Abstract
Aim
This study aimed to isolate moderately halophilic bacteria from salted goat skins, to characterize these microorganisms and to determine their industrially important enzymes such as amylase, catalase, oxidase, caseinase, cellulase, DNase, lipase, lecithinase, protease, pullulanase, urease, phospholipase, xylanase and β-galactosidase.
Methods
Enzymes of these bacteria, isolated from skin samples belonging to eight countries and identified using phenotypic and genotypic methods, were examined in agar media.
Results
Thirty-nine isolates were fairly similar to species of genera Staphylococcus, Bacillus, Salinicoccus, Gracilibacillus, Chromohalobacter and Halomonas. Various carbon sources were utilized, and all isolates produced enzyme. Enzyme-producing species were Staphylococcus saprophyticus subsp. saprophyticus, Staphylococcus arlettae, Bacillus pumilus, Gracilibacillus dipsosauri, Salinicoccus roseus, Bacillus licheniformis, Chromohalobacter beijerinckii, Staphylococcus xylosus, Halomonas eurihalina, Staphylococcus equorum subsp. equorum, Halomonas zhanjiangensis, Halomonas venusta and Chromohalobacter canadensis. Fairly high percentage of isolates produced protease (87%) and catalase (100%). While more than 50% of isolates produced lipase (64%), β-galactosidase (59%) and oxidase (56%), less than 50% of isolates produced urease (46%), caseinase (28%), amylase (26%), lecithinase (8%) and cellulase (5%).
Conclusion
We detected that moderately halophilic bacteria on skins produced important enzymes, which may be used in diverse industrial applications in leather, feed, detergent, paper, food, chemical, medical, pharmaceutical, textile industries.
Özet
Amaç
Bu çalışma, ılımlı halofil bakterileri tuzlanmış keçi derilerinden izole etmeyi, bu mikroorganizmaları karakterize etmeyi ve bunların amilaz, katalaz, oksidaz, kaseinaz, selülaz, DNaz, lipaz, lesitinaz, proteaz, pullulanaz, üreaz, fosfolipaz, ksilinaz ve β-galaktosidaz gibi endüstriyel olarak önemli enzimlerini saptamayı amaçlamıştır.
Yöntemler
Sekiz ülkeye ait olan deri örneklerinden izole edilerek fenotipik ve genotipik metodlara göre tanımlanan bu bakterilerin enzimleri, agar besiyerlerinde incelendi.
Bulgular
Staphylococcus, Bacillus, Salinicoccus, Gracilibacillus,Chromohalobacter ve Halomonas cinslerine ait 39 izolat bu cinslere ait türlere oldukça benzerdi. Çeşitli karbon kaynakları kullanıldı ve izolatların tümü enzim üretti. Enzim üreten türler Staphylococcus saprophyticus subsp. saprophyticus, Staphylococcus arlettae, Bacillus pumilus, Gracilibacillus dipsosauri, Salinicoccus roseus, Bacillus licheniformis, Chromohalobacter beijerinckii, Staphylococcus xylosus, Halomonas eurihalina, Staphylococcus equorum subsp. equorum, Halomonas zhanjiangensis, Halomonas venusta ve Chromohalobacter canadensis’di. İzolatların oldukça yüksek yüzdesi proteaz (87%) ve katalaz (100%) enzimlerini üretti. İzolatların yarısından fazlası lipaz (64%), β-galaktosidaz (59%) ve oksidaz (56%) enzimlerini üretirken, yarısından azı üreaz (46%), kazeinaz (28%), amilaz (26%), lesitinaz (8%) ve selülaz (5%) üretti.
Sonuç
Derilerdeki ılımlı halofil bakterilerin, deri, yem, deterjan, kâğıt, gıda, kimya, medikal, ilaç, tekstil gibi çeşitli endüstriyel uygulamalarda kullanılabilecek önemli enzimleri ürettiklerini saptadık.
Introduction
Enzymatic processes provide many advantages in terms of saving energy, water, raw materials and chemicals that cannot be achieved using conventional chemical processes. Due to enzymes’ biodegradable structure, obtaining high quality product, low energy consumption, low-cost material and less environmental pollution, enzymes are more preferred as economically and ecologically alternatives to chemicals in industrial applications [1]. The use of microbial enzymes is widespread in biotechnological applications as metabolic catalysts for centuries. Industrial enzymes have been used in food, feed, chemical and pharmaceutical, textile, biofuel, paper and pulp, detergent and leather industries as well as production of fish sauce and soy sauce, bioremediation, saline waste water, and oil field waste treatment [2].
Annual spending on enzymes is quite high worlwide. According to report by BBC Research (2014), globally the enzyme market is approximately estimated at 7.1 billion dollars by 2018 [3]. Due to the high demand for industrial enzymes, researchers have been focused on finding new industrial microorganisms producing different enzymes that can withstand extreme conditions. Especially moderate halophiles are considered to be potential sources for industrial enzymes such as cellulases, lipases, xylanases, proteases, amylases, nucleases, and esterases which are able to perform their functions under a wide range of salinity (optimally 3–15% NaCl), pH, and temperatures [1], [2], [4], [5], [6]. Furthermore, moderate halophiles present opportunities owing to easily growing on low-cost substrate and low water content [7]. In the previous studies, different moderately halophilic bacterial genera (Bacillus, Chromohalobacter, Halobacillus, Halomonas, Marinococcus, Oceanobacillus, Salibacillus, Salinicoccus, Salinivibrio and Thalassobacillus), isolated from natural saline environments, produced DNase, lipase, xylanase, amylase, inulinase, protease, cellulase, pullulanase, pectinase and caseinase [5], [8], [9].
Moderately halophilic bacteria have been isolated from hypersaline environments such as saline lakes, salterns, solar salt evaporation ponds, saline soils, salt mine soil, marine sediments and other saline habitats such as salted fish, fermented anchovy sauce, meat and hides [10], [11], [12], [13], [14], [15], [16]. Although characterization of moderately halophilic bacteria, isolated from different saline environments, using conventional and molecular techniques and detail experiments on their hydrolytic enzymes have been carried out by several researchers [5], [8], [9], investigation of moderately halophilic bacteria found on salted goat skins, using both conventional and molecular techniques and examination of their industrial enzymes have not been reported previously. Screening of enzyme producing moderately halophilic bacteria on goat skins cured with different countries’ salt will be important for determination of industrially potential microorganisms. Hence, the present study focused on the screening industrially important moderately halophilic bacteria producing amylase, catalase, oxidase, caseinase, cellulase, DNase, lipase, lecithinase, protease, pullulanase, urease, phospholipase, xylanase and β-galactosidase. In order to characterize moderately halophilic bacteria isolated from the skins and to understand their physiological and biochemical characteristics for enzyme production, phenotypic characteristics and comparative partial 16S rRNA gene sequence analysis of these microorganisms were also examined in this study.
Materials and methods
Sample collection
Twenty-three salted goat skin samples imported from Australia, Bulgaria, Israel, South Africa, Russia, China, France, and four salted goat skin samples preserved in Turkey were collected from different tanneries in the Leather Organized Tannery Region, Tuzla and Corlu, Turkey. The samples were then placed into sterile prelabeled translucent ziplock bags and transported in a sterile and cold container. The samples were named according to its origin: Australia (AL, AVT), Turkey (E, KE, T, YE), Bulgaria (B, BLG, BT), Israel (IRL), South Africa (KT, K, NE, GK), Russia (RU, RA), China (CCN, CN, CC), France (F, FR, FN, FS) (Table 1).
Phenotypic characteristics of the moderately halophilic bacterial isolates.
| Characteristics | Staphylococcus saprophyticus subsp. saprophyticus | Staphylococcus arlettae | Bacillus pumilus | Gracilibacillus dipsosauri | Salinicoccus roseus | Bacillus licheniformis | Chromohalobacter beijerinckii |
|---|---|---|---|---|---|---|---|
| Isolate numbers | 7 | 6 | 6 | 5 | 3 | 2 | 2 |
| Pigmentation | Yellow | Cream | White | White | Pink | White | Cream |
| Gram staining | + | + | + | + | + | + | − |
| Cell morphology | Cocci | Cocci | Rod | Rod | Cocci | Rod | Rod |
| NaCl range (%) | 3−12.5 | 0.5–15 | 3–15 | 0.5–20 | 0.5–25 | 3–12.5 | 3–25 |
| Optimum NaCl (%) | 10 | 10 | 10 | 10 | 10 | 10 | 7.5–10 |
| Temperature range (°C) | 20–40 | 20–40 | 20–40 | 20–45 | 20–40 | 20–45 | 4–40 |
| Optimum temperature (°C) | 37 | 37 | 37 | 37 | 37 | 37 | 30–37 |
| pH range | 6–8 | 6–8 | 6–8 | 5–9 | 5–9 | 6–10 | 5–9 |
| Optimum pH | 7.5 | 7.5 | 7.5 | 7.5 | 7.5 | 7 | 7.5 |
| Endospore formation | − | − | + | + | − | + | − |
| Flagella | − | − | + | + | − | + | + |
| Production of indole | − | − | − | − | − | − | − |
| Citrate utilization | − | − | + | − | − | + | + |
| Methyl-red test | + | + | − | + | − | + | + |
| Voges-Proskauer test | − | − | + | − | − | + | − |
| Production of H2S | − | − | − | − | − | − | − |
| Nitrate reduction | − | − | − | + | + | + | + |
| Production of N2 | − | − | − | − | + | − | − |
| Production of NH3 | + | − | + | + | + | + | + |
| Production of acid from different carbon energy sources | |||||||
| Sucrose | + | + | + | + | − | + | − |
| D-glucose | + | + | + | + | − | + | + |
| D-galactose | + | − | + | − | − | + | + |
| Fructose | + | + | + | + | − | + | − |
| D-trehalose | + | + | + | + | − | + | + |
| D-melibiose | − | + | − | + | − | + | + |
| D-mannose | − | + | + | + | − | + | + |
| D-xylose | − | + | + | + | − | + | + |
| Lactose | + | − | + | + | + | − | − |
| Maltose | + | + | − | + | − | − | − |
| L-arabinose | − | + | + | + | − | + | + |
| D-cellobiose | − | − | + | − | − | + | − |
| Hydrolytic activities | |||||||
| Catalase | + | + | + | + | + | + | + |
| Protease | + | + | + | + | + | + | − |
| Lipase | + | − | + | + | + | + | − |
| B-galactosidase | + | − | + | + | − | + | − |
| Oxidase | − | − | + | + | + | + | + |
| Urease | + | + | − | − | − | − | − |
| Caseinase | − | − | + | − | + | + | − |
| Amylase | − | − | − | + | + | + | − |
| Lecithinase | − | − | − | − | − | − | − |
| Cellulase | − | − | − | − | − | + | − |
| Pullulanase | − | − | − | − | − | − | − |
| Xylanase | − | − | − | − | − | − | − |
| Phospholipase | − | − | − | − | − | − | − |
| DNase | − | − | − | − | − | − | − |
| Staphylococcus xylosus | Halomonas eurihalina | Staphylococcus equorumsubsp. equorum | Halomonas zhanjiangensis | Halomonas venusta | Chromohalobacter canadensis | ||
| Isolate numbers | 2 | 2 | 1 | 1 | 1 | 1 | Positive isolates (%) |
| Pigmentation | Yellow | Cream | Cream | Yellow | Cream | White | |
| Gram staining | + | − | + | − | − | − | |
| Cell morphology | Cocci | Rod | Cocci | Rod | Rod | Rod | |
| NaCl range (%) | 3–12.5 | 0.5–25 | 0.5–15 | 3–20 | 3–15 | 3–20 | |
| Optimum NaCl (%) | 10 | 10 | 10 | 7.5–10 | 10 | 7.5–10 | |
| Temperature range (°C) | 20–40 | 4–45 | 20–40 | 4–40 | 20–40 | 20–45 | |
| Optimum temperature (°C) | 37 | 37 | 37 | 30 | 37 | 30–37 | |
| pH range | 6–8 | 5–9 | 6–8 | 6–10 | 6–10 | 5–9 | |
| Optimum pH | 7 | 7 | 7.5 | 7.5 | 7.5 | 7.5 | |
| Endospore formation | − | − | − | − | − | − | 33 |
| Flagella | − | + | − | + | + | + | 51 |
| Production of indole | − | − | − | − | − | + | 3 |
| Citrate utilization | − | − | − | + | + | − | 31 |
| Methyl-red test | + | + | + | − | + | + | 74 |
| Voges-Proskauer test | − | − | − | − | − | − | 21 |
| Production of H2S | − | + | − | − | − | − | 5 |
| Nitrate reduction | + | + | + | + | + | + | 51 |
| Production of N2 | − | − | − | − | − | − | 8 |
| Production of NH3 | + | + | + | + | + | + | 85 |
| Production of acid from different carbon energy sources | |||||||
| Sucrose | + | + | + | + | + | − | 85 |
| D-glucose | + | + | + | + | + | + | 92 |
| D-galactose | + | + | − | − | − | + | 56 |
| Fructose | + | − | − | + | − | − | 74 |
| D-trehalose | + | + | + | + | + | − | 90 |
| D-melibiose | − | − | + | − | − | + | 44 |
| D-mannose | + | + | + | + | + | + | 74 |
| D-xylose | + | − | + | + | + | + | 69 |
| Lactose | + | + | + | + | − | + | 72 |
| Maltose | + | + | − | + | − | + | 62 |
| L-arabinose | + | + | + | + | − | + | 72 |
| D-cellobiose | − | + | − | − | − | + | 28 |
| Hydrolytic activities | |||||||
| Catalase | + | + | + | + | + | + | 100 |
| Protease | + | + | + | − | − | − | 87 |
| Lipase | − | + | − | − | − | − | 64 |
| B-Galactosidase | + | − | + | − | − | − | 59 |
| Oxidase | − | + | − | + | + | − | 56 |
| Urease | + | + | + | − | − | − | 46 |
| Caseinase | − | − | − | − | − | − | 28 |
| Amylase | − | − | − | − | − | − | 26 |
| Lecithinase | + | − | + | − | − | − | 8 |
| Cellulase | − | − | − | − | − | − | 5 |
| Pullulanase | − | − | − | − | − | − | − |
| Xylanase | − | − | − | − | − | − | − |
| Phospholipase | − | − | − | − | − | − | − |
| DNase | − | − | − | − | − | − | − |
Isolation of the moderately halophilic bacteria
To isolate moderately halophilic bacteria, 20 g skin samples were separately soaked in flasks containing 180 mL of 10% NaCl. The flasks were placed into orbital shaker at 100 rpm for 4 h at 25°C. Sterile physiological saline solution containing 10% NaCl was used to dilute skin solutions. An aliquot of 0.1 mL of each direct and serial dilutions (from 10−1 to 10−6) of skin solutions was spread onto the surface of the agar plates containing Complex Medium I (CMI) supplemented with 0.5% (w/v) yeast extract with 10% final salt concentration (SW10, saline water) consisting of (w/v): 8.1% NaCl, 0.2% KCl, 0.7% MgCl2, 0.006% NaHCO3, 0.96% MgSO4, 0.0026% NaBr and 0.036% CaCl2 [17]. Yeast extract and all chemicals used in SW10 were from the same company (Merck, Darmstadt, Germany). The pH of the media was adjusted to 7.5 prior to autoclaving. The plates were incubated at 37°C during 24 h. After incubation period, bacterial cultures were selected by their different colony morphologies and re-streaked on CMI agar to obtain pure isolates. Since the selections of isolates were made according to their different colony morphologies, a few different species having similar colony morphologies might have been missed. Then, the phenotypic and genotypic analysis of the pure cultures were performed.
Amplification and sequencing of 16S rRNA genes
The genomic DNA of the isolates was extracted by QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s recommended method. The 16S rRNA genes were amplified using the universal primers 16F27 (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 16R1488 (5′-CGG TTA CCT TGT TAG GAC TTC ACC-3′). A PCR cycler (Techne, Staffordshire, UK) was used for amplification. Amplification reactions contained each primer (2.5 μL), dNTPs (10 mM, 8 μL), PCR buffer (5 μL), MgCl2 (25 mM, 2.5 μL), template DNA (1 μL), Taq DNA polymerase [(0.5 μL), (Thermo Fisher Scientific, Vilnius, Lithuania)], dH2O (28 μL), in a final volume of 50 μL [18]. The following conditions were used in the amplification of 16S rRNA gene: 95°C for 5 min, followed by 25 cycles of 94°C for 1 min, 50°C for 1 min and 72°C for 2 min, with final 10 min extension at 72°C. The PCR products were then checked on agarose gel with ethidium bromide staining and purified using QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). The 16S rRNA gene sequences were determined by IONTEK Laboratory (Turkey). 16S rRNA gene similarities between isolates and closely related species were determined further using ChromasPro (South Brisbane, Australia) and EzTaxon-e tool (Seoul, Korea) (Table 1) [19].
Phenotypic characterization of the isolates
The phenotypic characterization of bacterial cells was performed according to Gram reaction, pigmentation and cell morphology, growth at different salt concentrations (0, 0.5, 3, 5, 7.5, 10, 12.5, 15, 20 and 25%), growth at different pH (4, 5, 6, 7, 8, 9, 10, 11), growth at different temperatures (4, 28, 35, 37, 40, 45°C), citrate utilization, indole production, methyl red and Voges-Proskauer tests, H2S production, reduction of nitrate to nitrite, production of N2, production of NH3 from peptone, the ability to produce acid from different carbon sources. Acid production from different carbon sources was separately examined using 1% (w/v) lactose, sucrose, D-glucose, D-galactose, D-trehalose, D-melibiose, D-mannose, D-xylose, D-cellobiose, L-arabinose, fructose, maltose, 0.5% (w/v) yeast extract, and 0.001% (w/v) phenol red [9], [20]. All carbon sources and phenol red used in this experiment were obtained from same company (Merck, Darmstadt, Germany).
Enzymatic activities of the moderately halophilic isolates
Catalase activity (hydrogen-peroxide oxidoreductase, EC 1.11.1.6) was determined by adding 3% H2O2 to colonies grown on CMI agar medium. The immediate appearance of bubbles was accepted as a positive test result. Oxidase activity (ferrocytochrome-c, EC 1.9.3.1) was examined by transfering colony of the isolate with a sterile loop onto filter paper moistened with oxidase reagent (Merck, Darmstadt, Germany). Occurrence of color change from pink to dark purple in a few seconds indicated positive oxidase activity [9], [20], [21], [22]. Amylase activity (4-α-D-glucan glucanohydrolase, EC 3.2.1.1) was detected using CMI agar medium supplemented with 0.5% (w/v) soluble starch. After incubation, the plate was flooded with 0.3% I2–0.6% KI solution. Clear halos around the colonies indicated starch hydrolysis. The DNase test agar (Merck, Darmstadt, Germany) was used to determine DNase activity (deoxyribonuclease I, EC 3.1.21.1). After incubation, the plate was flooded with 1N HCl. Clear zones around the colonies showed hydrolysis of DNA [8], [21], [22]. The cellulose medium agar plate containing 0.2% (w/v) carboxymethyl cellulose was used to detect production of cellulase (4-β-D-glucan cellobiohydrolase, EC 3.2.1.91). After incubation, 0.1% congo red test reagent (Merck, Darmstadt, Germany) was flooded on the colonies and left for 30 min. Then, the colonies were washed with 1 M NaCl solution. Clear zones around the colonies showed cellulase activity [21], [22], [23]. Hydrolysis of casein was tested with the Plate Count Agar medium containing 2% skim milk. After incubation, clear zones around the colonies were interpreted as caseinase production (caseinolytic protease, EC 3.4.21.92) [9], [21], [22]. Lipase activity (triacylglycerol acylhydrolase, EC 3.1.1.3) was screened on Tween 80 agar medium containing 1% (w/v) Tween 80 (Merck, Darmstadt, Germany). After incubation, opaque zones around the colonies were accepted as evidence of lipase activity [21], [22], [23]. Due to the presence of fats on the skins, lipase activity of the isolates was also tested in agar medium containing 5% (w/v) butter. After incubation, 20% cupper-sulfate solution was flooded onto the plates. Positive bluish green colonies were interpreted as phospholipase activity (phosphatidylcholine 1-acylhydrolase, EC 3.1.1.32) [21], [22], [24], [25]. Protease activity (gelatinase A, EC 3.4.24.24) was screened on gelatin agar medium containing 2% gelatin (w/v). After incubation, the plate was flooded with Frazier solution. Clear zones around the colonies were interpreted as positive protease activities [8], [21], [22]. Urease activity (urea amidohydrolase, EC 3.5.1.5) was detected on Christensen Urea Agar (Difco, Detroit, USA). After growth was obtained, the tube was examined for pink or red color changes [21], [22], [26]. To detect pullulolytic (pullulan 6-α-glucanohydrolase, EC 3.2.1.41) and xylanolytic (4-β-D-xylan xylanohydrolase, EC 3.2.1.8) activities of the isolates, plates containing the chromogenic substrates such as azurine-cross-linked (AZCL)-pullulan and AZCL-xylan (Megazyme, Wicklow, Finland) were used. Clear zones around the colonies were accepted as positive pullulolytic and xylanolytic activities [8], [21], [22]. Lecithinase activity (phosphatidylcholine 2-acylhydrolase, EC 3.1.1.4) was investigated on lecithine agar plate containing 5% egg yolk (w/v). Opaque zones around the colonies showed lecithinase activity [21], [22], [24], [25]. Beta-galactosidase activity (β-D-galactoside galactohydrolase, EC 3.2.1.23) was detected in test tubes containing 1 mL of 10% NaCl (w/v) sterile distilled water and ONPG (ortho-nitrophenyl-β-galactoside) discs (Sigma-Aldrich, Buchs, Switzerland). The formation of yellow color was accepted as positive β-galactosidase activity [21], [22], [23]. The pH of all media was adjusted to 7.5. The salt mixture (SW10) was used in all biochemical test media.
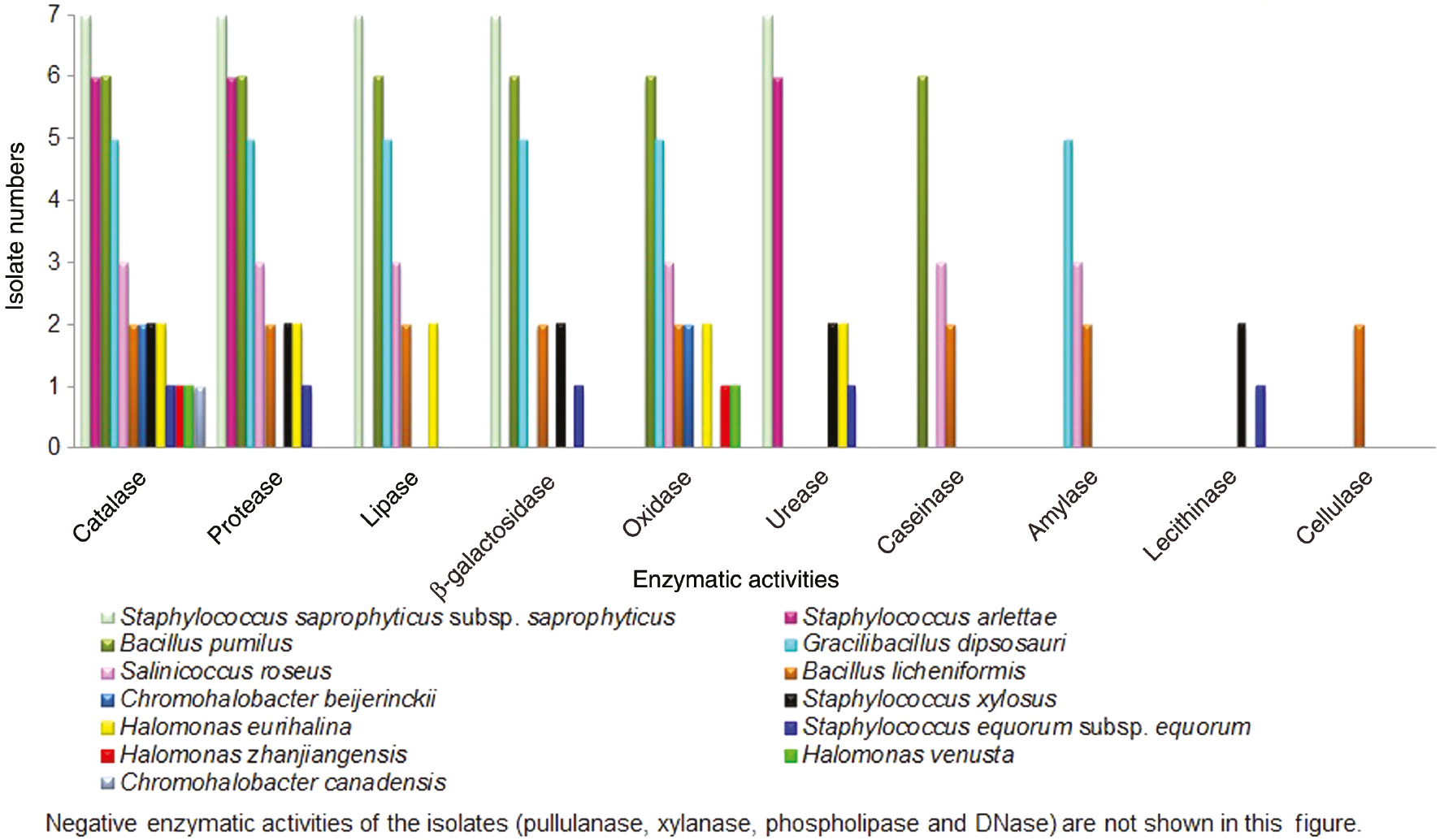
Comparison of enzymatic activities of moderately halophilic bacterial isolates.
Results
The 16S rRNA the pairwise sequence similarities of the isolates were found as 98.9–100% for the isolates belonging to Firmicutes and 99–99.9% for the isolates belonging to Proteobacteria [25]. While four different genera [Staphylococcus (16 isolates), Bacillus (8 isolates), Gracilibacillus (5 isolates) and Salinicoccus (3 isolates)] were determined in Firmicutes, two different genera [Halomonas (4 isolates) and Chromohalobacter (3 isolates)] were found in Proteobacteria (Table 1).
Goat skin samples used in this study were cured with each country’s preservation salt. Different species of moderately halophilic bacteria may be found in these salts. Hence, every country’s goat skin samples may have different moderately halophilic bacterial species. To verify this, we collected salted goat skin samples belonging to eight different countries. Our study demonstrated that the isolate numbers, presence and prevalence of different moderately halophilic bacterial species on the goat skin samples showed differences according to countries. While the goat skins belonging to Africa contained five different species, goat skin samples belonging to Israel and Russia contained only one species. The other skin samples contained a few species (Table 2).
The presence and prevalence of different moderately halophilic bacterial species on the goat skin samples.
| Isolate numbers | Phylogenetically similar to | Country origins and codes of the goat skin samples | Prevalence of different bacterial species on the skins | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Turkey * E * KE *T *YE | Australia * AL * AVT | Africa * KT * K *NE *GK | Bulgaria * B * BLG *BT | China *CCN *CN *CC | Israel * IRL | Russia *RU *RA | France *F *FR *FN *FS | |||
| 7 | S. saprophyticus subsp. saprophyticus | + | − | − | − | − | − | − | + | 2 |
| 6 | S. arlettae | − | − | + | + | + | − | − | − | 3 |
| 6 | B. pumilus | + | − | + | − | + | − | − | − | 3 |
| 5 | G. dipsosauri | − | − | + | − | − | − | + | − | 2 |
| 3 | S. roseus | − | + | − | − | − | − | − | + | 2 |
| 2 | B. licheniformis | − | − | − | − | + | + | − | − | 2 |
| 2 | S. xylosus | − | − | − | + | − | − | − | − | 1 |
| 2 | C. beijerinckii | − | − | − | + | − | − | − | − | 1 |
| 2 | H. eurihalina | − | + | − | − | − | − | − | − | 1 |
| 1 | S. equorum subsp. equorum | − | − | + | − | − | − | − | − | 1 |
| 1 | H. zhanjiangensis | − | − | + | − | − | − | − | − | 1 |
| 1 | H. venusta | + | − | − | − | − | − | − | − | 1 |
| 1 | C. canadensis | − | − | − | + | − | − | − | − | 1 |
| Total numbers of different species for each country | 3 | 2 | 5 | 4 | 3 | 1 | 1 | 2 | ||
While all isolates were catalase positive, more than half of the isolates were oxidase positive. It has been known that catalase and oxidase enzymes are related with aerobic microorganisms. A few bacteria produced indole from tryptophan; fermented glucose and produced acetoin and 2,3 butanediol in Voges-Proskauer test; formed H2S and N2 gases. More than half of the isolates catabolized glucose and produced acidic end products; used nitrate as a terminal electron acceptor and reduced nitrate to nitrite; produced NH3 from peptone broth. Ammonia odor released from the goat skin samples was related to protein catabolism. While 85%, 92%, 56%, 74%, 90%, 74%, 69%, 72%, 62% and 72% of the isolates respectively produced acid from sucrose, D-glucose, D-galactose, fructose, D-trehalose, D-mannose, D-xylose, lactose, maltose and L-arabinose, acid productions from D-melibiose and D-cellobiose were detected as 44% and 28%, respectively. Thirty one percent of the isolates utilized citrate as a sole carbon source for their energy needs (Table 1).
While all isolates produced catalase, 87%, 64%, 59%, 46%, 28%, 26%, 8%, 5% of the isolates produced protease, lipase, β-galactosidase, urease, caseinase, amylase, lecithinase and cellulase, respectively (Table 1).
Some of the isolates exhibited combined enzymatic activities. While 5%, 36% and 31% of isolates produced eight, six and five different enzymes, respectively, 15% of isolates produced three different enzymes. Furthermore, 10% and 3% of the isolates produced two and one enzymes (Figure 1). In the present study, isolates exhibiting most combined activities were belong to the genera Bacillus, Staphylococcus, Gracilibacillus, Salinicoccus and Halomonas. Among the isolates B. licheniformis produced the highest number of enzymes (Figure 1). In order to characterize these enzymes and determine their biochemical properties, more detailed investigation is currently under way.
Discussion
Production of lipase, pullulanase, amylase, protease, xylanase, inulinases, pectinase, cellulases and DNase enzymes by moderately halophilic bacteria (Salicola, Salinicoccus, Marinococcus, Salinivibrio, Virgibacillus, Oceanobacillus, Thalassobacillus, Halovibrio, Halobacillus, Piscibacillus, Halomonas, Gracilibacillus, Bacillus, Chromohalobacter) which were isolated from various saline environments in Spain and Howz Soltan playa (a hypersaline lake) were stated in the previous studies [5], [8]. The enzymatic activities of our test isolates were similar to the enzymatic activities of the same species previously identified [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44].
Catalase enzymes are used in textile industry as a bleaching agent and elimination of hydrogen peroxide in dairy industry [45], [46]. Our all isolates produced this enzyme. Proteases are used in removing hair from hides, leather processing, laundry, detergent production, cheese production, softening meat, improving wool quality [47]. The species of Bacillus licheniformis, Bacillus pumilus, Gracilibacillus dipsosauri, Halomonas eurihalina, Staphylococcus arlettae, Staphylococcus equorum subsp. equorum, Staphylococcus saprophyticus subsp. saprophyticus, Salinicoccus roseus and Staphylococcus xylosus were able to produce protease in this study. Lipases are used in dairy industry for hydrolysis of milk fat, removal of subcutaneous fat in leather industry, biosysnthesis of drugs in pharmaceutical industry [47]. Bacillus licheniformis,Bacillus pumilus, Gracilibacillus dipsosauri, Halomonas eurihalina,Salinicoccus roseus and Staphylococcus saprophyticus subsp. saprophyticus were able to produce lipase (Table 1). Proteases and lipases are notably important because they are mainly used in bating, soaking, degrasing, tanning and final stages of leather product. Hence, salt-tolerant enzymes produced by moderately halophilic bacteria are good candidates for leather industry [48]. The enzyme β-galactosidase, produced by Bacillus licheniformis, Bacillus pumilus, Gracilibacillus dipsosauri, Staphylococcus equorum subsp. equorum, Staphylococcus saprophyticus subsp. saprophyticus and Staphylococcus xylosus, in this study, may be used in the synthesis of galacto-oligosaccharides from lactose [49]. In the present study, moderately halophilic Halomonas eurihalina, Staphylococcus arlettae, Staphylococcus equorum subsp. equorum, Staphylococcus saprophyticus subsp. saprophyticus and Staphylococcus xylosus showed positive urease activity. Microbial ureases are used for wine production to remove urea [50]. Caseinase plays an essential role in degrading casein found in milk [26]. Caseinase activity was seen in Bacillus licheniformis, Bacillus pumilus and Salinicoccus roseus. Amylases play a vital role in starch hydrolysis, food, textile, paper and pulp industry, bread and baking, detergent, pharmaceutical [47], [51]. In the present study, Bacillus licheniformis, Gracilibacillus dipsosauri and Salinicoccus roseus produced amylase. Staphylococcus equorum subsp. equorum and Staphylococcus xylosus in the present study were capable of producing lecithinase enzyme that hydrolysis lecithine. In our study, only moderately halophilic Bacillus licheniformis produced cellulase enzyme which may be used in food, chemical, textile, feed, paper and detergent industries, biomedical science and agriculture [47]. In accordance with data from previous studies that investigated enzymatic studies, members of genera Bacillus, Gracilibacillus, Halomonas, Salinicoccus, Salinivibrio and Staphylococcus were known to secrete extracellular enzymes such as protease, lipase, amylase, urease [5], [8], [51]. None of our moderately halophilic isolates produced pullulanase, xylanase, phospholipase and DNase in the present study (Table 1).
In the present study, moderately halophilic bacteria were especially isolated from goat skin samples belonging to different countries. Presence of moderately halophilic bacteria on all salted goat skins was closely related with the preservation salt used in the curing of goat skin. It has been known that salted goat skin containing fats, proteins, carbohydrates and blood offers an ideal saline environment for growth of moderately halophilic bacteria and production of hydrolytic enzymes. Our biochemical test results proved that all isolates were able to utilize a wide variety of organic compounds and carbon sources. These isolates produced different enzymes such as protease, catalase, lipase, β-galactosidase, urease, caseinase, amylase, lecithinase and cellulase. These enzymes may have a wide range of potential applications in different industries such as baking, beverage, dairy, dry cleaning, feed, food, laundry, meat, paper, pharmaceutical, starch and textile.
Acknowledgments
This work was supported by the Scientific Research Project Commission of Marmara University, Project no. FEN C-DRP-040712-0281 belonging to PhD Study. We thank the Scientific Research Project Commission and the tanneries in Tuzla and Corlu Leather Organized Industrial Zones, Turkey for helping us during collection of skin samples.
Conflict of interest: There is no conflict of interest among authors.
References
1. Delgado-García M, Valdivia-Urdiales B, Aquilar-González CN, Contreras-Esquivel JC, Rodríguez-Herrera R. Halophilic hydrolases as a new tool for the biotechnological industries. J Sci Food Agric 2012;92:2575–80.10.1002/jsfa.5860Suche in Google Scholar PubMed
2. Gupta S, Sharma P, Dev K, Sourirajan A. Halophilic bacteria of Lunsu produce an array of industrially important enzymes with salt tolerant activity. Biochem Res Int 2016;2016:1–10.10.1155/2016/9237418Suche in Google Scholar PubMed PubMed Central
3. BBC Research, Global Markets for Enzymes in Industrial Applications, Wellesley, Massachusetts. Available at: http://www.bccresearch.com/pressroom/bio/global-market-industrial-enzymes-reach-nearly-$7.1-billion-2018, 2014. Accessed: Feb 2017.Suche in Google Scholar
4. Ventosa A, Nieto JJ, Oren A. Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev 1998;62:504–44.10.1128/MMBR.62.2.504-544.1998Suche in Google Scholar PubMed PubMed Central
5. Rohban R, Amoozegar MA, Ventosa A. Screening and isolation of halophilic bacteria producing extracellular hydrolyses from Howz Soltan Lake, Iran. J Ind Microbiol Biotechnol 2009;36:333–40.10.1007/s10295-008-0500-0Suche in Google Scholar PubMed
6. Kumar S, Karan R, Kapoor S, Singh SP, Khare SK. Screening and isolation of halophilic bacteria producing industrially important enzymes. Braz J Microbiol 2012;43:1595–603.10.1590/S1517-83822012000400044Suche in Google Scholar
7. Yin J, Chen JC, Wu Q, Chen GQ. Halophiles, coming stars for industrial biotechnology. Biotechnol Adv 2015;33:1433–42.10.1016/j.biotechadv.2014.10.008Suche in Google Scholar PubMed
8. Sánchez-Porro C, Martín S, Mellado E, Ventosa A. Diversity of moderately halophilic bacteria producing extracellular hydrolytic enzymes. J Appl Microbiol 2003;94:295–300.10.1046/j.1365-2672.2003.01834.xSuche in Google Scholar PubMed
9. Sánchez-Porro C, Yilmaz P, De la Haba RR, Birbir M, Ventosa A. Thalassobacillus pellis sp. nov., a moderately halophilic, Gram-positive bacterium isolated from salted hides. Int J Syst Evol Microbiol 2011;5:1206–10.10.1099/ijs.0.024778-0Suche in Google Scholar PubMed
10. Stuart LS, Frey RW, James LH. Microbiological studies of salt in relation to the reddening salted hides. Technical Bulletin 1933;383:1–24.Suche in Google Scholar
11. Kim SJ, Lee JC, Han SI, Whang KS. Halobacillus salicampi sp nov., a moderately halophilic bacterium isolated from solar saltern sediment. Anton Leeuw Int J G 2016;109:713–20.10.1007/s10482-016-0672-ySuche in Google Scholar PubMed
12. Ventosa A, Oren A, Yanhe M. Halophiles and hypersaline environments: current research and future trends. Berlin, Heidelberg: Springer-Verlag, 2011.10.1007/978-3-642-20198-1Suche in Google Scholar
13. Mehrshad M, Amoozegar MA, Didari M, Bagheri M, Fazeli SA, Schumann P, et al. Bacillus halosaccharovorans sp nov., a moderately halophilic bacterium from a hypersaline lake. Int J Syst Evol Microbiol 2013;63:2776–81.10.1099/ijs.0.046961-0Suche in Google Scholar PubMed
14. Wang YX, Xiao W, Dong MH, Zhao Q, Li ZY, Lai YH, et al. Halomonas qiaohouensis sp nov., isolated from salt mine soil in southwest China. Anton Leeuw Int J G 2014;106:253–60.10.1007/s10482-014-0189-1Suche in Google Scholar
15. Jung WY, Lee SH, Jin HM, Jeon CO. Lentibacillus garicola sp nov., isolated from myeolchi-aekjeot, a Korean fermented anchovy sauce. Anton Leeuw Int J G 2015;107:1569–76.10.1007/s10482-015-0450-2Suche in Google Scholar
16. Bailey DG, Birbir M. A study of the extremely halophilic microorganisms found on commercially brine-cured cattle hides. J Amer Leather Chem Assoc 1993;88:285–93.Suche in Google Scholar
17. Ventosa A, García MT, Kamekura M, Onishi H, Ruíz-Berraquero F. Bacillus halophilus sp. nov., a moderately halophilic Bacillus species. Syst Appl Microbiol 1989;12:162–6.10.1016/S0723-2020(89)80009-8Suche in Google Scholar
18. Mellado E, Nieto JJ, Ventosa A. Phylogenetic interferences and taxonomic consequences of 16S ribosomal DNA sequence comparison of Chromohalobacter marismortui, Volcaniella eurihalina and Deleya halophila and reclassification of V. eurihalina as Halomonas eurihalina comb. nov. Int J Syst Bacteriol 1995;45:712–6.10.1099/00207713-45-4-712Suche in Google Scholar PubMed
19. Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, et al. Introducing EzTaxon: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 2012;62:716–21.10.1099/ijs.0.038075-0Suche in Google Scholar PubMed
20. Arahal DR, Dewhirst FE, Paster BJ, Volcani BE, Ventosa A. Phylogenetic analyses of some extremely halophilic archaea isolated from dead sea water, determined on the basis of their 16S rRNA sequences. Appl Environ Microbiol 1996;62:3779–86.10.1128/aem.62.10.3779-3786.1996Suche in Google Scholar PubMed PubMed Central
21. Bairoch A. The enzyme database in 2000. Nucleic Acids Res 2000;28:304–5.10.1093/nar/28.1.304Suche in Google Scholar PubMed PubMed Central
22. Placzek S, Schomburg I, Chang A, Jeske L, Ulbrich M, Tillack J, et al. BRENDA in 2017: new perspectives and new tools in BRENDA. Nucleic Acids Res 2017;45:380–8.10.1093/nar/gkw952Suche in Google Scholar PubMed PubMed Central
23. Birbir M, Calli B, Mertoglu B, Elevi Bardavid R, Oren A, Ogmen MN, et al. Extremely halophilic archaea from Tuz Lake, Turkey, and the adjacent Kaldirim and Kayacik salterns. World J Microb Biot 2007;23:309–16.10.1007/s11274-006-9223-4Suche in Google Scholar
24. Bese M. Biochemical tests and media used in microbiology. Ankara, Turkey: Ankara University, 1974:298.Suche in Google Scholar
25. Caglayan P. Characterization of moderately halophilic bacteria found on the sheep and goat skins, Ph.D. Thesis, Marmara University, Istanbul, Turkey, 2015.Suche in Google Scholar
26. Johnson TR, Case CL. Laboratory Experiments in microbiology, 9th ed. San Francisco: Pearson Education, Inc., Pearson Benjamin Cummings, 2010.Suche in Google Scholar
27. Schleifer KH, Kloos WE. Isolation and characterization of Staphylococci from human skin, Amended descriptions of Staphylococcus epidermidis and Staphylococcus saprophyticus and descriptions of three new species: Staphylococcus cohnii, Staphylococcus haemolyticus, and Staphylococcus xylosus. Int J Syst Bacteriol 1975;25:50–61.10.1099/00207713-25-1-50Suche in Google Scholar
28. Schleifer KH, Kilpper-Bälz R, Devriese LA. Staphylococcus arlettae sp. nov., S. equorum sp. nov. and S. k1oosii sp. nov.: three new coagulase-negative, novobiocin-resistant species from animals. Syst Appl Microbiol 1985;5:501–9.10.1016/S0723-2020(84)80007-7Suche in Google Scholar
29. Kloos WE, George CG, Olgiate JS, Pelt LV, McKinnon ML, Zimmer BL, et al. Staphylococcus hominis subsp. novobiosepticus subsp. nov., a novel trehalose- and N-acetyl-D-glucosamine-negative, novobiocin- and multiple-antibiotic-resistant subspecies isolated from human blood cultures. Int J Syst Bacteriol 1998;49:799–812.10.1099/00207713-48-3-799Suche in Google Scholar
30. Casaburi A, Villani F, Toldra F, Sanz Y. Protease and esterase activity of staphylococci. Int J Food Microbiol 2006;112:223–9.10.1016/j.ijfoodmicro.2006.04.008Suche in Google Scholar
31. Sakinç T, Kleine B, Gatermann SG. Biochemical characterization of the surface-associated lipase of Staphylococcus saprophyticus. FEMS Microbiol Lett 2007;274:335–41.10.1111/j.1574-6968.2007.00857.xSuche in Google Scholar
32. Kim B-S, Kim C-T, Park BH, Kwon S, Cho Y-J, Kim N, et al. Draft genome sequence of Staphylococcus saprophyticus subsp. saprophyticus M1-1, isolated from the grills of a Korean rockfish, Sebates schlegeli Hilgendorf, after high hydrolytic pressure processing. J Bacteriol 2012;194:4441–2.10.1128/JB.00848-12Suche in Google Scholar
33. Soto-Ramírez N, Sánchez-Porro C, Rosas S, González W, Quiñones M, Ventosa A, et al. Halomonas avicenniae sp. nov., isolated from the salty leaves of the black mangrove Avicennia germinans in Puerto Rico. Int J Syst Evol Microbiol 2007;57:900–5.10.1099/ijs.0.64818-0Suche in Google Scholar
34. Kim KK, Jin L, Yang HC, Lee ST. Halomonas gomseomensis sp. nov., Halomonas janggokensis sp. nov., Halomonas salaria sp. nov. and Halomonas denitrificans sp. nov., moderately halophilic bacteria isolated from saline water. Int J Syst Evol Microbiol 2007;57:675–81.10.1099/ijs.0.64767-0Suche in Google Scholar
35. Chen Y-G, Zhang Y-Q, Huang H-Y, Klenk H-P, Tang S-K, Huang K, et al. Halomonas zhanjiangensis sp. nov., a halophilic bacterium isolated from a sea urchin. Int J Syst Evol Microbiol 2009;59:2888–93.10.1099/ijs.0.010173-0Suche in Google Scholar
36. Ventosa A, Márquez MC, Ruiz-Berraquero F, Kocur M. Salinicoccus roseus gen. nov., sp. nov., a new moderately halophilic Gram-positive coccus. Syst Appl Microbiol 1990;13:29–33.10.1016/S0723-2020(11)80177-3Suche in Google Scholar
37. Kämpfer P, Arun AB, Busse H-J, Young C-C, Lai W-A, Rekha PD, et al. Salinicoccus sesuvii sp. nov., isolated from the rhizosphere of Sesuvium portulacastrum. Int J Syst Evol Microbiol 2011;61:2348–52.10.1099/ijs.0.027524-0Suche in Google Scholar PubMed
38. Beutling DM, Peçonek J, Stan-Lotter H. Chromohalobacter beijerinckii: a psychrophilic, extremely halotolerant and enzymatically active microbe from salted food with the capacity for biogenic amine production. Eur Food Res Technol 2009;229:725–30.10.1007/s00217-009-1106-0Suche in Google Scholar
39. Arahal DR, García MT, Ludwig W, Schleifer KH, Ventosa A. Transfer of Halomonas canadensis and Halomonas israelensis to the genus Chromohalobacter as Chromohalobacter canadensis comb. nov. and Chromohalobacter israelensis comb. nov. Int J Syst Evol Microbiol 2001;51:1443–8.10.1099/00207713-51-4-1443Suche in Google Scholar PubMed
40. Wainø M, Tindall BJ, Schumann P, Ingvorsen K. Gracilibacillus gen. nov., with description of Gracilibacillus halotolerans gen. nov., sp. nov.; transfer of Bacillus dipsosauri to Gracilibacillus dipsosauri comb. nov., and Bacillus salexigens to the Genus Salibacillus gen. nov., as Salibacillus salexigens comb. nov. Int J Syst Bacteriol 1999;49:821–31.10.1099/00207713-49-2-821Suche in Google Scholar PubMed
41. Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, et al. Bergey’s manual of systematic bacteriology the firmicutes. New York: Springer-Verlag, 2009:163.Suche in Google Scholar
42. Shivaji S, Chaturvedi P, Suresh K, Reddy GS, Dutt CB, Wainwright M, et al. Bacillus aerius sp. nov., Bacillus aerophilus sp. nov., Bacillus stratosphericus sp. nov. and Bacillus altitudinis sp. nov., isolated from cryogenic tubes used for collecting air samples from high altitudes. Int J Syst Evol Microbiol 2006;56:1465–73.10.1099/ijs.0.64029-0Suche in Google Scholar PubMed
43. Parvathi A, Krishna K, Jose J, Nair S. Biochemical and molecular characterization of Bacillus pumilus isolated from coastal environment in cochin, China. Braz J Microbiol 2009;40:269–75.10.1590/S1517-83822009000200012Suche in Google Scholar
44. Seo JK, Park TS, Kwon IH, Piao MY, Lee CH, Ha JK. Characterization of cellulolytic and xylanolytic enzymes of Bacillus licheniformis JK7 isolated from the rumen of a native Korean goat. Asian-Aust J Anim Sci 2013;26:50–8.10.5713/ajas.2012.12506Suche in Google Scholar PubMed PubMed Central
45. Jegannathan KR, Nielsen PH. Environmental assessment of enzyme use in industrial production, a literature review. J Clean Prod 2013;42:228–40.10.1016/j.jclepro.2012.11.005Suche in Google Scholar
46. Kiehl K, Straube T, Opwis K, Gutmann J. Strategies for permanent immobilization of enzymes on textile carriers. Eng Life Sci 2015;15:622–6.10.1002/elsc.201400148Suche in Google Scholar
47. Amoozegar MA, Siroosi M. Hydrolytic enzymes in halophilic bacteria, properties and biotechnological potential, Halophiles, Biodiversity and Sustainable Exploitation, vol. 6, Switzerland: Springer International Publishing, 2015:355–78.10.1007/978-3-319-14595-2_13Suche in Google Scholar
48. De Souza FR, Gutterres M. Application of enzymes in leather processing: a comparison between chemical and coenzymatic. Braz J Chem Eng 2012;29:473–81.10.1590/S0104-66322012000300004Suche in Google Scholar
49. Karan R, Capes MD, DasSarma S. Function and biotechnology of extremophilic enzymes in low water activity. Aquat Biosyst 2012;8:1–15.10.1186/2046-9063-8-4Suche in Google Scholar PubMed PubMed Central
50. Liu J, Xu Y, Nie Y, Zhao GA. Optimization production of acid urease by Enterobacter sp. in an approach to reduce urea in Chinese rice wine. Bioprocess Biosyst Eng 2012;35:651–7.10.1007/s00449-011-0643-7Suche in Google Scholar PubMed
51. Souza PM, Magalhães PO. Application of microbial α-amylase in industry-a review. Braz J Microbiol 2010;41:850–61.10.1590/S1517-83822010000400004Suche in Google Scholar PubMed PubMed Central
©2018 Walter de Gruyter GmbH, Berlin/Boston
Artikel in diesem Heft
- Frontmatter
- Research Articles
- Comparative study of stirred and fluidized tank reactor for hydroxyl-kojic acid derivatives synthesis and their biological activities
- Synthesis and antimicrobial activities of some novel thiazole compounds
- Water based PHEMA hydrogels for controlled drug delivery
- Cultural conditions optimization for production of β-galactosidase from Bacillus licheniformis ATCC 12759 under solid-state fermentation
- A novel application of oriental beetle juvenile Anomala corpulenta to reflect Pb contamination in soil
- Purification and characterization of Rhizoctonia solani AG-4 strain ZB-34 α-amylase produced by solid-state fermentation using corn bran
- Grain amino acid composition of barley (Hordeum vulgare L.) cultivars subjected to selenium doses
- Microsatellite-based characterization of cotton genotypes for verticillium wilt and fiber quality traits
- Novel pectinase from Piriformospora indica, optimization of growth parameters and enzyme production in submerged culture condition
- Application of Bdellovibrio bacteriovorus for reducing fouling of membranes used for wastewater treatment
- Large unstained cells are correlated with inflammatory biomarkers in patients with invasive aspergillosis
- Detection of industrially potential enzymes of moderately halophilic bacteria on salted goat skins
- Strain improvement of newly isolated Lactobacillus acidophilus MS1 for enhanced bacteriocin production
- The effects of hydraulic calcium silicate containing endodontic materials on oxidative stress in erythrocytes and liver
Artikel in diesem Heft
- Frontmatter
- Research Articles
- Comparative study of stirred and fluidized tank reactor for hydroxyl-kojic acid derivatives synthesis and their biological activities
- Synthesis and antimicrobial activities of some novel thiazole compounds
- Water based PHEMA hydrogels for controlled drug delivery
- Cultural conditions optimization for production of β-galactosidase from Bacillus licheniformis ATCC 12759 under solid-state fermentation
- A novel application of oriental beetle juvenile Anomala corpulenta to reflect Pb contamination in soil
- Purification and characterization of Rhizoctonia solani AG-4 strain ZB-34 α-amylase produced by solid-state fermentation using corn bran
- Grain amino acid composition of barley (Hordeum vulgare L.) cultivars subjected to selenium doses
- Microsatellite-based characterization of cotton genotypes for verticillium wilt and fiber quality traits
- Novel pectinase from Piriformospora indica, optimization of growth parameters and enzyme production in submerged culture condition
- Application of Bdellovibrio bacteriovorus for reducing fouling of membranes used for wastewater treatment
- Large unstained cells are correlated with inflammatory biomarkers in patients with invasive aspergillosis
- Detection of industrially potential enzymes of moderately halophilic bacteria on salted goat skins
- Strain improvement of newly isolated Lactobacillus acidophilus MS1 for enhanced bacteriocin production
- The effects of hydraulic calcium silicate containing endodontic materials on oxidative stress in erythrocytes and liver

