Abstract
Background
Forkhead box P3 (FOXP3) plays a critical role in the pathogenesis of autoimmune disorders. In the present study, we genotyped three single-nucleotide polymorphisms, namely, rs2232365, rs3761548, and rs3761549, to determine the relationship between FOXP3 polymorphisms and neuromyelitis optica spectrum disorder (NMOSD) susceptibility among the Northern Chinese Han population.
Materials and methods
We genotyped single nucleotide polymorphisms at loci of the FOXP3 gene (rs2232365, rs3761548, and rs3761549136) in 136 NMOSD patients and 224 healthy subjects using the multiplex SNaPshot technique. Allele, genotype, and haplotype frequencies were compared. qPCR was used to analyze the mRNA expression levels of FOXP3 in the peripheral blood mononuclear cells of 63 NMOSD patients and 35 healthy subjects. Non-parametric tests were used to test the FOXP3 mRNA expression across the different groups.
Results
The minor allele frequency (MAF) of G in rs2232365 was markedly lower in the NMOSD group than in the control group (odds ratio [OR] = 0.57, 95% confidence interval [95% CI]: 0.41–0.79, p = 0.001). Using genetic (codominant, dominant, and recessive) models and performing haplotype analyses, the MAF of G in rs2232365 was shown to be associated with protection against NMOSD in this population. Furthermore, haplotype analysis revealed that the haplotype GCT and the rs2232365, rs3761548, and rs3761549 alleles predicted protection against NMOSD (OR = 0.63, 95% CI = 0.41–0.97, p = 0.038). The proportions of the three genotypes of rs2232365 (p = 0.001) were not significantly different between the moderate-to-severe (Expanded Disability Status Scale (EDSS) ≥ 3 points) and mild (EDSS < 3 points) groups. Evidently, the proportion of patients with the AA genotype (64.3%) among the rs2232365 patients was significantly greater in the moderate-to-severe group than in the mild group (36.4%). However, the proportion of patients with the GG genotype (15.2%) among the rs2232365 patients was significantly greater in the mild group than in the moderate-to-severe group (2.9%). The mRNA expression of FOXP3 was markedly greater in the NMOSD group than in the control group (p = 0.001). Nevertheless, acute non-treatment patients exhibited lower FOXP3 mRNA expression than healthy controls and patients in the remission group (p = 0.004 and 0.007, respectively).
Conclusion
FOXP3 polymorphisms and haplotypes are related to NMOSD susceptibility among the Han Chinese population. The minor allele G of FOXP3 rs2232365 and the haplotype GCT are associated with protection against NMOSD. The GG genotype may decrease the severity of NMOSD, whereas the AA genotype is related to moderate-to-severe NMOSD. FOXP3 mRNA expression is greater in patients with NMOSD than in healthy controls. However, it is decreased in acute non-treatment patients compared with healthy controls.
Abbreviations
- ANT
-
acute non-treatment group
- AOGC
-
acute long-term oral glucocorticoid group
- APQ4
-
anti-aquaporin-4
- CD
-
Crohn’s disease
- CNS
-
central nervous system
- FOXP3
-
Forkhead box P3
- IL
-
interleukin
- MAF
-
minor allele frequency
- mRNA
-
messenger RNA
- MS
-
multiple sclerosis
- NMOSD
-
neuromyelitis optica spectrum disorder
- PBMCs
-
peripheral blood mononuclear cells
- PCR
-
polymerase chain reaction
- RA
-
rheumatoid arthritis
- SLE
-
systemic lupus erythematosus
- SNPs
-
single nucleotide polymorphisms
- TGF-β
-
transforming growth factor-β
- Tregs
-
regulatory T cells
1 Background
Neuromyelitis optica spectrum disorder (NMOSD) is a rare neurologic autoimmune disease and a type of central nervous system (CNS) autoimmune disorder that may lead to inflammatory demyelinating lesions in the spinal cord or optic nerves [1,2]. The incidence of NMOSD in China per 100,000 person-years was 0.278. In the northern Chinese cities of Heilongjiang and Shanxi Provinces, the crude incidence rates of NMOSD per 100,000 person-years are 0.155 and 0.425, respectively [46]. Humoral immunity plays a crucial role in the pathogenesis of NMOSD because anti-aquaporin-4 (APQ4) immunoglobulin G (IgG) autoantibodies can bind to astrocytes and promote complement- and antibody-dependent cell-mediated cytotoxic effects [3,4]. Furthermore, the upregulation of proinflammatory factors in the cerebrospinal cord and serum, as well as the infiltration of local and systemic AQP4-specific lymphocytes and T cells in the brain, can considerably facilitate cerebral injury during NMOSD [5–12]. However, the mechanism through which endogenous immunity limits inflammatory injury in NMOSD patients has largely not been identified. Moreover, additional studies are warranted to understand the specific pathogenic mechanism and etiology of NMOSD, although several potential environmental and genetic factors affecting NMOSD susceptibility were recently identified. Studies have reported that gene polymorphisms in interleukin (IL)-1 receptor-associated kinase 1 [13], B-lymphoid tyrosine kinase [14], signal transducer and activator of transcription 4 [15], proline-rich coiled-coil 2A [16], and cholesterol 7α-hydroxylase [17] are associated with NMOSD susceptibility. However, compared with those of other immune disorders, such as systemic lupus erythematosus (SLE), multiple sclerosis (MS), autoimmune thyroiditis, and rheumatoid arthritis (RA), genetic studies on NMOSD are relatively rare.
Forkhead box P3 (FOXP3) is a transcription factor and a member of the fork-winged helix family. It is encoded by FOXP3 located on the X chromosome. FOXP3 deletion combined with loss of regulatory T cells (Tregs) can accelerate inflammatory and autoimmune syndromes [18–20]. Furthermore, ectopic FOXP3 expression results in the suppressive ability of CD4CD25+ T cells to prevent autoimmune gastritis and inflammatory bowel disease [19]. However, Tregs with FOXP3 deficiency exhibit decreased expression of marker genes such as cytotoxic T lymphocyte protein 4, Epstein‒Barr virus-induced gene 3, interleukin (IL)-10, and ectonucleoside triphosphate diphosphohydrolase 1; in contrast, T effector cytokine genes such as cytokine interferon gamma, tumor necrosis factor alpha, IL-4, and IL-17 are expressed [21–24]. In Scurfy mice, the FOXP3 protein, which lacks the forkhead domain, is expressed owing to a frameshift mutation in Foxp3 [25]. Several additional loss-of-function Foxp3 mutations have been discovered in patients who develop immune dysregulation, polyendocrinopathy, enteropathy, or X-linked inheritance syndrome [26,27]. Genetic mutations in Foxp3 generally occur along with functional Treg loss, resulting in different autoimmune disorders. Previous studies have suggested that NMOSD susceptibility is associated with defects in naive Tregs [28,29]. Moreover, the promoter region of the FOXP3 gene contains three well-documented single nucleotide polymorphisms (SNPs) (SNPrs2232365, rs3761548, and rs3761549), which have been confirmed to be associated with the development of various autoimmune diseases, including MS, Graves’ disease, cancer, vitiligo, and ulcerative colitis [44,45,47–49]. Notably, these FOXP3 promoter SNPs can directly lead to changes in the binding of transcription factors, affecting the transcriptional activity of FOXP3 and its protein expression [50–53], which in turn may have an impact on the onset of autoimmune diseases.
As a result, in the present study, we determined whether FOXP3 SNPs at the above loci (rs2232365, rs3761548, and rs3761549) can predict NMOSD susceptibility in the Han Chinese population. The relationships between FOXP3 alleles, genetic models, linkage disequilibrium (LD), and haplotypes and NMOSD were determined. Simultaneously, genotypes were stratified based on AQP4–IgG status and clinical manifestations. Finally, FOXP3 messenger RNA (mRNA) expression was detected in peripheral blood mononuclear cells (PBMCs) collected from patients with NMOSD and healthy Han Chinese people. To the best of our knowledge, our study is the first to explore the polymorphism of FOXP3 and its susceptibility to NMOSD in the northern Han Chinese population. The findings of this study increase the known spectrum of FOXP3-associated diseases and may provide some reference value for future gene modification treatments for NMOSD.
2 Materials and methods
2.1 Study participants
A total of 136 patients with NMOSD (11 men and 125 women) were recruited from the Department of Neurology, Second Hospital of Hebei Medical University, between December 2020 and March 2023. Furthermore, 224 normal individuals (207 women and 17 men) were recruited from a physical examination center. The participants were northern Han Chinese. NMOSD is a rare neurologic autoimmune disease. The incidence of NMOSD in China per 100,000 person-years was 0.278 [46]. Therefore, our study sample size is relatively limited. Patients with NMOSD who fulfilled the 2015 diagnostic criteria for NMOSD were recruited [30]. The exclusion criteria were as follows: had (a) other underlying autoimmune disorders, such as RA, SLE, or thyroid disorders, or (b) chronic disorders, such as tumors, hypertension, cerebrovascular disorders, or diabetes. Informed consent was obtained from all participants before starting the study. Patient information, including age, sex, onset age, AQP4 status, initial symptoms, clinical presentations, disease history, and medication history, was collected using a standard case report form.
2.2 Characteristics of the participants
Table S2 presents the characteristics of the participants. In total, 360 participants were enrolled for SNP analysis: 224 normal subjects (92.4% women) and 136 participants with NMOSD (91.9% women). The average ages of the participants in the NMOSD and control groups were 45.53 ± 14.15 and 43.77 ± 11.60 years, respectively; these findings suggest that there were no significant differences in age or sex between the two groups (p > 0.05). The average age of onset of NMOSD was 40.58 ± 12.96 years. In addition, 107 patients (78.68%) tested positive for serum anti-AQP4 antibodies. Optic neuritis was observed in 52 (38.24%) patients, acute myelitis in 57 (41.91%) patients, and brain and mixed attacks in 27 (19.85%) patients (Table S2 in the supplementary materials).
2.3 DNA extraction and genotyping
Briefly, 3–4 mL of peripheral venous blood was collected from patients with NMOSD and controls into ethylenediaminetetraacetic acid anticoagulation tubes. Thereafter, genomic DNA was extracted using a Blood Genomic DNA Extraction Kit (Gu De, Guangzhou, China) according to the specific instructions. DNA samples were stored at −80°C until subsequent analysis. Three FOXP3 genetic variants were chosen based on previous studies on additional autoimmune diseases (rs2232365, rs3761548, and rs3761549) and were identified via the PubMed database. Gene genotyping was performed using the SNaPshot SNP method, primarily involving polymerase chain reaction (PCR) amplification, followed by purification of PCR products. This mixture was subsequently subjected to an extension reaction, after which 1 µL of the extension product was taken, mixed with 9 µL of HiDi formamide, denatured at 95°C for 3 min, immediately placed in an ice bath, and finally loaded onto a sequencer for processing. Genotyping was performed using a Snapshot Kit (ABI, USA). Table S1 lists the sequences of primers used. Table S1 shows the detailed PCR conditions and representative peak charts used in the supplementary materials.
2.4 qPCR for FOXP3 mRNA
Fasting venous blood (5 mL) was obtained from each participant to isolate PBMCs. Total RNA was extracted using total RNApure reagent (Servicebio, Wuhan, China) according to the specific instructions. The Prime Script™ RT reagent Kit was used to prepare cDNA from the RNA samples by using gDNA Eraser (Servicebio) according to specific protocols. qPCR mix (Servicebio) was used, and qPCR was performed using an ABI 7300 system. The reaction system comprises 2× Universal Blue SYBR Green qPCR Master Mix (10.0 μL), forward primer (10 μm) (0.4 μL), reverse primer (10 μm) (0.4 μL), template (2 μL), followed by addition of nuclease-free water to make a total volume of 20 μL. The PCR amplification conditions were 10 min at 95°C, 15 s at 95°C, and 1 min at 60°C for 40 cycles; followed by a final extension for 10 min at 72°C. Upon obtaining the cycle threshold (CT) value, calculate 2–△△CT, representing the relative quantification value of the target gene expression. Standardize and proceed with non-parametric tests for statistical analysis. Primer information is provided in the supplementary materials.
2.5 Statistical analysis
Demographic, clinical, and laboratory data are presented as mean ± standard deviation; on the other hand, frequencies are presented as numbers and percentages. Differences in the average values were compared using the unpaired Student’s t-test for both groups, whereas the chi-square test and Fisher’s exact test were used to analyze categorical data. Furthermore, the chi-squared test was used to calculate the Hardy–Weinberg equilibrium (HWE). The SNPs that significantly deviated from HWE (p < 0.05) were excluded. Using diverse genetic models (codominant, dominant, and recessive), logistic regression analysis was conducted to analyze the relationships after adjusting for sex and age. Furthermore, SHEsis (http://analysis2.bio-x.cn/myAnalysis.php), an online software package, was used for the HWE test, LD, and haplotype analysis. The SNPStats tool (https://www.snpstats.net/start.htm) was used to construct the haplotypes while analyzing the relationships among the associated factors. FOXP3 mRNA expression was compared between the two groups using the non-parametric Mann–Whitney U test (non-normal distribution). In contrast, FOXP3 mRNA expression was compared across the three groups using the Kruskal–Wallis (non-normal) test and Dunn’s test. The statistical analysis was performed using SPSS 25.0 (IBM Corp., Armonk, NY, USA). A two‐tailed p < 0·05 was used to indicate statistical significance.
-
Ethical approval: The research related to human use has been complied with all the relevant national regulations, institutional policies and in accordance the tenets of the Helsinki Declaration, and has been approved by the authors’ institutional review board or equivalent committee. This study was approved by the Ethics Committee of the Second Hospital of Hebei Medical University (No. 2021-R513).
-
Informed consent: Informed consent has been obtained from all individuals included in this study.
3 Results
3.1 Genotype and allele frequency distributions of the FOXP3 polymorphisms in the NMOSD and normal groups
The distribution of FOXP3 allele frequencies conformed to HWE in the NMOSD and control groups (p > 0.05). Furthermore, the minor allele frequency (MAF) of the three FOXP3 SNPs was >5%. Table 1 presents the genotyping quality results.
Data from the quality evaluation for genotyping
| Test of HWE MAF (present‐study data) | ||||
|---|---|---|---|---|
| SNP groups | p‐value | Allele | Frequencies | |
| rs2232365 | NMOSD | 0.826238 | G | 0.290 |
| Control | 0.171883 | G | ||
| rs3761548 | NMOSD | 0.840396 | A | 0.217 |
| Control | 0.197898 | A | ||
| rs3761549 | NMOSD | 0.248432 | T | 0.154 |
| Control | 0.201864 | T | ||
Abbreviations: Hardy‒Weinberg equilibrium (HWE); MAF: minor allele frequency.
Table 2 displays the FOXP3 allele and genotype distributions of rs2232365, rs3761548, and rs3761549 in both groups. The NMOSD group was not significantly associated with rs3761548 or rs3761549 (p > 0.05). Nevertheless, the frequency of the rs2232365 genotype was significantly different between the NMOSD group and the control group. In addition, the MAF G in rs2232365 was lower in the NMOSD group (0.29) than in the control group (0.42); this difference was strongly associated with a lower NMOSD risk (odds ratio [OR] = 0.57, 95% confidence interval [CI]: 0.41–0.79, p = 0.001).
Genotypes and allele frequencies of the Foxp3 polymorphisms in the NMOSD patients and controls
| HCs | NMOSD | OR | ||||
|---|---|---|---|---|---|---|
| Gene SNP | Model | Genotype | n (%) | n (%) | 95% CI | p‐values |
| rs2232365(A/G) | Alleles | A | 261 (0.58) | 193 (0.71) | 1 | 0.001 |
| G | 187 (0.42) | 79 (0.29) | 0.57 (0.41–0.79) | |||
| Codominant | A/A | 81 (0.36) | 69 (0.51) | 1.00 | 0.004 | |
| A/G | 99 (0.44) | 55 (0.40) | 0.65 (0.41–1.03) | |||
| G/G | 44 (0.20) | 12 (0.09) | 0.32 (0.16–0.66) | |||
| Dominant | A/A | 81 (0.36) | 69 (0.51) | 1 | 0.007 | |
| A/G-G/G | 143 (0.64) | 67 (0.49) | 0.55 (0.35–0.85) | |||
| Recessive | A/A-A/G | 180 (0.80) | 124(0.91) | 1 | 0.005 | |
| G/G | 44 (0.20) | 12 (0.09) | 0.40 (0.20–0.79) | |||
| Overdominant | A/A-G/G | 125 (0.56) | 81 (0.60) | 1 | 0.470 | |
| A/G | 99 (0.44) | 55 (0.40) | 0.85 (0.55–1.32) | |||
| rs3761548(A/C) | Alleles | A | 102(0.23) | 59 (0.22) | 1 | 0.737 |
| C | 346 (0.77) | 213 (0.78) | 0.94 (0.65–1.35) | |||
| Codominant | C/C | 137 (0.61) | 83 (0.61) | 1 | 0.680 | |
| A/C | 72 (0.32) | 47 (0.35) | 1.08 (0.68–1.71) | |||
| A/A | 15 (0.07) | 6 (0.04) | 0.69 (0.26–1.87) | |||
| Dominant | C/C | 137 (0.61) | 83 (0.61) | 1 | 0.940 | |
| A/C-A/A | 87 (0.39) | 53 (0.39) | 1.02 (0.65–1.58) | |||
| Recessive | C/C-A/C | 209 (0.93) | 130 (0.96) | 1 | 0.420 | |
| A/A | 15 (0.07) | 6 (0.04) | 0.67 (0.25–1.79) | |||
| Overdominant | C/C-A/A | 152 (0.68) | 89 (0.65) | 1 | 0.640 | |
| A/C | 72 (0.32) | 47 (0.35) | 1.11 (0.71–1.76) | |||
| rs3761549(C/T) | Alleles | C | 363(0.81) | 230(0.85) | 1 | 0.228 |
| T | 85(0.19) | 42(0.15) | 1.28 (0.86–1.92) | |||
| Codominant | C/C | 150 (0.67) | 99 (0.73) | 1 | 0.500 | |
| C/T | 63 (0.28) | 32 (0.24) | 0.77 (0.47–1.26) | |||
| T/T | 11 (0.05) | 5 (0.04) | 0.70 (0.23–2.08) | |||
| Dominant | C/C | 150 (0.67) | 99 (0.73) | 1 | 0.240 | |
| C/T-T/T | 74 (0.33) | 37 (0.27) | 0.76 (0.47–1.21) | |||
| Recessive | C/C-C/T | 213 (0.95) | 131 (0.96) | 1 | 0.600 | |
| T/T | 11 (0.05) | 5 (0.04) | 0.75 (0.25–2.21) | |||
| Overdominant | C/C-T/T | 161 (0.72) | 104 (0.77) | 1 | 0.330 | |
| C/T | 63 (0.28) | 32 (0.24) | 0.78 (0.48–1.28) |
The genotypic frequency data were adjusted for age and sex using logistic regression analyses.
The results of genotypic frequencies using logistic regression analyses.
Abbreviations: 95% CI, 95% confidence interval; HCs, healthy controls; NMOSD, neuromyelitis optica spectrum disorders; OR, odds ratio.
The values in bold are considered to be significant.
The SNP distribution was examined using four genetic models (codominant, dominant, recessive, and overdominant). The GG genotype of rs2232365 markedly decreased NMOSD susceptibility in the codominant (G/G vs A/A, OR = 0.32, 95% CI = 0.16–0.66, p = 0.004), dominant (A/G + G/G vs A/A, OR = 0.55, 95% CI = 0.35–0.85, p = 0.007), and recessive (G/G vs A/G + A/A, OR = 0.40, 95% CI = 0.20–0.79, p = 0.005) models. These findings suggest the protective role of the GG phenotype against NMOSD. Additional details are presented in Table 2.
3.2 LD and haplotype analyses
Among the three SNPs investigated in this study, we determined LD based on D′ and r 2 values. rs2232365 and rs3761548 exhibited LD (D′ = 0.859, r 2 = 0.363) (Figure 1). Furthermore, rs2232365 exhibited LD with rs3761549 (D′ = 0.878, r 2 = 0.282). In addition, rs3761548 and rs3761549 exhibited LD (D′ = 0.774). Next, we elucidated FOXP3 haplotypes based on the LD among the three SNPs. The following haplotypes were constructed in the order of rs2232365, rs3761548, and rs3761549; haplotypes (ACT, AAT, GCC, GAT) with a frequency of <3.0% in both groups were excluded. Table 3 presents the remaining three haplotypes. The GCT haplotype was significantly different between the NMOSD group and the control group. In particular, the GCT haplotype with the minor G allele of rs2232365 may be associated with protection against NMOSD (OR = 0.63, 95% CI = 0.41–0.97, p = 0.038) and may have protected against NMOSD susceptibility.

Logistic chosen for haplotype analysis: rs2232365, rs3761548, and rs3761549. LD patterns in FOXP3. D′ (a) and r 2 (b) mean LD coefficients of the three SNPs. Each block represents the LD relationship between two SNPs. The rs2232365 and rs3761548 variants exhibited LD for D′ = 0.859 and r 2 = 0.363. The rs2232365 and rs3761549 variants exhibited LD for D′ = 0.878 and r 2 = 0.282. The rs3761548 and rs3761549 variants exhibited LD for D′ = 0.774.
Haplotype analysis of FOXP3 polymorphisms in NMOSD patients and controls
| Haplotype | Case (%) | HCs (%) | p‐value | OR (95% CI) |
|---|---|---|---|---|
| ACC | 0.6432 | 0.5804 | 1 | |
| GAC | 0.1641 | 0.2277 | 0.074 | 0.69 (0.46–1.04) |
| GCT | 0.1264 | 0.1875 | 0.038 | 0.63 (0.41–0.97) |
| Rare | * | * | 0.110 | 4.00 (0.75–21.36) |
Abbreviations: HCs, healthy controls; 95% CI, 95% confidence interval; OR, odds ratios; * represents rare haplotype.
3.3 Stratification analyses of FOXP3 polymorphisms based on the clinical characteristics of patients with NMOSD
Statistical analysis was performed to assess the role of FOXP3 SNPs in the analyzed clinical features (AQP4 status and onset of symptoms). Allele and genotype frequencies did not significantly differ after stratifying the clinical features (Table 4). Patients with NMOSD were divided into moderate-to-severe (Expanded Disability Status Scale [EDSS] ≥3 points) and mild (EDSS <3 points) groups [31], with average ages of 48.31 ± 14.42 and 44.59 ± 15.19 years, respectively. The two groups did not exhibit any significant difference in average age (p = 0.145), and the percentages of women in the two groups were 92.9 and 90.0%, respectively, with no significant difference in sex (p = 0.677). However, in the rs2232365 subgroup, the percentages were significantly different between the two groups (p = 0.001), with genotype AA accounting for 64.3% of the participants in the moderate-to-severe subgroup, which was markedly greater than that in the mild subgroup (36.4%). In contrast, in the mild group, the GG genotype accounted for a noticeably greater percentage (15.2%) of the population in the mild group than in the moderate-to-severe group (2.9%).
Association of FOXP3 rs2232365 rs3761548 and rs3761549 with clinical characteristics of patients with NMOSD
| rs2232365 | rs3761548 | rs3761549 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical characteristics | AA | AG | GG | p | CC | AC | AA | p | CC | CT | TT | p |
| AQP4‐IgG positive n (%) | 56 (52.3) | 44 (41.1) | 7 (6.5) | 0.24 | 69 (64.5) | 34 (31.8) | 4 (3.7) | 0.28 | 77 (72) | 26 (24.3) | 4 (3.7) | 0.91 |
| AQP4‐IgG negative n (%) | 13 (44.8) | 11 (37.9) | 5 (17.2) | 14 (48.3) | 13 (44.8) | 2 (6.9) | 22 (75.9) | 6 (20.7) | 1 (3.5) | |||
| OR (95%CI) | 1 | 1.08 (0.44–2.64) | 3.08 (0.84–11.25) | 1 | 1.88 (0.80–4.45) | 2.46 (0.41–14.79) | 1 | 0.81 (0.30–2.21) | 0.87 (0.09–8.24) | |||
| Onset syndromes, n (%) | ||||||||||||
| Optic neuritis | 29 (42.0) | 23 (41.8) | 3 (25.0) | 0.903 | 30 (36.1) | 21 (44.7) | 1 (16.7) | 0.553 | 40 (40.4) | 11 (34.4) | 1 (20.0) | 0.721 |
| Acute myelitis | 26 (37.7) | 21 (38.2) | 7 (58.3) | 37 (44.6) | 17 (36.2) | 3 (50.0) | 39 (39.4) | 14 (43.8) | 4 (80.0) | |||
| Brain attacks | 8 (11.6) | 7 (12.7) | 1 (8.3) | 9 (10.8) | 5 (10.6) | 2 (33.3) | 12 (12.1) | 4 (12.5) | 0 (0.0) | |||
| Mix attacks | 6 (8.7) | 4 (7.3) | 1 (8.3) | 7 (8.4) | 4 (8.5) | 0 (0.0) | 8 (8.1) | 3 (9.4) | 0 (0.0) | |||
| EDSS score EDSS ≥3 EDSS <3 | 45 (64.3) 24 | 23 (32.9) 32 | 2 (2.9) 10 | 0.001 | 49 (70.0) 34 | 19 (27.1) 28 | 2 (2.9) 4 | 0.083 | 54 (77.1) 45 | 15 (21.4) 17 | 1 (1.4) 4 | 0.269 |
| (36.4) | (48.5) | (15.2) | (51.5) | (42.4) | (6.1) | (68.2) | (25.8) | (6.1) | ||||
Abbreviations: 95% CI, 95% confidence interval; OR, odds ratios; NMOSD, neuromyelitis optica spectrum disorders; APQ4: anti-aquaporin-4; EDSS, Expanded Disability Status Scale.
3.4 FOXP3 mRNA expression in the NMOSD and control groups
A total of 63 NMOSD samples were collected to measure FOXP3 mRNA expression in PBMCs (3 men and 60 women; average age of 45.56 ± 13.34 years). Furthermore, 35 healthy control samples were randomly collected (3 men and 32 women; average age of 46.86 ± 14.33 years). No obvious differences in age or sex were noted between the two groups (p = 0.653 and 0.663, respectively). Compared with those in the control group, FOXP3 mRNA expression in the NMOSD group was markedly greater (p = 0.001) (Figure 2a). Moreover, the 63 NMOSD patients were classified into three groups: (1) the acute non-treatment group (ANT), in which six acute-phase patients were admitted with no previous medication before blood sampling or drug administration for 1 month before disease onset; (2) the acute long-term oral glucocorticoid group (AOGC), in which 20 acute-phase patients received routine long-term oral glucocorticoids before acute onset; and (3) the remission group, in which 37 stable-phase patients received long-term oral drugs (such as glucocorticoids, mycophenolate mofetil, and azathioprine). When comparing ANT patients with age- and sex-matched healthy controls, the ANT group had decreased FOXP3 mRNA expression (p = 0.004) (Figure 2b).

Relative mRNA expression of FOXP3 in the NMOSD, HCS, and ANT groups. FOXP3 mRNA levels in PBMCs. The p‐value was calculated by comparisons of individuals’ relative expression using the 2−ΔΔCt method. RT‐PCR was used to determine the relative quantification of the FOXP3 mRNA expression level in the PBMCs of HCs and NMOSD patients. (a) FOXP3 mRNA expression was increased in the total NMOSD patients (n = 63) compared with that in the healthy controls (n = 35) (p = 0.001). (b) FOXP3 mRNA expression was lower in the acute untreated group (n = 6) than in the healthy control group (n = 35) (p = 0.004). p < 0.05 according to the non-parametric Mann–Whitney U test (non‐normality). HCs, healthy controls; ANT, acute non‐treatment. Each p‐value was corrected for age and sex.
When the three groups were adjusted for age and sex, FOXP3 expression followed the order of ANT < AOGC < remission group, from the lowest to the highest. Moreover, FOXP3 mRNA expression was lower in the ANT group than in the other two groups (p = 0.009). In addition, the ANT group exhibited significant differences compared with the remission group (p = 0.007). However, compared with those in the remission group, the AOGC group did not exhibit any significant differences; nevertheless, FOXP3 mRNA expression was lower in the AOGC group (p = 0.763) (Figure 3). In addition, FOXP3 mRNA expression was not significantly different among the diverse genotypes of NMOSD (n = 61). Nevertheless, patients with NMOSD who carry the AA genotype may exhibit decreased FOXP3 expression compared with those who carry the AG and GG genotypes (p > 0.05) (Figure 4).
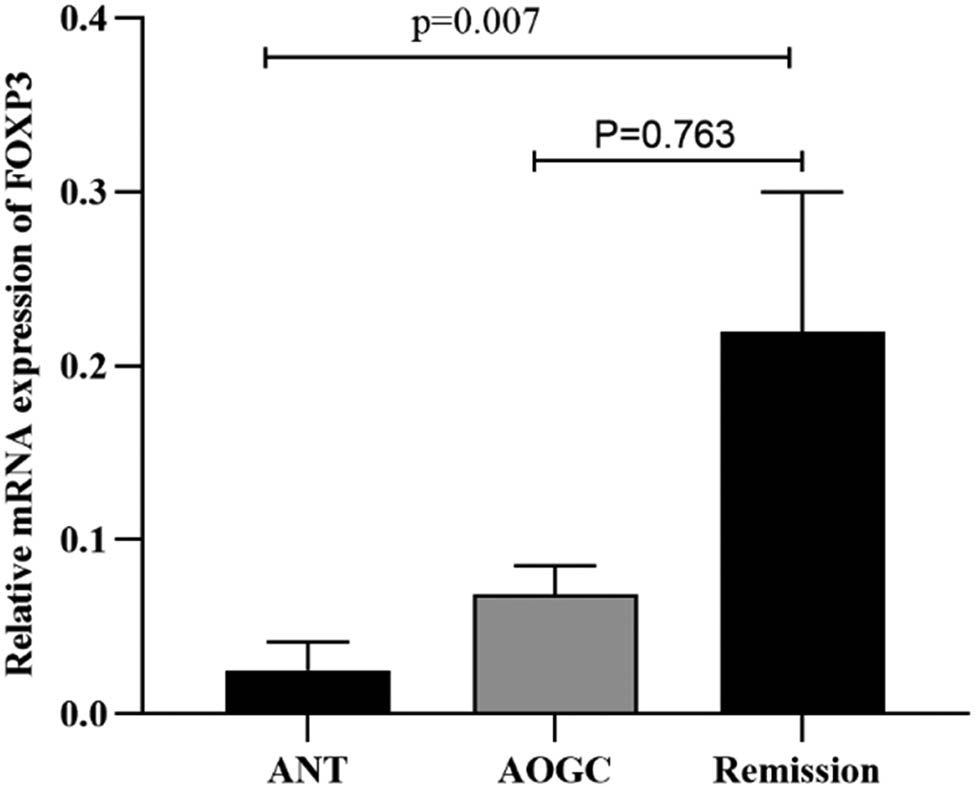
Relative mRNA expression of FOXP3 in the ANT, AOGC, and remission groups. The 63 NMOSD patients were divided into three groups: ANT (n = 6), AOGC (n = 20), and remission (n = 37). The expression levels of FOXP3 were arranged in ascending order among the three groups: ANT<AOGC<remission group. FOXP3 mRNA expression in the ANT was lower than that in the other two groups (p = 0.009). The proportion of patients in the ANT group was lower than that in the remission group (p = 0.007). There was no significant difference in the AOGC score compared to that in the remission group (p = 0.763). We used the Kruskal–Wallis test with Dunn’s correction (non‐normality) for three-group comparisons. All p values were corrected for age and sex.

FOXP3 mRNA expression levels in patients with different genotypes of rs2232365, rs3761548, and rs3761549 among the NMOSD patients. The data of 61 NMOSD patients, including rs2232365 (AA 25, AG 27, and GG 9), rs3761548 (AC 26, CC 34, and AA1), and rs3761549 (CC 44, CT 13, and TT 4), were analyzed. The Mann–Whitney U test (non‐normality) was applied for the statistical analysis. There was no observable significant variation in FOXP3 mRNA expression in NMOSD patients with different genotypes (p > 0.05). Each p-value was corrected for age and sex.
3.5 Gender analysis of FOXP3 polymorphisms
Analysis was carried out on 11 male (average age of 45.65 ± 13.88 years) and 125 female (average age of 44.09 ± 5.32 years) NMOSD patients. No obvious differences in age were noted between the two groups (p = 0.204). The ratios of the corresponding alleles rs2232365, rs3761548, and rs3761549 were not significantly different between the sexes (p = 0.201, p = 0.972, and p = 0.711, respectively). Figure 5). Specifically, 28.0% of the male NMOSD patients had the G allele of rs2232365, which was lower than the 40.9% ratio of female patients. Analysis of the rs2232365, rs3761548, and rs3761549 genotypes in NMOSD patients of different sexes revealed no statistically significant differences in the rs2232365, rs3761548, and rs3761549 genotypes between male and female NMOSD patients (p = 0.074, p = 0.092, and p = 0.007, respectively) (Figure 5). However, for rs2232365, the GG genotype had a prevalence of 7.1% in females, which was noticeably lower than the 27.3% prevalence in male patients. Similarly, there was a lower percentage of female patients (35.2%) with the CC genotype of rs3761548 than male patients (63.6%). Moreover, for rs3761549, 2.4% of the TT genotype was female, which was markedly lower than the 18.2% ratio in male patients.

Ratios of alleles and genotypes of rs2232365, rs3761548, and rs3761549 in male and female NMOSD patients. The alleles and genotypes of 11 male and 125 female NMOSD patients, including rs2232365 (A, G), rs3761548 (A, C), and rs3761549 (C, T), as well as rs2232365 (GG, AG, AA), rs3761548 (CC, AC, AA), and rs3761549 (CC, CT, TT), were analyzed. The chi-squared test was applied for the statistical analysis. There was no statistically significant difference in the ratios of the alleles or genotypes of rs2232365, rs3761548, or rs3761549 between male and female NMOSD patients (p > 0.05).
4 Discussion
In the present study, we elucidated the association between three SNPs (rs2232365, rs3761548, and rs3761549) in FOXP3 and NMOSD susceptibility in the northern Han Chinese population. We observed that there was no significant association between rs3761548 or rs3761549 and NMOSD risk; however, rs2232365 was significantly associated with decreased NMOSD risk. The MAF of G in rs2232365 was markedly decreased in the NMOSD group compared with the control group, suggesting that rs2232365 allele render protection to NMOSD in this population. In addition, we observed that the GCT haplotype carrying the minor allele G of rs2232365 exerted a protective effect.
FOXP3 is located on Xp11.23 and is a specific Treg marker. The activities and phenotypes of human Tregs are determined via FOXP3 expression [32]. During the immune response in humans, a minor CD4 T-cell subset may not lead to an immune response by suppressing additional T-cell functions when they recognize antigens. These cells are called natural Tregs or Tregs [33,34]. CD4CD25 Foxp3 Tregs exert important immunomodulatory effects by secreting immunosuppressive factors such as transforming growth factor-β (TGF-β), IL-10, IL-35, perforin, and granzyme or via cell contact-dependent mechanisms [33,35]. The ability of the immune system to tolerate self- or nonpathogenic non-self-structures can be regulated via immunosuppressive mechanisms. These data suggest that regulating FOXP3 expression can be a critical approach for controlling the immune response. Loss of functional FOXP3 leads to serious autoimmune disorders in patients with immune dysregulation [36,37]. Polymorphisms in FOXP3 can impair Treg function and result in autoimmune disorders in humans [38]. Abnormal numbers or functional defects of Tregs have been observed in individuals with NMOSD [39]. Therefore, FOXP3 polymorphisms may be associated with the occurrence of NMOSD via the phenotype and function of Tregs.
NMOSD is a type of CNS autoimmune disorder. However, its pathogenesis has not been fully elucidated, and it has complex genetic and polygenic inheritance patterns. Many genetic models have been examined to explore the relationship between FOXP3 polymorphisms and NMOSD to better understand the possible genetic patterns of the SNP loci of FOXP3 during NMOSD. Using codominant, dominant, and recessive genetic models, we observed that patients with the homozygous GG genotype of rs2232365 had a lower NMOSD risk than those with the wild-type AA genotype. Furthermore, the minor allele G of rs2232365 exerts a protective effect on NMOSD, consistent with the findings of Eftekharian et al., in which the FOXP3 rs2232365A/G polymorphism was detected in 410 individuals with MS and 446 healthy controls via PCR-based restriction fragment length polymorphism. They confirmed that FOXP3 rs2232365 G alleles can predict a markedly decreased risk of MS in the tested population [38]. In addition, Stadtlober et al. included 196 patients with SLE and 157 healthy controls and evaluated the association between rs2232365G/A and rs3761548C/A FOXP3 variants and SLE susceptibility and the SLE disease activity index; they noted that the G/C haplotype plays a protective role against SLE, whereas allele A in the above two variants may favor disease activity and autoimmunity. FOXP3 variants may affect Treg activity by inhibiting TGF-β1 generation and increasing disease activity and autoantibody levels in patients with SLE [40]. This finding is similar to that of our study in that the G haplotype (GCT) carrying the minor allele of rs2232365 may exert a protective effect. In the future, we should further investigate the specific mechanism underlying the minor allele GCT of rs2232365 in NMOSD. Interestingly, in another study that included 50 patients with Crohn’s disease (CD) and 154 healthy controls, the rs2232365 haplotype (dominant model) exerted a protective effect on 60% of patients with CD susceptibility and decreased the endoscopic severity index by 60% [41]. In addition, the rs2232365A/G FOXP3 variant is associated with prognostic outcomes and susceptibility to different autoimmune disorders, such as RA [42,43]. These findings support the findings of our study. These results revealed that FOXP3 polymorphisms are associated with the susceptibility of NMOSD in the Northern Chinese Han population.
FOXP3 exerts an important regulatory effect on FOXP3+ Tregs. Diverse single-nucleotide variants have been detected in the FOXP3 promoter region; these variants affect FOXP3 expression while impairing Treg differentiation and function, further affecting the occurrence and development of autoimmune diseases [44,45].
Subsequently, we stratified AQP4-IgG status and onset symptoms according to the rs2232365, rs3761548, and rs3761549 genotypes. However, these three SNPs were not associated with NMOSD onset symptoms or AQP4–IgG status. These results can shed novel insights into the relationship between FOXP3 polymorphisms and NMOSD.
However, the classification of patients with NMOSD into moderate-to-severe (EDSS ≥3 points) and mild (EDSS <3 points) groups for the three genotype frequencies of rs2232365 resulted in significant differences between the two groups (p = 0.001). In the moderate-to-severe group, the frequency of allele AA was 64.3%, which was significantly greater than that in the mild group (36.4%). In contrast, in the mild group, the frequency of the genotype GG was 15.2%, which was significantly greater than that in the moderate-to-severe group (2.9%). Therefore, the GG genotype may decrease NMOSD severity, whereas the AA genotype may be associated with NMOSD severity. This finding is similar to the findings of Stadtlober et al., who indicated that the allele A of rs2232365 may favor disease activity and autoimmunity [40]. In future studies, the sample size should increase, and the genotype should be further stratified to explore and identify the associations between disease severity and pathological mechanisms.
Next, we assessed the possible relationships between the three FOXP3 SNPs and mRNA expression. In our study, FOXP3 mRNA expression did not significantly differ among patients with NMOSD of diverse genotypes. However, FOXP3 mRNA expression was lower in patients with NMOSD and carrying the AA genotype of FOXP3 rs2232365 than in those carrying the AG or GG genotype; on the other hand, FOXP3 mRNA expression was greater in patients with the GG genotype than in those with the AG or AA genotype. The rs2232365 genotype was correlated with FOXP3 mRNA expression in the following order: GG > AG > AA. Furthermore, the results regarding the sex distribution of the FOXP3 polymorphism showed that for rs2232365 in females, the G allele and GG genotype were significantly lower than those in males, the CC genotype at rs3761548 was lower than that in males, and the TT genotype at rs3761549 was lower than that in males. These observations suggest that alterations in the alleles and genotypes of rs2232365, rs3761548, and rs3761549 may have greater potential to influence the expression level of female FOXP3 mRNA, which in turn may have an impact on the onset of NMOSD in female patients. Although the above experimental results showed no significant difference, other related studies have shown that gene polymorphisms might regulate mRNA expression through various mechanisms [56], and the degree of expression of SNPs correlates with the level of the inactive X chromosome [57]. Additionally, some genes with SNP–sex interactions exhibit sex-biased gene expression [58]. In future studies, the sample size should be expanded to further elucidate the relationships among FOXP3 polymorphisms, sex, and mRNA expression, as well as research into SNP functionality. Finally, we determined FOXP3 mRNA expression in 63 patients with NMOSD and 35 healthy controls and observed that FOXP3 mRNA expression was increased in the NMOSD group compared with the control group (p = 0.001). Among the 63 NMOSD patients, 57 underwent immunotherapy-related treatment, which could lead to an increase in FOXP3 mRNA and thus facilitate the repair of immune regulation. This finding is in line with the findings of Pitarokoili et al. [54,55], who suggested that FOXP3 mRNA expression can be upregulated after treatment with immunotherapy-related drugs. However, these findings contradict those of Brill et al. [29]. Therefore, the sample size needs to be increased to investigate the expression of FOXP3 mRNA further. Considering that there is a drug effect, patients in the acute phase were divided into three groups: the ANT, AOGC, and remission groups. FOXP3 mRNA expression was lower in the ANT group than in the control group (p = 0.004). Furthermore, compared with that in the AOGC and remission groups, FOXP3 mRNA expression was significantly lower in patients with NMOSD (p = 0.009). In addition, the ANT group exhibited significant differences compared with the remission group (p = 0.007). FOXP3 mRNA expression was increased in the AOGC group compared with the ANT group but decreased compared with that in the remission group, but the difference was not statistically significant. This may be due to the small sample size of our study. Subsequently, FOXP3 mRNA expression was markedly decreased in the ANT group. Therefore, FOXP3 mRNA expression has some significance for predicting disease activity and evaluating therapeutic effects. In the present study, we detected FOXP3 expression in PBMCs collected from patients with NMOSD and compared it between the ANT and control groups for the first time. Because the sample size of our study was small, future studies should be conducted with a larger sample size; in addition, additional mechanistic studies are warranted, and the effects of drug treatments should be considered.
5 Conclusion
We observed that FOXP3 polymorphisms and haplotypes are associated with NMOSD susceptibility in the Han Chinese population. The minor G allele of FOXP3 rs2232365 and the GCT haplotype (the rs2232365, rs3761548, and rs3761549 alleles) are associated with decreased NMOSD susceptibility and protection against NMOSD in this population. Furthermore, the GG genotype may decrease the severity of NMOSD, whereas the AA genotype may be associated with moderate NMOSD severity. FOXP3 mRNA expression was lower in the ANT group than in the control group. These results suggest that FOXP3 is a candidate mechanism-specific druggable target for NMOSD and has potential value for observing the dynamics of this disease. Nevertheless, our sample size is insufficient to achieve the ideal statistical power needed to detect correlations of small effect sizes; therefore, our findings should be further validated in larger cohorts. Moreover, we should determine the effect of rs2232365 on the pathogenesis of NMOSD, which is our future research direction.
Acknowledgements
The authors are grateful to all the patients and healthy volunteers for their participation, and all the colleagues who helped us to collect the blood samples.
-
Funding information: This study was supported by the Hebei Natural Science Foundation (No. H2022206483) and the S&T Program of Hebei (No. 22377711D).
-
Author contributions: Jing Liu: conceptualization, methodology, investigation, formal analysis, writing – original draft preparation; Gaoning Wang: software and investigation; Yulin Wang: methodology and formal analysis; Ruoyi Guo: data curation, writing – review and editing; Bin Li: conceptualization, project administration and funding acquisition. All the authors have read and approved the final manuscript.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Lucchinetti CF, Mandler RN, McGavern D, Bruck W, Gleich G, Ransohoff RM, et al. A role for humoral mechanisms in the pathogenesis of Devic’s neuromyelitis optica. Brain. 2002;125:1450–61. 10.1093/brain/awf151.Search in Google Scholar PubMed PubMed Central
[2] Jarius S, Wildemann B. AQP4 antibodies in neuromyelitis optica: diagnostic and pathogenetic relevance. Nat Rev Neurol. 2010;6:383–92. 10.1038/nrneurol.2010.72.Search in Google Scholar PubMed
[3] Lennon VA, Wingerchuk DM, Kryzer TJ, Pittock SJ, Lucchinetti CF, Fujihara K, et al. A serum autoantibody marker of neuromyelitis optica: distinction from multiple sclerosis. Lancet. 2004;364:2106–12. 10.1016/s0140-6736(04)17551-x.Search in Google Scholar PubMed
[4] Lennon VA, Kryzer TJ, Pittock SJ, Verkman AS, Hinson SR. IgG marker of optic-spinal multiple sclerosis binds to the aquaporin-4 water channel. J Exp Med. 2005;202:473–7. 10.1084/jem.20050304.Search in Google Scholar PubMed PubMed Central
[5] Kaneko K, Sato DK, Nakashima I, Ogawa R, Akaishi T, Takai Y, et al. CSF cytokine profile in MOG-IgG+ neurological disease is similar to AQP4-IgG+ NMOSD but distinct from MS: a cross-sectional study and potential therapeutic implications. J Neurol Neurosurg Psychiatry. 2018;89:927–36. 10.1136/jnnp-2018-317969.Search in Google Scholar PubMed PubMed Central
[6] Uzawa A, Mori M, Arai K, Sato Y, Hayakawa S, Masuda S, et al. Cytokine and chemokine profiles in neuromyelitis optica: significance of interleukin-6. Mult Scler. 2010;16:1443–52. 10.1177/1352458510379247.Search in Google Scholar PubMed
[7] Uzawa A, Mori M, Kuwabara S. Cytokines and chemokines in neuromyelitis optica: pathogenetic and therapeutic implications. Brain Pathol. 2014;24:67–73. 10.1111/bpa.12097.Search in Google Scholar PubMed PubMed Central
[8] Pohl M, Fischer MT, Mader S, Schanda K, Kitic M, Sharma R, et al. Pathogenic T cell responses against aquaporin 4. Acta Neuropathol. 2011;122:21–34. 10.1007/s00401-011-0824-0.Search in Google Scholar PubMed PubMed Central
[9] Varrin-Doyer M, Spencer CM, Schulze-Topphoff U, Nelson PA, Stroud RM, Cree BA, et al. Aquaporin 4-specific T cells in neuromyelitis optica exhibit a Th17 bias and recognize Clostridium ABC transporter. Ann Neurol. 2012;72:53–64. 10.1002/ana.23651.Search in Google Scholar PubMed PubMed Central
[10] Pohl M, Kawakami N, Kitic M, Bauer J, Martins R, Fischer MT, et al. T cell-activation in neuromyelitis optica lesions plays a role in their formation. Acta Neuropathol Commun. 2013;1:85. 10.1186/2051-5960-1-85.Search in Google Scholar PubMed PubMed Central
[11] Wang HH, Dai YQ, Qiu W, Lu ZQ, Peng FH, Wang YG, et al. Interleukin-17-secreting T cells in neuromyelitis optica and multiple sclerosis during relapse. J Clin Neurosci. 2011;18:1313–7. 10.1016/j.jocn.2011.01.031.Search in Google Scholar PubMed
[12] Chen T, Lennon VA, Liu YU, Bosco DB, Li Y, Yi MH, et al. Astrocyte-microglia interaction drives evolving neuromyelitis optica lesion. J Clin Invest. 2020;130:4025–38. 10.1172/jci134816.Search in Google Scholar PubMed PubMed Central
[13] Yan H, Guo R, Chen W, Xi X, Wang L, Ma J, et al. Associations of IRAK1 gene polymorphisms and mRNA expression with NMOSD risk in the Northern Chinese Han population. Front Neurol. 2021;12:661791. 10.3389/fneur.2021.661791.Search in Google Scholar PubMed PubMed Central
[14] Yin BW, Li B, Mehmood A, Yuan C, Song S, Guo RY, et al. BLK polymorphisms and expression level in neuromyelitis optica spectrum disorder. CNS Neurosci Ther. 2021;27:1549–60. 10.1111/cns.13738.Search in Google Scholar PubMed PubMed Central
[15] Shi Z, Zhang Q, Chen H, Lian Z, Liu J, Feng H, et al. STAT4 polymorphisms are associated with neuromyelitis optica spectrum disorders. Neuromol Med. 2017;19:493–500. 10.1007/s12017-017-8463-9.Search in Google Scholar PubMed
[16] Zhang J, Chen MJ, Zhao GX, Li HF, Wu L, Xu YF, et al. Common genetic variants in PRRC2A are associated with both neuromyelitis optica spectrum disorder and multiple sclerosis in Han Chinese population. J Neurol. 2021;268:506–15. 10.1007/s00415-020-10184-z.Search in Google Scholar PubMed
[17] Kim HJ, Park HY, Kim E, Lee KS, Kim KK, Choi BO, et al. Common CYP7A1 promoter polymorphism associated with risk of neuromyelitis optica. Neurobiol Dis. 2010;37:349–55. 10.1016/j.nbd.2009.10.013.Search in Google Scholar PubMed
[18] Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4:337–42. 10.1038/ni909.Search in Google Scholar PubMed
[19] Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–61. 10.1126/science.1079490.Search in Google Scholar PubMed
[20] Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–6. 10.1038/ni904.Search in Google Scholar PubMed
[21] Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277–84. 10.1038/ni1437.Search in Google Scholar PubMed
[22] Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–70. 10.1038/nature05479.Search in Google Scholar PubMed
[23] Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–40. 10.1038/nature06878.Search in Google Scholar PubMed PubMed Central
[24] Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–5. 10.1038/nature05543.Search in Google Scholar PubMed
[25] Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. 10.1038/83784.Search in Google Scholar PubMed
[26] Gambineri E, Torgerson TR, Ochs HD. Immune dysregulation, polyendocrinopathy, enteropathy, and X-linked inheritance (IPEX), a syndrome of systemic autoimmunity caused by mutations of FOXP3, a critical regulator of T-cell homeostasis. Curr Opin Rheumatol. 2003;15:430–5. 10.1097/00002281-200307000-00010.Search in Google Scholar PubMed
[27] Chatila TA, Blaeser F, Ho N, Lederman HM, Voulgaropoulos C, Helms C, et al. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest. 2000;106:R75–81. 10.1172/jci11679.Search in Google Scholar
[28] Cho EB, Cho HJ, Seok JM, Min JH, Kang ES, Kim BJ. The IL-10-producing regulatory B cells (B10 cells) and regulatory T cell subsets in neuromyelitis optica spectrum disorder. Neurol Sci. 2018;39:543–9. 10.1007/s10072-018-3248-y.Search in Google Scholar PubMed
[29] Brill L, Lavon I, Vaknin-Dembinsky A. Foxp3+ regulatory T cells expression in neuromyelitis optica spectrum disorders. Mult Scler Relat Disord. 2019;30:114–8. 10.1016/j.msard.2019.01.047.Search in Google Scholar PubMed
[30] Tan CT, Mao Z, Qiu W, Hu X, Wingerchuk DM, Weinshenker BG. International consensus diagnostic criteria for neuromyelitis optica spectrum disorders. Neurology. 2016;86:491–2. 10.1212/wnl.0000000000002366.Search in Google Scholar PubMed
[31] Wang L, Du L, Li Q, Li F, Wang B, Zhao Y, et al. Neuromyelitis optica spectrum disorder with anti-aquaporin-4 antibody: outcome prediction models. Front Immunol. 2022;13:873576. 10.3389/fimmu.2022.873576.Search in Google Scholar PubMed PubMed Central
[32] Roncador G, Brown PJ, Maestre L, Hue S, Martínez-Torrecuadrada JL, Ling KL, et al. Analysis of FOXP3 protein expression in human CD4+CD25+ regulatory T cells at the single-cell level. Eur J Immunol. 2005;35:1681–91. 10.1002/eji.200526189.Search in Google Scholar PubMed
[33] Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–87. 10.1016/j.cell.2008.05.009.Search in Google Scholar PubMed
[34] Li MO, Rudensky AY. T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat Rev Immunol. 2016;16:220–33. 10.1038/nri.2016.26.Search in Google Scholar PubMed PubMed Central
[35] Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–64. 10.1146/annurev.immunol.25.022106.141623.Search in Google Scholar PubMed PubMed Central
[36] Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27:18–20. 10.1038/83707.Search in Google Scholar PubMed
[37] Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–1. 10.1038/83713.Search in Google Scholar PubMed
[38] Eftekharian MM, Sayad A, Omrani MD, Ghannad MS, Noroozi R, Mazdeh M, et al. Single nucleotide polymorphisms in the FOXP3 gene are associated with increased risk of relapsing-remitting multiple sclerosis. Hum Antibodies. 2016;24:85–90. 10.3233/hab-160299.Search in Google Scholar PubMed
[39] Ma X, Qin C, Chen M, Yu HH, Chu YH, Chen TJ, et al. Regulatory T cells protect against brain damage by alleviating inflammatory response in neuromyelitis optica spectrum disorder. J Neuroinflammation. 2021;18:201. 10.1186/s12974-021-02266-0.Search in Google Scholar PubMed PubMed Central
[40] Stadtlober NP, Flauzino T, da Rosa Franchi Santos LF, Iriyoda TMV, Costa NT, Lozovoy MAB, et al. Haplotypes of FOXP3 genetic variants are associated with susceptibility, autoantibodies, and TGF-β1 in patients with systemic lupus erythematosus. Sci Rep. 2021;11:5406. 10.1038/s41598-021-84832-3.Search in Google Scholar PubMed PubMed Central
[41] Inoue CJ, Flauzino T, Gonçalves BP, Paula JCC, Galvão TC, Miyazaki PK, et al. FOXP3 variants are independently associated with transforming growth factor B1 plasma levels in female patients with inflammatory bowel disease. Clinics. 2022;77:100084. 10.1016/j.clinsp.2022.100084.Search in Google Scholar PubMed PubMed Central
[42] Paradowska-Gorycka A, Jurkowska M, Felis-Giemza A, Romanowska-Próchnicka K, Manczak M, Maslinski S, et al. Genetic polymorphisms of Foxp3 in patients with rheumatoid arthritis. J Rheumatol. 2015;42:170–80. 10.3899/jrheum.131381.Search in Google Scholar PubMed
[43] Hashemi V, Farrokhi AS, Tanomand A, Babaloo Z, Hojjat-Farsangi M, Anvari E, et al. patients with rheumatoid arthritis in Iranian population. Immunol Lett. 2018;204:16–22.10.1016/j.imlet.2018.10.001Search in Google Scholar PubMed
[44] Cheng Z, Guo Y, Ming L. Functional Foxp3 polymorphisms and the susceptibility to cancer: an update meta-analysis. Medicine. 2018;97:e11927. 10.1097/md.0000000000011927.Search in Google Scholar PubMed PubMed Central
[45] Giri PS, Patel S, Begum R, Dwivedi M. Association of FOXP3 and GAGE10 promoter polymorphisms and decreased FOXP3 expression in regulatory T cells with susceptibility to generalized vitiligo in Gujarat population. Gene. 2021;768:145295. 10.1016/j.gene.2020.145295.Search in Google Scholar PubMed
[46] Tian DC, Li Z, Yuan M, Zhang C, Gu H, Wang Y, et al. Incidence of neuromyelitis optica spectrum disorder (NMOSD) in China: a national population-based study. Lancet Reg Health West Pac. 2020;2:100021. 10.1016/j.lanwpc.2020.100021.Search in Google Scholar PubMed PubMed Central
[47] Li HN, Li XR, Du YY, Yang ZF, Lv ZT. The association between Foxp3 polymorphisms and risk of graves’ disease: a systematic review and meta-analysis of observational studies. Front Endocrinol (Lausanne). 2020 Jun 16;11:392. 10.3389/fendo.2020.00392. PMID: 32612577; PMCID: PMC7308555.47.Search in Google Scholar PubMed PubMed Central
[48] Zhang Y, Zhang J, Liu H, He F, Chen A, Yang H, et al. Meta-analysis of FOXP3 gene rs3761548 and rs2232365 polymorphism and multiple sclerosis susceptibility. Medicine. 2019 Sep;98(38):e17224. 10.1097/MD.0000000000017224. PMID: 31567981; PMCID: PMC6756718.Search in Google Scholar PubMed PubMed Central
[49] Xia SL, Ying SJ, Lin QR, Wang XQ, Hong WJ, Lin ZJ, et al. Association of ulcerative colitis with FOXP3 gene polymorphisms and its colonic expression in chinese patients. Gastroenterol Res Pract. 2019 Feb;2019:4052168. 10.1155/2019/4052168. PMID: 30918515; PMCID: PMC6409000.Search in Google Scholar PubMed PubMed Central
[50] Song P, Wang XW, Li HX, Li K, Liu L, Wei C, et al. Association between FOXP3 polymorphisms and vitiligo in a Han Chinese population. Br J Dermatol. 2013;169(3):571–8. 10.1111/bjd.12377.Search in Google Scholar PubMed
[51] Naderi-Mahabadi F, Zarei S, Fatemi R, Kamali K, Pahlavanzadeh Z, Jeddi-Tehrani M, et al. Association study of forkhead box P3 gene polymorphisms with unexplained recurrent spontaneous abortion. J Reprod Immunol. 2015;110:48–53. 10.1016/j.jri.2015.04.001.Search in Google Scholar PubMed
[52] Watkin RD, Nawrot T, Potts RJ, Hart BA. Mechanisms regulating the cadmium-mediated suppression of Sp1 transcription factor activity in alveolar epithelial cells. Toxicology. 2003;184(2–3):157–78. 10.1016/S0300-483X(02)00577-2.Search in Google Scholar
[53] Nie J, Li YY, Zheng SG, Tsun A, Li B. FOXP3(+) Treg cells and gender bias in autoimmune diseases. Front Immunol. 2015;6. 10.3389/fimmu.2015.00493.Search in Google Scholar PubMed PubMed Central
[54] Salehi N, Nourbakhsh M, Noori S, Rezaeizadeh H, Zarghi A. Tehranolid and artemisinin effects on ameliorating experimental autoimmune encephalomyelitis by modulating inflammation and remyelination. Mol Neurobiol. 2023 Oct;60(10):5975–86. 10.1007/s12035-023-03449-x Epub 2023 Jun 30. PMID: 37391648.Search in Google Scholar PubMed
[55] Pitarokoili K, Bachir H, Sgodzai M, Grüter T, Haupeltshofer S, Duscha A, et al. Induction of regulatory properties in the intestinal immune system by dimethyl fumarate in lewis rat experimental autoimmune neuritis. Front Immunol. 2019 Sep;10:2132. 10.3389/fimmu.2019.02132. PMID: 31552056; PMCID: PMC6746892 Search in Google Scholar PubMed PubMed Central
[56] Lu YH, Wang BH, Jiang F, Mo XB, Wu LF, He P, et al. Multi-omics integrative analysis identified SNP-methylation-mRNA: interaction in peripheral blood mononuclear cells. J Cell Mol Med. 2019 Jul;23(7):4601–10. 10.1111/jcmm.14315, Epub 2019 May 20. PMID: 31106970; PMCID: PMC6584519.Search in Google Scholar PubMed PubMed Central
[57] Cotton AM, Ge B, Light N, Adoue V, Pastinen T, Brown CJ. Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome. Genome Biol. 2013 Nov;14(11):R122. 10.1186/gb-2013-14-11-r122. PMID: 24176135; PMCID: PMC4053723.Search in Google Scholar PubMed PubMed Central
[58] Scholz M, Horn K, Pott J, Wuttke M, Kühnapfel A, Nasr MK, et al. X-chromosome and kidney function: evidence from a multi-trait genetic analysis of 908,697 individuals reveals sex-specific and sex-differential findings in genes regulated by androgen response elements. Nat Commun. 2024 Jan;15(1):586. 10.1038/s41467-024-44709-1. PMID: 38233393; PMCID: PMC10794254.Search in Google Scholar PubMed PubMed Central
© 2024 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Research Articles
- Brain expression profiles of two SCN1A antisense RNAs in children and adolescents with epilepsy
- Silibinin suppresses glioblastoma cell growth, invasion, stemness, and glutamine metabolism by YY1/SLC1A5 pathway
- Early exercise intervention promotes myelin repair in the brains of ischemic rats by inhibiting the MEK/ERK pathway
- Comparative analysis of CRASH and IMPACT in predicting the outcome of 340 patients with traumatic brain injury
- Association between FOXP3 polymorphisms and expression and neuromyelitis optica spectrum disorder risk in the Northern Chinese Han population
- Trehalose improves the movement ability of Aβarc Drosophila by restoring the damaged mitochondria
- The ACE2/Ang-(1-7)/MasR axis alleviates brain injury after cardiopulmonary resuscitation in rabbits by activating PI3K/Akt signaling
- Single cocaine exposure attenuates the intrinsic excitability of CRH neurons in the ventral BNST via Sigma-1 receptors
- Effect of dopamine on limbic network connectivity at rest in Parkinson’s disease patients with freezing of gait
- FT4-to-FT3 ratio is a novel prognostic marker in subacute combined spinal cord degeneration patients
- Suanzaoren decoction exerts its antidepressant effect via the CaMK signaling pathway
- Acute ischemic STROKE – from laboratory to the Patient’s BED (STROKELABED): A translational approach to reperfusion injury. Study Protocol
- Thyroid hormone T3 induces Fyn modification and modulates palmitoyltransferase gene expression through αvβ3 integrin receptor in PC12 cells during hypoxia
- Activating α7nAChR suppresses systemic inflammation by mitigating neuroinflammation of the medullary visceral zone in sepsis in a rat model
- Amelioration of behavioral and histological impairments in somatosensory cortex injury rats by limbal mesenchymal stem cell transplantation
- TTBK2 T3290C mutation in spinocerebellar ataxia 11 interferes with ciliogenesis
- In a rodent model of autism, probiotics decrease gut leakiness in relation to gene expression of GABA receptors: Emphasize how crucial the gut–brain axis
- A data science approach to optimize ADHD assessment with the BRIEF-2 questionnaire
- Cystatin C alleviates unconjugated bilirubin-induced neurotoxicity by promoting bilirubin clearance from neurocytes via exosomes, dependent on hepatocyte UGT1A1 activity
- Macrophage accumulation in dorsal root ganglion is associated with neuropathic pain in experimental autoimmune neuritis
- Identifying key biomarkers and therapeutic candidates for post-COVID-19 depression through integrated omics and bioinformatics approaches
- The hidden link: Investigating functional connectivity of rarely explored sub-regions of thalamus and superior temporal gyrus in Schizophrenia
- A pilot evaluation of the diagnostic accuracy of ChatGPT-3.5 for multiple sclerosis from case reports
- Review Articles
- Adaptation of the layer V supraspinal motor corticofugal projections from the primary (M1) and premotor (PM) cortices after CNS motor disorders in non-human primates: A survey
- Comorbidity in spinal cord injury in Iran: A narrative review
- Lipid-based nanoparticles for drug delivery in Parkinson’s disease
- Disgust sensitivity and psychopathic behavior: A narrative review
- Rapid Communications
- Long COVID elevated MMP-9 and release from microglia by SARS-CoV-2 Spike protein
- Internal consistency of the Mental Health Professional Culture Inventory: A pilot study in Romanian population
- Retraction
- Retraction of “Effect of C-phycocyanin on HDAC3 and miRNA-335 in Alzheimer’s disease”
- Corrigendum
- Corrigendum to “The ACE2/Ang-(1-7)/MasR axis alleviates brain injury after cardiopulmonary resuscitation in rabbits by activating PI3K/Akt signaling”
- Corrigendum to “Tongxinluo promotes axonal plasticity and functional recovery after stroke”
Articles in the same Issue
- Research Articles
- Brain expression profiles of two SCN1A antisense RNAs in children and adolescents with epilepsy
- Silibinin suppresses glioblastoma cell growth, invasion, stemness, and glutamine metabolism by YY1/SLC1A5 pathway
- Early exercise intervention promotes myelin repair in the brains of ischemic rats by inhibiting the MEK/ERK pathway
- Comparative analysis of CRASH and IMPACT in predicting the outcome of 340 patients with traumatic brain injury
- Association between FOXP3 polymorphisms and expression and neuromyelitis optica spectrum disorder risk in the Northern Chinese Han population
- Trehalose improves the movement ability of Aβarc Drosophila by restoring the damaged mitochondria
- The ACE2/Ang-(1-7)/MasR axis alleviates brain injury after cardiopulmonary resuscitation in rabbits by activating PI3K/Akt signaling
- Single cocaine exposure attenuates the intrinsic excitability of CRH neurons in the ventral BNST via Sigma-1 receptors
- Effect of dopamine on limbic network connectivity at rest in Parkinson’s disease patients with freezing of gait
- FT4-to-FT3 ratio is a novel prognostic marker in subacute combined spinal cord degeneration patients
- Suanzaoren decoction exerts its antidepressant effect via the CaMK signaling pathway
- Acute ischemic STROKE – from laboratory to the Patient’s BED (STROKELABED): A translational approach to reperfusion injury. Study Protocol
- Thyroid hormone T3 induces Fyn modification and modulates palmitoyltransferase gene expression through αvβ3 integrin receptor in PC12 cells during hypoxia
- Activating α7nAChR suppresses systemic inflammation by mitigating neuroinflammation of the medullary visceral zone in sepsis in a rat model
- Amelioration of behavioral and histological impairments in somatosensory cortex injury rats by limbal mesenchymal stem cell transplantation
- TTBK2 T3290C mutation in spinocerebellar ataxia 11 interferes with ciliogenesis
- In a rodent model of autism, probiotics decrease gut leakiness in relation to gene expression of GABA receptors: Emphasize how crucial the gut–brain axis
- A data science approach to optimize ADHD assessment with the BRIEF-2 questionnaire
- Cystatin C alleviates unconjugated bilirubin-induced neurotoxicity by promoting bilirubin clearance from neurocytes via exosomes, dependent on hepatocyte UGT1A1 activity
- Macrophage accumulation in dorsal root ganglion is associated with neuropathic pain in experimental autoimmune neuritis
- Identifying key biomarkers and therapeutic candidates for post-COVID-19 depression through integrated omics and bioinformatics approaches
- The hidden link: Investigating functional connectivity of rarely explored sub-regions of thalamus and superior temporal gyrus in Schizophrenia
- A pilot evaluation of the diagnostic accuracy of ChatGPT-3.5 for multiple sclerosis from case reports
- Review Articles
- Adaptation of the layer V supraspinal motor corticofugal projections from the primary (M1) and premotor (PM) cortices after CNS motor disorders in non-human primates: A survey
- Comorbidity in spinal cord injury in Iran: A narrative review
- Lipid-based nanoparticles for drug delivery in Parkinson’s disease
- Disgust sensitivity and psychopathic behavior: A narrative review
- Rapid Communications
- Long COVID elevated MMP-9 and release from microglia by SARS-CoV-2 Spike protein
- Internal consistency of the Mental Health Professional Culture Inventory: A pilot study in Romanian population
- Retraction
- Retraction of “Effect of C-phycocyanin on HDAC3 and miRNA-335 in Alzheimer’s disease”
- Corrigendum
- Corrigendum to “The ACE2/Ang-(1-7)/MasR axis alleviates brain injury after cardiopulmonary resuscitation in rabbits by activating PI3K/Akt signaling”
- Corrigendum to “Tongxinluo promotes axonal plasticity and functional recovery after stroke”

