05, Table 2). These results demonstrated that Lnc5q21.2 is a poor prognostic marker.
Univariate and multivariate analysis of Lnc5q21.2 expression with overall survival in CRC patients
| Clinical parameter | Univariate analysis |
Multivariate analysis | ||
|---|---|---|---|---|
| HR (95%CI) | P value | HR (95%CI) | P value | |
| Gender (male vs. female) | 0.580 (0.283,1.189) | 0.137 | ||
| Age (>50 vs. ≤50 years) | 0.651 (0.266,1.597) | 0.349 | ||
| Tumor size (>4 vs. ≤4 cm) | 0.899 (0.439,1.840) | 0.771 | ||
| Differentiation (low vs. high or middle differentiation) | 2.591 (1.257,5.342) | 0.010* | 2.598 (1.234,5.470) | 0.012* |
| TNM stage (III/IV vs. I/II) | 6.609 (2.694,16.213) | 0.000*** | 7.358 (1.486,36.425) | 0.014* |
| Lymph node metastasis (positive vs. negative) | 4.889 (2.169,11.020) | 0.000*** | 0.881 (0.203,3.812) | 0.865 |
| Lnc5q21.2 (high expression vs. low expression) | 7.501 (1.017,55.346) | 0.048* | 10.388 (1.396,77.319) | 0.022* |
| Smoking (yes vs. no) | 0.842 (0.385,1.840) | 0.666 | ||
| Drinking (yes vs. no) | 0.511 (0.155,1.685) | 0.270 | ||
| Inherited (yes vs. no) | 0.492 (0.117,2.068) | 0.333 | ||
HR: hazard ratio; CRC, colorectal cancer. *P < 0.05; ***P < 0.001.
The association of Lnc5q21.2 and microsatellite instability (MSI) was also evaluated by detecting mismatch repair genes (MMR) expression with immunohistochemistry, including MLH1, MSH2, MSH6 and PMS2. Deficiency of MMR (dMMR) was observed in 9.6% (12/125) of cases. KRAS mutation was found in 8% (10/125) and BRAF was mutated in 4.8% (6/125) of patients. No association was found between Lnc5q21.2 expression and dMMR, KRAS or BRAF mutation (all P > 0.05, Supplementary Table S5).
Lnc5q21.2 transcript is a new long non-coding RNA
First, the transcript Lnc5q21.2 was evaluated by Northern Blot, and then the full-length sequence was obtained by 5’ and 3’ RACE technique for 668nt (Supplementary Table S6), validating that Lnc5q21.2 is a novel long intergenic non-coding RNA with a poly (A) tail (Supplementary Figure S1A–C). Analyzing by nuclear- cytoplasmic separation and FISH assays, Lnc5q21.2 was displayed mainly located in the nucleus of HCT116 cells (Supplementary Figure S1D and E).
The potential coding ability for protein or peptic coding was estimated using Coding Potential Assessment Tool (CPAT) (http://lilab.research.bcm.edu/cpat/index.php), Coding Potential Calculator (CPC, https://cpc.gao-lab.org/),[22] LGC (https://ngdc.cncb.ac.cn/lgc/),[23] PLEK (https://sourceforge.net/projects/plek/)[24] and ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). Lnc5q21.2 was predicted possibly encoding two peptides according to ORF Finder software. No protein or peptide coding ability was found by other databases (Figure 1E and Supplementary Table S7). To further evaluate the possibility of peptide coding ability of Lnc5q21.2, the full-length sequence and two predicted peptide ORFs were cloned into pcDNA3.1-3xFlag expression vector. TMEM176A, encoding a short protein sequence, was used as the control. Supposing exists three possible translational start sites at the 5’ of Lnc5q21.2, various expressing constructs were built by selecting three different start site. These expression constructs include the full length Lnc5q21.2. sequence and the potential nucleotide sequences for predicting to coding peptides. No fusion protein was found by expressing these constructs in RKO cells (Figure 1E–F), excluding the protein/peptide coding ability of Lnc5q21.2.
Up-regulating Lnc5q21.2 by 6mA modification
The promoters of lncRNAs were reported almost as conservative as protein-coding genes and their expression regulation is also similar.[25] RNA transcription may be also regulated by N6-methyladenine (6mA) DNA modification.[26,27] While no CpG islands were discovered by analyzing the sequence around transcription start site (TSS), excluding promoter region methylation to regulate Lnc5q21.2 expression. Then, ChIP assay was employed to assess the possibility of histone modulation of Lnc5q21.2 expression. No apparent difference was observed for activating marker (H3K4me3) and inhibiting marker (H3K9me2) in Lnc5q21.2 highly expressed HCT116 and unexpressed RKO cells, indicating that Lnc5q21.2 was not regulated by histone modification (Supplementary Figure S2A).
DNA 6mA is related to transcriptional activation, and [G/C] AGG [C/T] is the most prevalent motif for 6mA modification.[28] Existing “AGG” sequence around the TSS of Lnc5q21.2 hints the possibility of the expression regulation by 6mA modification. By analyzing with ChIP assay, DNA 6mA was found in Lnc5q21.2 expressed HCT116 cells, while no 6mA was detected in Lnc5q21.2 unexpressed RKO cells, indicating 6mA regulation of its expression (Supplementary Figure S2B). DNA 6mA modification was mediated by methyltransferase N6AMT1.[28] Thereafter, siRNA was employed to knock down N6AMT1. DNA 6mA level was reduced after knockdown of N6AMT1 in HCT116 cells (Supplementary Figure S2C). The result further demonstrated that Lnc5q21.2 expression is regulated by 6mA modification.
Lnc5q21.2 promotes CRC growth
LncRNAs may play multifaceted function depending on their sequences and structure.[25,29] To understand the role of Lnc5q21.2 in CRC, the full-length sequence was cloned into Lentiviral vectors to obtain Lnc5q21.2 stably expressed RKO and LS180 cells. Lnc5q21.2 was observed to promote CRC cell proliferation and migration, increase S phase and reduce G2/M phase cells. Above results were validated by knockout of Lnc5q21.2 in HCT116 cells (Figure 2A-E and Supplementary Figure S3), suggesting the oncogenic role of Lnc5q21.2 in CRC cells.

The role of Lnc5q21.2 in CRC cell growth. A: The OD value in Lnc5q21.2 expressed and unexpressed CRC cells. Each experiment was repeated for three times and OD value was shown as mean ± SD. B: Colony formation assay shows the efficiency of Lnc5q21.2 on CRC cells. Each experiment was repeated three times. C: Cell cycle distribution in Lnc5q21.2 unexpressed and expressed CRC cells. Each experiment was repeated three times. D: Transwell assay shows the role of Lnc5q21.2 in cell migration and invasion. Each experiment was repeated three times. E: Western blots show the levels of cell cycle, invasion and migration related molecules in Lnc5q21.2 expressed and unexpressed CRC cells. F: Represents results for Lnc5q21.2 unexpressed and re-expressed RKO cell xenografts. G: Tumor growth curves and tumor weight of Lnc5q21.2 unexpressed and overexpressing RKO cells. H: Represents results for Lnc5q21.2 highly expressed and knockout HCT116 cell xenografts. I: Tumor growth curves and tumor weight of Lnc5q21.2 highly expressed and knockout HCT116 cell. *P < 0.05, **P < 0.01, ***P < 0.001. CRC, colorectal cancer; WT, wild type; KO, knockout.
To further estimate the effect of Lnc5q21.2 on CRC, RKO cell xenograft mice were utilized. The tumor volume was 176.3 ± 126.4 mm3 vs. 957.8 ± 209.9 mm3 in Lnc5q21.2 silenced and overexpressed xenografts, increasing the tumor volume by Lnc5q21.2 (P < 0.001, Figure 2F and 2G). The tumor weight was 0.09 ± 0.05 g vs. 0.47 ± 0.19 g in Lnc5q21.2 silenced and overexpressed xenografts, increasing the tumor weight by Lnc5q21.2 (P < 0.001, Figure 2G). The promoting role of Lnc5q21.2 in CRC growth was verified by CRISPR/Cas9 knockout technique. The tumor volume was 477.9 ± 132.7 mm3 vs. 64.4 ± 28.1 mm3 in Lnc5q21.2 highly expressed and deleted HCT116 cell xenografts. The tumor volume was decreased by depletion of Lnc5q21.2 (P < 0.001, Figure 2H and 2I). The tumor weight was 0.457 ± 0.110 g vs. 0.051 ± 0.039 g in Lnc5q21.2 highly expressed and deleted HCT116 cell xenografts. Reduced tumor weight was observed in Lnc5q21.2 depleted HCT116 cell xenografts (P < 0.001, Figure 2I). Lnc5q21.2 promotes CRC cell xenograft growth in mice.
Lnc5q21.2 was discovered to interact with HOXA10
To further understand the mechanism of Lnc5q21.2 in CRC, RNA pulldown and mass spectrometry techniques were employed. Two extra bands were obtained from the nuclear lysates of Lnc5q21.2 highly expressed HCT116 cells and analyzed by mass spectrometry (Figure 3A). Among Lnc5q21.2 binding proteins, HOXA10 was discovered and validated by RNA pulldown and western blot techniques (Figure 3B). Lnc5q21.2 binding to HOXA10 was reproduced by RIP assay using HOXA10 antibody in crosslinking treated and untreated HCT116 cell lysates (Figure 3C).
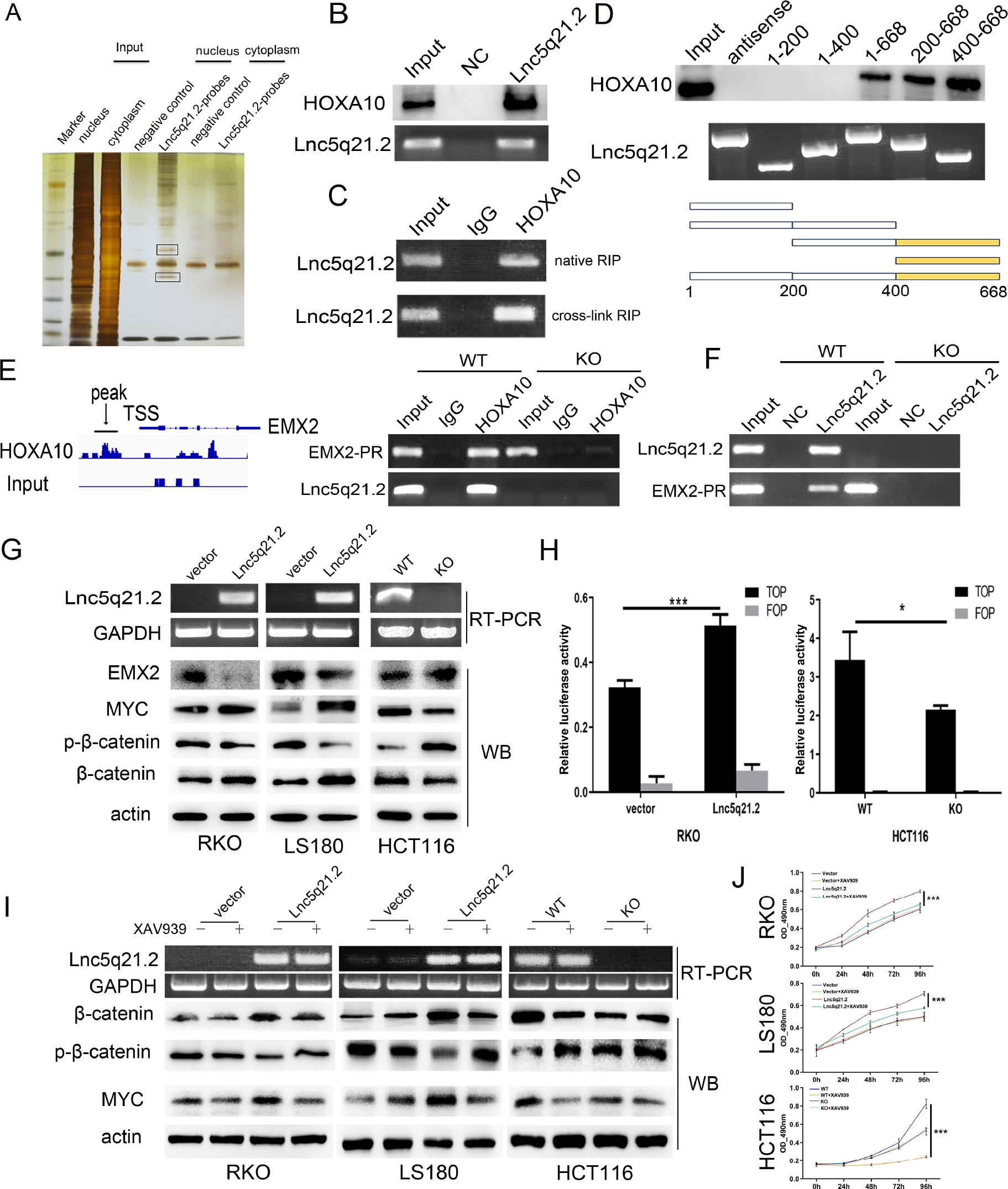
Lnc5q21.2 interacting with HOXA10 and promotes the Wnt signaling pathway. A: RNA pull-down assay shows the interacting of Lnc5q21.2 with proteins. The SDS-PAGE was stained with silver. The labeled bands were resected for mass spectrometry. B: Western blot validation for the specificity biding of Lnc5q21.2 and HOXA10 by HOXA10 antibody after RNA pull-down. C: Lnc5q21.2 enrichment in native and cross-linked RIP. D: One region in the 3’ end of Lnc5q21.2 is necessary to associate with HOXA10. Different Lnc5q21.2 fragments were used for RNA pull-down assay. The combination of Lnc5q21.2 fragment was revealed by western blot using HOXA10 antibody. E: ChIP-seq and ChIP-PCR results indicate HOXA10 binding to the promoter region of EMX2. F: ChIRP assay shows the promoter region of EMX2 existed in the complex obtained by Lnc5q21.2 probes, suggesting the interaction. G: Western blots show the effect of Lnc5q21.2 on the expression levels of EMX2, myc, β-catenin and p-β-catenin in RKO, LS180, and HCT116 cells. Semi-RT-PCR demonstrated that the expression of Lnc5q21.2 and GAPDH in CRC cells. vector, control vector; Lnc5q21.2, Lnc5q21.2 expression vector; actin, internal control; WT, Lnc5q21.2 high expression control; KO, Lnc5q21.2 expression knockout; GAPDH, internal control. H: Results of TCF/LEF luciferase reporter assay. I: Western blot shows the levels of myc, β-catenin and p-β-catenin in Lnc5q21.2 expressed and unexpressed CRC cells before and after XAV939 treatment. actin: internal control. J: Growth curves represent cell viability evaluated by MTT assay in CRC cells with or without XAV939(10 μmol/L) treatment. Each experiment was repeated for three times and OD value was shown as mean ± SD. *P < 0.05, ***P < 0.001. HOXA10, meobox A 10; ChIP, chromatin immunoprecipitation; CRC, colorectal cancer; PCR, polymerase chain reaction.
The evidence of Lnc5q21.2 binding to HOXA10 was further acquired by leveraging Lnc5q21.2 truncating fragments. The binding sequence of Lnc5q21.2 to HOXA10 was the fragment F400-668 (Figure 3D).
Lnc5q21.2 activates Wnt signaling by HOXA10 in CRC cells
To deeply understand the mechanism of Lnc5q21.2 interacting with HOXA10, the ChIP-sequence (ChIP-seq) and ChIP-PCR were borrowed. By using HOXA10 antibody, the combination of HOXA10 to the promoter region of EMX2 was discovered by ChIP-seq and validated by ChIP-PCR. At the same time, the sequence of Lnc5q21.2 was also discovered in the binding complex, suggesting that HOXA10 binds to both Lnc5q21.2 and the promoter region of EMX2 (Figure 3E). Labeled Lnc5q21.2 probes were used for ChIRP technique. The promoter region of EMX2 sequence was detected in the complexes of Lnc5q21.2 highly expressed HCT116 cells, while it did not emerge in Lnc5q21.2 unexpressed cells, further suggesting that Lnc5q21.2 binds to HOXA10 and EMX2 promoter region simultaneously (Figure 3F). The role of Lnc5q21.2 involves in EMX2 expression regulation was further validated by detecting the levels of EMX2 in Lnc5q21.2 highly expressed and unexpressed CRC cells (Supplementary Figure S4).
EMX2 mutation causes developmental defects in Drosophila melanogaster and homozygous mutants produced by intercrossing of heterozygotes died soon after birth in mice.[30] The transcription of EMX2 has been shown to be repressed by HOXA10 through binding its promoter region, supporting our above findings.[31]
Both HOXA10 and EMX2 have been displayed to regulate the Wnt signaling in cancers, independently.[32,33] The regulation of EMX2 by HOXA10 was reported previously.[31,34] Our above results demonstrated that HOXA10 binds to the promoter region of EMX2 gene. The regulation of Wnt signaling by HOXA10 was verified by siRNA knockdown technique. As shown in Supplementary Figure S5, reduced the total level of β-catenin and myc was displayed in HCT116 cells, while increased the level of p-β-catenin was observed after knockdown of HOXA10.
Thereafter, we explored the regulation role of Lnc5q21.2 in Wnt signaling. The key proteins of Wnt signaling were detected by western blot in CRC cells with or without expression of Lnc5q21.2. Increased the levels of β-catenin and myc were observed in Lnc5q21.2 re-expressed RKO and LS180 cells, while the levels of EMX2 and p-β-catenin were reduced. The levels of β-catenin and myc were reduced, and the levels of EMX2 and p-β-catenin were increased by knocking out Lnc5q21.2 in HCT116 cells (Figure 3G). Then, the role of Lnc5q21.2 in Wnt signaling was further determined by analyzing its effect on the activity of TCF/LEF transcription with dualluciferase reporter assay. Increased the activity was shown significantly in β-catenin and Lnc5q21.2 co-transfected RKO cells, while the activity was reduced by deletion of Lnc5q21.2 in HCT116 cells (Figure 3H).
For further verifying the role of Lnc5q21.2 in Wnt signaling, a Wnt signaling inhibitor (XAV939) was employed. CRC cells were divided into four groups, including empty vector, empty vector + XAV939, overexpression of Lnc5q21.2, overexpression of Lnc5q21.2 + XAV939 treatment. The impact of Lnc5q21.2 on Wnt signaling was evaluated by detecting β-catenin and MYC with western blot. The level of β-catenin and MYC was reduced by XAV939 in Lnc5q21.2 re-expressed RKO and LS180 cells. While no obvious changes were observed for β-catenin and MYC by deleting Lnc5q21.2 in HCT116 cells (Figure 3I). These results provide more evidence for Lnc5q21.2 taking part in Wnt signaling.
To further reveal Lnc5q21.2 promotion of CRC cell proliferation by activating Wnt signaling, MTT assay was employed. The OD values were 0.604 ± 0.014, 0.615 ± 0.047, 0.799 ± 0.017 and 0.654 ± 0.016 in untreated empty vector, XAV939 treated empty vector, untreated Lnc5q21.2 re-expressed and XAV939 treated Lnc5q21.2 re-expressed RKO cells, respectively. In LS180 cells, the OD values were 0.499 ± 0.040, 0.494 ± 0.009, 0.708 ± 0.019, and 0.579 ± 0.007 in untreated empty vector, XAV939 treated empty vector, untreated Lnc5q21.2 re-expressed and XAV939 treated Lnc5q21.2 re-expressed LS180 cells, respectively. No significant difference was displayed before and after treatment with XAV939 in Lncq5q21.2 unexpressed CRC cells (all P > 0.05, Figure 3J), while reduced the OD value was observed by XAV939 in Lncq5q21.2 re-expressing CRC cells (all P < 0.001, Figure 3J). These effects were verified by deleting Lnc5q21.2 in HCT116 cells. The OD values were 0.825 ± 0.051, 0.245 ± 0.010, 0.541 ± 0.017, and 0.523 ± 0.040 in Lnc5q21.2 highly expressed cells without treatment, XAV939 treatment, deletion of Lnc5q21.2, and deleting Lnc5q21.2 + XAV939, respectively. No significant changes were observed between Lnc5q21.2 deleting and Lnc5q21.2 deleting + XAV939 groups (P > 0.05). Nevertheless, the OD value was reduced significantly in the wild type HCT116 cells under the treatment of XAV939 (P < 0.001, Figure 3J). Above evidence further demonstrate that Lnc5q21.2 promotes CRC cell growth by activating Wnt signaling.
Lnc5q21.2 promotes ATR activity by activating Wnt signaling in CRC cells
It was noticed that HOXA10 is involved in homologous recombinant DNA repair (HR), and the Wnt signaling is also related to DDR.[35,36] It is possible that Lnc5q21.2 involves in HR. The key regulators of HR were examined utilizing western blots. No apparent changes were observed for total levels of ATM, ATR, CHK2, CHK1, and the levels of p-ATM and p-CHK2, whilst the levels of p-ATR and p-CHK1 were increased by re-expressing Lnc5q21.2 in RKO and LS180 cells under the low dose cisplatin treatment, indicating that Lnc5q21.2 activated ATR signaling (Figure 4A). Similar results were obtained in HCT116 cells. The total levels of ATM, ATR, CHK2, CHK1, p-ATM and p-CHK2 were not changed before and after deletion of Lnc5q21.2, the levels of p-ATR and p-CHK1 were decreased after deletion of Lnc5q21.2 (Figure 4A).

Lnc5q21.2 sensitives CRC cell to ATR inhibitor. A: Western blot shows the levels of ATM, ATR, p-ATM, p-ATR, CHK1, CHK2, p-CHK1 and p-CHK2 in Lnc5q21.2 expressed and unexpressed CRC cells before and after cisplatin treatment. Actin: internal control. B: The IC50 curve of VE-822 in CRC cells. C: Western blot shows the levels of ATR, p-ATR, CHK1, p-CHK1 and γH2AX in Lnc5q21.2 expressed and unexpressed CRC cells before and after cisplatin or VE-822 treatment. actin: internal control. D: Representative colony formation results to show the synthetic lethality between Lnc5q21.2 and ATR inhibitor. CRC, colorectal cancer. *P < 0.05, ***P < 0.001.
The role of Lnc5q21.2 in ATR signaling was further verified using VE- 822, an ATR inhibitor. Under low dose cisplatin treatment, the IC50 value of VE-822 was 1.09 ± 0.16 vs. 0.13 ± 0.04 (P < 0.001), and 2.22 ± 0.42 vs. 1.37 ± 0.14 (P < 0.05) μmol/L in Lnc5q21.2 silenced and reexpressed RKO and LS180 cells, respectively. The IC50 was reduced significantly by forced re-expressing Lnc5q21.2 (Figure 4B). The IC50 value was 2.10 ± 0.49 vs. 13.30 ± 2.41 μmol/L in wild type and Lnc5q21.2 deleted HCT116 cells, increasing the value by depletion of Lnc5q21.2 (P < 0.01, Figure 4B).
By utilizing AZD0156, an ATM inhibitor, the role of Lnc5q21.2 in ATM signaling was further tested. The IC50 of AZD0156 was 17.47 ± 0.58 vs. 16.96 ± 0.61 and 13.45 ± 0.93 vs. 14.32 ± 0.13 μmol/L in Lnc5q21.2 unexpressed and re-expressed RKO and LS180 cells, independently. The IC50 was 14.46 ± 0.48 vs. 15.04 ± 1.66 μmol/L in wild type and Lnc5q21.2 deleted HCT116 cells. No significant effect was observed by AZD0156 with or without Lnc5q21.2 expression (all P > 0.05, Supplementary Figure S6). These results further reflect that Lnc5q21.2 may drive ATR signaling, without influencing ATM signaling.
The role of Lnc5q21.2 in ATR signaling was further tested by using VE822. With the treatment of cisplatin, the levels of γ-H2AX were increased and the levels of p-CHK1 and p-ATR were reduced by VE822 in Lnc5q21.2 re-expressed RKO and LS180 cells. Accordingly, the levels of γ-H2AX were increased and the levels of p-CHK1 and p-ATR were reduced in Lnc5q21.2 highly expressed HCT116 cells, indicating that Lnc5q21.2 promotes ATR signaling (Figure 4C).
The influence of Lnc5q21.2 in ATR signaling was evaluated by colony formation. Lnc5q21.2 unexpressed and expressed cells were treated with cisplatin, VE-822 and cisplatin+VE-822 and the colony formation efficiency was normalized by untreated empty vector group. Under these three different treatments, the colony formation efficiency was 94.97% ± 2.53% vs. 96.51% ± 1.36% (P > 0.05), 71.71% ± 2.50% vs. 72.43% ± 22.40% (P > 0.05) and 78.45% ± 5.88% vs. 30.40% ± 4.01% (P < 0.05) in RKO cells without and with expression of Lnc5q21.2. It was 69.47% ± 4.52% vs. 64.87% ± 3.99% (P > 0.05), 62.70% ± 4.31% vs. 67.30% ± 0.64% (P > 0.05) and 58.33% ± 2.46% vs. 30.39% ± 3.73% (P < 0.001) in LS180 cells before and after re-expression of Lnc5q21.2. The efficiency was 84.69% ± 2.14% vs. 74.77% ± 6.22% (P > 0.05), 47.10% ± 9.24% vs. 55.26% ± 9.35% (P > 0.05) and 24.65% ± 2.73% vs. 47.60% ± 3.08% (P < 0.05) before and after deleting Lnc5q21.2 in HCT116 cells (Figure 4D). The results demonstrated that the colony formation efficiency was reduced by Lnc5q21.2 under cisplatin+VE-822 treatment. No significant difference was observed with cisplatin or VE-822 single reagent treatment group, with or without Lnc5q21.2 expression.
To further verify the sensitivity of Lnc5q21.2 to ATR inhibitor under cisplatin treatment, xenograft mouse models were used. Before and after re-expression of Lnc5q21.2, the tumor volumes were 213.38 ± 0.99 mm3 vs. 401.27 ± 73.92 mm3 (P < 0.05), 231.27 ± 75.07 mm3 vs. 265.23 ± 88.59 mm3 (P > 0.05), 217.26 ± 58.02 mm3 vs. 91.29 ± 20.68 mm3 (P < 0.05) in cisplatin, VE-822 and cisplatin plus VE-822 treatment, respectively. The tumor volume was reduced significantly in the combination of cisplatin and VE-822 treatment group by re-expressing Lnc5q21.2 (Figure 5A and 5B). In cisplatin, VE-822 and cisplatin plus VE-822 treatment groups, the tumor weights were 0.034 ± 0.012 g vs. 0.113 ± 0.027 g (P < 0.05), 0.044 ± 0.009 g vs. 0.050 ± 0.026 g (P > 0.05), 0.040 ± 0.015 g vs. 0.006 ± 0.003 g (P < 0.05) before and after re-expression of Lnc5q21.2, respectively. Reduced tumor weight was observed by cisplatin and VE- 822 treatment in Lnc5q21.2 re-expressed cell xenografts (Figure 5C). These results suggest that Lnc5q21.2 enforced the sensitivity of cisplatin treated CRC to VE- 822.
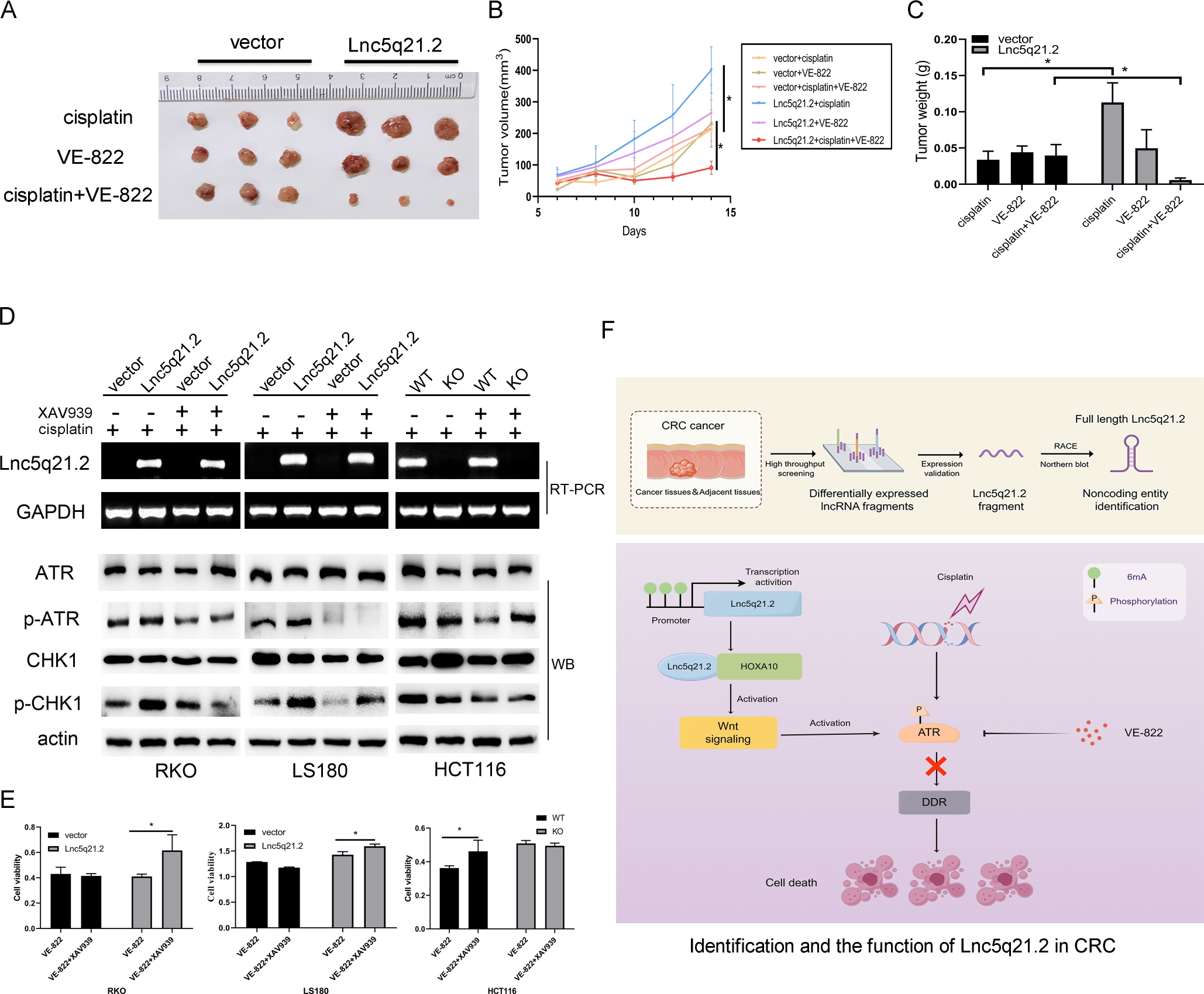
Lnc5q21.2 sensitives CRC cell to ATR inhibitor. A: Represents results for Lnc5q21.2 unexpressed and re-expressed RKO cell xenografts under various treatment. B: The growth curves of Lnc5q21.2 unexpressed and re-expressed RKO cell xenografts under various treatment. C: Tumor weight in Lnc5q21.2 unexpressed and re-expressed RKO cell xenografts under various treatment. D: The p-ATR and p-CHK1 was detected before and after XAV939 treatment under employing low dose cisplatin. XAV939, Wnt/β-catenin signaling inhibitor. E: Represent cell viability evaluated by MTT assay in CRC cells with or without XAV939 and VE-822 treatment. *P < 0.05. F: Identification and the function of Lnc5q21.2 in CRC. CRC, colorectal cancer.
To get more clues for Lnc5q21.2 promoting ATR signaling by activating Wnt signaling, XAV939 was employed. Under low dose cisplatin treatment, the total levels of ATR and CHK1 were not changed with or without Lnc5q21.2 expression. While the levels of p-ATR and p-CHK1 were decreased by XAV939 in Lnc5q21.2 expressed CRC cells (Figure 5D).
To gain more evidence of Lnc5q21.2 activating ATR signaling through Wnt signaling, XAV939 and VE-822 were applied under low dose cisplatin treatment. The OD value was 0.431 ± 0.05 vs. 0.415 ± 0.02 (P > 0.05), and 0.411 ± 0.02 vs. 0.616 ± 0.12 (P < 0.05) for VE-822 or VE-822 plus XAV939 treatment groups, in Lnc5q21.2 unexpressed or over-expressed RKO cells, respectively. It was 1.29 ± 0.00 vs. 1.18 ± 0.01 (P > 0.05), and 1.43 ± 0.06 vs. 1.59 ± 0.04 (P < 0.05) for VE-822 or VE-822 plus XAV939 treatment groups, in Lnc5q21.2 unexpressed or over-expressed LS180 cells, respectively. In HCT116 cells, the OD value was 0.36 ± 0.01 vs. 0.47 ± 0.06 (P < 0.05) and 0.51 ± 0.02 vs. 0.49 ± 0.02 (P > 0.05) for VE-822 or VE-822 plus XAV939 treatment groups, before and after deletion of Lnc5q21.2, respectively. In Lnc5q21.2 unexpressed CRC cells, no significant difference was shown for ATR between Wnt plus ATR inhibitor groups (Figure 5E). It was significantly different in Lnc5q21.2 expressed cells for the treatment using ATR inhibitor and both Wnt and ATR inhibitors. The results were further validated by siRNA knockdown of β-catenin, and ICG-001, another Wnt signaling inhibitor (Supplementary Figure S7 and S8). These data suggest that Lnc5q21.2 promotes ATR signaling through Wnt signaling.
Discussion
A recent study for the first time discovered that a lncRNA, which is overlapped with cortex gene, links to the evolution of a visible trait in animals, suggesting “the key regulator is RNA, not a protein”.[5] In human genome, roughly 80% of diseases-related phenotype loci was identified to be associated to noncoding genome regions. Nonetheless, the linkage between phenotype-related loci and lncRNA-associated diseases almost remains uncharacterized.[37] LncRNA has been recognized as important modulators in physiology and disease states, including carcinogenesis.[4] Grasping cancer-related lncRNAs and unlocking their regulatory-circuits may garner diagnostic markers and innovate therapeutic strategies. In this study, we identified a novel lncRNA and its full-length sequence is 668 nt. The lncRNA was designated as Lnc5q21.2, owing to the genomic coding region locating in Chromosome 5q21.2. The expression of Lnc5q21.2 is regulated by 6mA in the promoter region. The location of molecules in subcellular may roughly reflect their role in cells. Lnc5q21.2 was detected mainly in the nucleus, implying to join the regulating action. The irregular expression of LncRNA hints the role in carcinogenesis and progression. The expression of Lnc5q21.2 was increased with a progression tendency from noncancerous colorectal mucosa to adjacent tissue, adenoma and CRC, reminding the possible oncogenic function. The high levels of Lnc5q21.2 is associated with alcohol and may serve as an independent poor 5-year OS marker. LncRNAs may serve as oncogenes to promote cancer growth, or tumor suppressors to inhibit tumor progression depending on distinct tumor types.[38] Lnc5q21.2 promotes CRC cell proliferation, migration, invasion and cell cycle progression. It promotes tumor growth in CRC cell xenograft mice. These results provide some evidence for Lnc5q21.2 as an oncogenic molecule.
LncRNAs play their role by interacting with DNA, RNA and proteins.[4] The interaction of Lnc5q21.2 with HOXA10 was revealed and verified by RNA pulldown, RIP and western blot techniques. The binding sequence was identified to be located at 400–668nt of Lnc5q21.2. Given that HOXA10 was involved in cell fate determining signaling, including DDR, Wnt, Sonic hedgehog and Notch signaling pathways.[35,39,40,41] Next, we tried to find the target genes of HOXA10 through Lnc5q21.2. The combination of HOXA10, Lnc5q21.2 and the promoter region sequence of EMX2 was discovered by ChIP-seq and validated by ChIP-PCR and ChIRP assays using HOXA10 antibody and labeled nuclear probe from Lnc5q21.2. This interaction existed in Lnc5q21.2 highly expressed cells, without emerging in unexpressed cells. HOXA10 has been shown to regulate Wnt signaling via directly binding to the promoter region of EMX2.[31,34,42] HOXA10 has been reported to be associated with chemo-resistance through regulating HR pathway, and Wnt signaling was verified to modulate cell fate determination and DDR.[35,43,44] The role of Lnc5q21.2 in Wnt signaling was therefore explored. As expected, Lnc5q21.2 activated Wnt signaling through interacting with HOXA10 to modulate the expression of EMX2. To further understand the regulation network of Wnt signaling and HR, the mechanism of Lnc5q21.2 in HR was investigated. In CRC cells, Lnc5q21.2 enhanced ATR signaling by promoting Wnt signaling, while without influencing of ATM signaling.
With the deep insight into the mechanism of LncRNA, more attentions were paid for its application in cancer detection and therapeutics.[45] Targeting DDR has been innovated cancer therapy.[46,47] In CRC, Lnc5q21.2 increased the sensitivity of ATR inhibitor both in vitro and in vivo. In conclusions, Lnc5q21.2 is a novel long intergenic noncoding RNA. The expression of Lnc5q21.2 is regulated by 6mA modification. Highly expression of Lnc5q21.2 is associated with alcohol and serves as a potential independent poor prognostic marker. Lnc5q21.2 promotes ATR repair by activating Wnt signaling through interacting with HOXA10 via modulating EMX2 expression. Lnc5q21.2 plays an oncogenic role in CRC and serves as a sensitive marker for ATR inhibitor (Figure 5F).
Supplementary Information
Supplementary materials are only available at the official site of the journal (www.intern-med.com).
Funding statement: This work was supported by the National Key Research and Development Program of China (grant number 2020YFC2002705); National Science Foundation of China (grant number 82272632, 81672138); Youth Innovation Science Foundation of Chinese PLA general hospital (grant number 22QNCZ027, 22QNFC069); National Science Foundation of China (grant number 82403742).
Acknowledgements
We are grateful to Stephen B. Baylin for giving the colorectal cancer cell line DKO as a gift.
-
Author Contributions
M. Zhang: Conceptualization, Project administration, Data curation, Writing original draft. C. Zhu: Data curation, Formal analysis, Writing original draft, Validation. A. Gao: Data curation. J. G. Herman: Supervision. François Fuks: Supervision. J. Luo: Data curation. X. Su: Methodology. H. Cui: Supervision. R. Chen: Supervision. M. Guo: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing review& editing. All authors read and approved the final manuscript.
-
Ethical Approval
All tissue samples were obtained from the Chinese PLA General Hospital, under the guidelines approved by the institutional review board at the Chinese PLA General Hospital (NO.20090701-015). The animal experiments were approved by the Animal Ethics Committee of the Chinese PLA General Hospital (NO.2022-X18-119).
-
Informed Consent
Not applicable.
-
Conflict of Interest
The authors declare no competing interest.
-
Use of Large Language Models, AI and Machine Learning Tools
None declared.
-
Data Availability Statement
Correspondence and requests for materials should be addressed to Mingzhou Guo.
References
1 Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA 2021;325:669–685.10.1001/jama.2021.0106Search in Google Scholar PubMed
2 Chung DC, Gray DM 2nd, Singh H, Issaka RB, Raymond VM, Eagle C, et al. A Cell-free DNA Blood-Based Test for Colorectal Cancer Screening. N Engl J Med 2024;390:973–983.10.1056/NEJMoa2304714Search in Google Scholar PubMed
3 Rinn JL, Chang HY. Long Noncoding RNAs: Molecular Modalities to Organismal Functions. Annu Rev Biochem 2020;89:283–308.10.1146/annurev-biochem-062917-012708Search in Google Scholar PubMed
4 Winkler L, Dimitrova N. A mechanistic view of long noncoding RNAs in cancer. Wiley Interdiscip Rev RNA 2022;13:e1699.10.1002/wrna.1699Search in Google Scholar PubMed PubMed Central
5 Pennisi E. Surprise RNA paints colorful patterns on butterfly wings. Science 2024;383:1039–1040.10.1126/science.adp0471Search in Google Scholar PubMed
6 Ganesh VS, Riquin K, Chatron N, Yoon E, Lamar KM, Aziz MC, et al. Neurodevelopmental Disorder Caused by Deletion of CHASERR, a lncRNA Gene. N Engl J Med 2024;391:1511–1518.10.1056/NEJMoa2400718Search in Google Scholar PubMed PubMed Central
7 Balusu S, Horré K, Thrupp N, Craessaerts K, Snellinx A, Serneels L, et al. MEG3 activates necroptosis in human neuron xenografts modeling Alzheimer's disease. Science 2023;381:1176–1182.10.1126/science.abp9556Search in Google Scholar PubMed PubMed Central
8 Liao X, Zhang S, Li X, Qian W, Li M, Chen S, et al. Dynamic structural remodeling of LINC01956 enhances temozolomide resistance in MGMT-methylated glioblastoma. Sci Transl Med 2024;16:eado1573.10.1126/scitranslmed.ado1573Search in Google Scholar PubMed
9 Kilgas S, Syed A, Toolan-Kerr P, Swift ML, Roychoudhury S, Sarkar A, et al. NEAT1 modulates the TIRR/53BP1 complex to maintain genome integrity. Nat Commun 2024;15:8438.10.1038/s41467-024-52862-wSearch in Google Scholar PubMed PubMed Central
10 Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015;47:199–208.10.1038/ng.3192Search in Google Scholar PubMed PubMed Central
11 Pinkney HR, Ross CR, Hodgson TO, Pattison ST, Diermeier SD. Discovery of prognostic lncRNAs in colorectal cancer using spatial transcriptomics. NPJ Precis Oncol 2024;8:230.10.1038/s41698-024-00728-1Search in Google Scholar PubMed PubMed Central
12 Jin Y, Zuo Y, Li G, Liu W, Pan Y, Fan T, et al. Advances in spatial transcriptomics and its applications in cancer research. Mol Cancer 2024;23:129.10.1186/s12943-024-02040-9Search in Google Scholar PubMed PubMed Central
13 Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell 2015;28:529–540.10.1016/j.ccell.2015.09.006Search in Google Scholar PubMed PubMed Central
14 Arun G, Diermeier SD, Spector DL. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol Med 2018;24:257–277.10.1016/j.molmed.2018.01.001Search in Google Scholar PubMed PubMed Central
15 Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015;21:1253–1261.10.1038/nm.3981Search in Google Scholar PubMed
16 Rheinbay E, Nielsen MM, Abascal F, Wala JA, Shapira O, Tiao G, et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 2020;578:102–111.10.1038/s41586-020-1965-xSearch in Google Scholar PubMed PubMed Central
17 Gao A, Bai P, Zhang M, Yao Y, Herman JG, Guo M. RASSF1A promotes ATM signaling and RASSF1A methylation is a synthetic lethal marker for ATR inhibitors. Epigenomics 2023;15:1205–1220.10.2217/epi-2023-0306Search in Google Scholar PubMed
18 Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008;3:1101–1108.10.1038/nprot.2008.73Search in Google Scholar PubMed
19 Li H, Zhang M, Linghu E, Zhou F, Herman JG, Hu L, et al. Epigenetic silencing of TMEM176A activates ERK signaling in human hepatocellular carcinoma. Clin Epigenetics 2018;10:137.10.1186/s13148-018-0570-4Search in Google Scholar PubMed PubMed Central
20 Liu L, Yue H, Liu Q, Yuan J, Li J, Wei G, et al. LncRNA MT1JP functions as a tumor suppressor by interacting with TIAR to modulate the p53 pathway. Oncotarget 2016;7:15787–15800.10.18632/oncotarget.7487Search in Google Scholar PubMed PubMed Central
21 Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP). J Vis Exp 2012:3912.10.3791/3912Search in Google Scholar PubMed PubMed Central
22 Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 2007;35:W345–W349.10.1093/nar/gkm391Search in Google Scholar PubMed PubMed Central
23 Wang G, Yin H, Li B, Yu C, Wang F, Xu X, et al. Characterization and identification of long non-coding RNAs based on feature relationship. Bioinformatics 2019;35:2949–2956.10.1093/bioinformatics/btz008Search in Google Scholar PubMed
24 Li A, Zhang J, Zhou Z. PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinformatics 2014;15:311.10.1186/1471-2105-15-311Search in Google Scholar PubMed PubMed Central
25 Wu Z, Liu X, Liu L, Deng H, Zhang J, Xu Q, et al. Regulation of lncRNA expression. Cell Mol Biol Lett 2014;19:561–575.10.2478/s11658-014-0212-6Search in Google Scholar PubMed PubMed Central
26 Boulias K, Greer EL. Means, mechanisms and consequences of adenine methylation in DNA. Nat Rev Genet 2022;23:411–428.10.1038/s41576-022-00456-xSearch in Google Scholar PubMed PubMed Central
27 Li Y, Zhang XM, Luan MW, Xing JF, Chen J, Xie SQ. Distribution Patterns of DNA N6-Methyladenosine Modification in Non-coding RNA Genes. Front Genet 2020;11:268.10.3389/fgene.2020.00268Search in Google Scholar PubMed PubMed Central
28 Xiao CL, Zhu S, He M, Chen D, Zhang Q, Chen Y, et al. N<sup>6</sup>-Methyladenine DNA Modification in the Human Genome. Mol Cell 2018;71:306–318.10.1016/j.molcel.2018.06.015Search in Google Scholar PubMed
29 Herman AB, Tsitsipatis D, Gorospe M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol Cell 2022;82:2252–2266.10.1016/j.molcel.2022.05.027Search in Google Scholar PubMed PubMed Central
30 Yoshida M, Suda Y, Matsuo I, Miyamoto N, Takeda N, Kuratani S, et al. Emx1 and Emx2 functions in development of dorsal telencephalon. Development 1997;124:101–111.10.1242/dev.124.1.101Search in Google Scholar PubMed
31 Troy PJ, Daftary GS, Bagot CN, Taylor HS. Transcriptional repression of peri-implantation EMX2 expression in mammalian reproduction by HOXA10. Mol Cell Biol 2003;23:1–13.10.1128/MCB.23.1.1-13.2003Search in Google Scholar PubMed PubMed Central
32 Zhang Y, Cao G, Yuan QG, Li JH, Yang WB. Empty Spiracles Homeobox 2 (EMX2) Inhibits the Invasion and Tumorigenesis in Colorectal Cancer Cells. Oncol Res 2017;25:537–544.10.3727/096504016X14756640150695Search in Google Scholar PubMed PubMed Central
33 Wang C, Li Y, Yu K, Jiang Z, Wang Y, Yang G. HOXA10 inhibit the osteogenic differentiation of periodontal ligament stem cells by regulating β-catenin localization and DKK1 expression. Connect Tissue Res 2021;62:393–401.10.1080/03008207.2020.1756271Search in Google Scholar PubMed
34 Sarno JL, Kliman HJ, Taylor HS. HOXA10, Pbx2, and Meis1 protein expression in the human endometrium: formation of multimeric complexes on HOXA10 target genes. J Clin Endocrinol Metab 2005;90:522–528.10.1210/jc.2004-0817Search in Google Scholar PubMed
35 Kim JW, Kim JY, Kim JE, Kim SK, Chung HT, Park CK. HOXA10 is associated with temozolomide resistance through regulation of the homologous recombinant DNA repair pathway in glioblastoma cell lines. Genes Cancer 2014;5:165–174.10.18632/genesandcancer.16Search in Google Scholar PubMed PubMed Central
36 Karimaian A, Majidinia M, Bannazadeh Baghi H, Yousefi B. The crosstalk between Wnt/β-catenin signaling pathway with DNA damage response and oxidative stress: Implications in cancer therapy. DNA Repair (Amst) 2017;51:14–19.10.1016/j.dnarep.2017.01.003Search in Google Scholar PubMed
37 Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R, Luo X, Bhagat G, Green PH, et al. A long noncoding RNA associated with susceptibility to celiac disease. Science 2016;352:91–95.10.1126/science.aad0467Search in Google Scholar PubMed PubMed Central
38 Song H, Adu-Amankwaah J, Zhao Q, Yang D, Liu K, Bushi A, et al. Decoding long non‑coding RNAs: Friends and foes in cancer development (Review). Int J Oncol 2024;64:61.10.3892/ijo.2024.5649Search in Google Scholar PubMed PubMed Central
39 Li J, Chang J, Wang J, Xu D, Yang M, Jiang Y, et al. HOXA10 promote pancreatic cancer progression via directly activating canonical NF-κB signaling pathway. Carcinogenesis 2022;43:787–796.10.1093/carcin/bgac042Search in Google Scholar PubMed
40 Song C, Han Y, Luo H, Qin Z, Chen Z, Liu Y, et al. HOXA10 induces BCL2 expression, inhibits apoptosis, and promotes cell proliferation in gastric cancer. Cancer Med 2019;8:5651–5661.10.1002/cam4.2440Search in Google Scholar PubMed PubMed Central
41 Sengupta A, Banerjee D, Chandra S, Banerji SK, Ghosh R, Roy R, et al. Deregulation and cross talk among Sonic hedgehog, Wnt, Hox and Notch signaling in chronic myeloid leukemia progression. Leukemia 2007;21:949–955.10.1038/sj.leu.2404657Search in Google Scholar PubMed
42 Jimenez-García MP, Lucena-Cacace A, Otero-Albiol D, Carnero A. Empty spiracles homeobox genes EMX1 and EMX2 regulate WNT pathway activation in sarcomagenesis. J Exp Clin Cancer Res 2021;40:247.10.1186/s13046-021-02048-9Search in Google Scholar PubMed PubMed Central
43 Zhong Z, Virshup DM. Wnt Signaling and Drug Resistance in Cancer. Mol Pharmacol 2020;97:72–89.10.1124/mol.119.117978Search in Google Scholar PubMed
44 Kaur A, Lim JYS, Sepramaniam S, Patnaik S, Harmston N, Lee MA, et al. WNT inhibition creates a BRCA-like state in Wnt-addicted cancer. EMBO Mol Med 2021;13:e13349.10.15252/emmm.202013349Search in Google Scholar PubMed PubMed Central
45 Nemeth K, Bayraktar R, Ferracin M, Calin GA. Non-coding RNAs in disease: from mechanisms to therapeutics. Nat Rev Genet 2024;25:211–232.10.1038/s41576-023-00662-1Search in Google Scholar PubMed
46 Zhang M, Li X, Herman JG, Gao A, Wang Q, Yao Y, et al. Methylation of NRIP3 Is a Synthetic Lethal Marker for Combined PI3K and ATR/ATM Inhibitors in Colorectal Cancer. Clin Transl Gastroenterol 2024;15:e00682.10.14309/ctg.0000000000000682Search in Google Scholar PubMed PubMed Central
47 Gao A, Guo M. Epigenetic based synthetic lethal strategies in human cancers. Biomark Res 2020;8:44.10.1186/s40364-020-00224-1Search in Google Scholar PubMed PubMed Central
© 2025 Meiying Zhang, Cheng Zhu, Aiai Gao, James G. Herman, François Fuks, Jianjun Luo, Xiaomo Su, Hengmi Cui, Runsheng Chen, Mingzhou Guo, published by De Gruyter on behalf of Scholar Media Publishing
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Perspective
- Prehospital neuroprotective intervention: A critical imperative evidenced by the FRONTIER trial outcomes
- Artificial intelligence in rheumatology: A transformative perspective
- Dialysis adequacy revisited: Kt/V's blind spot for phosphorus and iodine
- Review Article
- The plakin family: Potential therapeutic targets for digestive system tumors
- Original Article
- Lnc5q21.2, a novel long intergenic RNA, sensitizes colorectal cancer cells to ATR inhibitor by activating Wnt pathway
- Effect of bone marrow mononuclear cells therapy on long-term survival of patients with liver cirrhosis: A 10-year retrospective cohort study using propensity score matching
- Associations between serum sex steroid hormone metabolites and gastric cancer and precancerous lesions in men: A 11.8-year prospective study
- Comprehensive investigation of cuproptosis-related genes in clinical features, biological characteristics, and immune microenvironment in B-cell Non-Hodgkin lymphoma
- Protocol
- A multicenter, prospective, randomized, open-label, blinded endpoint trial of intravenous thrombolysis with tenecteplase for acute non-large vessel occlusion in extended time window (OPTION): Rationale and design
- Rapid Communication
- Association between anemia and 1-year recurrence of paroxysmal atrial fibrillation after radiofrequency ablation
Articles in the same Issue
- Perspective
- Prehospital neuroprotective intervention: A critical imperative evidenced by the FRONTIER trial outcomes
- Artificial intelligence in rheumatology: A transformative perspective
- Dialysis adequacy revisited: Kt/V's blind spot for phosphorus and iodine
- Review Article
- The plakin family: Potential therapeutic targets for digestive system tumors
- Original Article
- Lnc5q21.2, a novel long intergenic RNA, sensitizes colorectal cancer cells to ATR inhibitor by activating Wnt pathway
- Effect of bone marrow mononuclear cells therapy on long-term survival of patients with liver cirrhosis: A 10-year retrospective cohort study using propensity score matching
- Associations between serum sex steroid hormone metabolites and gastric cancer and precancerous lesions in men: A 11.8-year prospective study
- Comprehensive investigation of cuproptosis-related genes in clinical features, biological characteristics, and immune microenvironment in B-cell Non-Hodgkin lymphoma
- Protocol
- A multicenter, prospective, randomized, open-label, blinded endpoint trial of intravenous thrombolysis with tenecteplase for acute non-large vessel occlusion in extended time window (OPTION): Rationale and design
- Rapid Communication
- Association between anemia and 1-year recurrence of paroxysmal atrial fibrillation after radiofrequency ablation

