Performance of Fujirebio Espline SARS-CoV-2 rapid antigen test for identifying potentially infectious individuals
-
Gian Luca Salvagno
To the Editor,
An important aspect, that has become clear several months since the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) outbreak has begun, is that diagnostic testing is critical for prevention, diagnosis, prognostication and clinical management of coronavirus disease 2019 (COVID-19) [1, 2]. Recent statistics attest that the dramatic demand for diagnostic testing has placed enormous pressure on clinical laboratories [3, 4], frequently plagued by shortage of technical and human resources [5]. The usage of rapid molecular or antigen diagnostic assays is a possible solution to overcome such ongoing limitation of SARS-CoV-2 test availability, enabling to process large numbers of samples in short time, with reasonable costs, even outside laboratory environments [6, 7]. In an official pronunciation, the World Health Organization (WHO) recently concluded that lateral flow immunoassays for SARS-CoV-2 antigen detection display lower sensitivity than molecular test, but have the advantages of providing rapid, relatively cheap and timely identification of most infectious COVID-19 cases [8]. The Fujirebio Espline SARS-CoV-2 is a rapid antigen test that has recently been commercialized. Since the performance of this test for diagnosing SARS-CoV-2 infection has been assessed in previous investigations [9], [10], [11], we focused on its potential effectiveness for detecting patients with high nasopharyngeal viral load, who may be responsible for the vast majority of community SARS-CoV-2 transmission [12, 13].
The study population consisted of a series of patients with laboratory confirmed SARS-CoV-2 infection determined by molecular testing at the Pederzoli Hospital (Peschiera del Garda, Verona, Italy) between 5 and 19 April, 2021. Upper respiratory specimens were collected in agreement with WHO guidelines [14]. A single swab (Virus swab UTM™, Copan, Brescia, Italy) was collected from each patient and concomitantly used for both antigen screening and molecular testing, within 1 h from collection.
The Fujirebio Espline SARS-CoV-2 (Fujirebio Inc., Tokyo, Japan) is an immunochromatographic test for SARS-CoV-2 nucleocapsid (N) antigen detection by means of enzyme immunoassay principle. The method, encompassing a simple manual procedure without dedicated instrumentation, uses 20 μL of samples and provides final result in 30 min. Briefly, 20 µL of specimen are mixed with 10 μL of concentrated treatment solution for 5 min and then added to the testing cassette in correspondence of the sample window [9], where SARS-CoV-2 nucleocapsid antigens eventually present in the sample bind to anti-SARS-CoV-2 nucleocapsid antibodies conjugated with alkaline phosphatase (ALP) and migrate towards the interpretation window area. The nucleocapsid antigen-antibody complexes are then captured by anti-SARS-CoV-2 antibodies immobilized on the SARS-CoV-2 test line. The ALP enzyme reacts with the substrate, forming a colored test line within the interpretation window, while excess ALP-labeled antibodies continue their migration up to the reference window, generating another colored line. The appearance of a colored line within the interpretation window reflects sample positivity for SARS-CoV-2 nucleocapsid antigens.
Altona Diagnostics RealStar® SARSCoV-2 RT-PCR Kit (Altona Diagnostics GmbH, Hamburg, Germany) is a real-time reverse transcription polymerase chain reaction (rRT-PCR) test intended for detection of SARS-CoV-2 RNA. The method encompasses two different amplifications and detections, the former deigned to target SARS-CoV-2 E gene (specific for lineage B-betacoronavirus, thus including SARS-CoV-2), and the latter targeting a specific sequence within the SARS-CoV-2 S gene. RNA was extracted using AltoStar® Purification Kit 1.5 (Altona Diagnostics), and the assay was carried out on a Bio-Rad CFX96™ Deep Well Dx Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). The final test results were reported as quantitative and qualitative measures, respectively as cycle threshold (Ct) values or by mirroring higher risk of sample infectivity with Ct values of both E and S genes <29.5 [15].
The diagnostic efficiency of Fujirebio Espline SARS-CoV-2 was assessed by constructing a receiver operating characteristic (ROC) curve and calculating the diagnostic accuracy, sensitivity and specificity at the diagnostic threshold for potential sample infectivity, with Analyse-it software (Analyse-it Software Ltd, Leeds, UK). Results were presented as mean and standard deviation (SD). The investigation was carried out as part of routine clinical laboratory operations, using pre-existing specimens collected for systematic SARS-CoV-2 diagnostic screening (with antigen immunoassay) and molecular testing at the local facility, and thereby patient informed consent and Ethical Committee approval were unnecessary. All test results were anonymized prior to statistical analysis. The study was conducted in accordance with the Declaration of Helsinki, under the terms of relevant local legislation.
The final study population consisted of 174 patients (mean age, 43 ± 19 years; 80 women, 46%) with positive SARS-CoV-2 molecular test at our institution during the study period. The Ct values of these positive samples were 30.5 ± 7.2 for the SARS-CoV-2 S gene and 30.9 ± 7.0 for the SARS-CoV-2 E gene, respectively. A high Spearman’s correlation was found between the Ct values of the two genes (r=0.97; 95% CI, 0.97–0.98; p<0.001). A total number of 47/174 samples tested positive with Fujirebio Espline SARS-CoV-2, thus accounting for 27% positive percent agreement (95% CI, 21–34%).
A total number of 59 nasopharyngeal samples (33.9%) displayed Ct values of both S and E genes <29.5 and were hence associated with higher risk of infectivity [15]. The diagnostic efficiency of Fujirebio Espline SARS-CoV-2 for identifying these samples is shown in Figure 1. The AUC was 0.89 (95% CI, 0.83–0.94; p<0.001), with 0.92 accuracy (95% CI, 0.87–0.96), 0.78 sensitivity (95% CI, 0.65–0.88, corresponding to 13 false negative test results) and 0.99 specificity (95% CI, 0.95–1.00, corresponding to 1 false positive test result). The mean Ct value of the 13 negative samples was 27.6 (range, 24.6–29.2) for the S gene and 28.3 (range, 25.9–29.4) for the E gene, respectively.
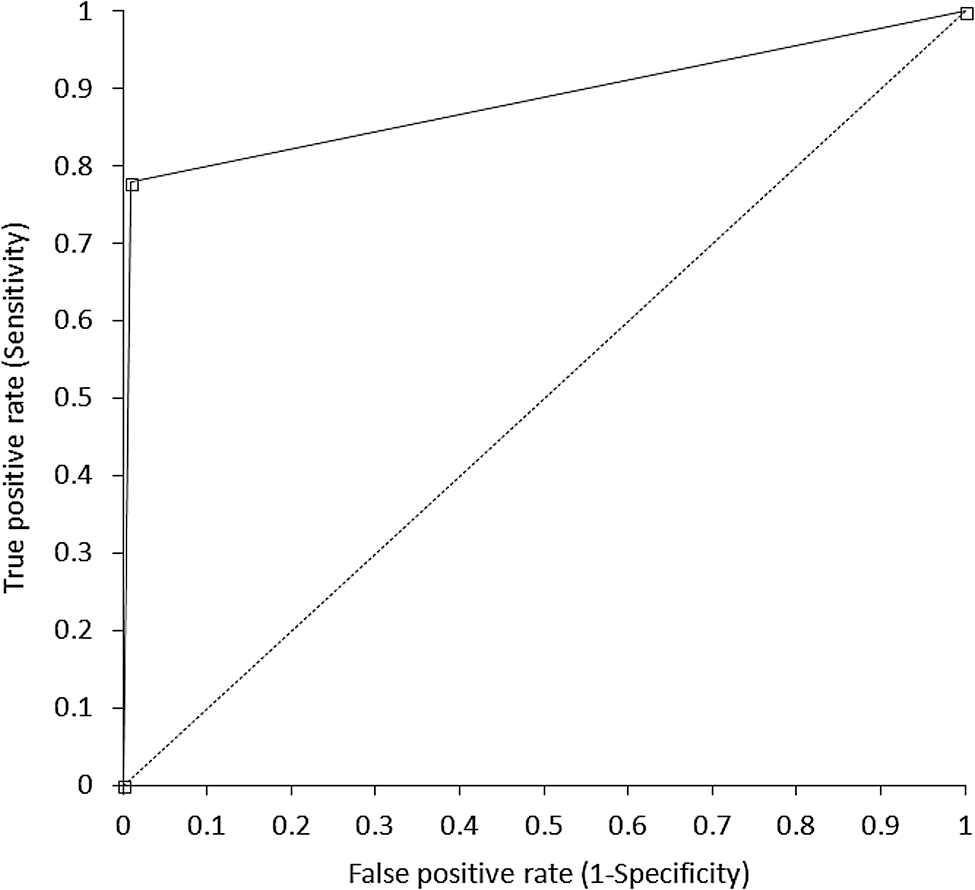
Receiver operating characteristic (ROC) curve Fujirebio Espline SARS-CoV-2 rapid antigen test for identifying potentially infectious individuals (i.e., cycle threshold <29.5).
Unlike previous investigations [9], [10], [11, 16], we undertook this study to explore whether Fujirebio Espline SARS-CoV-2 may have sufficient diagnostic performance for identifying subjects with higher nasopharyngeal viral load, who may also be potential high risk sources of contagion, rather than more specifically addressing its performance for diagnosing COVID-19. This is in line with the recent recommendations of the Task Force on COVID-19 of the International Federation of Clinical Chemistry and Laboratory Medicine (IFCC), which endorse the use of such tests for screening rather than for diagnosing SARS-CoV-2 infection [17].
Taken together, our results demonstrate that Fujirebio Espline SARS-CoV-2 has remarkably high specificity, close to 100%, for assessing potential subject infectivity from nasopharyngeal specimens, such that a positive test result would allow to identify with near perfect certainty patients with high nasopharyngeal viral load, i.e., those most likely to spread the virus in the community. On the other hand, the sensitivity we found in our subjects was 78%, thus reflecting that a meaningful number of infective patients may be missed with this test. Nonetheless, no Ct values <24.6 were observed for either SARS-CoV-2 gene in these false negative cohort, which thus reflects a rather limited risk (i.e., between 2 and 3%) of being associated with a positive viral culture according to the data earlier published by Gniazdowski et al. [15].
In conclusion, this study demonstrates that the Fujirebio Espline SARS-CoV-2 rapid antigen test has excellent specificity for identifying patients at high risk of being active sources of contagion, whilst its sensitivity would not allow to completely rule out the risk that subjects testing negative carry low infective risk. Therefore, this rapid antigen test would find ideal placing for rapid, inexpensive and decentralized screening of subjects with high SARS-CoV-2 viral load in high prevalence areas, who shall be timely isolated and/or treated (when becoming symptomatic).
-
Research funding: The authors received no funding for this work.
-
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
-
Competing interests: Authors state no conflict of interest.
-
Informed consent: Not applicable.
-
Ethical approval: Not applicable.
References
1. Schnitzler, L, Janssen, LMM, Evers, SMAA, Jackson, LJ, Paulus, ATG, Roberts, TE, et al.. The broader societal impacts of COVID-19 and the growing importance of capturing these in health economic analyses. Int J Technol Assess Health Care 2021;37:e43. https://doi.org/10.1017/s0266462321000155.Search in Google Scholar PubMed
2. Lippi, G, Plebani, M. The critical role of laboratory medicine during coronavirus disease 2019 (COVID-19) and other viral outbreaks. Clin Chem Lab Med 2020;58:1063–9. https://doi.org/10.1515/cclm-2020-0240.Search in Google Scholar PubMed
3. Our World in Data. Daily COVID-19 tests. Available from: https://ourworldindata.org/grapher/full-list-covid-19-tests-per-day [Accessed 4 Aug 2021].Search in Google Scholar
4. Lippi, G, Mattiuzzi, C, Santos de Oliveira, MH, Henry, BM. Clinical predictors of SARS-CoV-2 testing pressure on clinical laboratories: a multinational study analyzing google trends and over 100 million diagnostic tests. Lab Med 2021;52:311–4. https://doi.org/10.1093/labmed/lmab013.Search in Google Scholar PubMed PubMed Central
5. American Association of Clinical Chemistry. Coronavirus testing survey. Available from: https://www.aacc.org/science-and-research/covid-19-resources/aacc-covid-19-testing-survey [Accessed 4 Aug 2021].Search in Google Scholar
6. Mattiuzzi, C, Henry, BM, Lippi, G. Making sense of rapid antigen testing in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) diagnostics. Diagnosis (Berl) 2020;8:27–31. https://doi.org/10.1515/dx-2020-0131.Search in Google Scholar PubMed
7. Dinnes, J, Deeks, JJ, Berhane, S, Taylor, M, Adriano, A, Davenport, C, et al.. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev 2021;3:CD013705. https://doi.org/10.1002/14651858.CD013705.Search in Google Scholar PubMed PubMed Central
8. World Health Organization. Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays; 2020.Search in Google Scholar
9. Aoki, K, Nagasawa, T, Ishii, Y, Yagi, S, Kashiwagi, K, Miyazaki, T, et al.. Evaluation of clinical utility of novel coronavirus antigen detection reagent, Espline® SARS-CoV-2. J Infect Chemother 2021;27:319–32. https://doi.org/10.1016/j.jiac.2020.11.015.Search in Google Scholar PubMed PubMed Central
10. Basso, D, Aita, A, Padoan, A, Cosma, C, Navaglia, F, Moz, S, et al.. Salivary SARS-CoV-2 antigen rapid detection: a prospective cohort study. Clin Chim Acta 2021;517:54–9. https://doi.org/10.1016/j.cca.2021.02.014.Search in Google Scholar PubMed PubMed Central
11. Ishii, T, Sasaki, M, Yamada, K, Kato, D, Osuka, H, Aoki, K, et al.. Immunochromatography and chemiluminescent enzyme immunoassay for COVID-19 diagnosis. J Infect Chemother 2021;27:915–8. https://doi.org/10.1016/j.jiac.2021.02.025.Search in Google Scholar PubMed PubMed Central
12. Rao, SN, Manissero, D, Steele, VR, Pareja, J. A systematic review of the clinical utility of cycle threshold values in the context of COVID-19. Infect Dis Ther 2020;9:573–86. https://doi.org/10.1007/s40121-020-00324-3.Search in Google Scholar PubMed PubMed Central
13. Singanayagam, A, Patel, M, Charlett, A, Lopez Bernal, J, Saliba, V, Ellis, J, et al.. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill 2020;25:2001483. https://doi.org/10.2807/1560-7917.es.2020.25.32.2001483.Search in Google Scholar
14. World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases. Interim guidance; 2020.Search in Google Scholar
15. Gniazdowski, V, Morris, CP, Wohl, S, Mehoke, T, Ramakrishnan, S, Thielen, P, et al.. Repeat COVID-19 molecular testing: correlation of SARS-CoV-2 culture with molecular assays and cycle thresholds. Clin Infect Dis 2021;73:e860–9. https://doi.org/10.1093/cid/ciaa1616.Search in Google Scholar PubMed PubMed Central
16. Menchinelli, G, Bordi, L, Liotti, FM, Palucci, I, Capobianchi, MR, Sberna, G, et al.. Lumipulse G SARS-CoV-2 Ag assay evaluation using clinical samples from different testing groups. Clin Chem Lab Med 2021;59:1468–76. https://doi.org/10.1515/cclm-2021-0182.Search in Google Scholar PubMed
17. Bohn, MK, Lippi, G, Horvath, AR, Erasmus, R, Grimmler, M, Gramegna, M, et al.. IFCC interim guidelines on rapid point-of-care antigen testing for SARS-CoV-2 detection in asymptomatic and symptomatic individuals. Clin Chem Lab Med 2021;59:1507–15. https://doi.org/10.1515/cclm-2021-0455.Search in Google Scholar PubMed
© 2021 Walter de Gruyter GmbH, Berlin/Boston
Articles in the same Issue
- Frontmatter
- Editorial
- From Camille Nούς to Apollonian and the Dionysian scientists
- Review
- The role of D-dimer in periprosthetic joint infection: a systematic review and meta-analysis
- Mini Reviews
- Updated picture of SARS-CoV-2 variants and mutations
- Systematic review and cumulative meta-analysis of the diagnostic accuracy of glial fibrillary acidic protein vs. S100 calcium binding protein B as blood biomarkers in observational studies of patients with mild or moderate acute traumatic brain injury
- Opinion Papers
- The 6C model for accurately capturing the patient’s medical history
- Webside manner: maskless communication
- Original Articles
- Ways that nurse practitioner students self-explain during diagnostic reasoning
- Diagnostic reasoning: relationships among expertise, accuracy, and ways that nurse practitioner students self-explain
- Perspectives on the current state of pre-clerkship clinical reasoning instruction in United States medical schools: a survey of clinical skills course directors
- Use of a structured approach and virtual simulation practice to improve diagnostic reasoning
- Analyzing diagnostic errors in the acute setting: a process-driven approach
- Morning report goes virtual: learner experiences in a virtual, case-based diagnostic reasoning conference
- Stroke hospitalization after misdiagnosis of “benign dizziness” is lower in specialty care than general practice: a population-based cohort analysis of missed stroke using SPADE methods
- Discrepancy between emergency department admission diagnosis and hospital discharge diagnosis and its impact on length of stay, up-triage to the intensive care unit, and mortality
- Automated capture-based NGS workflow: one thousand patients experience in a clinical routine framework
- Short Communication
- Characterizing the relationship between diagnostic intensity and quality of care
- Case Reports – Lessons in Clinical Reasoning
- Lessons in clinical reasoning ‒ pitfalls, myths, and pearls: a case of confusion, disequilibrium, and “picking at the air”
- Hickam’s dictum, Occam’s razor, and Crabtree’s bludgeon: a case of renal failure and a clavicular mass
- Letters to the Editor
- Three learning concepts to improve diagnosis and enhance the practice of medicine
- Distributed cognition: a framework for conceptualizing telediagnosis in teams
- Performance of Fujirebio Espline SARS-CoV-2 rapid antigen test for identifying potentially infectious individuals
Articles in the same Issue
- Frontmatter
- Editorial
- From Camille Nούς to Apollonian and the Dionysian scientists
- Review
- The role of D-dimer in periprosthetic joint infection: a systematic review and meta-analysis
- Mini Reviews
- Updated picture of SARS-CoV-2 variants and mutations
- Systematic review and cumulative meta-analysis of the diagnostic accuracy of glial fibrillary acidic protein vs. S100 calcium binding protein B as blood biomarkers in observational studies of patients with mild or moderate acute traumatic brain injury
- Opinion Papers
- The 6C model for accurately capturing the patient’s medical history
- Webside manner: maskless communication
- Original Articles
- Ways that nurse practitioner students self-explain during diagnostic reasoning
- Diagnostic reasoning: relationships among expertise, accuracy, and ways that nurse practitioner students self-explain
- Perspectives on the current state of pre-clerkship clinical reasoning instruction in United States medical schools: a survey of clinical skills course directors
- Use of a structured approach and virtual simulation practice to improve diagnostic reasoning
- Analyzing diagnostic errors in the acute setting: a process-driven approach
- Morning report goes virtual: learner experiences in a virtual, case-based diagnostic reasoning conference
- Stroke hospitalization after misdiagnosis of “benign dizziness” is lower in specialty care than general practice: a population-based cohort analysis of missed stroke using SPADE methods
- Discrepancy between emergency department admission diagnosis and hospital discharge diagnosis and its impact on length of stay, up-triage to the intensive care unit, and mortality
- Automated capture-based NGS workflow: one thousand patients experience in a clinical routine framework
- Short Communication
- Characterizing the relationship between diagnostic intensity and quality of care
- Case Reports – Lessons in Clinical Reasoning
- Lessons in clinical reasoning ‒ pitfalls, myths, and pearls: a case of confusion, disequilibrium, and “picking at the air”
- Hickam’s dictum, Occam’s razor, and Crabtree’s bludgeon: a case of renal failure and a clavicular mass
- Letters to the Editor
- Three learning concepts to improve diagnosis and enhance the practice of medicine
- Distributed cognition: a framework for conceptualizing telediagnosis in teams
- Performance of Fujirebio Espline SARS-CoV-2 rapid antigen test for identifying potentially infectious individuals

