Abstract
Over 50 years have elapsed since the clinical definition of non-A, non-B hepatitis and 36 years since the unveiling of hepatitis C virus (HCV) and the availability of specific serological assays, but few advances in the serological diagnosis of HCV infection have occurred. Testing for anti-HCV is still based on the detection of reactivity towards the structural Core region of HCV, which appears to be dominant throughout the different phases of infection, plus antibodies towards antigens expressed by several non-structural regions. Unlike testing for other viral diseases, antibodies towards the envelope region are not detectable by the first line assays employed for screening or diagnosis and are scarcely represented in the supplementary assays employed to confirm the reactivity by screening assays. Clinical laboratories are often confronting issues on samples that give discrepant results among assays and are not confirmed by supplemental testing. Results obtained on those samples are usually labelled as ‘indeterminate’ and are often considered as false positive – though a true reference to define anti-HCV positivity is still lacking. The diagnostic answer is then deprived of clinical significance and affects patient management and counselling. The only advance, though not recent, has been the availability of assays detecting the HCV core antigen, which is considered as a surrogate of HCV RNA, with lesser sensitivity but bearing some operational and economic advantages in diagnosis and population screening, and lately of assays combining HCV antigen and anti-HCV detection. This paper summarizes the history of HCV serology and provides some insights on its limitations and potential developments.
Introduction: hepatitis C
Viral hepatitis is still a major public health challenge worldwide. The two more severe forms, hepatitis B and C, cause around 3,500 deaths per day, and mortality is increasing [1]. While hepatitis B is still the most diffused form of chronic viral hepatitis, 56.8 million viraemic HCV infections were estimated globally at the beginning of 2020 [1]. Although this number represents a decrease from 2015, forecasts from the Polaris Observatory HCV Collaborators suggest that we are not currently on track to achieve global elimination targets by 2030 [2], since only 36 % of people living with hepatitis C had been diagnosed and only 20 % of those had received curative treatment. The course of HCV infection is variable: acute hepatitis is often asymptomatic and up to 85 % of patients with acute HCV infection develop a persistent infection that may resolve spontaneously in about 20–30 % cases or evolve to chronicity; chronic hepatitis C is a major cause of liver cirrhosis, hepatocellular carcinoma and liver transplantation (Figure 1) [3], [4], [5], [6], [7]. On the bright side, tremendous advances have been made on therapy: hepatitis C became the first curable, chronic viral infection after the introduction of direct antiviral agents (DAAs) that revolutionized antiviral treatment, leading to viral eradication in more than 95 % of all patients infected with HCV treated by those drugs [8]. This breakthrough shall increase chances to meet the WHO global strategy to eliminate viral hepatitis by 2030, and more specifically reducing new infections and deaths by 90 and 65 %, respectively [4] and highlights the need of more efforts to identify infected individuals and opportunity for better linkage between diagnosis and provision of care. The first step is then to identify all subjects that have been exposed to HCV, and the logical strategy on this purpose is to test for anti-HCV antibodies. In the following paragraphs we aim to describe the history, advantages and limitations of HCV serology.
The discovery of hepatitis C virus
In 1972 a nosocomial outbreak of parenterally transmitted hepatitis affected both marrow transplant patients and normal platelet donors in an oncology unit [9]. Because of the characteristics of the clinical illness and the lack of serological evidence of hepatitis B virus (HBV) the outbreak was attributed to hepatitis A, but when stored sera were tested by the newly developed tests for hepatitis A virus (HAV) all were negatives and no patient seroconverted afterwards. Other similar episodes were reported and by the mid-seventies of last century clinicians became aware that a significant number of patients affected by acute or chronic hepatitis of presumed viral origin could not be diagnosed as being infected by HAV or HBV. Those cases were also unrelated to other known viruses which occasionally affect the liver and occurred in a variety of non-transfusion epidemiologic settings paralleling those implicated in the transmission of HBV. Chronic hepatitis could be seen in 30–40 % of patients and epidemiologic evidence favoured the existence of an asymptomatic chronic carrier state. That nosographic entity was then named in 1974 non-A,non-B hepatitis (nAnBH) [10], [11]) and starting from there researchers throughout the world tried to identify the potential causative agent(s) of that disease. The quest for the culprit took all of 15 years before the target was reached in April 1989 by a research team that included scientists from Chiron Corporation and the US National Institute of Health [12]. With an unusual approach, they constructed a random-primed complementary DNA library from plasma containing the still uncharacterized nAnBH agent, screened that library with serum from a patient diagnosed with nAnBH and isolated a complementary DNA clone, labelled 5-1-1-, that was shown to encode an antigen associated specifically with that infection (Figure 2) [13]. That clone derived from an RNA molecule of at least 10,000 nucleotides, positive-stranded and consistent with the infectious agent being an RNA virus similar to the Togaviridae or Flaviviridae. Further characterization by overlapping cDNA clones allowed to establish that the HCV RNA sequence (9,379 nucleotides) has a single large open reading frame that can encode a viral polyprotein precursor of 3011 amino acids (Figure 3). While there is little overall amino acid and nucleotide sequence homology with other viruses, the 5′ HCV nucleotide sequence upstream of this large open reading frame has substantial similarity to the 5′ terminus of pestiviral genomes. The polyprotein also has significant sequence similarity to helicases encoded by animal Pestiviruses, plant Potyviruses, and – as postulated before–human Flaviviruses. A basic, presumed nucleocapsid domain is located at the N terminus upstream of a region containing numerous potential N-linked glycosylation sites. These HCV domains are in the same relative position as observed in the Pestiviruses and Flaviviruses and the hydrophobic profiles of all three viral polyproteins are similar. These combined data indicate that HCV is an unusual virus that is most related to the Pestiviruses, such as the hog cholera and bovine diarrhoea viruses [13]. Of note, significant genome diversity within the putative 5′ structural gene region of different HCV isolates were apparent, suggesting the presence of closely related but distinct viral genotypes [14].
![Figure 2:
Schematic representation of the isolation of a first HCV-related antigen. Total nucleic acid content from a chimpanzee plasma infected with nAnBH was expressed as cDNA libraries and challenged with serum from a nAnBH patient as a putative source of antibodies to the nAnBH agent. After discarding interferent clones, a single small clone of about 150 base pairs, named 5-1-1, was identified as truly derived from the putative HCV genome. Modified from ref. [13].](/document/doi/10.1515/cclm-2025-0501/asset/graphic/j_cclm-2025-0501_fig_002.jpg)
Schematic representation of the isolation of a first HCV-related antigen. Total nucleic acid content from a chimpanzee plasma infected with nAnBH was expressed as cDNA libraries and challenged with serum from a nAnBH patient as a putative source of antibodies to the nAnBH agent. After discarding interferent clones, a single small clone of about 150 base pairs, named 5-1-1, was identified as truly derived from the putative HCV genome. Modified from ref. [13].
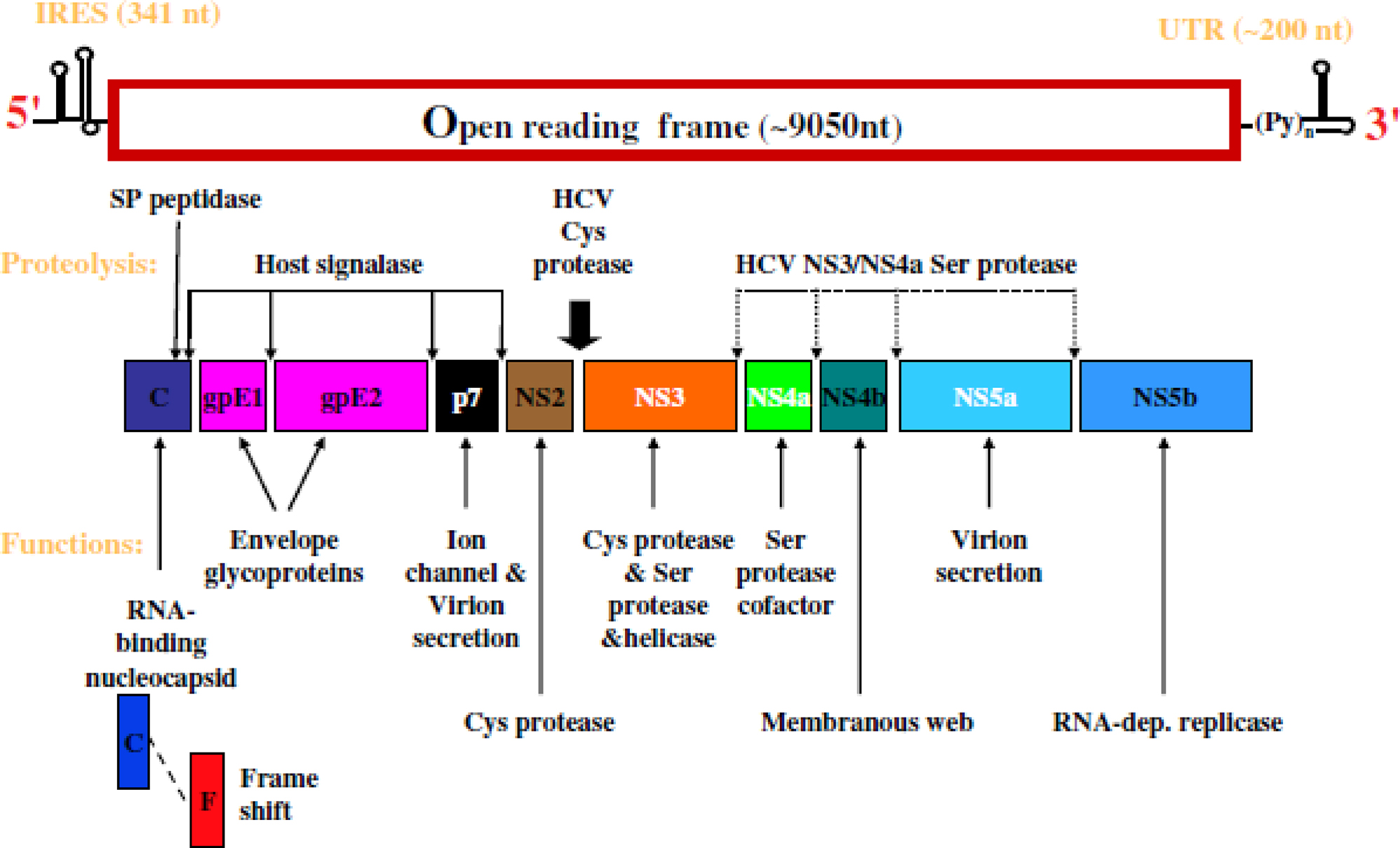
Genome structure of the hepatitis C virus (HCV). IRES, internal ribosome entry site (RNA sequence that helps ribosomes attachment); UTR, untranslated region; cys, cysteine; ser, serine.
The early quest for nAnBH biomarkers
Even before HCV was identified intensive research was conducted to try and develop diagnostic tools for nAnBH. Shirachi et al. [15] found evidence for a new hepatitis-specific antigen from double immunodiffusion assays between acute and convalescent sera obtained from patients with nAnB post-transfusion hepatitis (nAnB PTH). The antigen was found in the acute phase sera of all 13 nAnB PTH patients with longer incubation and duration periods but only transiently in 4 out of 10 sera from patients with shorter incubation and duration periods and was also detected in few specimens from acute hepatitis patients who had not received a blood-transfusion. That antigen, named HC, was distinct from those of hepatitis A and B (surface and core) as it migrated in the serum beta-globulin region and had a molecular weight between 100,000 and 300,000 daltons [15]. Antibodies against HC were found in only 30 % of long-incubation PTH and did not persist for long. In 1979 Tabor et al. [16] were able to detect by counterelectrophoresis an antigen in serum samples from six of seven chimpanzees during the acute phase of experimentally induced non-A, non-B hepatitis using antiserum from a chimpanzee convalescent from human non-A, non-B hepatitis. That antigen was detected in serum samples obtained from three humans with chronic nAnBH who transmitted the infection to other humans and to chimpanzees by experimental inoculation. In addition, the antigen was detected in serum obtained retrospectively from 11 to 31 former blood donors whose blood had transmitted post transfusion nAnBH several years previously to recipients of a single unit of their blood. Unlike the results from Shirachi, antibody to this antigen was detected in convalescent serum samples from all seven chimpanzees studied, in convalescent serum from the nurse infected by accidental needlestick, and in serum from a haemodialysis patient convalescent from nAnBH. Similar results have been obtained by Kabiri et al. [17]: convalescent serum from one infected chimpanzee reacted, in indirect immunofluorescent tests, with some of the hepatocyte nuclei in sections of autologous liver biopsy specimens and specimens from eight of the other chimpanzees and serum from a convalescent patient reacted in the same way. These positive sera did not react with liver sections from uninfected chimpanzees. While those cumulative results and further studies suggested that there may be at least two antigen/antibody systems correlated to nAnBH [18], [19], [20] those data were not conclusive and uncertainty on the serological biomarkers of nAnBH, coupled with the frustrating unavailability of testing systems that could enable routine testing, persisted. Difficulties encountered in the development of serologic tests for nAnB antigens were likely related to a lack of potent antibodies coupled with low concentrations of the infectious agent [21]. Also, as keenly pointed out by Dienstag et al. [22], circulating immune complexes could contain and mask viral antigens and this may explain the failure of conventional immunological techniques to detect virus antigens. All hypotheses on putative nAnB-related antigens and antibodies were hampered by the lack of identification of the causative agent and further studies carried out over the following years still yielded conflicting results [21], 23].
The first assay for HCV diagnosis
The breakthrough in the diagnostic field for hepatitis C was strictly linked to the identification of the hepatitis C virus (HCV) thanks to the identification of a complementary DNA clone that was shown to encode an antigen associated specifically with NANBH infections [12]. The same research group was able to develop a specific assay for a blood-borne nAnB virus in which a polypeptide synthesized in recombinant yeast clones of the hepatitis C virus (HCV) was used to capture circulating viral antibodies [24]. HCV antibodies were detected in six of seven human sera that were shown previously to transmit NANBH to chimpanzees. Assays of 10 blood transfusions in the United States that resulted in chronic nAnBH revealed that there was at least one positive blood donor in nine of these cases and that all 10 recipients seroconverted during their illnesses. About 80 % of chronic nAnB PTH patients from Italy and Japan had circulating HCV antibody while a much lower frequency (15 %) was observed in acute, resolving infections. In addition, 58 % of nAnB patients from the United States with no identifiable source of parenteral exposure to the virus were also positive for HCV antibody. The Authors then concluded that the newly discovered HCV was a major cause of nAnBH throughout the world and that their serological test was able to identify a large share of infected cases. This was soon confirmed with the same technique by further research. Alter et al. [25] measured an antibody to HCV by radioimmunoassay in prospectively followed transfusion recipients and their donors. Of 15 patients with chronic non-A, non-B hepatitis documented by liver biopsy, all seroconverted for the antibody and 3 out of 5 with acute resolving non-A, non-B also seroconverted. The development of anti-HCV was delayed (mean delay, 21.9 weeks after transfusion, or 15 weeks after the onset of clinical hepatitis) and took approximately one year in one patient. Anti-HCV persisted in 14 of the 15 patients with chronic disease with a mean follow-up of 6.9 years but disappeared in the three patients with acute resolving disease after a mean of 4.1 years. Anti-HCV was detected in samples of donor serum given to 14 of the 16 anti-HCV-positive patients (87.5 %) for whom all donor samples were available. Those data suggested that being HCV the predominant agent of transfusion-associated nAnBH, the screening of donors for anti-HCV could prevent majority of cases of the disease, while ‘surrogate’ assays such as anti-HBc and ALT would have detected approximately only half of the anti-HCV-positive donors involved in the transmission of hepatitis. Those conclusions were soon reinforced by another study [26] in which antibodies reacting with the antigen discovered by Choo et al. [12] were looked for in 643 sera from 23 patients with PTH and their corresponding donors and were detected in 15 out of 17 (88.2 %) nAnB PTH among patients whose sera over time displayed multiple alanine aminotransferase (ALT) peaks. In general, the antibody was detected after several peaks of serum ALT elevations and once detected it persisted for years. In contrast to the patients of chronic hepatitis, the antibody was barely detected in patients with a single episode of ALT elevation (1 out of 6). Of the 15 well-defined cases of nAnB PTH that showed multiple ALT peaks and HCV seroconversion, 11 (73.3 %) were shown to be transfused with at least one unit of blood positive for the antibody. The retrospective analysis showed that all tested donor blood found to be positive for the antibody had been transfused to recipients who afterwards developed nAnBH. These data confirmed further that the cloned cDNA originated from an etiological agent of nAnBH, the hepatitis C virus, and had the screening been done with an anti-HCV assay, 12/23 cases of nAnB-PTH, and notably 11 out of 17 cases of chronic post-transfusion hepatitis, would have been prevented.
First generation of commercial anti-HCV assays
The brilliant pieces of research described so far paved the way to blood screening for HCV infection. This fundamental step in disease prevention needed a further development, i.e., the availability of laboratory assays able to process thousands of blood units within a timeframe compatible with the availability of blood products. The first enzyme linked immunoassay (EIA) for the detection of anti-HCV antibody was released in the spring of 1990 [24] and after a few months another assay was commercially available [27]. Both assays were indirect EIAs in which the solid phase was coated with the recombinant c100-3, expressed from the NS4 region of the putative HCV genome like the original 5-1-1 protein. The impact of those assays on blood donations screening was huge, as demonstrated by a large study carried out in 1990 [27]. In that study a first generation enzyme immunoassay (EIA) was evaluated for efficacy in the detection of anti-HCV in a panel of human specimens known to contain the infectious agent of nAnBH and by testing 6,118 serum specimens from volunteer blood donors supposedly at low risk of infection. On the latter the prevalence of antibodies to HCV was 0.36 %, while it was 10.08 % among 3,718 commercial plasma donors, highlighting the relevant risk linked to blood products obtained from that group which is not fully investigated for high-risk behaviors.
A major step: second generation assays
Just one year after the release of first generation assays the same manufacturers released the ‘second generation’ of HCV antibody assays. The format was the same (indirect EIA with recombinant antigens anti-IgG conjugate), but the coating of the solid phase was enriched substantially by adding antigens expressed by the putative NS3 and Core region [28]. The latter, being a structural region near the 5′ terminus of the putative HCV genome, was thought to raise specific antibodies earlier and with longer persistence than those directed towards non-structural antigens. The advantages of the second generation anti-HCV assays for the diagnosis of HCV infection were demonstrated soon: of 115 patients in whom post-transfusion non-A, non-B hepatitis developed, the first-generation EIAs detected anti-HCV in 51 (46 %), and the second-generation assay detected anti-HCV in an additional 16 (14 %), for a total rate of anti-HCV positivity of 60 %. Furthermore, by a lookback analysis of 1,247 patients who underwent transfusion and 1,235 matched controls a 3 % rate of non-A, non-B, non-C hepatitis was found. Extrapolating that rate to the transfusion recipients, only 74 of the 111 cases of hepatitis were attributable to the transfusion and 67 of the 74 PTH cases (91 %) were caused by HCV [29]. Other experiences confirmed those early findings. By studying a cohort of 210 patients with bleeding disorders Hatzakis et al. [28] found an anti-HCV prevalence of 87.1 % by a first generation anti-HCV EIA and 93.8 % by a second generation test (p=0.0026). Furthermore, at follow-up of 29 out of 111 patients (26.1 %) became anti-HCV negative by the first generation test whereas none of the 121 tested seroreverted by the second generation method. The increased sensitivity of second generation anti-HCV assays was mainly due to the addition of core-derived antigens on the solid phase. Antibodies to distinct HCV core peptides were detected at the onset of acute non-A, non-B hepatitis in 15 and 14 of 20 patients, compared to a positivity to C-100-3 in only 9 patients. In addition, in 186 patients with chronic non-A, non-B liver disease, anti-core peptides antibodies were detected in 170 patients (91 %) and anti-C100-3 only in 138 patients (74 %) [30]. In another study, 55 % of 97 well-characterized cases NANBH hepatitis were recognized as anti-HCV positive by both the c100-3 and the c22-3 (core) assays, whereas 21 additional cases (22 %) were detected only by the c22-3 assay [31]. Notably, the c22-3 assay appeared to be significantly better in detecting anti-HCV in NANB cases that were acquired by presumed non-parenteral routes, whereas the tests were nearly identical in their detection of anti-HCV in parenterally acquired cases. A more massive antigenic exposure in parenterally acquired, especially transfusion-related, cases and a subsequent stronger immune response, detectable even by the less-sensitive assay, could explain those findings. To summarize a notable number of papers that compared first- and second-generation anti-HCV assays, the latter detected anti-HCV antibodies in approximately 20 % more cases of acute transfusion-associated or sporadic nAnBH and in up to 10 % more cases of chronic NANBH hepatitis, in which anti-HCV prevalence was already very high by first generation assays [32], [33]. While second generation anti-HCV assays represented a huge advance in the detection of HCV carriers and the diagnosis of HCV-related chronic hepatitis, those assays were not considered the ultimate solution to HCV diagnosis: antibodies to HCV were not detectable for several weeks after the onset of an acute hepatitis while viral genome (HCV-RNA) appears in the serum early during the incubation period [3]. More sensitive antibody assays and reliable, suitable methods for the detection of HCV-RNA were then needed.
‘Third’ generation anti-HCV assays: myth and reality
In 1993 another supposed ‘breakthrough’ in HCV serology was announced with the commercial availability of the ‘third generation’ tests for HCV antibodies. The assay format was like the previous versions – indirect enzyme immunoassay to detect specific IgG antibodies – but the solid phase of the third gen commercial assay in some instances lacked NS4-derived antigens – whose utility had been questioned by some authors [34] – and in all cases included recombinant antigens coded by the 3′ terminal NS5 region of the HCV genome, along with the already employed antigens coded by the Core and NS3 regions [35]. The initial claim for those methods was an increased sensitivity due to the inclusion of that new antigen and better specificity compared to the second-generation tests. Indeed, in a first retrospective study on 203 sera from blood donors the positive predictive value increased to 0.52 by a third-generation screening assay compared to 0.23 or 0.37 by prior assays. On the other side, antibody response to the newly added NS5 antigen was not as prevalent as to the other antigens and had only a minor influence on sensitivity and the few samples that were reactive only for anti-NS5 could not be confirmed by supplemental assays and were negative for HCV-RNA [35]. An early detection of anti-HCV seroconversion by a third generation screening test was due to an earlier detection of the immunological response to the NS3 antigen and not to the additional NS5 [35], [36]. The additional value of NS5 antigens was questioned soon thereafter by other Authors [37], [38], [39] who outlined the relatively low percentage of samples reactive to those antigens compared to the other ones, especially NS3 and Core, in patients with a chronic HCV infection and the negligible relevance in seroconversion. In a conclusive analysis of 3,832 samples reactive for anti-HCV out of 624,910 blood donations screened, anti-NS5 antibodies were less frequently detected compared to anti-Core, anti-NS3 and anti-NS4 in samples confirmed as anti-HCV positive, most of which were also positive for HCV-RNA, but were represented in 380/945 (40.2 %) of all ‘indeterminates’, i.e., samples showing only a single anti-HCV reactivity, and in none of those 380 was positive for HCV-RNA [40].
In conclusion, many studies comparing different generations (Figure 4) and different versions of anti-HCV antibody assays allowed to reach a consensus on the major role of the humoral immune response raised towards Core and NS3- derived antigens both in the early stages of HCV infection and in chronic infections while the antigens coded by the other non-structural regions of the putative HCV genome are relegated to an ancillary role. Nowadays, NS5 antigens are included only in a minority of commercial assays (Table 1) and all currently available first-level tests for anti-HCV are labelled as ‘third generation’ irrespective of the antigens employed to capture specific anti-HCV antibodies (Figure 5) because they have been subjected to extensive adjustments over time to increase the ability to detect anti-HCV in the early phases of HCV infection and to increase specificity and robustness to HCV genome diversity [41]. Unfortunately, despite all efforts in this sense – focused mainly on a better expression of antigens derived from one structural (Core) and a couple of non-structural (NS3, NS4) coding regions of the HCV genome – the improvements in terms of sensitivity and specificity have not been outstanding.
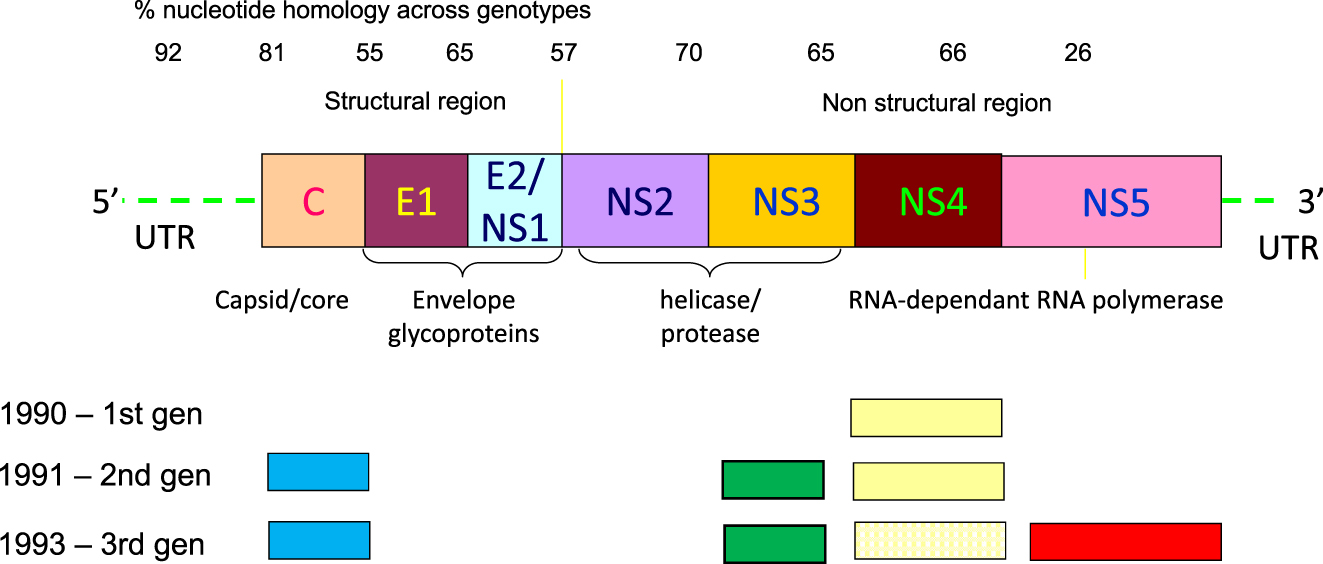
Schematic representation of the HCV genome and of the antigens included in the different generations of anti-HCV screening assays. The NS4 antigen(s) representation in third generation is dotted because not all assays included those epitopes. The dimension of the different HCV genome regions and derived antigens are exemplary and do not represent the real extension of those RNA or proteic compounds.
Characteristics of 11 commercial anti-HCV first level immunoassays.
| Assay name | Manufacturer | Assay principle | Solid phase | HCV antigens |
|---|---|---|---|---|
| ARCHITECT Anti- HCV | Abbott Laboratories | Chemiluminescent microparticle immunoassay | Paramagnetic particles | Core, NS3, NS4 |
| Alinity Anti-HCV | Abbott Laboratories | Chemiluminescent microparticle immunoassay | Paramagnetic particles | Core, NS3, NS4 |
| Murex Liaison XL HCV Ab | Diasorin | Chemiluminescent immunoassay | Paramagnetic particles | Core, NS3, NS4 |
| Vitros Anti-HCV | Ortho Clinical Diagnostics | Chemiluminescent immunoassay | Microwell | Core, NS3, NS4, NS5 |
| Elecsys Anti-HCV | Roche Diagnostics | Electrochemiluminescent immunoassay | Paramagnetic particles | Core, NS3, NS4 |
| Advia Centaur HCV Assay | Siemens Healthineers | Chemiluminescent immunoassay | Paramagnetic particles | Core, NS3, NS4, NS5 |
| Access HCV Ab Plus | Bio-Rad Laboratories | Chemiluminescent immunoassay | Paramagnetic particles | Core, NS3, NS4, NS5 |
| Lumipulse-Ab | Fujirebio | Chemiluminescent enzyme immunoassay | Ferrite particles | Core, NS3, NS5 |
| HCV AIA-Ab | Tosoh | Fluorescent enzyme immunoassay | Magnetic microparticles | Core, NS3, NS4 |
| Access Anti-HCV | Beckman Coulter | Chemiluminescent immunoassay | Magnetic particles | Core, NS3, NS4 |
| HISCL Anti-HCV | Sysmex Corporation | Chemiluminescent immunoassay | Magnetic particles | Core, NS3, NS4 |
-
The antigen composition reflects only the HCV coding regions, recombinant antigens or syntetic peptides derived from thse regions may vary according to the different manufacturers. Antigens expressed by the Core and NS3 regions of the putative HCV genome are included in all assays and NS4 antigens in all but one, whereas antigens from the NS5 region are coated on the solid phase in only 4/11 assays. Note: some assays are not available in all countries.
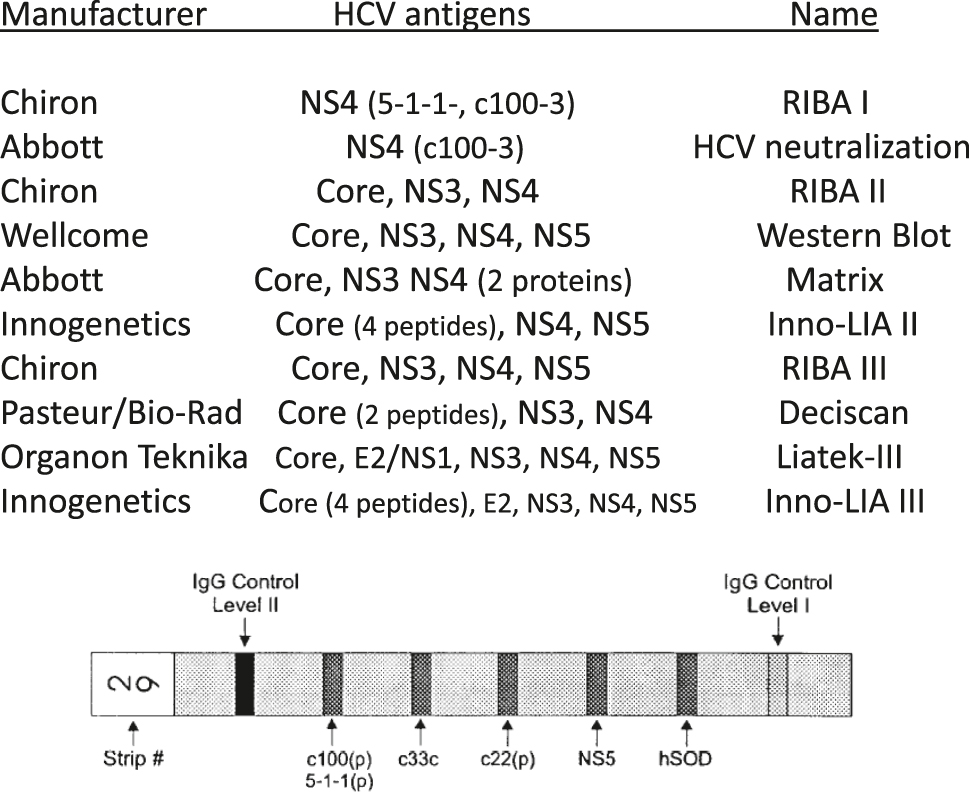
The most frequently employed supplemental assays for the confirmation of anti-HCV positivity available over time in most countries. The HCV antigens employed are named by coding regions of the HCV genome to allow a comparison among assays, while the actual composition and vectors employed to express the recombinant antigens are different across assays. In some instances the recombinant antigens employed combined sequences from two HCV regions. The drawing at the bottom depicts the solid phase of the most frequently used ‘third generation’ supplemental assay (Chiron RIBA III). That assay included four HCV recombinant antigens and two internal performance control (IgG level I and II), plus the SOD (superoxide dysmutase) band to check specificity.
HCV IgM as an alternative option
Commercial anti-HCV assay, regardless of the generation, detect only the specific IgG response. An early study demonstrated that IgM antibodies to HCV core were detected in 13 of 15 patients with post-transfusion hepatitis C and IgM to NS3 antigen in one of the 13; in nine of those patients IgM Core were transient and limited to the acute phase and were detectable coincidentally (nine cases) or earlier (four cases) than the IgG anti-HCV core response [42]. The average duration IgM anti-HCV core was 8.1 weeks but was considerably shorter in some patients and thus IgM detectability may be missed in acute-phase patients if specimens are not collected at short intervals. A typical IgM antibody response in nearly all patients with acute hepatitis C was confirmed also by other studies [43], [44] but since the IgM response usually did not precede the IgG response, the detection of IgM was deemed unlikely to narrow the window of seronegative infectivity. A potential role of HCV IgM response in chronic HCV infection, alike the IgM Core response in chronic hepatitis B [45], was then explored. In this context the initial results were quite consonant, despite the use of different non-commercial methods to detect specific HCV IgM and differences in the clinical conditions explored. Chen et al. [44] were able to demonstrate an IgM reactivity only in 28 % of chronic hepatitis C patients with mild disease activity compared to 45 % in patients with acute exacerbations; the IgM response was at low levels in nearly all patients. Low titers of IgM anti-HCV were detectable in 50–80 % of cases with chronic hepatitis C also in another relevant study [43]. IgM anti-HCV reactivity was typically found during acute exacerbation of chronic hepatitis C but also many patients with chronic active hepatitis C without acute exacerbation showed an IgM anti-HCV response and a correlation was found between the titer of IgM anti-HCV and the biochemical parameters of liver disease. Furthermore, in case of a sustained remission of liver disease due to alpha interferon therapy, positivity for IgM anti-HCV disappeared in more than 70 % of cases. In contrast, patients who do not respond to therapy rarely lose IgM anti-HCV. In conclusion, serum IgM antibodies to HCV appeared as a reliable markers of active HCV-induced liver disease both in acute and in chronic HCV infection and were useful as an adjunct in the clinical assessment of patients with chronic hepatitis C. Since post-treatment hepatitis C exacerbations showed the same sequence of events seen as in hepatitis B exacerbations (increases of viraemia followed by those of ALT and IgM anti-‘core’) [46] the findings of those early monitoring studies underscored the diagnostic and prognostic usefulness of monitoring anti-HCV-positive patients with quantitative assays for HCV markers [47]. However, despite those promising results, testing for specific IgM never attained routine use in HCV diagnostics. Several reasons shall explain the lack of adoption of such a simple and affordable tool. First, studies on HCV IgM have been carried out by commercial Core IgM tests or by in-house tests, some of which detected IgM to various HCV antigens; no real ‘standardization’ (or at least harmonization) of results was achieved, making it quite hard to compare results across different studies. Second, while a correlation between IgM response and active HCV infection, i.e., HCV-RNA positivity, was clear, a relationship between HCV IgM titers and HCV-RNA levels could not be found [48]. Truly quantitative IgM methods were not available while many quantitative assays for HCV-RNA were developed, making the latter a more reliable tool for HCV monitoring after treatment and to predict reactivation. After a few years the few commercial tests for HCV-IgM were dismissed and the interest for this serological biomarker disappeared soon.
The conundrum of anti-HCV confirmation
While anti-HCV commercial assays have attained a good sensitivity in chronically infected subjects, specificity has always been an issue. The low positive predictive value (PPV) of any assay when employed to test low prevalence populations, coupled with changes on HCV epidemiology with a progressive decrease of HCV incidence and prevalence in most countries [2] represent a challenge in reporting anti-HCV results. As an example, in the initial screening of 6,118 low risk US donors reported by Dawson et al. [27] only 22 out of 47 specimens reactive by the anti-HCV screening assays were confirmed as positive by supplemental testing, for a PPV lower than 50 %. To check specificity, ‘confirmatory’ assays are then needed: in the case of anti-HCV that problem has been dealt with already with the first generation assays by designing a Recombinant ImmunoBlot Assay (RIBA) in which two different antigens from the NS4 region (the afore mentioned c100-3 and 5-1-1) were coated on a nitrocellulose strip along with a control band (superoxide dismutase, SOD) to check interferences with the yeast vector in which the antigens were expressed [49]. The results by that method were scored as positive if both c100-3 and 5-1-1 bands were visible, indeterminate if only one viral band was detected and/or the SOD band was visible. It’s easy now to criticize such an approach, but at that time the algorithm worked quite well and allowed to increase the predictive value of a positive anti-HCV result by screening assays. Another confirmatory option was provided by the availability of a neutralization assay, based on the c100 antigen, that was able to confirm a high percentage of samples that resulted indeterminate by RIBA [50]. Supplemental assays paralleled the evolution of second generation screening assays: a new version of RIBA, including the same antigens expressed by the NS4 region already included in RIBA-I with the addition of core- and NS-3 derived recombinant antigens was available after just one year [51], [52] and the addition of those two antigens, as expected, increased the ability of that assay to ‘confirm’ reactivity by screening assays compared to the prior generation [53]. Other supplemental assays based on the same principle were soon available [40], [54], [55], [56], [57], [58], [59]: an overview of the more often used anti-HCV ‘confirmatory’ assays over time is provided in Figure 5. However, all those assays failed to meet the established criteria for a confirmatory assay, i.e., a different technique – those were still indirect immunoassays – and different antigen source, as the antigens employed were similar, and often the same, employed for screening assays. Moreover, the criteria to establish a ‘true’ anti-HCV positivity are still a matter of debate: translating the concept from immunoblots already in use to confirm the antibody reactivity towards the human immunodeficiency virus (HIV) [60], samples are usually confirmed as positive if a reactivity towards antigens expressed by at least two different portions of the HCV genome is detectable. However, the differences between HIV and HCV immunoblots are huge: HIV immunoblots have evolved over time from viral lysates and are designed to represent all major antigens encoded from the gag, pol and env region of the virus, so while the confirmation criteria may vary according to different guidelines issued by Institution or Scientific Societies the technical and scientific roots for positivity criteria are sound [61]. In the case of HCV, a viral lysate is still not available and all supplemental assay ‘mimic’ the first line assays employing the same or similar, recombinant antigens or synthetic peptides, so there’s no true reference. Additionally, by some methods a sample may be deemed as positive by a reactivity to two or more portions of the antigen expressed by the core region, which may make sense since that reactivity is the most frequent and the one showing longer persistence but leads to an increase of discordant results among assays and efforts to ‘standardize’ anti-HCV confirmation criteria were not really pursued. An Italian researcher stated at a national microbiology congress in the early 2000s, ‘Cuius EIA eius est RIBA’, i.e., each supplemental assay was more suited to ‘confirm’ anti-HCV reactivities by the screening test provided by the same company, which is not surprising since first- and second-line assays were developed in parallel.
To conclude, the whole setting of anti-HCV ‘confirmation’ resembles a closed system, and entropy increases as by the second principle of thermodynamics. As we have seen, some of the issues related to anti-HCV ‘confirmatory’ tests are inherent to the configuration of the supplemental assays, but the biological behaviour of the target analyte also plays a significant role, which we shall elucidate in the next section.
An alternative approach to anti-HCV confirmation is the so-called ‘orthogonal’ testing strategy, by which clinical samples are initially assayed by a first-line anti-HCV test and reactive specimens are then tested again by a second anti-HCV test to increase the specificity of the first result. In a recent evaluation of two broadly employed automated anti-HCV immunoassays [61], 516 anti-HCV reactive specimens out of 76,442 (0.67 %) routine samples assayed by the chemiluminescent anti-HCV assay A were tested by a second method (B) employing a similar technology, by which only 363 were reactive and 327 of those were positive by RIBA. In that low prevalence population, the PPV of anti-HCV was then increased from 64.1 to 90.1 %, and it increased from 55.2 to 83.9 % when the same algorithm was employed reverting the order of the first-level assays: 56 samples out of 87 reactive by method B were reactive also by method A, and 47 were positive also by RIBA. Those results indicate that the two-assay testing strategy may significantly reduce false positives in anti-HCV testing and identify inactive HCV infection in low seroprevalence settings. The drawback for this approach is that not all laboratories may be equipped with multiple first level anti-HCV assays and that the value of ‘orthogonal’ testing is not generally recognized yet.
Anti-HCV detection: not early, not forever
The first serological results on HCV acute infections already hinted the antibodies directed towards any of the antigens encoded by the structural Core region and the non-structural NS3, NS4 and NS5 regions cannot be detected earlier than some weeks – up to eight–after the onset of the clinical signs and symptoms [62]. The subsequent refinement of ‘third generation’ anti-HCV tests allowed to reduce that gap, but even by the most recent immunoassay the initial ‘window’ phase of several weeks after the onset of acute hepatitis C persists (Figure 6) [33], [41], [63], [64], [65]. This lack of sensitivity in the initial phase of an HCV infection may be due to biological factors, i.e., to a limited exposure of HCV antigens to the host immune system during an early infection that leads to a delayed antibody response, or to a gap in assay design. For the latter, it’s noteworthy that all first-line assays do not detect antibodies to HCV envelope antigens, which appear first compared to antibodies to other structural regions in most viral diseases [60], [66]. The HCV envelope coding region is just upstream to the Core region (Figure 3) and is divided in two subregions (E1 and E2). Antigens expressed by E2 have been cloned and expressed in Chinese hamster ovary cells and a purified E2 glycoprotein was used to construct an enzyme immunoassay for HCV E2 antibodies. The presence of those antibodies was positively correlated (97.3 %) to the presence of HCV RNA and highly concordant with a second generation anti-HCV enzyme immunoassay. Of note, E2 antibody was the first serological marker to appear in 3/5 HCV seroconversion panels and 42.4 % of core and 15.4 % of NS3 indeterminate specimens also contained antibodies to E2, suggesting that HCV infection had occurred in these individuals [67]. In another paper the effect of E2 hypervariability on the anti-E2 assay was studies by genotyping 49 samples positive by a third gen RIBA assay, all of which tested positive for anti-E2 [68]. This study confirmed the added value of testing for E2 antibodies, that were detectable in 8 of 12 RIBA indeterminate, HCV RNA positive samples, but also in 9 of 30 RIBA indeterminate, HCV RNA negative samples. Anti-E2 antibodies were also detectable in 9 out of 85 haemodialyzed patients who tested negative by a third generation EIA, and six of those were also positive for HCV-RNA [69]. Despite those promising data, difficulties to express recombinant E2 antigens in a suitable vector hampered their inclusion in commercial screening assays and nowadays only a few anti-HCV supplemental tests include E2 synthetic peptides along with recombinant antigens or peptides from other HCV regions (Figure 5). Two other relevant topics in anti-HCV serology are the persistence of antibodies over time and their protective effect towards HCV infection or reinfection. A spontaneous clearance of HCV infection (Figure 1) has been observed already in an early population study [5] and in a quite conclusive study by Ayoub et al. [6]. Applying a mathematical model developed to describe HCV transmission and clearance in two different populations, HCV clearance may be as high as 30–40 %, according to the risk characteristics of the population.
![Figure 6:
Estimates of the average window period for HCV detection after the clinical onset by HCV first level (screening) assays according to the evolution of commercially available tests. First, second and third generation refer to assays detecting anti-HCV, fourth generation assays combine anti-HCV and HCV core antigen detection. The data refer to immunocompetent individuals and were derived from references [32], [40] and [62], [63], [64]. In immunocompromised subjects this ‘window’ period may be extended by several weeks [64].](/document/doi/10.1515/cclm-2025-0501/asset/graphic/j_cclm-2025-0501_fig_006.jpg)
Estimates of the average window period for HCV detection after the clinical onset by HCV first level (screening) assays according to the evolution of commercially available tests. First, second and third generation refer to assays detecting anti-HCV, fourth generation assays combine anti-HCV and HCV core antigen detection. The data refer to immunocompetent individuals and were derived from references [32], [40] and [62], [63], [64]. In immunocompromised subjects this ‘window’ period may be extended by several weeks [64].
The early studies on HCV-infected people did not include a follow-up long enough to ascertain antibody persistence, but already in 1992 Alter et al. [5] demonstrated a rate of anti-HCV loss of 0.6 % per 100 person-year. In another longitudinal study on 178 multitransfused patients followed up for 8 years, 5 out of the 30 infected had a partial or full seroreversion [70] and in the early 2000s two independent studies carried out on open populations in Italy confirmed that. Kondili et al. [71] tested for anti-HCV by third generation screening and confirmatory assays 3,884 randomly selected individuals finding an initial prevalence of 2.4 %. After a median follow up of 7 years the incidence rate was of 1.4 cases per 10,000 person years but of the 36 individuals confirmed as anti-HCV positive at enrolment, seven (19.4 %) showed complete seroreversion. In the study by Mazzeo et al. [72], carried out on 1,996 individuals from the general population of a small town in North Italy, the initial anti-HCV prevalence (3.46 %) and the HCV incidence over a 10 year span (5.3 per 10,000 person years) were higher but the rate of complete seroreversion was quite similar, as 11 out of 65 (16.9 %) subjects spontaneously cleared HCV-Ab. Likewise, in a study on 63 HIV-infected men who have sex with men (MSM) followed up for 4 years [65] the 36 subjects who cleared HCV showed a significant decrease in anti-HCV levels and anti-HCV became undetectable during follow-up in eight, for a cumulative incidence of seroreversion of 37 %.
Anti-HCV seroreversion bears two relevant consequences: first, any serological survey based on antibody detection will underestimate the real prevalence of HCV infection over time in any given population. Second, this sustained rate of anti-HCV disappearance over time explains better than a supposed lack of specificity of the anti-HCV tests the frequent finding of low-level anti-HCV [70], often ‘not confirmed’ by supplemental testing and with no evidence of ongoing viral replication [65], [70], [71], [72]. Even a long-time persistence of anti-HCV antibodies does not appear to guarantee a ‘protective’ immunity. That issue is still debated, but HCV reinfections are quite common in people prone to high-risk behaviours, such as injecting drug use and subjects indulging to promiscuous sex relationship [73]. On the former [74] the overall rate of HCV reinfection three years after sustained viral response was 1.2 new cases per 100 person years at a median follow-up of 15.9 months but a much higher reinfection rate of 9 per 100 person years has been found in a similar study [75], possibly due to demographic differences. For the latter, men who have sex with men (MSM) have been shown to present reinfection rates ranging from 1.89 [76] up to 7.3 [77] per 100 person years. HCV reinfection is diagnosed by a positivity for HCV-RNA in people with sustained negativity after clearing a prior HCV infection [73] and by a new appearance on anti-HCV antibodies. An increase of anti-HCV levels if antibodies were still detectable has also been demonstrated and monitoring anti-HCV levels might therefore be an effective alternative for diagnosis of HCV reinfection [65].
The other way around: testing for HCV antigens
Already in the initial stages of nAnBH serology the presence of circulating viral antigens has been postulated and then demonstrated by different research groups [16], [22]. The low amount of circulating antigen(s) and the relative low sensitivity of then-available EIA techniques hampered the development of suitable assays for the detection of HCV antigens until 1999, when Komatsu et al. [78], employing monoclonal antibodies against hepatitis C virus (HCV) core protein, detected that HCV antigen (HCVAg) in 130 out of 490 anti-HCV positive sera (26.5 %), all positive also for HCV-RNA. The HCVAg was detectable in all 71 patients with chronic active hepatitis, cirrhosis and hepatocellular carcinoma and only in 18 of 32 patients with chronic inactive hepatitis. The correlation between HCVAg and quantitative HCV-RNA was significant (0.86, p<0.01) though HCVAg was negative in 14 of 144 sera positive for HCV-RNA. Similar results were obtained in an Italian study in which a commercial EIA was employed [79]: that assay showed a very good specificity (99.4 %) and correlated well with HCV-RNA. Though not as sensitive as the molecular test (the HCVAg detection limit was around 80,000–100,000 international units of HCV-RNA per mL), due to the rapid ramp-up of viremia and high levels of viral RNA during an acute HCV infection the HCVAg test reduced the time to a first positive result in acute hepatitis C by an average of 33.4 days (range: 19–71 days) compared to the anti-HCV assays, with an average delay of less than a full day from HCV-RNA detection. At that time, it was concluded that the measurement of total HCV core antigen showed a strong dynamic correlation with HCV RNA and could therefore serve as an alternative direct marker of viral infection. Also, a rapid, reliable assay for changes in HCV load may permit more frequent patient assessment and tailoring of the therapeutic regimen [80]. This opportunity for an easier, quicker and potentially cheaper diagnostic tool was hampered by the fact that then-available HCVAg assays could not readily detect that antigen with sufficient sensitivity beyond the early stages of infection, because the development of the antibody response led to the formation of immune complexes with anti-core antibodies. This hurdle was cleared almost a decade later, when a first commercial HCVAg immunoassays with an automated pretreatment apt to release the antigen from circulating virions and to disrupt antigen-antibody complexes became available [81]. This enhancement allowed to detect HCVAg throughout all phases of an active HCV infection, with increased sensitivity compared to the previous tests. Many studies on this new solution have been published since, and the general consensus, highlighted by guidelines from both the WHO [82] and the European Association for the Study of the Liver (EASL) [83] is that testing for the HCV core antigen is an adequate tool to ascertain an active HCV infection [84], [85] and to assess the sustained virological response after treatment in chronic hepatitis C [80], [86], [87]. Utilizing the HCVAg test as a reflex on samples positive for anti-HCV shall guarantee both economic and operations advantages and is cost-effective in population screening, where the reduced sensitivity compared to HCV-RNA is fully compensated by a reduction in people lost to follow-up when a two-tier strategy is adopted [88], [89]. HCVAg based diagnostics also offer improvements in time and equipment requirements since testing requires less pre-processing before analysis due to the lack of amplification steps and generally only requires a single readout instrument [90]. Unfortunately, outside of the Far East there are no other HCVAg assays available but the afore mentioned automated one, and this still hampers a wider adoption of this diagnostic solution.
Two better than one? HCV ‘combo’ assays
As we have seen, anti-HCV tests are unable to close the initial ‘window’ period of a primary HCV infection to less than a few weeks but testing for the HCV core antigen will significantly reduce that gap. To increase the sensitivity of HCV serological assays in the early stages of infection some manufacturers then choose to replicate the same approach used in HIV serology, combining the detection of anti-HCV and of the HCV core antigen in the same test [60]. This approach was aimed to reduce substantially the window phase of a primary HCV infection and thus ameliorate blood safety by adopting that device to screen blood donations [62] where HCV-RNA screening was not in place. The initial versions of those so-called ‘fourth generation’ HCV assays were indeed able to reduce significantly the initial ‘window’ phase: a French study demonstrated that the Monolisa HCV Ag/Ab ULTRA detected HCV 21.6 days earlier than anti-HCV Ab assays based on data from 23 hemodialysis patients [91]. Additionally, a study involving 1,251 routine clinical specimens from Germany, Netherlands, and South Africa found that a recently developed HCV combination immunoassay (Elecsys HCV Duo) had high sensitivity and specificity for detecting HCV infections and shortened the window period by 2.2–21.9 days relative to comparator Ab−only assays [92]. Therefore, fourth-generation HCV assays have the potential to improve early detection of HCV. Unfortunately, those fourth generation HCV assays have reduced sensitivity for the HCV core antigen compared to HCVAg immunoassays [92], [93] and the detection of that viral component after seroconversion to anti-HCV, in the lack of sample pretreatment, is hindered by binding of host antibodies to HCVAg, resulting in steric hindrance of immune complexes and/or competition for the antibodies used in the immunoassay [94].
Conclusions
Almost 50 years have elapsed since the identification and definition of hepatitis C and over that period we have witnessed many advances in the diagnosis and treatment of that disease (Figure 7). The availability of serological assays for the detection of specific antibodies was the breakthrough for the diagnosis of HCV infection. Unfortunately, no outstanding improvement on those diagnostic tools has been made over the last 30 years: still today, anti-HCV antibodies are detectable only some weeks after the infection and the development of a clinical illness, thus hampering their use in the early diagnosis of HCV infection. In addition, they do not last forever, which leads to an underestimation of HCV prevalence over time. Furthermore, while antibody levels appear to correlate with an active infection [65], [95], those levels do not represent a proxy to immunity against HCV and their employ in the evaluation of potential HCV vaccines [96], [97] is thus limited (Table 2).

The most significant steps in the diagnosis and clinical management of hepatitis C virus infection on a time scale. The green circles indicate four crucial clinical developments and the light blue circles indicate the main developments in HCV diagnostics. The two major breakthroughs (one diagnostic and one clinical) are highlighted in yellow.
A condensed view of the main aspects, features and limitations of HCV serology.
|
In our opinion there are two foreseeable improvements for HCV serology:
from a technology standpoint, enhancement in signal generation such as the use of electrochemical biosensors has been proposed and is actively investigated. Those biosensors work by employing a biorecognition element that, when bound to a desired analyte, causes a change in the electrochemical properties of the system, which can then be quantified. This approach may lead to an increased sensitivity and to a potential quantification of the antibody response [88].
As far as biologicals are concerned, the inclusion of well-constructed envelope antigens on the solid phase of screening and confirmatory assays becomes a dire need. This improvement on assay design may help an earlier identification of the humoral immune response to HCV during a primary infection and a more accurate long-term identification of subjects who cleared the infection. Studies will then be needed to eventually establish a ‘protective’ effect of those antibodies either after spontaneous or treatment-induced clearance or after vaccination [13], [96], [97], [98].
-
Research ethics: Not applicable.
-
Informed consent: Not applicable.
-
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and approved its submission. All authors contributed to the conceptualization, writing and revision of this paper.
-
Use of Large Language Models, AI and Machine Learning Tools: None declared.
-
Conflict of interest: The authors state no conflict of interest.
-
Research funding: None declared.
-
Data availability: Not applicable.
References
1. Global hepatitis report 2024: action for access in low- and middle-income countries. Geneva: World Health Organization; 2024. Licence: CC BY-NC-SA 3.0 IGO.Search in Google Scholar
2. Polaris Observatory HCV Collaborators, Terrault, NA, Tacke, F, Gamkrelidze, I, Craxi, A, Tanaka, J, et al.. Global change in hepatitis C virus prevalence and cascade of care between 2015 and 2020: a modelling study. Lancet Gastroenterol Hepatol 2022;7:396–415. https://doi.org/10.1016/s2468-1253-21-00472-6.Search in Google Scholar
3. Hoofnagle, JH. Hepatitis C: the clinical spectrum of disease. Hepatology 1997;26:15S–20S. https://doi.org/10.1002/hep.510260703.Search in Google Scholar PubMed
4. World Health Organization. Hepatitis C; 2023. Online. https://www.who.int/news-room/fact-sheets/detail/hepatitisc [Accessed 21 Mar 2025].Search in Google Scholar
5. Alter, MJ, Margolis, HS, Krawczynski, K, Judson, FN, Mares, A, Alexander, WJ, et al.. The natural history of community-acquired hepatitis C in the United States. N Engl J Med 1992;327:1899–905. https://doi.org/10.1056/nejm199212313272702.Search in Google Scholar PubMed
6. Ayoub, HH, Chemaitelly, H, Omori, R, Abu-Raddad, LJ. Hepatitis C virus infection spontaneous clearance: has it been underestimated? Int J Infect Dis 2018;75:60–6. https://doi.org/10.1016/j.ijid.2018.07.013.Search in Google Scholar PubMed
7. Martinello, M, Solomon, SS, Terrault, NA, Dore, GJ. Hepatitis C. Lancet 2023;402:1085–96. https://doi.org/10.1016/s0140-6736-23-01320-x.Search in Google Scholar
8. Manns, MP, Maasoumy, B. Breakthroughs in hepatitis C research: from discovery to cure. Nat Rev Gastroenterol Hepatol 2022;19:533–50. https://doi.org/10.1038/s41575-022-00608-8.Search in Google Scholar PubMed PubMed Central
9. Meyers, JD, Dienstag, JL, Purcell, RH, Thomas, ED, Holmes, KK. Parenterally transmitted non-A, non-B hepatitis: an epidemic reassessed. Ann Intern Med 1977;87:57–9. https://doi.org/10.7326/0003-4819-87-1-57.Search in Google Scholar PubMed
10. Prince, AM, Brotman, B, Grady, GF, Kuhns, WJ, Hazzi, C, Levine, RW, et al.. Long-incubation post-transfusion hepatitis without serological evidence of exposure to hepatitis B virus. Lancet 1974;2:241–6. https://doi.org/10.1016/s0140-6736-74-91412-3.Search in Google Scholar
11. Feinstone, SM, Kapikian, AZ, Purcell, RH, Alter, HJ, Holland, PV. Transfusion-associated hepatitis not due to viral hepatitis type A or B. N Engl J Med 1975;292:767–70. https://doi.org/10.1056/nejm197504102921502.Search in Google Scholar
12. Choo, QL, Kuo, G, Weiner, AJ, Overby, LR, Bradley, DW, Houghton, M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 1989;244:359–62. https://doi.org/10.1126/science.2523562.Search in Google Scholar PubMed
13. Houghton, M. The long and winding road leading to the identification of the hepatitis C virus. J Hepatol 2009;51:939–48. https://doi.org/10.1016/j.jhep.2009.08.004.Search in Google Scholar PubMed
14. Choo, QL, Richman, KH, Han, JH, Berger, K, Lee, C, Dong, C, et al.. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA 1991;15:2451–5. https://doi.org/10.1073/pnas.88.6.2451.Search in Google Scholar PubMed PubMed Central
15. Shirachi, R, Shiraishi, H, Tateda, A, Kikuchi, K, Ishida, N. Hepatitis “C” antigen in non-A, non-B post-transfusion hepatitis. Lancet 1978;21:853–6. https://doi.org/10.1016/s0140-6736-78-91567-2.Search in Google Scholar
16. Tabor, E, Mitchell, FD, Goudeau, AM, Gerety, RJ. Detection of an antigen-antibody system in serum associated with human non-A, non-B hepatitis. J Med Virol 1979;4:161–9. https://doi.org/10.1002/jmv.1890040302.Search in Google Scholar PubMed
17. Kabiri, M, Tabor, E, Gerety, RJ. Antigen-antibody system associated with non-A, non-B hepatitis detected by indirect immunofluorescence. Lancet 1979;4:221–4. https://doi.org/10.1016/s0140-6736-79-90237-x.Search in Google Scholar
18. Chircu, LV, Pezzella, M, Lacava, V, Ricci, G. Post-transfusion hepatitis: antigen/antibody systems correlated with non-A, non-B hepatitis. J Med Virol 1980;6:147–51. https://doi.org/10.1002/jmv.1890060207.Search in Google Scholar PubMed
19. Trépo, C, Vitvitski, L, Hantz, O, Chevallier, P, Lehman, H, Schlaak, M, et al.. Detection by immunofluorescence of a new “core-like” Ag/Ab system in liver and serum of patients with NANB hepatitis. Liver 1981;1:191–200. https://doi.org/10.1111/j.1600-0676.1981.tb00033.x.Search in Google Scholar PubMed
20. Spertini, O, Frei, PC. Demonstration of a single antigen-antibody system in 26 patients with non-A, non-B viral hepatitis. Lancet 1982;23:899–0. https://doi.org/10.1016/s0140-6736-82-90867-4.Search in Google Scholar
21. Bradley, DW, Maynard, JE. Non-A, non-B hepatitis: research progress and current perspectives. Dev Biol Stand 1983;54:463–73.Search in Google Scholar
22. Dienstag, JL, Bhan, AK, Alter, HJ, Feinstone, SM, Purcell, RH. Circulating immune complexes in non-A, non-B hepatitis. Possible masking of viral antigen. Lancet 1979;16:1265–7.10.1016/S0140-6736(79)92228-1Search in Google Scholar PubMed
23. Allain, JP, Duermeijer, W, Hellings, JA, Gazengel, C, Laurian, Y, Verroust, F. Non-A, non-B hepatitis in hemophilic patients with inhibitor treated with activated prothrombin complex concentrates: lack of correlation with an antigen possibly related to non-A, non-B hepatitis. Vox Sang 1984;47:47–53. https://doi.org/10.1111/j.1423-0410.1984.tb01560.x.Search in Google Scholar PubMed
24. Kuo, G, Choo, QL, Alter, HJ, Gitnick, GL, Redeker, AG, Purcell, RH, et al.. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 1989;244:362–4. https://doi.org/10.1126/science.2496467.Search in Google Scholar PubMed
25. Alter, HJ, Purcell, RH, Shih, JW, Melpoder, JC, Houghton, M, Choo, QL, et al.. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med 1989;321:1494–500. https://doi.org/10.1056/nejm198911303212202.Search in Google Scholar
26. Miyamura, T, Saito, I, Katayama, T, Kikuchi, S, Tateda, A, Houghton, M, et al.. Detection of antibody against antigen expressed by molecularly cloned hepatitis C virus cDNA: application to diagnosis and blood screening for posttransfusion hepatitis. Proc Natl Acad Sci USA 1990;87:983–7. https://doi.org/10.1073/pnas.87.3.983.Search in Google Scholar PubMed PubMed Central
27. Dawson, GJ, Lesniewski, RR, Stewart, JL, Boardaway, KM, Gutierrez, RA, Pendy, L, et al.. Detection of antibodies to hepatitis C virus in U.S. blood donors. J Clin Microbiol 1991;29:551–6. https://doi.org/10.1128/jcm.29.3.551-556.1991.Search in Google Scholar PubMed PubMed Central
28. Hatzakis, A, Polychronaki, H, Miriagou, V, Yannitsiotis, A, Chrispeels, J, Troonen, H, et al.. Antibody responses to hepatitis C virus by second-generation immunoassays in a cohort of patients with bleeding disorders. Vox Sang 1992;63:204–9. https://doi.org/10.1111/j.1423-0410.1992.tb05101.x.Search in Google Scholar PubMed
29. Aach, RD, Stevens, CE, Hollinger, FB, Mosley, JW, Peterson, DA, Taylor, PE, et al.. Hepatitis C virus infection in post-transfusion hepatitis. An analysis with first- and second-generation assays. N Engl J Med 1991;325:1325–9. https://doi.org/10.1056/nejm199111073251901.Search in Google Scholar
30. Okamoto, H, Tsuda, F, Machida, A, Munekata, E, Akahane, Y, Sugai, Y, et al.. Antibodies against synthetic oligopeptides deduced from the putative core gene for the diagnosis of hepatitis virus infection. Hepatology 1992;15:180–6. https://doi.org/10.1002/hep.1840150203.Search in Google Scholar PubMed
31. Brown, J, Dourakis, S, Karayannis, P, Goldin, R, Chiba, J, Ohba, H, et al.. Seroprevalence of hepatitis C virus nucleocapsid antibodies in patients with cryptogenic liver disease. Hepatology 1992;15:175–9.10.1002/hep.1840150202Search in Google Scholar PubMed
32. Jeffers, LJ, Hasan, F, De Medina, M, Reddy, R, Parker, T, Silva, M, et al.. Prevalence of antibodies to hepatitis C virus among patients with cryptogenic chronic hepatitis and cirrhosis. Hepatology 1992;15:187–90. https://doi.org/10.1002/hep.1840150204.Search in Google Scholar PubMed
33. Alter, HJ. New kit on the block: evaluation of second-generation assays for detection of antibody to the hepatitis C virus. Hepatology 1992;15:350–3. https://doi.org/10.1002/hep.1840150228.Search in Google Scholar PubMed
34. Zaaijer, HL, Cuypers, HTM, Reesink, HW, Lelie, PN. Should the c100 antigen be removed from HCV antibody assays? Vox Sang 1994;66:150. https://doi.org/10.1159/000462496.Search in Google Scholar
35. Uyttendaele, S, Claeys, H, Mertens, W, Verhaert, H, Vermylen, C. Evaluation of third-generation screening and confirmatory assays for HCV antibodies. Vox Sang 1994;66:122–9. https://doi.org/10.1111/j.1423-0410.1994.tb00293.x.Search in Google Scholar PubMed
36. Courouce, AM, Bouchardeau, F, Girault, A, Le Marrec, N, Vermylen, C. Significance of NS3 and NS5 antigen in screening for HCV antibody. Lancet 1994;343:853–4. https://doi.org/10.1016/s0140-6736-94-92054-0.Search in Google Scholar
37. Vernelen, K, Claeys, H, Verhaert, H, Volckaerts, A, Vermylen, C, Courouce, AM, et al.. Significance of NS3 and NS5 antigens in screening for HCV antibody. Lancet 1994;343:853. https://doi.org/10.1016/s0140-6736-94-92054-0.Search in Google Scholar
38. Goffin, E, Pirson, Y, Cornu, C, Jadoul, M, van Ypersele de Strihou, C. Significance of NS3 and NS5 antigens in screening for HCV antibody. Lancet 1994;343:854.10.1016/S0140-6736(94)92054-0Search in Google Scholar PubMed
39. Courouce, AM, Le Marrec, N, Girault, A, Ducamp, S, Simon, N. Anti-hepatitis C virus (anti-HCV) seroconversion in patients undergoing haemodialysis: comparison of second and third generation anti-HCV assays. Transfusion 1994;34:790–5. https://doi.org/10.1046/j.1537-2995.1994.34994378281.x.Search in Google Scholar PubMed
40. Dow, BC, Buchanan, I, Munro, H, Follett, EAC, Davidson, F, Prescott, LE, et al.. Relevance of RIBA-3 supplementary test to HCV PCR positivity and genotypes for HCV confirmation of blood donors. J Med Virol 1996;49:132–6. https://doi.org/10.1002/-sici-1096-9071-199606-49-2-132-aid-jmv10-3.0.co-2-g.Search in Google Scholar
41. Ponnuvel, S, Ali, H, Prakash, A, Steve, RJ, Singh, B, Cherian, AG, et al.. Superior performance of newly developed alinity Anti-HCV next assay in the diagnosis of HCV infection. J Med Virol 2025;97:e70307. https://doi.org/10.1002/jmv.70307.Search in Google Scholar PubMed
42. Clemens, JM, Taskar, S, Chau, K, Vallari, D, Shih, JW-K, Alter, HJ, et al.. IgM antibody response in acute hepatitis C viral infection. Blood 1992;9:169–72. https://doi.org/10.1182/blood.v79.1.169.bloodjournal791169.Search in Google Scholar
43. Brillanti, S, Masci, C, Ricci, P, Miglioli, M, Barbara, L. Significance of IgM antibody to hepatitis C virus in patients with chronic hepatitis C. Hepatology 1992;15:998–1001. https://doi.org/10.1002/hep.1840150604.Search in Google Scholar PubMed
44. Chen, P-J, Wand, J-T, Hwang, L-H, Yang, Y-H, Hsieh, C-L, Kao, J-H, et al.. Transient immunoglobulin M antibody response to hepatitis C virus capsid antigen in posttransfusion hepatitis C: putative serological marker for acute viral infection. Proc Nat Acad Sci USA 1992;89:5971–5. https://doi.org/10.1073/pnas.89.13.5971.Search in Google Scholar PubMed PubMed Central
45. Sjogren, MH, Lemon, SM. Low molecular weight lgM antibody to hepatitis B core antigen in chronic hepatitis B virus infection. J Infect Dis 1983;148:445–51. https://doi.org/10.1093/infdis/148.3.445.Search in Google Scholar PubMed
46. Brunetto, MR, Torrani Cerenzia, M, Oliveri, F, Piantino, P, Randone, A, Calvo, P-L, et al.. Monitoring the natural course and response to therapy of chronic hepatitis B with an automated semi-quantitative assay for IgM anti-HBc. J Hepatol 1993;19:431–6. https://doi.org/10.1016/s0168-8278-05-80554-9.Search in Google Scholar
47. Martinelli, AL, Brown, D, Braun, HB, Michel, G, Dusheiko, GM. Quantitative assessment of hepatitis C virus RNA and IgM antibodies to hepatitis C core in chronic hepatitis C. J Hepatol 1996;24:21–6. https://doi.org/10.1016/s0168-8278-96-80181-4.Search in Google Scholar
48. Chen, M, Sonnerborg, A, Sallberg, M. Levels of hepatitis C virus (HCV) RNA in serum and their relationship to levels of immunoglobulin M and G antibodies against HCV core protein. J Clin Microbiol 1995;33:778–80. https://doi.org/10.1128/jcm.33.3.778-780.1995.Search in Google Scholar PubMed PubMed Central
49. Zuck, TF, Rose, GA, Dumaswala, UJ, Geer, NJ. Experience with a transfusion recipient education program about hepatitis C. Transfusion 1990;325:1325–9.10.1046/j.1537-2995.1990.30891020339.xSearch in Google Scholar PubMed
50. Chaudary, RK, Frenette, S, Mo, T. Evaluation of hepatitis C virus kits. J Clin Microbiol 1991;29:2616–7. https://doi.org/10.1128/jcm.29.11.2616-2617.1991.Search in Google Scholar PubMed PubMed Central
51. Van der Poel, CL, Cuypers, HT, Reesink, HW, Weiner, AJ, Quan, S, Di Nello, R, et al.. Confirmation of hepatitis C virus infection by new four-antigen recombinant immunoblot assay. Lancet 1991;337:317–9. https://doi.org/10.1016/0140-6736-91-90942-i.Search in Google Scholar
52. Marcellin, P, Martinot-Peignoux, M, Boyer, N, Pouteau, M, Aumont, P, Erlinger, S, et al.. Second generation (RIBA) test in diagnosis of chronic hepatitis C. Lancet 1991;337:551–2. https://doi.org/10.1016/0140-6736-91-91335-r.Search in Google Scholar
53. Couroucé, AM, Janot, C. Recombinant immunoblot assay first and second generations on 732 blood donors reactive for antibodies to hepatitis C virus by ELISA. The Hepatitis study group of the French society of blood transfusion. Vox Sang 1991;61:177–80. https://doi.org/10.1111/j.1423-0410.1991.tb00943.x.Search in Google Scholar PubMed
54. Polito, AJ, DiNello, RK, Quan, S, Andrews, W, Rose, J, Lee, F, et al.. New-generation RIBA hepatitis C strip immunoblot assays. Beitr Infusionsther 1992;30:17–33.Search in Google Scholar
55. Lavanchy, D, Mayerat, C, Morel, B, Schneider, P, Zufferey, C, Gonvers, J-J, et al.. Evaluation of third-generation assays for detection of anti-hepatitis C virus (HCV) antibodies and comparison with presence of HCV RNA in blood donors reactive to c1OO-3 antigen. J Clin Microbiol 1994;32:2272–5. https://doi.org/10.1128/jcm.32.9.2272-2275.1994.Search in Google Scholar PubMed PubMed Central
56. Kuhns, M, de Medina, M, McNamara, A, Jeffers, LJ, Reddy, KR, Silva, M, et al.. Detection of hepatitis C virus RNA in hemodialysis patients. J Am Soc Nephrol 1994;4:1491–7. https://doi.org/10.1681/asn.v471491.Search in Google Scholar
57. Couroucé, AM, Noel, L, Barin, F, Elghouzzi, MH, Lunel, F, North, ML, et al.. A comparative evaluation of the sensitivity of five anti-hepatitis C virus immunoblot assays. Vox Sang 1998;74:217–24. https://doi.org/10.1046/j.1423-0410.1998.7440217.x.Search in Google Scholar
58. Maertens, G, Dekeyser, F, Van Geel, A, Sablon, E, Bosman, F, Zrein, M, et al.. Confirmation of HCV antibodies by the line immunoassay INNO-LIA HCV Ab III. Methods Mol Med 1999;19:11–25. https://doi.org/10.1385/0-89603-521-2-11.Search in Google Scholar
59. Kodani, M, Martin, M, Landgraf de Castro, V, DrobeniucJ, KS, Kamili, S. An automated immunoblot method for detection of IgG antibodies to hepatitis C virus: a potential supplemental antibody confirmatory assay. J Clin Microbiol 2019;57:e01567–18. https://doi.org/10.1128/jcm.01567-18.Search in Google Scholar PubMed PubMed Central
60. Branson, BM. HIV diagnostics. Current recommendations and opportunities for improvement. Infect Dis Clin North Am 2019;33:611–8.10.1016/j.idc.2019.04.001Search in Google Scholar PubMed
61. Huang, Y, Pan, H, Gao, Q, Lv, P, Xu, X, Zhao, Z. The role of a two-assay serological testing strategy for anti-HCV screening in low-prevalence populations. Sci Rep 2021;11:8689. https://doi.org/10.1038/s41598-021-88138-2.Search in Google Scholar PubMed PubMed Central
62. Kleinmann, SH, Lelie, N, Busch, MP. Infectivity of human immunodeficiency virus-1, hepatitis C virus, and hepatitis B virus and risk of transmission by transfusion. Transfusion 2009;49:2454–89. https://doi.org/10.1111/j.1537-2995.2009.02322.x.Search in Google Scholar PubMed
63. Schreiber, GB, Busch, MP, Kleinman, SH, Korelitz, JJ, For the Retrovirus Epidemiology Study Group. The risk of transfusion-transmitted viral infections. N Engl J Med 1996;334:1685–90. https://doi.org/10.1056/nejm199606273342601.Search in Google Scholar PubMed
64. Netski, DM, Mosbruger, T, Depla, E, Maertens, G, Ray, SC, Hamilton, RG, et al.. Humoral immune response in acute hepatitis C virus infection. Clin Infect Dis 2005;41:667–75. https://doi.org/10.1086/432478.Search in Google Scholar PubMed
65. Vanhommerig, JW, Thomas, XV, van der Meer, JTM, Geskus, RB, Bruisten, SM, Molenkamp, R, et al.. Hepatitis C virus (HCV) antibody dynamics following acute HCV infection and reinfection among HIV-infected men who have sex with men. Clin Infect Dis 2014;59:1678–85. https://doi.org/10.1093/cid/ciu695.Search in Google Scholar PubMed
66. Murin, CD, Wilson, IA, Ward, AB. Antibody responses to viral infections: a structural perspective across three different enveloped viruses. Nat Microbiol 2019;4:734–47. https://doi.org/10.1038/s41564-019-0392-y.Search in Google Scholar PubMed PubMed Central
67. Lesniewski, R, Okasinski, G, Carrick, R, Van Sant, C, Desai, S, Johnson, R, et al.. Antibody to hepatitis C virus second envelope (HCV-E2) glycoprotein: a new marker of HCV infection closely associated with viremia. J Med Virol 1995;45:415–22. https://doi.org/10.1002/jmv.1890450411.Search in Google Scholar PubMed
68. Chaudhary, RRK, Burres, E. Detection of antibody to envelope (E2) antigen of hepatitis C virus. Can J Infect Dis 1997;8:229–32. https://doi.org/10.1155/1997/386268.Search in Google Scholar PubMed PubMed Central
69. Lee, DS, Lesniewski, RR, Sung, YC, Min, WK, Park, SG, Lee, KH, et al.. Significance of anti-E2 in the diagnosis of HCV infection in patients on maintenance hemodialysis: Anti-E2 is frequently detected among anti-HCV antibody-negative patients. J Am Soc Nephrol 1996;7:2409–13. https://doi.org/10.1681/asn.v7112409.Search in Google Scholar
70. Lefrère, JJ, Guiramand, S, Lefrère, F, Mariotti, M, Aumont, P, Lerable, J, et al.. Full or partial seroreversion in patients infected by hepatitis C virus. J Infect Dis 1997;175:316–22. https://doi.org/10.1093/infdis/175.2.316.Search in Google Scholar PubMed
71. Kondili, LA, Chionne, P, Costantino, A, Villano, U, Lo Noce, C, Pannozzo, F, et al.. Infection rate and spontaneous seroreversion of anti-hepatitis C virus during the natural course of hepatitis C virus infection in the general population. Gut 2002;50:693–6. https://doi.org/10.1136/gut.50.5.693.Search in Google Scholar PubMed PubMed Central
72. Mazzeo, C, Azzaroli, F, Giovanelli, S, Dormi, A, Festi, D, Colecchia, A, et al.. Ten year incidence of HCV infection in northern Italy and frequency of spontaneous viral clearance. Gut 2003;52:1030–4. https://doi.org/10.1136/gut.52.7.1030.Search in Google Scholar PubMed PubMed Central
73. Martinello, M, Carson, JM, Van Der Valk, M, Rockstroh, JK, Ingiliz, P, Hellard, M, et al.. Reinfection incidence and risk among people treated for recent hepatitis C virus infection: the REACT study. AIDS 2023;37:1883–90. https://doi.org/10.1097/qad.0000000000003651.Search in Google Scholar
74. Chacón, F, Morano, L, Navarro, J, Granados, R, Llibre, JM, Ryan, P, et al.. Rate of hepatitis C reinfection after successful direct-acting antivirals treatment among people who inject drugs in Spain: the LIVERate study. BMC Pub Health 2024;24:3167. https://doi.org/10.1186/s12889-024-20625-3.Search in Google Scholar PubMed PubMed Central
75. Read, P, Tang, BZH, Silins, E, Doab, A, Cornelisse, VJ, Gilliver, R. Hepatitis C (HCV) reinfection and risk factors among clients of a low-threshold primary healthcare service for people who inject drugs in Sydney, Australia. Viruses 2024;16:957. https://doi.org/10.3390/v16060957.Search in Google Scholar PubMed PubMed Central
76. Ingiliz, P, Martin, TC, Rodger, A, Stellbrink, H-J, Mauss, S, Boesecke, C, et al.. HCV reinfection incidence and spontaneous clearance rates in HIV-positive men who have sex with men in Western Europe. J Hepatol 2017;66:282–7. https://doi.org/10.1016/j.jhep.2016.09.004.Search in Google Scholar PubMed
77. Ingiliz, P, Wehmeyer, MH, Boesecke, C, Schulze Zur Wiesch, J, Schewe, K, Lutz, T, et al.. Reinfection with the hepatitis C virus in men who have sex with men after successful treatment with direct-acting antivirals in Germany: current incidence rates, compared with rates during the interferon era. Clin Infect Dis 2020;71:1248–54. https://doi.org/10.1093/cid/ciz949.Search in Google Scholar PubMed
78. Komatsu, F, Takasaki, T. Determination of serum hepatitis C virus (HCV) core protein using a novel approach for quantitative evaluation of HCV viremia in anti-HCV positive patients. Liver 1999;19:375–80. https://doi.org/10.1111/j.1478-3231.1999.tb00065.x.Search in Google Scholar PubMed
79. Icardi, G, Ansaldi, F, Bruzzone, BM, Durando, P, Lee, S, De Luigi, C, et al.. Novel approach to reduce the hepatitis C virus (HCV) window period: clinical evaluation of a new enzyme-linked immunosorbent assay for HCV core antigen. J Clin Microbiol 2001;39:3110–4. https://doi.org/10.1128/jcm.39.9.3110-3114.2001.Search in Google Scholar PubMed PubMed Central
80. Zanetti, AR, Romanò, L, Brunetto, M, Colombo, M, Bellati, G, Tackney, C. Total HCV core antigen assay: a new marker of hepatitis C viremia for monitoring the progress of therapy. J Med Virol 2003;70:27–30. https://doi.org/10.1002/jmv.10355.Search in Google Scholar PubMed
81. Morota, K, Fujinami, R, Kinukawa, H, Machida, T, Ohno, K, Saegusa, H, et al.. A new sensitive and automated chemiluminescent microparticle immunoassay for quantitative determination of hepatitis C virus core antigen. J Virol Methods 2009;157:8–14. https://doi.org/10.1016/j.jviromet.2008.12.009.Search in Google Scholar PubMed
82. World Health Organization. Guidelines on hepatitis B and C testing. Geneva: World Health Organization; 2017. Licence: CC BY-NC-SA 3.0 IGO.Search in Google Scholar
83. European Association for the Study of the Liver, Negro, F, Aghemo, A, Berenguer, M, Dalgard, O, Dusheiko, G, et al.. EASL recommendations on treatment of hepatitis C: final update of the series. J Hepatol 2020;73:1170–218. https://doi.org/10.1016/j.jhep.2020.08.018.Search in Google Scholar PubMed
84. Vanhommerig, JW, van de Laar, TJ, Koot, M, van Rooijena, MS, Schinkel, J, Speksnijder, AG, et al.. Evaluation of a hepatitis C virus (HCV) antigen assay for routine HCV screening among men who have sex with men infected with HIV. J Virol Methods 2015;213:147–50. https://doi.org/10.1016/j.jviromet.2014.11.026.Search in Google Scholar PubMed
85. Adee, M, Zhong, H, Reipold, EI, Zhuo, Y, Shilton, S, Chhatwal, J. Cost-effectiveness of a core antigen-based rapid diagnostic test for hepatitis C. Value Health 2022;25:1107–15. https://doi.org/10.1016/j.jval.2022.01.004.Search in Google Scholar PubMed
86. Galli, C, Julicher, P, Plebani, M. HCV core antigen comes of age: a new opportunity for the diagnosis of hepatitis C virus infection. Clin Chem Lab Med 2018;56:880–8. https://doi.org/10.1515/cclm-2017-0754.Search in Google Scholar PubMed
87. Martínez, AM, Núñez Serrano, P, Fernández de Cañete Camacho, JC, Moreno Planas, JM. Hepatitis C virus core antigen as an alternative to RNA in the assessment of response to treatment with direct-acting oral antivirals. Hepat Mon 2021;21:e118579. https://doi.org/10.5812/hepatmon.118579.Search in Google Scholar
88. Juelicher, P, Chulanov, VP, Pimenov, NN, Chirkova, E, Yankina, A, Galli, C. Streamlining the screening cascade for active hepatitis C in Russia: a cost-effectiveness analysis. PLoS One 2019;14:e0219687. https://doi.org/10.1371/journal.pone.0219687.Search in Google Scholar PubMed PubMed Central
89. Marcellusi, A, Mennini, FS, Ruf, M, Galli, C, Aghemo, A, Brunetto, MR, et al.. Optimizing diagnostic algorithms to advance hepatitis C elimination in Italy: a cost effectiveness evaluation. Liver Int 2022;42:26–37. https://doi.org/10.1111/liv.15070.Search in Google Scholar PubMed PubMed Central
90. Baber, AS, Suganthan, B, Ramasamy, RP. Current advances in hepatitis C diagnostics. J Biol Eng 2024;18:48. https://doi.org/10.1186/s13036-024-00443-2.Search in Google Scholar PubMed PubMed Central
91. Laperche, S, Le Marrec, N, Girault, A, Bouchardeau, F, Servant-Delmas, A, Maniez-Montreuil, M, et al.. Simultaneous detection of hepatitis C virus (HCV) core antigen and anti-HCV antibodies improves the early detection of HCV infection. J Clin Microbiol 2005;43:3877–83. https://doi.org/10.1128/jcm.43.8.3877-3883.2005.Search in Google Scholar PubMed PubMed Central
92. Laperche, S, Nubling, CM, Stramer, SL, Brojer, E, Grabarczyk, P, Yoshizawa, H, et al.. Sensitivity of hepatitis C virus core antigen and antibody combination assays in a global panel of window period samples. Transfusion 2015;55:2489–98. https://doi.org/10.1111/trf.13179.Search in Google Scholar PubMed PubMed Central
93. Majchrzak, M, Bronner, K, Laperche, S, Riester, E, Bakker, E, Bollhagen, R, et al.. Multicenter performance evaluation of the elecsys HCV duo immunoassay. J Clin Virol 2022;156:105293. https://doi.org/10.1016/j.jcv.2022.105293.Search in Google Scholar PubMed
94. Bui, TI, Brown, AP, Brown, M, Lawless, S, Roemmich, B, Anderson, NW, et al.. Comparison of a dual antibody and antigen HCV immunoassay to standard of care algorithmic testing. J Clin Microbiol 2024;62:e0083224. https://doi.org/10.1128/jcm.00832-24.Search in Google Scholar PubMed PubMed Central
95. Kee, KM, Wang, JH, Hung, CH, Chen, CH, Lee, CM, Chang, KC, et al.. Decreased anti-HCV titer and associated factors in chronic hepatitis C patients after sustained virological response: a prospective study. J Gastroenterol Hepatol 2012;27:1106–11. https://doi.org/10.1111/j.1440-1746.2011.06946.x.Search in Google Scholar PubMed
96. Houghton, M, Abrignani, S. Prospects for a vaccine against the hepatitis C virus. Nature 2005;436:961–6. https://doi.org/10.1038/nature04081.Search in Google Scholar PubMed
97. Johnson, J, Freedman, H, Logan, M, Alexander, J, Wong, J-X, Hockman, D, et al.. A recombinant hepatitis C virus genotype 1a E1/E2 envelope glycoprotein vaccine elicits antibodies that differentially neutralize closely related 2a strains through interactions of the N-Terminal hypervariable region 1 of E2 with scavenger receptor B1. J Virol 2019;93:e00810–9. https://doi.org/10.1128/jvi.00810-19.Search in Google Scholar
98. Soerensen, A, Popovic, F, Olesen, CH, Mendez, BL, Lohse, B, Chen, Z, et al.. Selection and characterization of a broadly neutralizing class of HCV anti-E2 VH1-69 antibodies. PLoS Pathog 2025;21:e1012428. https://doi.org/10.1371/journal.ppat.1012428.Search in Google Scholar PubMed PubMed Central
© 2025 Walter de Gruyter GmbH, Berlin/Boston
Articles in the same Issue
- Frontmatter
- Editorials
- Challenging the dogma: why reviewers should be allowed to use AI tools
- Multivariate approaches to improve the interpretation of laboratory data
- Review
- Interference of therapeutic monoclonal antibodies with electrophoresis and immunofixation of serum proteins: state of knowledge and systematic review
- Opinion Papers
- Urgent call to the European Commission to simplify and contextualize IVDR Article 5.5 for tailored and precision diagnostics
- The importance of laboratory medicine in the management of CKD-MBD: insights from the KDIGO 2023 controversies conference
- Supplementation of pyridoxal-5′-phosphate in aminotransferase reagents: a matter of patient safety
- HCV serology: an unfinished agenda
- From metabolic profiles to clinical interpretation: multivariate approaches to population-based and personalized reference intervals and reference change values
- Genetics and Molecular Diagnostics
- A multiplex allele specific PCR capillary electrophoresis (mASPCR-CE) assay for simultaneously analysis of SMN1/SMN2/NAIP copy number and SMN1 loss-of-function variants
- General Clinical Chemistry and Laboratory Medicine
- From assessment to action: experience from a quality improvement initiative integrating indicator evaluation and adverse event analysis in a clinical laboratory
- Evaluation of measurement uncertainty of 11 serum proteins measured by immunoturbidimetric methods according to ISO/TS 20914: a 1-year laboratory data analysis
- Assessing the harmonization of current total vitamin B12 measurement methods: relevance and implications
- The current status of serum insulin measurements and the need for standardization
- Method comparison of plasma and CSF GFAP immunoassays across multiple platforms
- Cerebrospinal fluid leptin in Alzheimer’s disease: relationship to plasma levels and to cerebrospinal amyloid
- Verification of the T50 Calciprotein Crystallization test: bias estimation and interferences
- An innovative immunoassay for accurate aldosterone quantification: overcoming low-level inaccuracy and renal dysfunction-associated interference
- Oral salt loading combined with postural stimulation tests for confirming and subtyping primary aldosteronism
- Evaluating the performance of a multiparametric IgA assay for celiac disease diagnosis
- Clinical significance of anti-mitochondrial antibodies and PBC-specific anti-nuclear antibodies in evaluating atypical primary biliary cholangitis with normal alkaline phosphatase levels
- Reference Values and Biological Variations
- Establishment of region-, age- and sex-specific reference intervals for aldosterone and renin with sandwich chemiluminescence immunoassays
- Validation of a plasma GFAP immunoassay and establishment of age-related reference values: bridging analytical performance and routine implementation
- Comparative analysis of population-based and personalized reference intervals for biochemical markers in peri-menopausal women: population from the PALM cohort study
- Hematology and Coagulation
- Evaluation of stability and potential interference on the α-thalassaemia early eluting peak and immunochromatographic strip test for α-thalassaemia --SEA carrier screening
- Cardiovascular Diseases
- Analytical and clinical evaluation of an automated high-sensitivity cardiac troponin I assay for whole blood
- Diabetes
- Method comparison of diabetes mellitus associated autoantibodies in serum specimens
- Letters to the Editor
- Permitting disclosed AI assistance in peer review: parity, confidentiality, and recognition
- Response to the editorial by Karl Lackner
- Hemolysis detection using the GEM 7000 at the point of care in a pediatric hospital setting: does it affect outcomes?
- Estimation of measurement uncertainty for free drug concentrations using ultrafiltration
- Cryoglobulin pre-analysis over the weekend
- Accelerating time from result to clinical action: impact of an automated critical results reporting system
- Recent decline in patient serum folate test levels using Roche Diagnostics Folate III assay
- Kidney stones consisting of 1-methyluric acid
- Congress Abstracts
- 7th EFLM Conference on Preanalytical Phase
- Association of Clinical Biochemists in Ireland Annual Conference
- Association of Clinical Biochemists in Ireland Annual Conference
- 17th Congress of the Portuguese Society of Clinical Chemistry, Genetics and Laboratory Medicine
Articles in the same Issue
- Frontmatter
- Editorials
- Challenging the dogma: why reviewers should be allowed to use AI tools
- Multivariate approaches to improve the interpretation of laboratory data
- Review
- Interference of therapeutic monoclonal antibodies with electrophoresis and immunofixation of serum proteins: state of knowledge and systematic review
- Opinion Papers
- Urgent call to the European Commission to simplify and contextualize IVDR Article 5.5 for tailored and precision diagnostics
- The importance of laboratory medicine in the management of CKD-MBD: insights from the KDIGO 2023 controversies conference
- Supplementation of pyridoxal-5′-phosphate in aminotransferase reagents: a matter of patient safety
- HCV serology: an unfinished agenda
- From metabolic profiles to clinical interpretation: multivariate approaches to population-based and personalized reference intervals and reference change values
- Genetics and Molecular Diagnostics
- A multiplex allele specific PCR capillary electrophoresis (mASPCR-CE) assay for simultaneously analysis of SMN1/SMN2/NAIP copy number and SMN1 loss-of-function variants
- General Clinical Chemistry and Laboratory Medicine
- From assessment to action: experience from a quality improvement initiative integrating indicator evaluation and adverse event analysis in a clinical laboratory
- Evaluation of measurement uncertainty of 11 serum proteins measured by immunoturbidimetric methods according to ISO/TS 20914: a 1-year laboratory data analysis
- Assessing the harmonization of current total vitamin B12 measurement methods: relevance and implications
- The current status of serum insulin measurements and the need for standardization
- Method comparison of plasma and CSF GFAP immunoassays across multiple platforms
- Cerebrospinal fluid leptin in Alzheimer’s disease: relationship to plasma levels and to cerebrospinal amyloid
- Verification of the T50 Calciprotein Crystallization test: bias estimation and interferences
- An innovative immunoassay for accurate aldosterone quantification: overcoming low-level inaccuracy and renal dysfunction-associated interference
- Oral salt loading combined with postural stimulation tests for confirming and subtyping primary aldosteronism
- Evaluating the performance of a multiparametric IgA assay for celiac disease diagnosis
- Clinical significance of anti-mitochondrial antibodies and PBC-specific anti-nuclear antibodies in evaluating atypical primary biliary cholangitis with normal alkaline phosphatase levels
- Reference Values and Biological Variations
- Establishment of region-, age- and sex-specific reference intervals for aldosterone and renin with sandwich chemiluminescence immunoassays
- Validation of a plasma GFAP immunoassay and establishment of age-related reference values: bridging analytical performance and routine implementation
- Comparative analysis of population-based and personalized reference intervals for biochemical markers in peri-menopausal women: population from the PALM cohort study
- Hematology and Coagulation
- Evaluation of stability and potential interference on the α-thalassaemia early eluting peak and immunochromatographic strip test for α-thalassaemia --SEA carrier screening
- Cardiovascular Diseases
- Analytical and clinical evaluation of an automated high-sensitivity cardiac troponin I assay for whole blood
- Diabetes
- Method comparison of diabetes mellitus associated autoantibodies in serum specimens
- Letters to the Editor
- Permitting disclosed AI assistance in peer review: parity, confidentiality, and recognition
- Response to the editorial by Karl Lackner
- Hemolysis detection using the GEM 7000 at the point of care in a pediatric hospital setting: does it affect outcomes?
- Estimation of measurement uncertainty for free drug concentrations using ultrafiltration
- Cryoglobulin pre-analysis over the weekend
- Accelerating time from result to clinical action: impact of an automated critical results reporting system
- Recent decline in patient serum folate test levels using Roche Diagnostics Folate III assay
- Kidney stones consisting of 1-methyluric acid
- Congress Abstracts
- 7th EFLM Conference on Preanalytical Phase
- Association of Clinical Biochemists in Ireland Annual Conference
- Association of Clinical Biochemists in Ireland Annual Conference
- 17th Congress of the Portuguese Society of Clinical Chemistry, Genetics and Laboratory Medicine

![Figure 1:
Natural history of hepatitis C virus infection. Due to the broad clinical spectrum of disease presentation and outcomes the percentages reported are indicative and have been derived from references [3], [4], [5], [6], [7].](/document/doi/10.1515/cclm-2025-0501/asset/graphic/j_cclm-2025-0501_fig_001.jpg)
