Abstract
Millions of people worldwide are chronically exposed to environmental arsenic through drinking water, increasing their risk of various adverse cardiometabolic outcomes. To understand the inter-individual variation in arsenic susceptibility, this systematic review explores all epidemiological evidence on interactions between single nucleotide polymorphisms (SNPs) and arsenic exposure in relation to cardiometabolic health. Five electronic databases were searched until April 2023. From 42,202 retrieved publications, 18 candidate gene-environment (cGxE) studies were included, and no genome-wide association studies were found. Of 676 SNPs in 148 genes tested, 40 SNPs in 24 genes, 4 haplotypes and combined SNPs in MCP-1/APOE, were reported to statistically significantly interact with arsenic exposure. These genes were involved in arsenic metabolism, oxidative stress or defence, DNA damage repair, endothelial (dys) function, inflammation or immune function, tumour suppressor activity, or were previously implicated in cardiometabolic disease pathways. Most studies did not explore the same SNPs (or strong proxies), and none of the identified SNP-arsenic interactions were replicated for the same arsenic species and cardiometabolic outcome. Whilst some SNPs are suggestive of influencing susceptibility to arsenic for various cardiometabolic outcomes, further research is needed to understand the interplay between arsenic and genetic variants, identify at-risk populations, and improve risk assessment.
Introduction
Environmental exposure to arsenic, one of the most abundant elements in the earth’s crust, is increasingly recognised to elevate risk of adverse cardiovascular disease, hypertension and diabetes, with evidence being most conclusive at moderate to high levels of arsenic exposure [1], [2], [3]. Inter-individual variation in susceptibility to adverse arsenic-related health effects is influenced by the arsenic species (i.e., inorganic or organic, valency), concentration, and metabolism efficiency. The latter is shaped by biological factors (e.g., age, sex), lifestyle factors (e.g., nutritional status, physical activity) and potentially various genetic polymorphisms [4].
Several genetic studies – including genome-wide association [GWA] [5], linkage [6], and candidate gene studies [7] - have reported significant effects of variants near the arsenic methyltransferase (AS3MT) gene on arsenic metabolism and subsequent non-cardiometabolic disease outcomes, such as skin lesions and cancers [8]. Further candidate gene, linkage and exome-wide association studies suggest that additional genetic variants affecting arsenic metabolism and arsenic-related health outcomes likely exist, including genes critical for carbon-folate metabolism (e.g., forminotransferase cyclodeaminase [FTCD]) [9], genes involved in the classic arsenic methylation pathway (e.g., glutathione S-transferase omega [GSTO] [10], purine nucleoside phosphorylase [PNP] [11]), and genes involved in DNA damage repair (e.g., MPO [myeloperoxidase] [12], and NBS1 [nibrin]) [13].
However, whilst one previous narrative review explored arsenic exposure, arsenic metabolism, and their interaction with genetic (and epigenetic) factors in relation to cancer outcomes [4], thus far, no study has systematically summarised the totality of evidence for the influence of genetic variations on arsenic-related cardiometabolic outcome susceptibility. Therefore, this systematic review aims to i) summarise all available epidemiological evidence exploring whether associations between environmental arsenic exposure and risk of adverse cardiometabolic outcomes change in the presence of common genetic variations, and ii) support the current understanding of the biomolecular mechanisms that may underpin arsenic-related cardiometabolic outcomes, based on the genetic variants proposed to be involved.
Methods
The study protocol was prospectively registered with PROSPERO (CRD42019146144). Findings are reported following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) (Supplementary Table 1) and Joanna Briggs Institute (JBI) guidelines [14], 15].
Search strategy and definitions
Five electronic databases were systematically searched using English search terms: PubMed, Web of Science, Scopus, EMBASE, and TOXLINE via TOXnet. Databases were searched from inception until 24 July 2019, with an updated search on 10 April 2023. Note, TOXLINE was excluded in the updated search as the database was retired in December 2019. Keywords used included indexed MeSH terms as well as free text terms related to ‘genetic polymorphism’, ‘genetic’, ‘arsenic’ and ‘arsenicals’ and were informed by previous publications [1], 16], and a medical information specialist (full search strategy in Supplementary Table 2). No restriction on the language or date was applied. Bibliographies of relevant publications (e.g., reviews, grey literature) and all included studies in the full-text screening were cross-referenced (forward and backward) to identify any additional eligible studies. The GWAS Catalog (https://www.ebi.ac.uk/gwas/) and Google Scholar were searched for additional studies fulfilling the inclusion criteria.
The exposure of interest was single nucleotide polymorphisms (SNPs): a single nucleotide change in the DNA sequence, comprising one of the most abundant types of genetic variation. No specific molecular processes, genes or SNPs were selected as a focus a priori, enabling the synthesis of all available evidence on common genetic variation(s) that may interact with arsenic exposure to impact arsenic-related cardiometabolic outcomes. Other variations in the DNA sequence, including other genetic variants (e.g., indels, microsatellites, repetitive elements), structural variation, and epigenetic variation were excluded. Haplotypes of linked SNPs within a chromosomal region were included.
This study included arsenic-related cardiometabolic diseases such as cardiovascular diseases (CVD, including coronary heart diseases [CHD], cerebrovascular diseases [CeD], peripheral arterial diseases [PAD], deep vein thrombosis [DVT]), diabetes mellitus (including type 1 [T1DM], type 2 [T2DM] and gestational [GDM]), hypertension, obesity, and metabolic syndrome. Subclinical cardiometabolic diseases (carotid atherosclerosis, arterial stiffness, ECG abnormality), and diagnostic markers (blood pressure [systolic, diastolic, pulse], blood glucose levels, carotid intima-media thickness [CIMT], HbA1c% levels) as continuous outcomes were included. Predictive biomarkers (e.g., asymmetric dimethylarginine (ADMA), fatty acid-binding protein 4 (FABP4) for CVDs) were excluded. Standard definitions following commonly used guidelines for the different cardiometabolic outcomes were applied (e.g., ACC/AHA [17] and ESC/ESH [18] guidelines for hypertension, or WHO [19], ADA [20], and NICE [21] guidelines for T2DM). Other diseases, such as skin lesions or cancer, were excluded, as were studies that only assessed the impact of SNPs on arsenic metabolism.
Study selection
Studies that met the inclusion criteria were: i) peer-reviewed quantitative epidemiological studies, ii) studies exploring whether one or more SNPs affects susceptibility to adverse cardiometabolic outcomes related to arsenic (i.e., whether a genetic variant is associated with higher or lower cardiometabolic risk when a person is exposed to arsenic), and iii) studies including arsenic species to which the general population is commonly environmentally exposed (i.e., inorganic arsenic [iAs] exposure or its metabolites [MMA, DMA], see Supplementary Table 3). Studies were excluded if they were studies that i) did not include a quantitative measure of arsenic exposure, ii) focused on forms of arsenic exposure uncommon in the general population such as medical therapy (e.g., melarsoprol treatment for parasitic infections, arsenic trioxide for leukemia), or intentional poisoning (homicide, suicide), ii) were non-human (i.e., in vitro, animal studies), iii) had a secondary study design (e.g., reviews, meta-analyses), or that reported on data previously reported elsewhere, iv) were conference proceedings, case reports or case series, v) lacked an available full-text (after request of full-text from the authors), or vi) had overlapping study populations with another included study within this review (with exception of those studying a different cardiometabolic outcome and partially overlapping study populations). Studies on arsenic-exposed populations that did not include SNP-related arsenic susceptibility or SNP-arsenic interaction analyses were also excluded.
After the duplicates were removed in Endnote, abstracts and titles were blindly double-screened using Rayyan (https://rayyan.ai/). In this stage, all studies on SNPs in relation to arsenic-related disease or arsenic metabolism were included. Ambiguity in the inclusion and exclusion criteria was checked by conflict piloting after the first 500 records were screened, resulting in an inter-rater reliability kappa score of 98.6 %. In the second stage, full-texts were retrieved and double-screened, focusing only on arsenic-related adverse cardiometabolic outcomes as previously defined. Any disagreements between two reviewers were discussed until consensus was reached. To minimise bias, non-English records were included and translated/reviewed by a speaker of the language (i.e., Bengali, Dutch, French, Spanish, German, Serbian). However, few non-English studies were retrieved for full-text screening.
Data extraction
A data extraction tool was developed and pilot-tested on five randomly selected studies, and refined accordingly. Data were extracted by KvD, and verified by MJ. Data extracted included: first author, year of publication, study design, participant characteristics (country, age, sex, ethnicity and/or genetic ancestry), number of participants, years of follow-up (cohort studies), gene(s) and SNP(s) assessed (including their allele/genotype/haplotype frequencies), genotyping method, type of arsenic exposure (e.g., water, urinary or blood arsenic), method of arsenic exposure assessment, adverse cardiometabolic outcome (e.g., CVD, metabolic syndrome), outcome ascertainment method, measures of the combined effects (or interaction) of the SNPs and arsenic exposure on negative cardiometabolic outcomes (e.g., odds ratio [OR] estimates for interaction terms, joint ORs of the combined SNP-arsenic effect, relative excess risk due to the interaction [RERI], attributable proportion due to the interaction [AP], synergy index [SI], p-values for interaction [Pint]), variables adjusted for in the main statistical analyses, and an open field to record additional information. When both p-values and Q-values (p-values adjusted for multiple comparisons) were reported, both were extracted, and results were discussed based on Q-values. When both unadjusted and adjusted measures of effect were reported, both were extracted, but the models that adjusted for the greatest number of confounders were discussed in the information synthesis. This approach was taken even when models adjusted for different variables across studies considering the adjusted effect estimates are more likely to be representative of the “true” effect than crude models.
To create an overview of all genes and SNPs assessed within the studies included review, an overview table was generated listing the corresponding gene, rsID, alleles, and consequence of the variation for each individual SNP (i.e., missense variant, synonymous variant, intron variant, noncoding transcript variant, 5′ UTR variant, 3′ UTR variant, 2KB upstream variant, 500B downstream variant) based on information in the included studies, the dbSNP database (https://www.ncbi.nlm.nih.gov/snp/) and the integrative database GeneCards (https://www.genecards.org/).
Individual study quality assessment
To allow for the incorporation of genetic and environmental components in individual study quality assessment, methodological quality was assessed using study design adapted versions of the quality assessment tool developed by Moon et al. 2017 for studies on environmental arsenic [1], 22] – and the Q-genie tool for the quality assessment of genetic association studies [23]. The tool was pilot-tested on five randomly selected studies, and refined accordingly. No overall score (e.g., good, fair, poor) was given as this can oversimplify important differences in bias, confounding, and overall quality of the different studies. Almost none of the studies explored the same SNP and cardiometabolic outcomes (Table 2, 3), therefore no overall assessment of the certainty of the body of evidence using Grading of Recommendations, Assessments, Development and Evaluation (GRADE) was performed.
Information synthesis
Due to the heterogeneity of included studies [i.e., genes/SNPs assessed, outcome assessed (e.g., CVD, DM, hypertension), and arsenic species assessed], data were descriptively synthesised, and no meta-analyses were performed. Studies were grouped and synthesised by the outcome of interest, and, where possible, further discussed by biochemical processes relevant genes are involved in. Due to the limited number of (heterogenous) studies included, publication bias could not be assessed.
Results
42,202 publications were retrieved from the databases (Figure 1). After removing duplicates, 18,922 studies were screened by abstract and title, and 261 in full text. A total of 18 studies met the inclusion criteria, with summary characteristics reported in Table 1, individual study results in Table 2, and summary results in Table 3. Studies excluded in full-text screening and reason for exclusion are listed in Supplementary Table 4. Most studies were conducted in Taiwan (n=7) [24], [25], [26], [27], [28], [29], [30] and Bangladesh (n=4) [31], [32], [33], [34], with others conducted in China (n=3) [35], [36], [37], Spain (n=2) [38], 39], Mexico [40], and the United States [41]. Included studies had a cross-sectional (n=7) [24], 33], [36], [37], [38], [39], [40], case-control (n=5) [26], [27], [28], [30], 35], cohort (n=4) [25], 29], 31], 41], or case-cohort (n=1) [34], design and included data from, among others, the HEALS [31], 33], 34], SHFS [41], Hortega [38], 39] and VALCAR [39] studies.

Study selection process using the PRISMA diagram.
Study characteristics of studies exploring the association of single nucleotide polymorphisms (SNPs) with arsenic-induced cardio-metabolic disease.
| Study | N | Women% (n) | Age | Location | Study design | Outcome | Outcome assessment | Arsenic assessment | Gene(s) | Genotyping | Study aim |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chen et al. 2012a | 240 P: 36 C: 204 |
60 % (145) |
56 ± 16.3 yr | 12 villages in Nantou County, Taiwan |
Cross-sectional Resident area ≥1 yr TP: NR |
Hypertension | Resting BP (20-min) using sphygmomanometer SBP ≥130 DBP ≥85, HTN history treated with anti-HTN drugs |
Urinary (spot morning) arsenic [As] (GF-AAS) |
SOD2, OGG1 | PCR-RFLP | Investigate whether individuals with different SOD2 and OGG1genotypes have increased arsenic related hypertension risk. |
| Chen et al. 2012b | 247 P: 111 C: 136 |
57 % (141) |
No MetS 60.4 ± 9.0 yr MetS 64.3 ± 7.9 yr |
Putai village, Taiwan |
Cohort TP: 1990–2002/3 |
Metabolic syndrome | ≥3 or more: FPG ≥110 mg/dL Trig ≥150 mg/dL HDL ≤40 mg/dL M ≤50 mg/dL W SBP ≥130 DBP ≥85 WG ≥90 cm M ≥80 cm W |
Urinary (spot) arsenic [PMI, SMI, MMA%, DMA%] (HPLC-ICP-MS) |
AS3MT, GSTO1, GSTO2 (Note, results of publication only focus on GSTO1) |
PCR-RFLP TaqMan SNP genotyping assay |
Evaluate the association among arsenic methylation, genetic polymorphisms and MetS occurrence. |
| Drobná et al. 2012 | 255 P: NR C: NR |
67 % (171) |
34 ± 18 yr | Zimapan and Lagunera regions, Mexico |
Cross-sectional Resident area ≥2 yr TP: NR |
Diabetes mellites type 2 | OGTT 2h-PBG ≥200 mg/dl, FBG ≥126 mg/dl, physician diagnosis, diabetes treatment |
Water arsenic [iAs] (HG-AAS) |
AS3MT | TaqMan SNP genotyping assay | Assess the interaction between AS3MT polymorphism and iAs exposure on diabetes development. |
| Fan et al. 2022 | 938 P: 319 C: 619 |
66 % (620) |
487 <60 yr 451 ≥60 yr |
Lv Liang City, Shanxi Province, China | Case-control TP: 2018 |
Diabetes mellites type 2 | HbA1c ≥6.5 % | Urinary (spot) arsenic [tAs] (LC-AFS) |
KEAP1 | Multiplex PCR | Explore whether T2DM is related to the low-to-moderate level arsenic exposure and KEAP1 rs11545829 SNP. |
| Farzan et al. 2016 | 1,137 P: 432 C: 705 |
49 % (559) |
Normal 37.4 ± 10.4 (Pre) HTN 43.1 ± 10.7 |
Araihazar, Bangladesh |
Cohort (HEALS) Resident area ≥5 yrs TP: 2002/4–2007/9 |
Blood pressure (SBP, DBP, pulse pressure) | Resting BP (5-min) using automated sphygmomanometer |
Water arsenic [tAs] (HR-ICP-MS) |
APOE, AS3MT, CBS, CYBA, GSTM1, GSTO1, GSTP1, GSTT1, HMOX1, ICAM1, IL6, MTHFR, NOS3, PNP, S1PR1, SOD2, TNF, VCAM1 | Illumina GoldenGate asay | Examine genetic susceptibility loci that may contribute to arsenic related increases in blood pressure. |
| Grau-Perez et al. 2017 | 395 | 19 % (77) |
≤ 4 μg/L blood As 25.8 ± 6.6 yr > 4 μg/L 26.4 ± 5.8 yr |
Zhongshan, Guangdong, China | Cross-sectional No change in living ≥1 yr TP: 2013/4 |
(Elevated) blood pressure (SBP, DBP) | Resting BP (no further details provided) | Blood arsenic [As] (ICP-MS) Urinary (spot) arsenic (HG-AFS) |
CCM3 | imLDR method | Investigate interaction effects between blood arsenic and CCM3 polymorphism on blood pressure. |
| Grau-Perez et al. 2017 | 1,838 P: 255 C: 1,583 |
60 % 1,122 |
36 (24–47) yr | Arizona, Oklahoma, North Dakota, South Dakota, United States |
Cohort (SHFS) TP: 1998/99–2006/9 |
Diabetes mellitus type 2 Insulin resistance |
FBG ≥126 mg/dL physician diagnosis, diabetes treatment IFG (100–126 mg/dl) and NFG <100 mg/dl), HOMA2-IR |
Urinary (morning spot) arsenic [tAs] (ICP-MS) [iAs, MMA, DMA] (HPLC-ICP-MS) |
AS3MT | Illumina Cardio-Metabo DNA Analysis BeadChip | Study potential effect modification of the arsenic related diabetes association by AS3MT polymorphisms. |
| Grau-Perez et al. 2018 | 1,451 P: 1,331 C: 120 |
51 % (740) | Overall 49.7 ± 0.2 yr No DM 48.5 ± 0.2 yr DM 67.1 ± 1.4 yr |
Valladolid, Spain | Cross-sectional (Hortega study) TP: 2001/3 |
Diabetes mellitus type 2 | FBG ≥126 mg/dL HbA1c ≥6.5 % physician diagnosis, diabetes treatment |
Urinary (spot) arsenic [tAs] (AEC-ICP-MS) |
ABCA1, ACE, ADIPOQ, ADRB2, AGT, AGTR1, AGTRAP, APOA5, ATXN2L, BCS1L, CAT, CDKAL1, CETP, COX5A, COX5B, COX6B1, COX6B2, COX7A1, CRP, CYBA, CYP11B1, EDN1, EDN2, EDN3, EDNRA, EDNRB, ETV5, FABP2, FADS1, FADS2, FAIM2, FTO, GCLC, GCLM, GDF9, GNPDA2, GPX4, GPX6, GSR, GSS, IGF2BP2, IL10, IL10RA, IL10RB, IL18, IL18R1, IL18RAP, IL1RL1, IL6, IL6R, IL6ST, IL8, IL8RA, IL8RB, INSIG2, KCTD15, LAMA1, LIPC, LPL, MAF, MC4R, MPO, MSRA, MSRB2, MTCH2, NCF2, NCF4, NDUFS1, NDUFS2, NDUFS3, NDUFS4, NDUFS6, NDUFS7, NDUFS8, NEGR1, NOS1, NOS2A, NOX3, NOX5, NR3C2, OGG1, PON1, PPARA, PPARG, PPARGC1A, PRL, RAC2, REN, SCNN1A, SDHB, SDHD, SH2B1, SLC30A8, SOD1, SOD2, SOD3, TCF7L2, TNFA, TNFRSF1A, TNFRSF1B, TXN, TXN2, TXNRD1, TXNRD2, UCP1, UCP2, UCP3, UQCRB, WWOX, XDH | Oligo-ligation-assay SNPlex | Evaluate the interaction of arsenic and diabetes-related candidate polymorphisms as a potential determinant of T2DM. |
| Hsieh et al. 2007 | 479 P: 235 C: 244 |
53 % (255) |
96: ≤55 yr 201: 55–65 yr 182: ≥65 yr |
Lanyang Basin, Ilan County, Taiwan | Nested case-control TP: 1991/4–1997/8 |
Carotid atherosclerosis | Intima media thickness >1 mm plus one of: plaque score≥1, stenosis of the ECCA >50 % | Water arsenic [As, CAE] (HG-AAS) |
MCP-1, APOE | PCR-RFLP | Investigate the joint effects of arsenic exposure through drinking water and genetic polymorphism of APOE and MCP-1 on carotid atherosclerosis risk. |
| Hsieh et al. 2011 | 863 P: 384 C: 479 |
53 % (455) |
Reference 59.2 ± 0.4 yr Cases 65.1 ± 0.4 yr |
Lotung, Ilan County, Taiwan | Nested case-control TP: 1991/4–1997/8 |
Carotid atherosclerosis | Intima media thickness >1 mm plus one of: plaque score ≥1, stenosis of the ECCA >50 % | Water arsenic [As] (HG-AAS) |
AS3MT, GSTO1, GSTO2, PNP, PNP/GSTO haplotype(s) | PCR-RFLP | Identify association between risk genotypes and development of carotid atherosclerosis in arsenic-exposed population. |
| Hsueh et al. 2005 | 292 P: 79 C: 213 |
61 % (177) |
No HTN 46.2 ± 0.7 yr HTN 54.2 ± 0.8 yr |
Homei, Fuhsing, & Hsinming, Taiwan | Nested case-control Resident area >6 months TP: 1989 - NR |
Hypertension | Health examination (not further details); or antihypertensive drug use SBP ≥140 DBP≥ 90 |
Water arsenic [CAE] (previous reports) [42] |
CYBA, SOD2, NOS3, CAT | PCR-RFLP | Assess the association of 4 genetic polymorphisms with arsenic-related hypertension. |
| Liang et al. 2023 | 385 P: 86 C: 299 |
100 % (385) |
Overall 29 (26–33) yr Non-GDM 29 (26–32) yr GDM 31 (28–34) yr |
Beichen district, Tianjin, China | Cross-sectional Resident area ≥6 yr TP: 2017/18 |
Gestational diabetes mellitus | 75 g OGTT at 24–28 weeks FBG ≥5.1 mmol/L or 1hPG ≥10 mmol/L or 2hPG ≥8.5 mmol/L |
Urinary (spot) arsenic [iAs%, MMA%, DMA%] (HPLC-ICP-MS) | AS3MT, N6AMT1 | Hi-SNP genotyping (3-round multiplex PCR with NGS) | Explore the individual and combined effects of N6AMT1 and AS3MT SNPs with arsenic metabolism on GDM. |
| Liao et al. 2009 | 121 P: 79 C: 42 |
61 % (74) |
ECG normal 62 ± 7.3 yr ECG abnormal 64.9 ± 8.7 yr |
Homei, Fuhsin and Hsinming, Taiwan | Cohort Live ≥5 days a week in area TP: 1993 -2002 |
ECG abnormality | 12-Lead ECG (MI or ischemia, conduction defect, arrhythmias, atrial enlargement or ventricular hypertrophy, prolonged ventricular repolarization) | Water arsenic [CAE] (previous reports) |
AS3MT, GSTO1, GSTO2, PON1, PON2 | TaqMan SNP Genotyping Assay; PCR-RFLP | Investigate possible contribution of genetic factors, and long-term arsenic exposure to CVD development. |
| Martíz-Barquero et al. 2015 | 1,970 P: 468 C: 1,502 |
51 % (1,006) |
VALCAR 46.4 ± 14.9 yr Hortega 54.4 ± 19.3 yr |
Valencia & Valladolid, Spain | Cross-sectional (VALCAR Hortega study) TP: 2001/3 |
Obesity, BMI |
BMI value was ≥30 kg/m2 | Blood plasma arsenic [As] (ICP-MS) |
EDN1, EDN2, EDN3, EDNRA, EDNRB | Oligo-ligation-assay SNPlex | Assess whether arsenic modulates the effect of EDNRB genetic polymorphisms in obesity development. |
| Pan et al. 2013 | 919 P: 83 C: 836 |
39 % (361) |
No DM 33.5 ± 11.7 yr DM 39.8 ± 10.8 yr |
Pabna district, Bangladesh | Follow-up of a skin-lesion case control TP: 2001/3–2009/11 |
Diabetes mellitus type 2 % HbA1c levels |
HbA1c ≥6.5 % | Water arsenic [As] (ICP-MS) |
ADAMTS9, BCL11A, CDC123, CDKN2A, CDKN2B, CENTD2, IDE, KCNQ1, KMGA2, LGR5, NOTCH2, PRC1, SLC30A8, TCF2, THADA, TSPAN8, WFS1 | Sequenome MassARRAY iPLEX | Evaluate interaction between SNPs in diabetes genes and water arsenic exposure T2DM risk. |
| Wang et al. 2007 | 605 P: 279 C: 326 |
52 % (316) |
110≤55 yr 258: 55–65 yr 237: ≥65 yr |
Lanyang Basin, Ilan County, Taiwan | Case-control TP: 1991/4–1997/98 |
Carotid atherosclerosis | Intima media thickness >1 mm, plaque score ≥1, stenosis of the ECCA >50 % | Water arsenic (HG-AAS) |
GSTM1, GSTT1, GSTP1, P53 | PCR-RFLP | Explore the joint effects water arsenic exposure and genetic polymorphisms on risk of carotid atherosclerosis. |
| Wu et al. 2014 | 1,078 | 60.9 % | 38 (27–52) yr | Araihazar, Bangladesh |
Cross-sectional (HEALS) Resident area ≥5 yr TP: 2000/2 – 2012 |
Carotid intima-media thickness | Carotid sonography using the SonoSite MicroMaxx ultrasound machine | Water arsenic [tAs] (HPLC-ICP-MS) Urinary (spot) arsenic [tAs] (GF-AAS) |
GSTM1, GSTT1, GSTO1,
GSTP1, MTHFR, CBS, PNPAS3MT, HMOX1, NOS3, SOD2, CYBA, APOE, TNF, IL6, ICAM1, S1PR1, VCAM1 |
Illumina GoldenGate asay | Evaluate whether the association between arsenic exposure and cIMT differs by 207 SNPs. |
| Wu et al. 2015 | 2,225 CVD: 447 CHD: 238 Str: 165 C: 1,375 |
53 % (1,181) |
Sub-cohort 38.6 ± 9.7 yr CVD 47.4 ± 9.7 yr CHD 45.4 ± 9.2 yr Stroke 50.8 ± 8.9 yr |
Araihazar, Bangladesh |
Case-cohort (HEALS) Resident area≥5 yr TP: 2000/2 - 2012 |
CVD, CHD, Stroke (mortality) | Validated verbal autopsy | Water arsenic [tAs] (ICP-MS) Urinary (spot) arsenic [tAs] (GFAAS) [As3+, As5+, MMA, DMA] (HPLC-ICP-MS) |
APOE, AS3MT, CBS, CYBA, GSTM1, GSTO1, GSTP1, GSTT1, HMOX1, ICAM1, IL6, MTHFR, NOS3, PNP, S1PR1, SOD2, TNF, VCAM1 | Illumina GoldenGate asay | Investigate whether associations between arsenic exposure and CVD risk are modified by genetic polymorphism in 18 genes. |
-
AAS, atomic absorption spectroscopy; BP, blood pressure; CHD, coronary heart disease; CVD, cardiovascular disease; DBP, diastolic blood pressure; MetS, metabolic syndrome; DM, diabetes mellitus; ECCA, extracranial carotid artery; ECG, electrocardiogram; FBG, fasting blood glucose; FPG, fasting plasma glucose; GDM, gestational diabetes mellitus; GFAAS, graphite furnace atomic absorption; HDL, high density lipoprotein; HEALS, health effects of arsenic longitudinal study; HG-AAS, hydride generation atomic absorption spectroscopy; HOMA2-IR, homeostasis model assessment insulin resistance; HPLC-ICP-MS, high-performance liquid chromatography ICP-MS; HR-ICP-MS, high-performace ICP-MS; HTN, hypertension; ICP-MS, inductively coupled plasma mass spectrometry; IFG, impaired fasting glycaemia; MetS, metabolic syndrome; MI, myocardial infarction; NR, not clearly reported; OGTT, oral glucose tolerance test; PCR-RFLP, polymerase chain reaction restriction fragment length polymorphism; SBP, systolic blood pressure; SHFS, strong heart family study; SNP, single nucleotide polymorphism; T2DM, Type 2 diabetes mellitus; TP, time period; Trig, triglycerides; WG, waist girth. Note, abbreviations of relevant genes can be found in Supplementary Table 4.
Summary results of studies exploring the association of single nucleotide polymorphisms (SNPs) with arsenic-related cardiometabolic outcomes (i.e., studies assessing environmental arsenic-SNP interaction with cardiometabolic outcomes).
| Study | Gene | Genotype | Arsenic | MoA 95 % CI | P (Q) | Ca/Co | Comments | |
|---|---|---|---|---|---|---|---|---|
| Chen et al. 2012a Hypertension, (HTN) Urinary arsenic (μg/g creatinine) |
SOD2 (rs4880) |
Val-Val | ≤ 8 μg/g | OR | 1.0 (ref.) | 51/69 | Both SOD2 and OGG1 genotypes may be prone to increased arsenic-related hypertension risk. Adjusted for age, gender, urinary Pb levels, renal disease history and SOD2 genotype (only OGG1). |
|
| Val-Val | > 8 μg/g | OR | 3.1 (1.2–7.9) | <0.05 | 15/11 | |||
| Val-Ala / Ala-Ala | ≤ 8 μg/g | OR | 1.2 (0.6–2.4) | – | 26/34 | |||
| Val-Ala / Ala-Ala | > 8 μg/g | OR | 4.2 (1.7–10.3) | <0.01 | 22/12 | |||
| OGG1 (rs1052133) |
Ser-Cys/Ser-Ser | ≤ 8 μg/g | OR | 1.0 (ref.) | 48/63 | |||
| Ser-Cys/Ser-Ser | > 8 μg/g | OR | 3.2 (1.4–7.4) | <0.01 | 25/17 | |||
| Cys-Cys | ≤ 8 μg/g | OR | 1.0 (0.5–2.0) | – | 29/40 | |||
| Cys-Cys | > 8 μg/g | OR | 3.4 (1.1–10.7) | <0.05 | 12/6 | |||
| Chen et al. 2012b Metabolic syndrome (MetS) Urinary arsenic metabolites (PMI, SMI, MMA%, DMA%) |
GSTO1 (rs4925) |
AA | PMI high | OR | 1.0 (ref.) | 24/48 | Findings suggest that decreasing MMA% and PMI and increasing DMA% and SMI are associated with increased MetS risk, which may be marginally modified by the GSTO1 genotype. Adjusted by age and betel nut chewing. Study did not report on GSTO2 or AS3MT polymorphisms results. High and low PMI, SMI, MMA% and DMA% were not clearly defined. |
|
| AD + DD | PMI high | OR | 1.32 (0.61–2.90) | 0.48 | 20/30 | |||
| AA | PMI low | OR | 2.03 (1.03–4.01) | 0.04 | 42/44 | |||
| AD + DD | PMI low | OR | 4.00 (1.65–9.71) | 0.002 | 24/13 | |||
| AA | SMI low | OR | 1.0 (ref.) | – | 20/57 | |||
| AD + DD | SMI low | OR | 2.91 (1.29–6.57) | 0.01 | 22/23 | |||
| AA | SMI high | OR | 4.65 (2.22–9.73) | <0.001 | 46/35 | |||
| AD + DD | SMI high | OR | 3.87 (1.62–9.26) | 0.002 | 22/20 | |||
| AA | MMA% high | OR | 1.0 (ref.) | 22/55 | ||||
| AD + DD | MMA% high | OR | 2.35 (1.07–5.15) | 0.03 | 23/24 | |||
| AA | MMA% low | OR | 3.36 (1.66–6.79) | <0.001 | 44/37 | |||
| AD + DD | MMA% low | OR | 3.24 (1.37–7.66) | 0.007 | 21/19 | |||
| AA | DMA% low | OR | 1.0 (ref.) | 23/57 | ||||
| AD + DD | DMA% low | OR | 1.86 (0.83–4.18) | 0.13 | 17/22 | |||
| AA | DMA% high | OR | 3.44 (1.69–6.99) | <0.001 | 43/35 | |||
| AD + DD | DMA% high | OR | 3.54 (1.59–7.87) | 0.002 | 27/21 | |||
| Drobná et al. 2012 Diabetes mellites type 2 (T2DM) Water arsenic, ppb |
AS3MT (rs11191439) |
TT | <52 ppb | OR | 1.0 (ref.) | – | Carriers of rs11191439 (greater than additive) and rs17881215 (approximately multiplicative) genotypes may be more likely to develop arsenic-related T2DM. Little indication of consistent association with any of the other 4 (rs3740393, rs3740390, rs10748835, VNTR variants) genotypes and T2DM. Adjusted for age, sex, hypertension, and obesity. 95 % CI not clearly reported for all categories, nor number of cases and controls in each group, or the P value. Cursive 95 % CI estimated based on Figure 1.3 |
|
| TC + CC | <52 ppb | OR | 3.47 (0.8–17.0) | – | – | |||
| TT | ≥52 ppb | OR | 5.68 (1.95–17.0) | – | – | |||
| TC + CC | ≥52 ppb | OR | 11.4 (2.2–58.8) | – | – | |||
| AS3MT (rs17881215) |
GG | <52 ppb | OR | 1.0 (ref.) | – | |||
| GC + CC | <52 ppb | OR | 1.97 (0.23–12.5) | – | – | |||
| GG | ≥52 ppb | OR | 4.60 (1.6–15.0) | – | – | |||
| GC + CC | ≥52 ppb | OR | 8.8 (1.6–47.3) | – | – | |||
| Fan et al. 2022 Diabetes mellites type 2 (T2DM) Urinary arsenic (μg/g creatinine) |
KEAP1 (rs11545829) |
GG + GA | μg/g (cont) | OR | 10.12 (2.89–35.41) | <0.05 | 300/548 | There was no statistically significant multiplicative nor addictive interaction between rs11545829 and urinary arsenic on T2DM risk. Adjusted for cigarette smoking, alcohol drinking, gender, age, hypertension, and BMI. |
| AA | μg/g (cont) | OR | 10.67 (0.20–564.83) | >0.05 | 19/71 | |||
| RERI | −4.06 (−17.52–1.61) | Pint 0.73 | ||||||
| AP | −0.74 (−3.10–0.14) | |||||||
| Farzan et al. 2016 Systolic blood pressure (SBP) Water arsenic, μg/l |
APOE (rs429358) |
Dominant model | μg/Ll (cont) | βint | 0.26 (−0.08–0.59) | 0.14 (0.78) | – | From the 235 SNPS, 44 SNPs showed an interaction with water arsenic for one or more blood pressure outcomes. The CYBA (rs3794624) polymorphism was the only one statistically significant after FDR adjustment. Adjusted for sex, age at enrolment, BMI, smoking status, educational attainment, and diabetes at baseline. Coefficients are in relation to each SNP, time and well arsenic concentration in the different genetic models. P-values are for interaction. Three models were run for dominant, recessive, and additive – additive models were not shown as they were similar to dominant models. Number of study participants for each model were not reported. |
| Recessive model | βint | 2.05 (0.57–3.54) | 0.01 (0.34) | – | ||||
| CBS (rs234709) |
Dominant model | βint | 0.34 (0.06–0.62) | 0.02 (0.54) | – | |||
| Recessive model | βint | 0.49 (−1.00–1.97) | 0.52 (0.94) | – | ||||
| CBS (rs2014564) |
Dominant model | βint | 0.28 (0.01–0.56) | 0.04 (0.67) | – | |||
| Recessive model | βint | 0.17 (−0.12–0.46) | 0.25 (0.86) | – | ||||
| CBS (rs2124459) |
Dominant model | βint | 0.40 (0.12–0.68) | 0.01 (0.34) | – | |||
| Recessive model | βint | 0.41 (−1.12–1.95) | 0.60 (0.96) | – | ||||
| CBS (rs234701) |
Dominant model | βint | 0.29 (0.02–0.57) | 0.04 (0.67) | – | |||
| Recessive model | βint | 0.12 (−0.18–0.41) | 0.44 (0.93) | – | ||||
| CYBA (rs3794624) |
Dominant model | βint | 0.02 (−0.29–0.33) | 0.90 (0.99) | – | |||
| Recessive model | βint | 2.43 (0.78–4.08) | 0.004 (0.34) | – | ||||
| GSTM1 (rs4147567) |
Dominant model | βint | 0.60 (0.17–1.03) | 0.01 (0.34) | – | |||
| GSTP1 (rs749174) |
Dominant model | βint | 0.29 (0.02–0.52) | 0.04 (0.67) | – | |||
| Recessive model | βint | 0.43 (−1.11–1.97) | 0.58 (0.96) | – | ||||
| GSTT1 (rs4630) |
Dominant model | βint | 0.10 (−0.33–0.52) | 0.66 (0.98) | – | |||
| Recessive model | βint | 1.07 (0.11–2.03) | 0.03 (0.67) | – | ||||
| IL6 (rs1524107) |
Dominant model | βint | 0.34 (0.08–0.60) | 0.01 (0.47) | – | |||
| Recessive model | βint | 0.10 (−0.81–1.01) | 0.83 (0.98) | – | ||||
| NOS3 (rs743506) |
Dominant model | βint | −0.10 (−0.36–0.16) | 0.45 (0.93) | – | |||
| Recessive model | βint | 0.72 (0.04–1.40) | 0.04 (0.67) | – | ||||
| S1PR1 (rs3753194) |
Dominant model | βint | −0.04 (−0.29–0.22) | 0.78 (0.98) | – | |||
| Recessive model | βint | 0.45 (0.02–0.88) | 0.04 (0.67) | – | ||||
| SOD2 (rs5746123) |
Dominant model | βint | 0.33 (0.02–0.64) | 0.04 (0.67) | – | |||
| Recessive model | βint | 0.18 (−0.75–1.12) | 0.70 (0.98) | – | ||||
| Diastolic blood pressure, (DBP) Water arsenic, μg/l |
APOE (rs429358) |
Dominant model | βint | 0.06 (−0.15–0.27) | 0.56 (0.99) | – | ||
| Recessive model | βint | 0.95 (0.03–1.88) | 0.04 (0.82) | – | ||||
| AS3MT (rs9527) |
Dominant model | βint | 0.22 (0.02–0.42) | 0.03 (0.74) | – | |||
| Recessive model | βint | −1.30 (−2.65–0.04) | 0.06 (0.88) | – | ||||
| AS3MT (rs4290163) |
Dominant model | βint | 0.26 (0.03–0.49) | 0.03 (0.74) | – | |||
| Recessive model | βint | −0.60 (−1.63–0.42) | 0.25 (0.88) | – | ||||
| GSTM1 (rs4147567) |
Dominant model | βint | 0.36 (0.09–0.63) | 0.01 (0.74) | – | |||
| GSTP1 (rs6591256) |
Dominant model | βint | 0.24 (0.08–0.40) | 0.003 (0.70) | – | |||
| Recessive model | βint | 0.20 (−0.06–0.46) | 0.14 (0.88) | – | ||||
| GSTP1 (rs1695) |
Dominant model | βint | 0.30 (0.04–0.57) | 0.02 (0.74) | – | |||
| HMOX1 (rs5755718) |
Dominant model | βint | 0.26 (0.04–0.48) | 0.02 (0.74) | – | |||
| Recessive model | βint | 0.16 (−0.11–0.43) | 0.24 (0.88) | – | ||||
| HMOX1 (rs8139532) |
Dominant model | βint | 0.29 (0.07–0.52) | 0.01 (0.74) | – | |||
| HMOX1 (rs16995662) |
Dominant model | βint | 0.25 (0.03–0.48) | 0.03 (0.74) | – | |||
| HMOX1 (rs2269534) |
Dominant model | βint | 0.25 (0.03–0.48) | 0.03 (0.74) | – | |||
| IL6 (rs2069840) |
Dominant model | βint | −0.11 (−0.29–0.07) | 0.24 (0.88) | – | |||
| Recessive model | βint | 0.59 (0.07–1.10) | 0.02 (0.74) | – | ||||
| IL6 (rs13306435) |
Dominant model | βint | 0.19 (0.01–0.36) | 0.04 (0.82) | – | |||
| Recessive model | βint | −0.37 (−1.12–0.37) | 0.33 (0.92) | – | ||||
| IL6 (rs1554606) |
Dominant model | βint | 0.28 (0.04–0.52) | 0.02 (0.74) | – | |||
| Recessive model | βint | −0.60 (−1.63–0.42) | 0.25 (0.88) | – | ||||
| IL6 (rs1474347) |
Dominant model | βint | 0.19 (0.03–0.34) | 0.02 (0.74) | – | |||
| Recessive model | βint | 0.02 (−0.46–0.49) | 0.94 (0.99) | – | ||||
| NOS3 (rs3918198) |
Dominant model | βint | 0.25 (0.02–0.47) | 0.03 (0.74) | – | |||
| NOS3 (rs1799983) |
Dominant model | βint | 0.30 (0.07–0.52) | 0.01 (0.74) | – | |||
| PNP (rs17882836) |
Dominant model | βint | 0.04 (−0.13–0.21) | 0.65 (0.99) | – | |||
| Recessive model | βint | 0.21 (0.02–0.40) | 0.03 (0.74) | – | ||||
| TNF (rs4248159) |
Dominant model | βint | 0.21 (0.02–0.40) | 0.03 (0.74) | – | |||
| Pulse pressure, PP Water arsenic, μg/L |
APOE (rs429358) |
Dominant model | βint | 0.22 (−0.01–0.44) | 0.06 (0.62) | – | ||
| Recessive model | βint | 1.08 (0.11–2.06) | 0.03 (0.52) | – | ||||
| AS3MT (rs4290163) |
Dominant model | βint | 0.34 (0.10–0.58) | 0.01 (0.45) | – | |||
| Recessive model | βint | −0.30 (−1.39–0.78) | 0.58 (0.90) | – | ||||
| AS3MT (rs10509761) |
Dominant model | βint | 0.11 (−0.05–0.28) | 0.18 (0.79) | – | |||
| Recessive model | βint | 0.32 (0.06–0.58) | 0.02 (0.49) | – | ||||
| CBS (rs6586281) |
Dominant model | βint | 0.23 (0.05–0.40) | 0.01 (0.46) | – | |||
| Recessive model | βint | −0.10 (−0.31–0.12) | 0.38 (0.88) | – | ||||
| CBS (rs234701) |
Dominant model | βint | 0.21 (0.03–0.39) | 0.02 (0.49) | – | |||
| Recessive model | βint | 0.06 (−0.13–0.25) | 0.54 (0.90) | – | ||||
| CBS (rs706208) |
Dominant model | βint | 0.19 (0.01–0.38) | 0.04 (0.57) | – | |||
| Recessive model | βint | −0.06 (−0.27–0.15) | 0.59 (0.91) | – | ||||
| CBS (rs3788050) |
Dominant model | βint | 0.12 (−0.10–0.34) | 0.27 (0.85) | – | |||
| Recessive model | βint | 0.69 (0.19–1.18) | 0.007 (0.45) | – | ||||
| CBS (rs234709) |
Dominant model | βint | 0.23 (0.05–0.41) | 0.01 (0.47) | – | |||
| Recessive model | βint | 0.32 (−0.69–1.33) | 0.53 (0.90) | – | ||||
| CBS (rs2124459) |
Dominant model | βint | 0.26 (0.07–0.44) | 0.01 (0.45) | – | |||
| Recessive model | βint | 0.17 (−0.88–1.21) | 0.76 (0.95) | – | ||||
| CBS (rs2849727) |
Dominant model | βint | 0.20 (0.02–0.37) | 0.03 (0.53) | – | |||
| Recessive model | βint | 0.53 (−0.47–1.53) | 0.30 (0.86) | – | ||||
| CBS (rs9983620) |
Dominant model | βint | 0.01 (−0.16–0.17) | 0.95 (0.98) | – | |||
| Recessive model | βint | 0.39 (0.01–0.76) | 0.04 (0.57) | – | ||||
| CYBA (rs3794624) |
Dominant model | βint | 0.01 (−0.19–0.21) | 0.90 (0.98) | – | |||
| Recessive model | βint | 2.10 (1.01–3.20) | <0.001 (0.05) | – | ||||
| CYBA (rs13306296) |
Dominant model | βint | 0.03 (−0.21–0.15) | 0.74 (0.94) | – | |||
| Recessive model | βint | 1.02 (0.22–1.81) | 0.01 (0.47) | – | ||||
| GSTM1 (rs4147567) |
Dominant model | βint | 0.30 (0.01–0.58) | 0.04 (0.57) | – | |||
| GSTP1 (rs749174) |
Dominant model | βint | 0.20 (0.02–0.38) | 0.03 (0.49) | – | |||
| Recessive model | βint | 0.49 (−0.54–1.52) | 0.35 (0.88) | – | ||||
| IL6 (rs2069845) |
Dominant model | βint | 0.18 (0.004–0.35) | 0.04 (0.57) | – | |||
| Recessive model | βint | −0.05 (−0.89–0.78) | 0.90 (0.98) | – | ||||
| IL6 (rs1524107) |
Dominant model | βint | 0.26 (0.08–0.43) | 0.004 (0.45) | – | |||
| Recessive model | βint | −0.02 (−0.63–0.58) | 0.94 (0.98) | – | ||||
| IL6 (rs1554606) |
Dominant model | βint | 0.29 (0.04–0.54) | 0.02 (0.49) | – | |||
| Recessive model | βint | −0.32 (−1.40–0.76) | 0.56 (0.90) | – | ||||
| MTHFR (rs9651118) |
Dominant model | βint | −0.04 (−0.21–0.14) | 0.69 (0.93) | – | |||
| Recessive model | βint | 0.48 (0.03–0.94) | 0.04 (0.55) | – | ||||
| NOS3 (rs743506) |
Dominant model | βint | −0.03 (−0.20–0.14) | 0.72 (0.94) | – | |||
| Recessive model | βint | 0.49 (0.04–0.94) | 0.03 (0.53) | – | ||||
| NOS3 (rs1808593) |
Dominant model | βint | 0.22 (0.03–0.40) | 0.02 (0.49) | – | |||
| PNP (rs17882836) |
Dominant model | βint | 0.10 (−0.09–0.29) | 0.30 (0.86) | – | |||
| Recessive model | βint | 0.28 (0.08–0.48) | 0.01 (0.45) | – | ||||
| PNP (rs1617940) |
Dominant model | βint | −0.05 (−0.22–0.12) | 0.58 (0.90) | – | |||
| Recessive model | βint | 0.47 (0.01–0.92) | 0.04 (0.57) | – | ||||
| SOD2 (rs10370) |
Dominant model | βint | 0.04 (−0.13–0.22) | 0.61 (0.91) | – | |||
| Recessive model | βint | 0.32 (0.06–0.58) | 0.02 (0.47) | – | ||||
| SOD2 (rs12526686) |
Dominant model | βint | 0.23 (0.05–0.40) | 0.01 (0.46) | – | |||
| Recessive model | βint | −0.16 (−0.38–0.07) | 0.17 (0.78) | – | ||||
| TNF (rs1800630) |
Dominant model | βint | 0.26 (0.08–0.43) | 0.01 (0.45) | – | |||
| Recessive model | βint | −0.08 (−0.34–0.17) | 0.52 (0.90) | – | ||||
| TNF (rs3179060) |
Dominant model | βint | 0.20 (0.02–0.38) | 0.03 (0.53) | – | |||
| VCAM1 (rs3176877) |
Dominant model | βint | 0.22 (0.04–0.39) | 0.02 (0.47) | – | |||
| Recessive model | βint | −0.13 (−0.34–0.09) | 0.26 (0.85) | – | ||||
| VCAM1 (rs3783617) |
Dominant model | βint | −0.02 (−0.20–0.15) | 0.78 (0.95) | – | |||
| Recessive model | βint | 0.48 (0.03–0.94) | 0.04 (0.55) | – | ||||
| VCAM1 (rs3917016) |
Dominant model | βint | 0.24 (0.03–0.46) | 0.02 (0.49) | – | |||
| Recessive model | βint | −0.20 (−0.95–0.56) | 0.60 (0.91) | – | ||||
| Gao et al. 2018 Systolic blood pressure, SBP Blood arsenic, μg/l |
CCM3 (rs9818496) |
CC | 0–4 μg/l | OR | 1.0 (ref.) | – | Interactions between the rs9818496, rs3804610 and rs6784267 CCM3 genotypic variants and blood arsenic increased the hazard of increased SBP. Adjusted for age, gender, BMI, smoking status, alcohol use status, education level, BG, TG, TC, HDL, LDL, and CRP. Number of study participants in each category not reported. |
|
| CC | >4 μg/L | OR | 1.62 (0.53–2.82) | – | – | |||
| CT + TT | 0–4 μg/l | OR | 1.35 (0.75–2.57) | – | – | |||
| CT + TT | >4 μg/L | OR | 2.68 (1.35–5.55) | – | – | |||
| Interaction (multiplicative) | ORint | 1.50 (1.15–1.95) | 0.003 | |||||
| CCM3 (rs3804610) |
TT | 0–4 μg/l | OR | 1.0 (ref.) | – | |||
| TT | >4 μg/L | OR | 1.62 (0.53–2.82) | – | – | |||
| CT + CC | 0–4 μg/l | OR | 1.35 (0.75–2.57) | – | – | |||
| CT + CC | >4 μg/L | OR | 2.68 (1.35–5.55) | – | – | |||
| Interaction (multiplicative) | ORint | 1.50 (1.15–1.95) | 0.003 | |||||
| CCM3 (rs6784267) |
CC | 0–4 μg/l | OR | 1.0 (ref.) | – | |||
| CC | >4 μg/L | OR | 1.46 (0.87–2.41) | – | – | |||
| CT + TT | 0–4 μg/l | OR | 1.29 (0.71–2.23) | – | – | |||
| CT + TT | >4 μg/L | OR | 1.82 (1.09–2.44) | – | – | |||
| Interaction (multiplicative) | ORint | 1.31 (1.08–1.59) | 0.006 | |||||
| Diastolic blood pressure, DBP Blood arsenic, μg/l |
CCM3 (rs9818496) |
CC | 0–4 μg/l | OR | 1.0 (ref.) | – | ||
| CC | >4 μg/L | OR | 1.32 (0.72–2.95) | – | – | |||
| CT + TT | 0–4 μg/l | OR | 1.27 (0.42–3.84) | – | – | |||
| CT + TT | >4 μg/L | OR | 1.57 (0.93–2.88) | – | – | |||
| Interaction (multiplicative) | ORint | 1.31 (0.87–1.90) | 0.067 | |||||
| CCM3 (rs3804610) |
TT | 0–4 μg/l | OR | 1.0 (ref.) | – | |||
| TT | >4 μg/L | OR | 1.32 (0.72–2.95) | – | – | |||
| CT + CC | 0–4 μg/l | OR | 1.27 (0.42–3.84) | – | – | |||
| CT + CC | >4 μg/L | OR | 1.57 (0.93–2.88) | – | – | |||
| Interaction (multiplicative) | ORint | 1.31 (0.87–1.90) | 0.067 | |||||
| CCM3 (rs6784267) |
CC | 0–4 μg/l | OR | 1.0 (ref.) | – | |||
| CC | >4 μg/L | OR | 1.40 (0.86–2.59) | – | – | |||
| CT + TT | 0–4 μg/l | OR | 1.33 (0.91–2.40) | – | – | |||
| CT + TT | >4 μg/L | OR | 1.54 (0.96–2.98) | – | – | |||
| Interaction (multiplicative) | ORint | 1.28 (1.06–1.55) | 0.052 | |||||
| Grau-Perez et al. 2017 Diabetes mellites type 2 (T2DM) Urinary arsenic (μg/g creatinine) & metabolism markers (iAs%, MMA%, DMA%) HOMA2-IR Urinary arsenic metabolism markers |
AS3MT (rs12768205) | GG | As 75th v 25th | HR | 1.14 (0.86–1.51) | Pint 0.79 | 121/809 | Results found no interaction between genetic polymorphisms, iAs exposure, arsenic metabolism with diabetes risk. Adjusted for gender, age, education, BMI, smoking status, WC, GFR, estimated vitamin B2, vitamin B6 and folate, AS3MT genotype and FG levels at baseline. GMRs were estimated per 5 % increase in arsenic metabolism markers. The association between iAs% and DMA% with HOMA2-IR differed by the AS3MT genetic variant. Adjusted for sex, age, education, BMI, smoking status, WC, GFR, ΣAs concentration, estimated vitamin B2, vitamin B6, and folate, AS3MTgenotype and FG levels at baseline. |
| GA | As 75th v 25th | HR | 1.14 (0.86–1.49) | 111/639 | ||||
| AA | As 75th v 25th | HR | 1.39 (0.79–2.44) | 20/138 | ||||
| AS3MT (rs12768205) | GG | iAs% v MMA% | GMR | 0.98 (0.76–1.26) | Pint 0.82 | 120/776 | ||
| GA | iAs% v MMA% | GMR | 1.02 (0.81–1.28) | 108/616 | ||||
| AA | iAs% v MMA% | GMR | 0.88 (0.49–1.57) | 20/134 | ||||
| AS3MT (rs12768205) | GG | MMA% v DMA% | GMR | 1.05 (0.87–1.27) | Pint 0.77 | 120/776 | ||
| GA | MMA% v DMA% | GMR | 1.04 (0.82–1.31) | 108/616 | ||||
| AA | MMA% v DMA% | GMR | 0.84 (0.47–1.51) | 20/134 | ||||
| AS3MT (rs12768205) | GG | DMA% v iAs% | GMR | 0.97 (0.84–1.12) | Pint 0.76 | 120/776 | ||
| GA | DMA% v iAs% | GMR | 0.95 (0.84–1.09) | 108/616 | ||||
| AA | DMA% v iAs% | GMR | 1.08 (0.77–1.54) | 20/134 | ||||
| AS3MT (rs12768205) | GG | iAs% v MMA% | GMR | 1.12 (1.04–1.20) | P int 0.03 | 775 | ||
| GA | iAs% v MMA% | GMR | 1.04 (0.98–1.12) | 616 | ||||
| AA | iAs% v MMA% | GMR | 1.04 (0.92–1.16) | 134 | ||||
| AS3MT (rs12768205) | GG | MMA% v DMA% | GMR | 0.93 (0.89–0.99) | Pint 0.15 | 775 | ||
| GA | MMA% v DMA% | GMR | 0.90 (0.84–0.97) | 616 | ||||
| AA | MMA% v DMA% | GMR | 0.83 (0.73–0.92) | 134 | ||||
| AS3MT (rs12768205) | GG | DMA% v iAs% | GMR | 0.98 (0.95–1.02) | P int 0.03 | 775 | ||
| GA | DMA% v iAs% | GMR | 1.02 (0.99–1.06) | 616 | ||||
| AA | DMA% v iAs% | GMR | 1.06 (0.98–1.14) | 134 | ||||
| Grau-Perez et al. 2018 Diabetes mellites type 2 (T2DM) Urinary arsenic (μg/g creatinine) |
IL8RA (rs1008563) Additive |
TT | 80th v 20th 80th v 20th 80th v 20th |
OR | 2.26 (2.18–2.34) | P int 0.004 | 40/329 | The study indicates suggestive evidence for a differential association of arsenic related T2DM and genetic polymorphisms in IL8RA, TXN, NR3C2, COX5A and GCLC. Tests for interaction were obtained from linear regression models with arsenic as a continuous variable. Only includes genes with top interaction P-values. Note, no polymorphism was significant after correcting for multiple testing (Bonferroni significance level 0.002) Adjusted for age, sex, education, urinary cotinine levels, smoking status, alcohol consumption, fish consumption, residence place and arsenobetaine. |
| TC | OR | 3.38 (3.29–3.46) | 46/608 | |||||
| CC | OR | 5.04 (4.69–5.42) | 27/313 | |||||
| IL8RA (rs1008562) Additive |
CC | 80th v 20th 80th v 20th 80th v 20th |
OR | 2.48 (2.40–2.56) | P int 0.01 | 49/451 | ||
| CG | OR | 3.48 (3.39–3.58) | 44/602 | |||||
| GG | OR | 4.89 (4.48–5.33) | 20/207 | |||||
| TXN (rs4135168) Dominant |
AA | 80th v 20th 80th v 20th |
OR | 4.19 (4.01–4.38) | P int 0.004 | 71/732 | ||
| AG + GG | OR | 2.48 (2.38–2.58) | 47/566 | |||||
| NR3C2 (rs13117325) Additive |
GG | 80th v 20th 80th v 20th 80th v 20th |
OR | 3.87 (3.73–4.02) | P int 0.007 | 48/572 | ||
| GA | OR | 2.82 (2.76–2.88) | 46/554 | |||||
| AA | OR | 2.06 (1.93–2.19) | 18/138 | |||||
| NR3C2 (rs2137335) Dominant |
TT | 80th v 20th 80th v 20th |
OR | 2.70 (2.63–2.76) | P int 0.01 | 82/779 | ||
| TC + CC | OR | 4.65 (4.30–5.03) | 33/492 | |||||
| GCLC (rs11415624) Additive |
DD | 80th v 20th 80th v 20th 80th v 20th |
OR | 3.80 (3.67–3.93) | P int 0.01 | 58/585 | ||
| DA | OR | 2.73 (2.67–2.79) | 54/574 | |||||
| AA | OR | 1.96 (1.84–2.09) | 8/164 | |||||
| COX5A (rs1133322) Recessive |
TC + TT | 80th v 20th 80th v 20th |
OR | 3.44 (3.36–3.53) | P int 0.01 | 96/978 | ||
| CC | OR | 2.03 (1.90–2.17) | 21/323 | |||||
| Hsieh et al. 2007 Carotid atherosclerosis Water arsenic, μg/L Cumulative water arsenic, mg/l/yr |
MCP-1 (rs1024611) APOE (rs429358, rs7412) |
0 | ≤10 μg/L | OR | 1.0 (ref.) | 1/5 | Those with two risk genotypes of APOE (AG or GG) and MCP-1 (ε4 allele) and high arsenic exposure have strikingly highest risk for the development carotid atherosclerosis, showing significant joint effect of arsenic exposure and risk genotypes of APOE and MCP-1. 0 = non-risk polymorphisms of the 2 genes, 1=at least one risk polymorphism, 2=two risk polymorphisms. Adjusted for age and gender. |
|
| 1 | ≤ 10 μg/L | OR | 2.7 (0.3–28.1) | – | 13/24 | |||
| 2 | ≤ 10 μg/L | OR | 5.1 (0.3–89.5) | – | 3/2 | |||
| 0 | > 10 μg/L | OR | 4.2 (0.4–41.5) | – | 37/53 | |||
| 1 | > 10 μg/L | OR | 5.8 (0.6–54.7) | – | 140/135 | |||
| 2 | > 10 μg/L | OR | 10.3 (1.0–102.5) | <0.05 | 41/25 | |||
| MCP-1 (rs1024611) APOE (rs429358, rs7412) |
0 | ≤ 0.2 mg/L/yr | OR | 1.0 (ref.) | 1/7 | |||
| 1 | ≤ 0.2 mg/L/yr | OR | 6.4 (0.7–59.0) | 0.05–0.1 | 22/25 | |||
| 2 | ≤ 0.2 mg/L/yr | OR | 10.1 (0.6–173.1) | – | 3/2 | |||
| 0 | > 0.2 mg/L/yr | OR | 7.0 (0.8–62.1) | 0.05–0.1 | 37/50 | |||
| 1 | > 0.2 mg/L/yr | OR | 8.6 (1.0–73.4) | <0.05 | 131/134 | |||
| 2 | > 0.2 mg/L/yr | OR | 15.7 (1.7–141.2) | <0.05 | 41/25 | |||
| Hsieh et al. 2011 Carotid atherosclerosis Water arsenic, μg/l |
PNP (rs1049564) (rs1130650) |
ht1GC/ht3AC | ≤50 μg/L | OR | 1.0 (ref.) | – | PNP, AS3MT, and GSTO polymorphisms may exacerbate the formation of atherosclerosis in individuals with high levels of arsenic concentration in well water (>50 μg/L). The polymorphism of the two PNP gene loci are listed in this order: rs1049564, rs1130650. The polymorphisms of the three GSTO gene loci are listed in this order: rs4925, rs2297235 and rs156697. Adjusted for age, gender, cigarette smoking, alcohol consumption, HTN, cholesterol, FG, and BMI. Number of study participants in each category not reported. |
|
| ht2AT | ≤50 μg/L | OR | 0.85 (0.31–2.35) | 0.76 (0.76) | – | |||
| ht1GC/ht3AC | >50 μg/L | OR | 1.38 (0.99–1.91) | 0.06 (0.09) | – | |||
| ht2AT | >50 μg/L | OR | 2.08 (1.32–3.30) | 0.002 (0.006) | – | |||
| AS3MT (rs11191439) | TT | ≤50 μg/L | OR | 1.0 (ref.) | – | |||
| TC | ≤50 μg/L | OR | 1.49 (0.16–14.17) | 0.73 (0.073) | – | |||
| TT | >50 μg/L | OR | 1.57 (0.99–2.49) | 0.05 (0.075) | – | |||
| TC | >50 μg/L | OR | 2.63 (1.00–6.92) | 0.05 (0.075) | – | |||
| GSTO (rs4925) GSTO2 (rs2297235, rs156697) |
ht1CAA/ht3CAG | ≤50 μg/L | OR | 1.0 (ref.) | – | |||
| ht2AGG | ≤50 μg/L | OR | 1.01 (0.40–2.56) | 0.98 (0.98) | – | |||
| ht1CAA/ht3CAG | >50 μg/L | OR | 1.54 (1.11–2.14) | 0.01 (0.03) | – | |||
| ht2AGG | >50 μg/L | OR | 1.62 (1.05–2.50) | 0.03 (0.045) | – | |||
| Hsueh et al. 2005 Hypertension (HTN) Water (cumulative) arsenic (mg/l-yr) |
CYBA (rs4673) |
CC | <10.5 mg/l-yr | OR | 1.0 (ref.) | 7/73 | Results suggest that the CYBA, SOD2, and NOS3 polymorphism may play a role in arsenic related hypertension, but no CAT polymorphisms. Adjusted for age, gender, triglycerides and LDL-cholesterol |
|
| CC | ≥10.5 mg/l-yr | OR | 3.8 (1.5–9.6) | <0.05 | 47/76 | |||
| CT + TT | <10.5 mg/l-yr | OR | 3.0 (0.5–19.2) | >0.05 | 2/8 | |||
| CT + TT | ≥10.5 mg/l-yr | OR | 3.7 (0.9–15.6) | >0.05 | 7/7 | |||
| SOD2 (rs4880) |
TT | <10.5 mg/l-yr | OR | 1.0 (ref.) | 3/57 | |||
| TT | ≥10.5 mg/l-yr | OR | 5.7 (1.6–20.9) | <0.05 | 33/56 | |||
| TC + CC | <10.5 mg/l-yr | OR | 4.5 (1.0–21.4) | >0.05 | 6/24 | |||
| TC + CC | ≥10.5 mg/l-yr | OR | 9.0 (2.3–35.0) | <0.05 | 21/25 | |||
| CAT (rs1001179) |
CC | <10.5 mg/l-yr | OR | 1.0 (ref.) | 8/73 | |||
| CC | ≥10.5 mg/l-yr | OR | 3.5 (1.4–8.6) | <0.05 | 51/75 | |||
| CT | <10.5 mg/l-yr | OR | 1.5 (0.2–14.9) | >0.05 | 1/8 | |||
| CT | ≥10.5 mg/l-yr | OR | 2.4 (0.5–11.6) | >0.05 | 3/8 | |||
| NOS3 (rs1799983) |
GG | <10.5 mg/l-yr | OR | 1.0 (ref.) | 8/65 | |||
| GG | ≥10.5 mg/l-yr | OR | 2.7 (1.1–6.7) | <0.05 | 44/69 | |||
| GT + TT | <10.5 mg/l-yr | OR | 0.4 (0.1–4.1) | >0.05 | 1/14 | |||
| GT + TT | ≥10.5 mg/l-yr | OR | 3.7 (1.2–11.7) | <0.05 | 10/13 | |||
| Liang et al. 2023 Gestational diabetes mellitus, GDM Urinary arsenic metabolites (iAs%, MMA%, DMA%) |
N6AMT1 (rs1997605) |
GA + AA | >19.37 iAs% | OR | 1.0 (ref.) | – | Results suggest that polymorphisms in N6AMT1 (rs1997605, rs1003671) could contribute to the differential arsenic susceptibility of individuals to GDM. The additive interactions between N6AMT1 rs1997605 GG genotypes and lower iAs% or higher DMA% were statistically significant. Additive interactions were also found between N6AMT1 rs1003671 GG genotypes and lower iAs% or higher DMA%. No significant combined effects were observed between AS3MT SNPs and urinary As metabolism indicators on GDM risk. Adjusted for age, ethnicity, education, pre-pregnancy body mass index, serum folate, B12, homocysteine (Hcy), urinary total arsenic, urinary arsenobetaine (AsB), and AS3MT. |
|
| GA + AA | ≤19.37 iAs% | OR | 1.04 (0.52–2.08) | 0.917 | – | |||
| GG | >19.37 iAs% | OR | 1.58 (0.72–3.43) | 0.253 | – | |||
| GG | ≤19.37 iAs% | OR | 3.23 (1.42–7.39) | 0.005 | – | |||
| RERI | 1.62 (−0.81–4.05) | – | – | |||||
| AP | 0.50 (0.01–0.99) | – | – | |||||
| SI | 3.64 (0.34–39.51) | – | – | |||||
| N6AMT1 (rs1997605) |
GA + AA | >1.11 MMA% | OR | 1.0 (ref.) | – | |||
| GA + AA | ≤1.11 MMA% | OR | 1.07 (0.54–2.13) | 0.839 | – | |||
| GG | >1.11 MMA% | OR | 1.66 (0.77–3.57) | 0.193 | – | |||
| GG | ≤1.11 MMA% | OR | 3.30 (1.43–7.61) | 0.005 | – | |||
| RERI | 1.56 (−0.95–4.07) | – | – | |||||
| AP | 0.47 (−0.03–0.98) | – | – | |||||
| SI | 3.13 (0.40–24.71) | – | – | |||||
| N6AMT1 (rs1997605) |
GA + AA | ≤79.62 DMA% | OR | 1.0 (ref.) | – | |||
| GA + AA | >79.62 DMA% | OR | 1.04 (0.52–2.08) | 0.917 | – | |||
| GG | ≤79.62 DMA% | OR | 1.56 (0.71–3.39) | 0.265 | – | |||
| GG | >79.62 DMA% | OR | 3.30 (1.45–7.51) | 0.004 | – | |||
| RERI | 1.71 (−0.76–4.17) | – | – | |||||
| AP | 0.52 (0.04–0.99) | – | – | |||||
| SI | 3.87 (0.34–44.42) | – | – | |||||
| N6AMT1 (rs1003671) |
GA + AA | >19.37 iAs% | OR | 1.0 (ref.) | – | |||
| GA + AA | ≤19.37 iAs% | OR | 1.14 (0.61–2.13) | 0.672 | – | |||
| GG | >19.37 iAs% | OR | 1.51 (0.42–5.47) | 0.532 | – | |||
| GG | ≤19.37 iAs% | OR | 5.40 (1.50–19.46) | 0.010 | – | |||
| RERI | 3.75 (−3.11–10.61) | – | – | |||||
| AP | 0.70 (0.19–1.20) | – | – | |||||
| SI | 6.76 (0.21–213.06) | – | – | |||||
| N6AMT1 (rs1003671) |
GA + AA | >1.11 MMA% | OR | 1.0 (ref.) | – | |||
| GA + AA | ≤1.11 MMA% | OR | 1.28 (0.70–2.34) | 0.421 | – | |||
| GG | >1.11 MMA% | OR | 3.01 (0.89–10.24) | 0.077 | – | |||
| GG | ≤1.11 MMA% | OR | 2.96 (0.82–10.63) | 0.097 | – | |||
| RERI | −0.34 (−5.37–4.70) | – | – | |||||
| AP | −0.11 (−1.91–1.69) | – | – | |||||
| SI | 0.85 (0.08–9.27) | – | – | |||||
| N6AMT1 (rs1003671) |
GA + AA | ≤79.62 DMA% | OR | 1.0 (ref.) | – | |||
| GA + AA | >79.62 DMA% | OR | 1.16 (0.62–2.13) | 0.646 | – | |||
| GG | ≤79.62 DMA% | OR | 1.52 (0.42–5.50) | 0.528 | – | |||
| GG | >79.62 DMA% | OR | 5.43 (1.51–19.55) | 0.010 | – | |||
| RERI | 3.76 (−3.13–10.66) | – | – | |||||
| AP | 0.69 (0.18–1.20) | – | – | |||||
| SI | 6.62 (0.23–194.10) | – | – | |||||
| AS3MT (rs1046778) |
TC + CC | >19.37 iAs% | OR | 1.0 (ref.) | – | |||
| TC + CC | ≤19.37 iAs% | OR | 1.68 (0.84–3.34) | 0.140 | – | |||
| TT | >19.37 iAs% | OR | 1.57 (0.71–3.51) | 0.267 | – | |||
| TT | ≤19.37 iAs% | OR | 1.56 (0.58–4.18) | 0.380 | – | |||
| RERI | −0.70 (−2.63–1.24) | – | – | |||||
| AP | −0.45 (−1.90–1.00) | – | – | |||||
| SI | 0.44 (0.04–5.49) | – | – | |||||
| AS3MT (rs1046778) |
TC + CC | >1.11 MMA% | OR | 1.0 (ref.) | – | |||
| TC + CC | ≤1.11 MMA% | OR | 1.72 (0.87–3.40) | 0.118 | – | |||
| TT | >1.11 MMA% | OR | 1.63 (0.74–3.60) | 0.225 | – | |||
| TT | ≤1.11 MMA% | OR | 1.58 (0.56–4.45) | 0.390 | – | |||
| RERI | −0.78 (−2.79–1.24) | – | – | |||||
| AP | −0.49 (−2.03–1.04) | – | – | |||||
| SI | 0.43 (0.03–5.53) | – | – | |||||
| AS3MT (rs1046778) |
TC + CC | ≤79.62 DMA% | OR | 1.0 (ref.) | – | |||
| TC + CC | >79.62 DMA% | OR | 1.68 (0.85–3.33) | 0.133 | – | |||
| TT | ≤79.62 DMA% | OR | 1.58 (0.71–3.51) | 0.264 | – | |||
| TT | >79.62 DMA% | OR | 1.55 (0.58–4.16) | 0.380 | – | |||
| RERI | −0.71 (−2.64–1.23) | – | – | |||||
| AP | −0.45 (−1.91–1.00) | – | – | |||||
| SI | 0.44 (0.04–5.45) | – | – | |||||
| AS3MT (rs1191453) |
TC + CC | >19.37 iAs% | OR | 1.0 (ref.) | – | |||
| TC + CC | ≤19.37 iAs% | OR | 1.07 (0.48–2.38) | 0.864 | – | |||
| TT | >19.37 iAs% | OR | 0.92 (0.44–1.93) | 0.825 | – | |||
| TT | ≤19.37 iAs% | OR | 1.85 (0.81–4.24) | 0.144 | – | |||
| RERI | 0.86 (−0.40–2.12) | – | – | |||||
| AP | 0.47 (−0.10–1.03) | – | – | |||||
| SI | −102.09 (NaN-NaN) | – | – | |||||
| AS3MT (rs1191453) | TC/CC | >1.11 MMA% | OR | 1.0 (ref.) | – | |||
| TC/CC | ≤1.11 MMA% | OR | 1.64 (0.73–3.70) | 0.233 | – | |||
| TT | >1.11 MMA% | OR | 1.41 (0.66–2.99) | 0.370 | – | |||
| TT | ≤1.11 MMA% | OR | 1.91 (0.81–4.54) | 0.140 | – | |||
| RERI | −0.13 (−1.75–1.48) | – | – | |||||
| AP | −0.07 (−0.93–0.79) | – | – | |||||
| SI | 0.87 (0.18–4.30) | – | – | |||||
| AS3MT (rs1191453) | TC + CC | ≤79.62 DMA% | OR | 1.0 (ref.) | – | |||
| TC + CC | >79.62 DMA% | OR | 1.20 (0.54–2.67) | 0.651 | – | |||
| TT | ≤79.62 DMA% | OR | 1.03 (0.49–2.16) | 0.937 | – | |||
| TT | ≤79.62 DMA% | OR | 1.87 (0.81–4.30) | 0.140 | – | |||
| RERI | 0.64 (−0.68–1.96) | – | – | |||||
| AP | 0.34 (−0.28–0.97) | – | – | |||||
| SI | 3.75 (0.01–940.13) | – | – | |||||
| Liao et al. 2009 ECG abnormality Water (cumulative) arsenic (ppm-yr) |
PON1 (rs662, rs705379) PON2 (rs7493) |
PON R-C-S - | ≤14.7 ppm-yr | OR | 1.0 (ref.) | 3/22 | Participants with high cumulative arsenic exposure and carrying the R-C-S haplotype was linked to increased ECG abnormality risk (synergistic effect). PON R-C-S refers to the PON1 (rs662), PON1 (rs705379), PON2 (rs7493) haplotype. Note, none of the eight included polymorphism reached statistical significance in univariate models. Adjusted by age, gender and cigarette smoking. |
|
| PON R-C-S + | ≤ 14.7 ppm-yr | OR | 1.57 (0.19–13.00) | 0.319 | 2/8 | |||
| PON R-C-S - | > 14.7 ppm-yr | OR | 4.27 (0.83–22.08) | 0.632 | 11/17 | |||
| PON R-C-S + | > 14.7 ppm-yr | OR | 19.19 (1.86–197.76) | 0.014 | 4/2 | |||
| Martíz-Barquero et al. 2015 Obesity Blood plasma arsenic (nmol/L) |
EDNRB (rs5351) |
GG | ≥50 nmol/L | OR | 1.0 (ref.) | 42/115 | From the 8 polymorphisms analysed, an association for two EDNRB polymorphism with obesity was found. A positive association for the two SNPs with obesity risk in those with higher arsenic exposure. Haplotype analysis indicates that there was a lower risk for obesity in individuals with higher arsenic levels, but no association for medium or low arsenic levels. Haplotype construction uses rs5351, rs3759475. Adjusted for age and sex. Number of study participants in each haplotype category not reported. |
|
| AG + AA | ≥50 nmol/L | OR | 0.51 (0.32–0.82) | 0.005 | 54/265 | |||
| GG | 20–50 nmol/L | OR | 1.0 (ref.) | 37/133 | ||||
| AG + AA | 20–50 nmol/L | OR | 0.70 (0.43–1.13) | 0.15 | 49/256 | |||
| GG | ≤20 nmol/L | OR | 1.0 (ref.) | 29/123 | ||||
| AG + AA | ≤20 nmol/L | OR | 0.70 (0.41–1.20) | 0.20 | 46/272 | |||
| EDNRB (rs3759475) |
CC | ≥50 nmol/L | OR | 1.0 (ref.) | 44/120 | |||
| CT + TT | ≥50 nmol/L | OR | 0.53 (0.33–0.85) | 0.009 | 54/256 | |||
| CC | 20–50 nmol/L | OR | 1.0 (ref.) | 35/132 | ||||
| CT + TT | 20–50 nmol/L | OR | 0.77 (0.47–1.25) | 0.30 | 52/259 | |||
| CC | ≤20 nmol/L | OR | 1.0 (ref.) | 29/126 | ||||
| CT + TT | ≤20 nmol/L | OR | 0.70 (0.41–1.21) | 0.20 | 46/268 | |||
| EDNRB (rs5351, rs3759475) |
GC | ≥50 nmol/L | OR | 1.0 (ref.) | – | |||
| AT | ≥50 nmol/L | OR | 0.53 (0.37–0.76) | <0.001 | – | |||
| AC | ≥50 nmol/L | OR | 0.66 (0.13–3.28) | 0.62 | – | |||
| GT | ≥50 nmol/L | OR | 0.78 (0.09–6.72) | 0.82 | – | |||
| GC | 20–50 nmol/L | OR | 1.0 (ref.) | – | ||||
| AT | 20–50 nmol/L | OR | 0.84 (0.59–1.20) | 0.34 | – | |||
| AC | 20–50 nmol/L | OR | 0.69 (0.08–6.05) | 0.74 | – | |||
| GT | 20–50 nmol/L | OR | 2.00 (0.49–8.11) | 0.33 | – | |||
| GC | ≤20 nmol/L | OR | 1.0 (ref.) | – | ||||
| AT | ≤20 nmol/L | OR | 0.82 (0.56–1.19) | 0.29 | – | |||
| AC + GT | ≤20 nmol/L | OR | 0.30 (0.04–2.52) | 0.27 | – | |||
| Pan et al. 2013 Diabetes mellites type 2 (T2DM) Water arsenic, μg/l |
ADAMTS9 | rs17070905 | P-value for interaction in the study population | 0.076 (0.063) | 957 adults, 83 T2DM |
Three SNPs showed a significant interaction with arsenic increasing T2DM risk. When using a subset of the population with exposure <148 μg/L water arsenic only the gene-environment interaction with NOTCH2 rs699780 remained significant. Adjusted for age, sex, BMI, smoking, skin lesions, using piece-wise regression models. Q-values are calculated using FDR adjustment for the P-value of interaction. |
||
| rs17070967 | 0.096 (0.063) | |||||||
| rs6766801 | 0.113 (0.063) | |||||||
| BCL11A | rs2058703 | 0.101 (0.063) | ||||||
| CDC123 | rs1051055 | 0.008 (0.033) | ||||||
| rs12126 | 0.017 (0.052) | |||||||
| CDKN2A | rs3088440 | 0.088 (0.063) | ||||||
| CDKN2B | rs1063192 | 0.138 (0.067) | ||||||
| rs3217986 | 0.078 (0.063) | |||||||
| rs3217992 | 0.938 (0.319) | |||||||
| CENTD2 | rs11603334 | 0.024 (0.061) | ||||||
| IDE | rs4646954 | 0.136 (0.067) | ||||||
| KCNQ1 | rs1057128 | 0.107 (0.063) | ||||||
| rs10798 | 0.093 (0.063) | |||||||
| rs8234 | 0.144 (0.067) | |||||||
| KMGA2 | rs343092 | 0.123 (0.065) | ||||||
| LGR5 | rs17109924 | 0.090 (0.063) | ||||||
| NOTCH2 | rs1043964 | 0.438 (0.168) | ||||||
| rs699779 | 0.096 (0.063) | |||||||
| rs699780 | 0.003 (0.021) | |||||||
| rs7527186 | 0.059 (0.063) | |||||||
| rs835575 | 0.115 (0.063) | |||||||
| rs835576 | 0.044 (0.063) | |||||||
| PRC1 | rs12911192 | 0.243 (0.109) | ||||||
| rs14280 | 0.960 (0.319) | |||||||
| rs7601 | 0.465 (0.168) | |||||||
| SLC30A8 | rs10282940 | 0.348 (0.146) | ||||||
| rs11558471 | 0.553 (0.194) | |||||||
| rs2466293 | 0.317 (0.138) | |||||||
| TCF2 | rs1058166 | 0.454 (0.168) | ||||||
| rs10962 | 0.115 (0.063) | |||||||
| rs2688 | 0.003 (0.021) | |||||||
| THADA | rs1549723 | 0.046 (0.063) | ||||||
| rs17031056 | 0.384 (0.156) | |||||||
| TSPAN8 | rs1051334 | 0.052 (0.063) | ||||||
| WFS1 | rs1801208 | 0.111 (0.063) | ||||||
| rs1801212 | 0.110 (0.063) | |||||||
| rs734312 | 0.449 (0.168) | |||||||
| % HbA1c levels Water arsenic, μg/l |
ADAMTS9 | rs17070905 | P-value for interaction in the study population | 0.091 (0.449) | No significant interactions were shown for SNPs with arsenic on T2DM risk. Adjusted for age, sex, BMI, smoking, skin lesions, using piece-wise regression models. Q-values are calculated using FDR adjustment for the P-value of interaction. |
|||
| rs17070967 | 0.136 (0.449) | |||||||
| rs6766801 | 0.144 (0.449) | |||||||
| BCL11A | rs2058703 | 0.553 (0.68) | ||||||
| CDC123 | rs1051055 | 0.354 (0.621) | ||||||
| rs12126 | 0.786 (0.745) | |||||||
| CDKN2A | rs3088440 | 0.681 (0.698) | ||||||
| CDKN2B | rs1063192 | 0.498 (0.677) | ||||||
| rs3217986 | 0.931 (0.793) | |||||||
| rs3217992 | 0.992 (0.816) | |||||||
| CENTD2 | rs11603334 | 0.707 (0.698) | ||||||
| IDE | rs4646954 | 0.417 (0.621) | ||||||
| KCNQ1 | rs1057128 | 0.248 (0.621) | ||||||
| rs10798 | 0.086 (0.449) | |||||||
| rs8234 | 0.017 (0.353) | |||||||
| KMGA2 | rs343092 | 0.161 (0.457) | ||||||
| LGR5 | rs17109924 | 0.29 (0.621) | ||||||
| NOTCH2 | rs1043964 | 0.326 (0.621) | ||||||
| rs699779 | 0.541 (0.353) | |||||||
| rs699780 | 0.023 (0.68) | |||||||
| rs7527186 | 0.376 (0.621) | |||||||
| rs835575 | 0.093 (0.449) | |||||||
| rs835576 | 0.078 (0.449) | |||||||
| PRC1 | rs12911192 | 0.571 (0.68) | ||||||
| rs14280 | 0.861 (0.792) | |||||||
| rs7601 | 0.714 (0.698) | |||||||
| SLC30A8 | rs10282940 | 0.905 (0.793) | ||||||
| rs11558471 | 0.655 (0.698) | |||||||
| rs2466293 | 0.123 (0.449) | |||||||
| TCF2 | rs1058166 | 0.365 (0.621) | ||||||
| rs10962 | 0.645 (0.698) | |||||||
| rs2688 | 0.491 (0.677) | |||||||
| THADA | rs1549723 | 0.385 (0.621) | ||||||
| rs17031056 | 0.587 (0.68) | |||||||
| TSPAN8 | rs1051334 | 0.137 (0.449) | ||||||
| WFS1 | rs1801208 | 0.411 (0.621) | ||||||
| rs1801212 | 0.351 (0.621) | |||||||
| rs734312 | 0.939 (0.793) | |||||||
| Wang et al. 2007 Carotid atherosclerosis Water arsenic (μg/L) |
GSTP1 (rs1695) | Ile/Ile | ≤50 μg/L | OR | 1.0 (ref.) | – | Results of this study suggest that there is a joint effect of arsenic exposure and GSTP1 and P53 (synergistic effect) genotypic variants on the risk of carotid atherosclerosis. Adjusted for age and gender. Number of study participants (patients/controls) in each category not reported. |
|
| Ile/Val + Val/Val | ≤50 μg/L | OR | 1.9 (0.9–4.0) | > 0.05 | – | |||
| Ile/Ile | >50 μg/L | OR | 2.7 (1.6–4.5) | <0.01 | – | |||
| Ile/Val + Val/Val | >50 μg/L | OR | 6.0 (3.3–10.7) | <0.001 | – | |||
| TP53 (rs1042522) | Arg/Arg | ≤50 μg/L | OR | 1.0 (ref.) | – | |||
| Arg/Pro + Pro/Pro | ≤50 μg/L | OR | 0.9 (0.5–1.9) | > 0.05 | – | |||
| Arg/Arg | >50 μg/L | OR | 1.4 (0.7–3.0) | > 0.05 | – | |||
| Arg/Pro + Pro/Pro | >50 μg/L | OR | 3.1 (1.7–5.7) | <0.001 | – | |||
| No. variant genotypes of GSTP1 and P53 | 0 | ≤50 μg/L | OR | 1.0 (ref.) | – | |||
| 1 | ≤50 μg/L | OR | 1.0 (0.4–2.2) | > 0.05 | – | |||
| 2 | ≤50 μg/L | OR | 1.9 (0.7–5.3) | > 0.05 | – | |||
| 0 | >50 μg/L | OR | 1.4 (0.6–3.3) | > 0.05 | – | |||
| 1 | >50 μg/L | OR | 2.8 (1.3–6.0) | <0.01 | – | |||
| 2 | >50 μg/L | OR | 6.1 (2.7–13.9) | <0.001 | – | |||
| Wu et al. 2014 Carotid intima-media thickness Water arsenic (μg/L) |
APOE (rs7256173) | Dominant | μg/L (cont) | βint | 49.6 (21.6, 77.6) | 0.0005 | – | Nine SNPs (APOE, AS3MT, PNP, TNF) had a nominally statistically significant interaction with well-water (or urinary) arsenic in cIMT. Coefficient of multiplicative interaction between a 1-standard-deviation increase (96.7 μg/L) in arsenic and each SNP. Adjusted for sex, age at cIMT measurement, BMI, smoking status, educational attainment, SBP, diabetes status at baseline, and change in urinary arsenic level between visits. Number of study participants (patients/controls) not reported. |
| AS3MT (rs10883790) | Additive | μg/L (cont) | βint | 2.0 (−6.1, 10.1) | 0.63 | – | ||
| Dominant | μg/L (cont) | βint | −2.1 (−11.9, 7.8) | 0.68 | – | |||
| Recessive | μg/L (cont) | βint | 21.0 (1.0, 41.0) | 0.039 | – | |||
| AS3MT (rs11191442) | Additive | μg/L (cont) | βint | 2.9 (−5.1, 10.9) | 0.47 | – | ||
| Dominant | μg/L (cont) | βint | −1.6 (−11.4, 8.2) | 0.75 | – | |||
| Recessive | μg/L (cont) | βint | 23.8 (4.4, 43.2) | 0.016 | – | |||
| AS3MT (rs3740392) | Additive | μg/L (cont) | βint | 2.8 (−5.2, 10.9) | 0.49 | – | ||
| Dominant | μg/L (cont) | βint | −1.8 (−11.7, 8.1) | 0.73 | – | |||
| Recessive | μg/L (cont) | βint | 23.6 (4.2, 43.1) | 0.017 | – | |||
| AS3MT (rs4919694) | Additive | μg/L (cont) | βint | 8.5 (−2.8, 19.8) | 0.14 | – | ||
| Dominant | μg/L (cont) | βint | 6.3 (−6.8, 19.4) | 0.34 | – | |||
| Recessive | μg/L (cont) | βint | 40.4 (3.4, 77.4) | 0.032 | – | |||
| PNP (rs17886095) | Dominant | μg/L (cont) | βint | 35.0 (4.1, 65.9) | 0.027 | – | ||
| PNP (rs17882804) | Additive | μg/L (cont) | βint | 8.9 (−0.8, 18.6) | 0.071 | – | ||
| Dominant | μg/L (cont) | βint | 13.5 (1.6, 25.4) | 0.026 | – | |||
| Recessive | μg/L (cont) | βint | −0.4 (−27.6, 26.9) | 0.98 | – | |||
| TNF (rs3790064) | Dominant | μg/L (cont) | βint | 24.7 (0.6, 48.7) | 0.044 | – | ||
| TNF (rs3093661) | Additive | μg/L (cont) | βint | −4.0 (−15.2, 7.2) | 0.48 | – | ||
| Dominant | μg/L (cont) | βint | −0.9 (−13.4, 11.7) | 0.89 | – | |||
| Recessive | μg/L (cont) | βint | −47.3 (−90.4, −4.2) | 0.032 | – | |||
| Carotid intima-media thickness Water arsenic (μg/L) |
APOE (rs7256173) | CC | < 40.4 μg/L | βint | 1.0 (ref.) | 504 | Adjusted for sex, age at cIMT measurement, BMI, smoking status, educational attainment, SBP, diabetes status at baseline, and change in urinary arsenic level between visits. Only total number of participants in each category was reported. |
|
| CC | ≥ 40.4 μg/L | βint | 8.7 (−1.4, 18.8) | – | 504 | |||
| CT + TT | < 40.4 μg/L | βint | −8.1 (−40.4, 24.1) | – | 25 | |||
| CT + TT | ≥ 40.4 μg/L | βint | 46.0 (10.7, 81.3) | – | 21 | |||
| AS3MT (rs10883790) | CA + AA | < 40.4 μg/L | βint | 1.0 (ref.) | 490 | |||
| CA + AA | ≥ 40.4 μg/L | βint | 8.0 (−2.3, 18.3) | – | 483 | |||
| CC | < 40.4 μg/L | βint | −5.4 (−32.2, 21.4) | – | 37 | |||
| CC | ≥ 40.4 μg/L | βint | 35.8 (9.2, 62.4) | – | 38 | |||
| AS3MT (rs11191442) | TT + TA | < 40.4 μg/L | βint | 1.0 (ref.) | 488 | |||
| TT + TA | ≥ 40.4 μg/L | βint | 7.8 (−2.5, 18.1) | – | 479 | |||
| AA | < 40.4 μg/L | βint | −5.8 (−32.5, 21.0) | – | 37 | |||
| AA | ≥ 40.4 μg/L | βint | 38.3 (12.1, 64.5) | – | 39 | |||
| AS3MT (rs3740392) | AA + AG | < 40.4 μg/L | βint | 1.0 (ref.) | 489 | |||
| AA + AG | ≥ 40.4 μg/L | βint | 7.2 (−3.1, 17.5) | – | 487 | |||
| GG | < 40.4 μg/L | βint | −5.1 (−31.6, 21.3) | – | 38 | |||
| GG | ≥ 40.4 μg/L | βint | 40.9 (14.4, 67.5) | – | 38 | |||
| Carotid intima-media thickness Urinary arsenic (μg/g creatinine) |
APOE (rs7256173) | Dominant | μg/g (cont) | βint | 50.3 (7.5, 93.2) | 0.021 | – | Coefficient of multiplicative interaction between a 1-standard-deviation increase (345.2 μg/g creatinine) in arsenic and each SNP. Adjusted for sex, age at cIMT measurement, BMI, smoking status, educational attainment, SBP, diabetes status at baseline, and change in urinary arsenic level between visits. Number of study participants (patients/controls) not reported. |
| AS3MT (rs10883790) | Additive | μg/g (cont) | βint | 6.3 (−3.8, 16.5) | 0.22 | – | ||
| Dominant | μg/g (cont) | βint | 2.6 (−9.3, 14.5) | 0.67 | – | |||
| Recessive | μg/g (cont) | βint | 40.9 (10.6, 71.3) | 0.008 | – | |||
| AS3MT (rs11191442) | Additive | μg/g (cont) | βint | 6.9 (−3.3, 17.0) | 0.18 | – | ||
| Dominant | μg/g (cont) | βint | 3.5 (−8.5, 15.4) | 0.57 | – | |||
| Recessive | μg/g (cont) | βint | 40.4 (10.2, 70.6) | 0.009 | – | |||
| AS3MT (rs3740392) | Additive | μg/g (cont) | βint | 6.3 (−4.0, 16.7) | 0.23 | – | ||
| Dominant | μg/g (cont) | βint | 2.4 (−9.6, 14.3) | 0.69 | – | |||
| Recessive | μg/g (cont) | βint | 47.6 (14.9, 80.2) | 0.004 | – | |||
| AS3MT (rs4919694) | Additive | μg/g (cont) | βint | 12.2 (−4.7, 29.0) | 0.16 | – | ||
| Dominant | μg/g (cont) | βint | 10.5 (−9.0, 29.9) | 0.29 | – | |||
| Recessive | μg/g (cont) | βint | 50.4 (−7.2, 108.0) | 0.086 | – | |||
| PNP (rs17886095) | Dominant | μg/g (cont) | βint | −6.7 (−34.6, 21.3) | 0.64 | – | ||
| PNP (rs17882804) | Additive | μg/g (cont) | βint | 9.6 (−0.9, 20.1) | 0.072 | – | ||
| Dominant | μg/g (cont) | βint | 8.3 (−2.6, 19.1) | 0.14 | – | |||
| Recessive | μg/g (cont) | βint | 65.4 (6.2, 124.6) | 0.030 | – | |||
| TNF (rs3790064) | Dominant | μg/g (cont) | βint | 0.6 (−22.2, 23.4) | 0.96 | – | ||
| TNF (rs3093661) | Additive | μg/g (cont) | βint | 0.6 (−17.2, 18.4) | 0.95 | – | ||
| Dominant | μg/g (cont) | βint | 0.3 (−17.9, 18.6) | 0.97 | – | |||
| Recessive | μg/g (cont) | βint | 14.8 (−126.4, 156.0) | 0.84 | – | |||
| Carotid intima-media thickness Urinary arsenic (μg/g creatinine) |
APOE (rs7256173) | CC | < 183 μg/g | β | 1.0 (ref.) | 508 | Adjusted for sex, age at cIMT measurement, BMI, smoking status, educational attainment, SBP, diabetes status at baseline, and change in urinary arsenic level between visits. Only total number of participants in each category was reported. |
|
| CC | ≥ 183 μg/g | β | 7.9 (−2.2, 18.1) | – | 514 | |||
| CT + TT | < 183 μg/g | β | 7.5 (−24.2, 39.2) | – | 26 | |||
| CT + TT | ≥ 183 μg/g | β | 27.9 (−6.6, 62.4) | – | 22 | |||
| AS3MT (rs10883790) | CA + AA | < 183 μg/g | β | 1.0 (ref.) | 486 | |||
| CA + AA | ≥ 183 μg/g | β | 5.9 (−4.4, 16.2) | – | 501 | |||
| CC | < 183 μg/g | β | −2.9 (−27.2, 21.3) | – | 46 | |||
| CC | ≥ 183 μg/g | β | 40.7 (11.6, 69.9) | – | 31 | |||
| AS3MT (rs11191442) | TT + TA | < 183 μg/g | β | 1.0 (ref.) | 483 | |||
| TT + TA | ≥ 183 μg/g | β | 5.7 (−4.6, 16.0) | – | 498 | |||
| AA | < 183 μg/g | β | −0.2 (−24.1, 23.8) | – | 47 | |||
| AA | ≥ 183 μg/g | β | 40.4 (11.4, 69.5) | – | 31 | |||
| AS3MT (rs3740392) | AA + AG | < 183 μg/g | β | 1.0 (ref.) | 484 | |||
| AA + AG | ≥ 183 μg/g | β | 5.7 (−4.5, 16.0) | – | 506 | |||
| GG | < 183 μg/g | β | 0.4 (−23.3, 24.2) | – | 48 | |||
| GG | ≥ 183 μg/g | β | 44.1 (14.6, 73.6) | – | 30 | |||
| Wu et al. 2015 Cardiovascular disease (CVD) Water arsenic, μg/l |
ICAM1 (rs281432) | CG + CC | < 45 μg/L | HR | 1.0 (ref.) | Pint 0.40 | 143/500 | From 170 SNPs explored, multiplicative interactions were statistically significant for CVD after multiple testing corrections for ICAM1 (rs2781432) and VCAM1 (rs3176867). Adjusted for sex, age, BMI, smoking status, educational attainment, SBP, diabetes status, and change in urinary arsenic between visits. |
| GG | < 45 μg/L | HR | 1.35 (0.84–2.18) | 43/173 | ||||
| CG + CC | ≥ 45 μg/L | HR | 1.67 (1.14–2.43) | 185/520 | ||||
| GG | ≥ 45 μg/L | HR | 2.98 (1.87–4.77) | 73/151 | ||||
| VCAM1 (rs3176867) | TC + TT | < 45 μg/L | HR | 1.0 (ref.) | Pint 0.04 | 166/594 | ||
| CC | < 45 μg/L | HR | 0.89 (0.56–1.41) | 16/65 | ||||
| TC + TT | ≥ 45 μg/L | HR | 1.30 (0.83–2.05) | 222/609 | ||||
| CC | ≥ 45 μg/L | HR | 2.13 (1.37–3.31) | 27/42 | ||||
| Cardiovascular disease (CVD) Water arsenic, μg/L |
APOE rs405509 | AA vs. AC + CC | μg/L (cont) | HR | 1.09 (0.78, 1.53) | 0.021 (0.287) | – | Coefficient of multiplicative interaction between a 1-SD increase (101.3 μg/L) in arsenic and each SNP. Adjusted for sex, age, BMI, smoking status, educational attainment, SBP, diabetes status, and change in urinary arsenic between visits. P-values from tests for multiplicative interactions between a 1-SD increase well-water arsenic and SNPs. |
| rs7259620 | GG vs. AG + AA | μg/L (cont) | HR | 1.35 (0.96, 1.89) | 0.041 (0.324) | – | ||
| AS3MT rs1046778 | TC + CC vs. TT | μg/L (cont) | HR | 1.22 (0.87, 1.72) | 0.038 (0.324) | – | ||
| rs10748839 | TC + CC vs. TT | μg/L (cont) | HR | 1.34 (0.91, 2.00) | 0.046 (0.324) | – | ||
| rs10786719 | AG + GG vs. AA | μg/L (cont) | HR | 1.28 (0.87, 1.90) | 0.046 (0.324) | – | ||
| rs11191454 | AG + GG vs. AA | μg/L (cont) | HR | 1.27 (0.87, 1.84) | 0.016 (0.287) | – | ||
| rs12573221 | AC + CC vs. AA | μg/L (cont) | HR | 1.18 (0.81, 1.71) | 0.040 (0.324) | – | ||
| rs4290163 | GT + TT vs. GG | μg/L (cont) | HR | 1.25 (0.85, 1.84) | 0.036 (0.324) | – | ||
| rs9527 | GG vs. GA + AA | μg/L (cont) | HR | 1.00 (0.63, 1.61) | 0.041 (0.324) | – | ||
| CBS rs1005585 | AG + GG vs. AA | μg/L (cont) | HR | 1.03 (0.67, 1.58) | 0.006 (0.274) | – | ||
| rs3788050 | GT + TT vs. GG | μg/L (cont) | HR | 1.10 (0.73, 1.66) | 0.015 (0.287) | – | ||
| rs8132811 | CT + TT vs. CC | μg/L (cont) | HR | 1.01 (0.71, 1.45) | 0.002 (0.141) | – | ||
| GSTO1 rs1147611 | CA + AA vs. CC | μg/L (cont) | HR | 1.65 (1.16, 2.36) | 0.026 (0.296) | – | ||
| rs11509438 | GA + AA vs. GG | μg/g (cont) | HR | 1.70 (1.19, 2.41) | 0.024 (0.295) | – | ||
| rs2282326 | AC + CC vs. AA | μg/L (cont) | HR | 1.66 (1.17, 2.36) | 0.041 (0.324) | – | ||
| ICAM1 rs281432 | GG vs. CG + CC | μg/L (cont) | HR | 1.82 (1.31, 2.54) | 9.4 × 10 −7 (0.0002) | – | ||
| NOS3 rs1800783 | TA + AA vs. TT | μg/L (cont) | HR | 1.06 (0.75, 1.50) | 0.022 (0.287) | – | ||
| rs6951150 | CT + TT vs. CC | μg/L (cont) | HR | 1.04 (0.73, 1.48) | 0.012 (0.287) | – | ||
| SOD2 rs2758331 | CA + AA vs. CC | μg/L (cont) | HR | 1.07 (0.72, 1.59) | 0.021 (0.287) | – | ||
| rs2758334 | TC + CC vs. TT | μg/L (cont) | HR | 0.96 (0.66, 1.41) | 0.009 (0.287) | – | ||
| rs8031 | TA + AA vs. TT | μg/L (cont) | HR | 1.02 (0.68, 1.51) | 0.046 (0.324) | – | ||
| VCAM1 | ||||||||
| rs3176867 | CC vs. TC + TT | μg/L (cont) | HR | 1.34 (0.95, 1.87) | 0.0004 (0.035) | – | ||
| rs3176871 | GG vs. GA + AA | μg/L (cont) | HR | 0.55 (0.32, 0.95) | 0.018 (0.287) | – | ||
| rs3765685 | AA vs. AG + GG | μg/L (cont) | HR | 1.20 (0.83, 1.74) | 0.014 (0.287) | – | ||
| Coronary heart disease, (CHD) Water arsenic, μg/L |
AS3MT (rs1046778) | TC+CC vs. TT | μg/L (cont) | HR | 1.25 (0.84–1.85) | 0.043 (0.315) | – | Coefficient of multiplicative interaction between a 1-SD increase (101.3 μg/L) in arsenic and each SNP. Adjusted for sex, age, BMI, smoking status, educational attainment, SBP, diabetes status, and change in urinary arsenic between visits. P-values from tests for multiplicative interactions between a 1-SD increase well-water arsenic and SNPs. |
| (rs11191454) | AG+GG vs. AA | μg/L (cont) | HR | 1.33 (0.88–2.01) | 0.036 (0.290) | – | ||
| (rs12573221) | AC+CC vs. AA | μg/L (cont) | HR | 1.17 (0.76–1.78) | 0.007 (0.257) | – | ||
| CBS (rs1005585) |
AG+GG vs. AA | μg/L (cont) | HR | 0.93 (0.55–1.58) | 0.009 (0.257) | – | ||
| (rs11700748) | TC+TT vs. CC | μg/L (cont) | HR | 1.04 (0.68–1.58) | 0.018 (0.257) | – | ||
| (rs2124459) | TC+CC vs. TT | μg/L (cont) | HR | 1.08 (0.67–1.73) | 0.050 (0.321) | – | ||
| (rs2849727) | TC+TT vs. CC | μg/L (cont) | HR | 1.08 (0.72–1.61) | 0.050 (0.321) | – | ||
| (rs3788050) | GT+TT vs. GG | μg/L (cont) | HR | 0.96 (0.58–1.58) | 0.018 (0.257) | – | ||
| (rs706208) | TC+CC vs. TT | μg/L (cont) | HR | 1.02 (0.67–1.56) | 0.031 (0.275) | – | ||
| GSTO1 (rs1147611) |
CA+AA vs. CC | μg/L (cont) | HR | 1.72 (1.13–2.60) | 0.028 (0.275) | – | ||
| (rs11509438) | GA+AA vs. GG | μg/L (cont) | HR | 2.07 (1.36–3.15) | 0.028 (0.275) | – | ||
| (rs2282326) | AC+CC vs. AA | μg/L (cont) | HR | 1.74 (1.15–2.63) | 0.036 (0.290) | – | ||
| ICAM1 (rs281432) |
GG vs. CG+CC | μg/L (cont) | HR | 1.56 (1.04–2.34) | 0.007 (0.257) | – | ||
| IL6 (rs2069835) |
TC+CC vs. TT | μg/L (cont) | HR | 1.27 (0.77–2.12) | 0.031 (0.275) | – | ||
| MTHFR (rs12121543) | CA+AA vs. CC | μg/L (cont) | HR | 0.92 (0.61–1.39) | 0.016 (0.257) | – | ||
| (rs17421462) | GG vs. GA+AA | μg/L (cont) | HR | 0.80 (0.47–1.36) | 0.029 (0.275) | – | ||
| (rs1801131) | AC+CC vs. AA | μg/L (cont) | HR | 0.92 (0.59–1.43) | 0.015 (0.257) | – | ||
| NOS3 (rs1800783) |
TA+AA vs. TT | μg/L (cont) | HR | 0.94 (0.63–1.39) | 0.015 (0.257) | – | ||
| (rs6951150) | TC+TT vs. CC | μg/L (cont) | HR | 0.96 (0.65–1.44) | 0.014 (0.257) | – | ||
| SOD2 (rs2758334) |
TC+CC vs. TT | μg/L (cont) | HR | 0.96 (0.61–1.51) | 0.042 (0.315) | – | ||
| VCAM1 (rs1409419) |
CC vs. TC+TT | μg/L (cont) | HR | 0.83 (0.56–1.24) | 0.027 (0.275) | – | ||
| (rs2209627) | AA vs. AG+GG | μg/L (cont) | HR | 0.70 (0.40–1.23) | 0.008 (0.257) | – | ||
| (rs3176867) | CC vs. TC+TT | μg/L (cont) | HR | 1.51 (1.01–2.26) | 0.027 (0.275) | – | ||
| (rs3176871) | GG vs. GA+AA | μg/L (cont) | HR | 0.48 (0.27–0.88) | 0.011 (0.257) | – | ||
| (rs3176878) | CC vs. TC+TT | μg/L (cont) | HR | 0.78 (0.45–1.35) | 0.014 (0.257) | – | ||
| (rs3917014) | GG vs. AG+AA | μg/L (cont) | HR | 1.55 (0.87–2.73) | 0.003 (0.260) | – | ||
| Stroke Water arsenic (μg/L) |
AS3MT | Coefficient of multiplicative interaction between a 1-SD increase (101.3 μg/L) in arsenic and each SNP. Adjusted for sex, age, BMI, smoking status, educational attainment, SBP, diabetes status, and change in urinary arsenic between visits. P-values from tests for multiplicative interactions between a 1-SD increase well-water arsenic and SNPs. |
||||||
| (rs10786719) | AG+GG vs. AA | μg/L (cont) | HR | 1.16 (0.64–2.08) | 0.050 (0.440) | – | ||
| (rs11191439) | TT vs. TC+CC | μg/L (cont) | HR | 0.82 (0.38–1.76) | 0.044 (0.440) | – | ||
| CBS | ||||||||
| (rs8132811) | CT+TT vs. CC | μg/L (cont) | HR | 0.97 (0.57–1.64) | 0.021 (0.440) | – | ||
| GSTO1 | ||||||||
| (rs1147611) | CA+AA vs. CC | μg/L (cont) | HR | 1.64 (0.99–2.73) | 0.027 (0.440) | – | ||
| ICAM1 | ||||||||
| (rs281432) | GG vs. CG+CC | μg/L (cont) | HR | 1.85 (1.14–3.01) | 8.3 × 10 −5 (0.14) | – | ||
| NOS3 | ||||||||
| (rs1800779) | AG+GG vs. AA | μg/L (cont) | HR | 1.36 (0.84–2.20) | 0.034 (0.440) | – | ||
| (rs1800783) | TA+AA vs. TT | μg/L (cont) | HR | 1.10 (0.69–1.76) | 0.013 (0.440) | – | ||
| (rs3793342) | CT+TT vs. CC | μg/L (cont) | HR | 1.29 (0.79–2.12) | 0.023 (0.440) | – | ||
| (rs3918169) | AG+GG vs. AA | μg/L (cont) | HR | 1.18 (0.73–1.92) | 0.031 (0.440) | – | ||
| (rs6951150) | CT+TT vs. CC | μg/L (cont) | HR | 1.01 (0.63–1.62) | 0.002 (0.193) | – | ||
| VCAM1 | ||||||||
| (rs3176867) | CC vs. TC+TT | μg/L (cont) | HR | 1.23 (0.76–1.99) | 0.007 (0.377) | – | ||
| Cardiovascular disease, (CVD) Urinary arsenic (μg/g creatinine) |
AS3MT | Coefficient of multiplicative interaction between a 1-SD increase (322 μg/g) in arsenic and each SNP. Adjusted for sex, age, BMI, smoking status, educational attainment, SBP, diabetes status, and change in urinary arsenic between visits. P-values from tests for multiplicative interactions between a 1-SD increase well-water arsenic and SNPs. |
||||||
| (rs12573221) | AC+CC vs. AA | μg/g (cont) | HR | 0.99 (0.64–1.54) | 0.011 (–) | – | ||
| (rs4290163) | GT+TT vs. GG | μg/g (cont) | HR | 1.11 (0.72–1.70) | 0.011 (–) | – | ||
| CBS | ||||||||
| (rs1005585) | AG+GG vs. AA | μg/g (cont) | HR | 1.02 (0.63–1.64) | 4.3 × 10−7 (–) | – | ||
| (rs3788050) | GT+TT vs. GG | μg/g (cont) | HR | 1.09 (0.69–1.73) | 1.1 × 10−6 (–) | – | ||
| (rs8132811) | CT+TT vs. CC | μg/g (cont) | HR | 1.00 (0.65–1.54) | 0.002 (–) | – | ||
| ICAM1 | ||||||||
| (rs281432) | GG vs. CG+CC | μg/g (cont) | HR | 1.68 (1.12–2.52) | 0.014 (–) | – | ||
| SOD2 | ||||||||
| (rs2758331) | CA+AA vs. CC | μg/g (cont) | HR | 0.97 (0.63–1.50) | 0.010 (–) | – | ||
| (rs2758334) | TC+CC vs. TT | μg/g (cont) | HR | 0.86 (0.56–1.31) | 0.005 (–) | – | ||
| (rs8031) | TA+AA vs. TT | μg/g (cont) | HR | 0.90 (0.59–1.38) | 0.008 (–) | – | ||
| Coronary heart disease (CHD) Urinary arsenic (μg/g creatinine) |
AS3MT | |||||||
| (rs12573221) | AC+CC vs. AA | μg/g (cont) | HR | 1.03 (0.63–1.69) | 0.035 (–) | – | ||
| CBS | ||||||||
| (rs1005585) | AG+GG vs. AA | μg/g (cont) | HR | 1.01 (0.57–1.79) | 0.0003 (–) | – | ||
| (rs3788050) | GT+TT vs. GG | μg/g (cont) | HR | 1.02 (0.59–1.79) | 0.0009 (–) | – | ||
| VCAM1 | ||||||||
| (rs1409419) | CC vs. TC+TT | μg/g (cont) | HR | 0.84 (0.54–1.31) | 0.002 (–) | – | ||
| (rs2209627) | AA vs. AG+GG | μg/g (cont) | HR | 0.57 (0.29–1.14) | 0.043 (–) | – | ||
| (rs3176878) | CC vs. TC+TT | μg/g (cont) | HR | 0.62 (0.31–1.25) | 0.050 (–) | – | ||
| Stroke Urinary arsenic (μg/g creatinine) |
CBS | |||||||
| (rs8132811) | CT+TT vs. CC | μg/g (cont) | HR | 0.89 (0.49–1.63) | 0.003 (–) | – | ||
| ICAM1 | ||||||||
| (rs281432) | GG vs. CG+CC | μg/g (cont) | HR | 1.60 (0.87–2.93) | 0.005 (–) | – | ||
-
Most studies only presented the results of statistically significant SNPs and thus these have been included here. Q-values are adjusted p-values after FDR correction. A, adenine; AP, attributable proportion due to the interaction; BG, blood glucose; βint, coefficient for interaction; BMI, body mass index; C, cytosine; Ca, cases; CHD, coronary heart disease; CI, confidence interval; cIMT, carotid intima-media thickness; Co, controls; CRP, C-reactive protein; CVD, cardiovascular disease; DBP, diastolic blood pressure; DM, diabetes mellitus; DMA, dimethylated arsenic; ECG, electrocardiogram; FDR, false discovery rate; G, guanine; GMR, geometric mean ratio; HDL, high-density lipoprotein; HOMA2-IR, homeostatic model assessment insulin resistance; HR, heart rate; HTN, hypertension; LDL, low-density lipoprotein; MoA, measure of association; MMA, monomethylated arsenic; OR, odds ratio; ORint, odds ratio for interaction; Pb, lead; Pint, p-value for interaction; PMI, primary methylation index; Ppb, parts per million; RERI, relative excess risk due to interactions; SBP, systolic blood pressure; SD, standard deviation; SI, synergy index; SMI, secondary methylation index; SNP, single nucleotide polymorphism; SNV, small nucleotide variant; T, thymine; TC, total cholesterol; TG, triglycerides; T2DM, Type 2 diabetes mellitus; WC, waist circumference. Abbreviations of relevant genes can be found in Supplementary Table 5. The bold MAO and P/Q values are considered statistically significant.
Single nucleotide polymorphisms (SNPs) associated with arsenic-related cardiometabolic disease.
| Significant SNP-arsenic | Arsenic and disease | Risk(b) | Non-significant results reported(c) | Gene (product) function(d) |
|---|---|---|---|---|
|
AS3MT
rs17881215 |
W + DM [40] |
↑ |
– |
Arsenic metabolism; transfer of methyl group from SAM to arsenical |
| rs11191439 | W + DM [40] | ↑ | W + CA [0.05 (0.075)] [27] W + CVD/CHD/Stroke [0.044 (0.440)] [34] (a) CW + ECG ab [29] W/U + CIMT [33] U + MetS [25] |
|
| rs12768205 | U[iAs% vs. MMA%] + IR [41] U[DMA% vs. iAs%] + IR [41] |
↑ | U[iAs% vs. MMA%] + DM [41] U[DMA% vs. iAs%] + DM [41] |
|
| rs10883790 | W/U + CIMT [33] | ↑ | W + CVD/CHD/Stroke [34] | |
| rs11191442 | W/U + CIMT [33] | ↑ | W + CVD/CHD/Stroke [34] | |
| rs3740392 | W/U + CIMT [33] | ↑ | W + CVD/CHD/Stroke [34] | |
| rs4919694 | W/U + CIMT [33] | ↑ | W + CVD/CHD/Stroke [34] | |
|
|
||||
|
APOE
rs7256173 |
W/U + CIMT [33] |
↑ |
W + CVD/CHD/Stroke [34] |
Inflammation/endothelial function; catabolism of triglyceride-rich lipoprotein constituent |
|
|
||||
|
CCM3
rs9818496 |
W + SBP [36] |
↑ |
W + DBP [36] |
Cell apoptosis; vascular development / angiogenesis; implicated in influencing cardiovascular-related physiological response |
| rs3804610 | W + SBP [36] | ↑ | W + DBP [36] | |
| rs6784267 | W + SBP [36] | ↑ | W + DBP [36] | |
|
|
||||
|
CDC123
rs1051055 |
W + DM [32] |
↑ |
W + % HbA1c [32] |
Cell division cycle; implicated in T2DM |
|
|
||||
|
COX5A
rs1133322 |
U + DM [38] |
↑ |
– |
Oxidative phosphorylation; component of cytochrome C oxidase; implicated in T2DM |
|
|
||||
|
CYBA
rs3794624 |
W + PP [31] |
↑ |
W + SBP [P 0.004 (0.34)](a) [31] W + CVD/CHD/Stroke [34] W/U + CIMT [33] |
Oxidative stress/defence; encodes the light, alpha subunit of Cytochrome B |
| rs4673 | CW + HTN [28] | ↑ | – | |
|
|
||||
|
EDNRB
rs3759475 |
B + Ob [39] |
↓ |
U + T2DM [38] |
Inflammation/endothelial function; located primarily in vascular endothelial cells with a role in vasoconstriction, vasodilation, and cell proliferation |
| rs5351 | B + Ob [39] | ↓ | U + T2DM [38] | |
|
|
||||
|
GCLC
rs11415624 |
U + DM [38] |
↑ |
– |
Oxidative stress/defence; glutathione synthesis; first rate limiting enzyme of glutathione synthesis |
|
|
||||
|
GSTO1
rs4925 |
U (PMI, SMI, MMA%, DMA%) + MetS [25] W + CA [27] |
↑ |
CW + ECG ab [29] W + CVD/CHD/Stroke [34] W/U + CIMT [33] |
Arsenic metabolism; glutathione-dependent thiol transferase and dehydroascorbate reductase activities; reduces MMA and DMA |
|
|
||||
|
GSTP1
rs1695 |
W + CA [30] |
↑ |
W + DBP [P 0.02 (0.74)](a) [31] W + CVD/CHD/Stroke [34] |
Arsenic metabolism; conjugation of reduced glutathione to a wide number of hydrophobic electrophiles; |
|
|
||||
|
ICAM1
rs281432 |
W + CVD/Stroke [34] |
↑ |
W + CHD [P 0.007 (0.257)](a) [34] U + CVD/Stroke/CHD [34] |
Inflammation/endothelial function; encodes cell adhesion molecules |
|
|
||||
|
IL8RA
rs1008563 |
U + DM [38] |
↑ |
– |
Implicated in T2DM; receptor to interleukin-8; activation of neutrophils |
| rs1008562 | U + DM [38] | ↑ | – | |
|
|
||||
|
NOS3
rs1799983 |
CW + HTN [28] |
↑ |
W + DBP [P 0.01 (0.74)](a) [31] |
Oxidative stress/defence; production of nitric oxide (NO) which is implicated in vascular smooth muscle relaxation, angiogenesis, blood clotting |
|
|
||||
|
NOTCH2
rs699780 |
W + DM [32] |
↑ |
W + % HbA1c [32] |
Cell fate decision; development; immune function |
|
|
||||
| NR3C2 rs13117325 |
U + DM [38] |
↑ |
– |
Cellular sodium ion homeostasis; implicated in T2DM; encodes the mineralocorticoid receptor |
| rs2137335 | U + DM [38] | ↑ | – | |
|
|
||||
|
N6AMT1
rs1997605 |
U (iAs%, MMA%, DMA% + GDM [37] |
↑ |
– |
Arsenic metabolism; methyltransferase; involved in converting MMA to DMA |
| rs1003671 | U (iAs%, MMA%, DMA% + GDM [37] | ↑ | – | |
|
|
||||
| OGG1 rs1052133 |
U + HTN [24] |
↑ |
U + T2DM [38] |
DNA repair; glycosylase removing premutagenic 8-oxo-G – a base by-product of ROS exposure |
|
|
||||
| PNP rs17886095 |
W/U + CIMT [33] |
↑ |
W + CVD/CHD/Stroke [34] |
Arsenic metabolism; catalyses the phosphorolysis of purine nucleosides; arsenic reductase |
| rs17882804 | W/U + CIMT [33] | ↑ | W + CVD/CHD/Stroke [34] | |
| rs3790064 | W/U + CIMT [33] | ↑ | W + CVD/CHD/Stroke [34] | |
|
|
||||
|
TP53
rs1042522 |
W + CA [30] |
↑ |
– |
Tumour suppressor; protects vascular smooth muscle from apoptosis; implicated in atherosclerosis |
|
|
||||
|
SOD2
rs4880 |
U + HTN [24] CW + HTN [28] |
↑ ↑ |
U + T2DM [38] |
Oxidative stress/defence; ROS scavenger in mitochondria; binds to superoxide by-products of oxidative phosphorylation |
|
|
||||
|
TCF2
rs2688 |
W + DM [32] |
↑ |
W + % HbA1c [32] |
Nephron development, embryonic pancreas development; implicated in T2DM |
|
|
||||
|
TNF
rs3093661 rs3790064 |
W + CIMT [33] W + CIMT [33] |
↑ ↑ |
W + CVD/CHD/Stroke [34] U + CIMT [33] U + CIMT [33] |
Inflammation/endothelial function; encodes multifunctional proinflammatory cytokine |
|
|
||||
|
TXN
rs4135168 |
U + DM [38] |
↑ |
– |
Oxidative stress/defence; cellular redox homeostasis; |
|
|
||||
|
VCAM1
rs3176867 |
W + CVD [34] |
↑ |
W + CHD [P 0.027 (0.275)](a) /Stroke [P 0.007 (0.377)](a) [34] U + CIMT [33] |
Inflammation/endothelial function; mediates leukocyte-endothelial cell adhesion and signal transduction; may have a role in CA |
|
|
||||
| Haplotypes | ||||
|
|
||||
| PON R-C-S haplotype [PON1 (rs662), PON1 (rs705379), PON2 (rs7493)] |
CW + ECG ab [29] |
↑ |
– |
Serum esterase/lactonase with antioxidant action on HDL; protective atherosclerosis |
|
|
||||
| PNP A-T haplotype (rs1049564, rs1130650) | W + CA [27] |
↑ |
– |
Arsenic metabolism; catalyses the phosphorolysis of purine nucleosides; arsenic reductase |
|
|
||||
| GST A-G-G haplotype [GSTO1 (rs925), GSTO2 (rs2297235, rs156697)] |
W + CA [27] |
↑ |
– |
Arsenic metabolism; glutathione-dependent thiol transferase and dehydroascorbate reductase activities; reduces MMA and DMA |
|
|
||||
| EDRNB A-T haplotype (rs5351, rs3759475) |
B + Ob [39] |
↓ |
– |
Inflammation/endothelial function; located primarily in vascular endothelial cells with a role in vasoconstriction, vasodilation, and cell proliferation |
|
|
||||
|
MCP-1
(rs1024611),
APOE
(rs429358, rs7412) |
W + CA [26] |
↑ |
– |
Inflammation/endothelial function; catabolism of triglyceride-rich lipoprotein constituent (APOE); chemotactic response and mobilization of intracellular calcium (MCP-1) |
-
B, blood; CA, carotid atherosclerosis; CHD, coronary heart disease; CVD, cardiovascular disease; CW, cumulative water arsenic; DBP, diastolic blood pressure; DM, diabetes mellitus; DMA, dimethylated arsenic; ECG, electrocardiogram; HDL, high-density lipoprotein; HTN, hypertension; IR, insulin resistance; MetS, metabolic syndrome; Ob, obesity; PMI, primary methylation index; ROS, reactive oxygen species; SAM, S-Adenosyl methionine; SBP, systolic blood pressure; SMI, secondary methylation index; U, urine, W, water. (a) Here the P (Q) values are presented with Q referring to the p-value adjusted for multiple testing. (b)Refers to whether there is a positive or negative association between the arsenic-SNV interaction and disease of interest. (c) Here, studies are listed that investigated a same SNV for which a significant interaction effect was reported in one study – but that reported non-significant interaction effects (before and/or after adjustment for multiple testing). (d)Information described in this column is based on information provided in the referenced included articles as well as the dbSNP database (https://www.ncbi.nlm.nih.gov/snp/) and the integrative database GeneCards (https://www.genecards.org/). ↓ Arsenic exposed individuals had lower disease risk in those carrying the variant compared to those carrying the wildtype. ↑ Arsenic exposed individuals had higher disease risk in those carrying the variant compared to the wildtype. – Indicates that there was no other study that explored this unique SNP. Details on each individual SNP can be found in supplementary Table 5.
Study samples were relatively small ranging from 121 participants [29] up to 2,225 participants [34]. Most had gender parity in participants (i.e., 45–55 % women) [26], 27], 30], 31], 34], 38], 39], or included predominantly women (i.e., >55 % participants were women) [24], 25], 28], 29], 33], 35], 37], 40], 41], whilst two included predominantly men [32], 36]. All studies were conducted in adults, and were conducted as early as 1989 [28] up to 2018 [35].
Cardiometabolic outcomes
Studies reported on different cardiometabolic outcomes, including: hypertension or blood pressure (SBP, DBP, PP) [24], 28], 31], 36], metabolic syndrome [25], obesity [39], T2DM or insulin resistance [32], 35], 38], 40], 41], GDM [37], carotid atherosclerosis or CIMT [26], 27], 30], 33], CVD [34], CHD [34], stroke [34] or ECG abnormalities consistent with MI or ischemia, conduction defect, arrhythmia, arterial enlargement or ventricular hypertrophy or prolonged ventricular repolarization (Table 1) [29].
Hypertension and blood pressure (i.e., systolic, diastolic, pulse pressure) were assessed using a mercury or automated sphygmomanometer [24], 28], 31]. Hypertension was defined as SBP≥130 mmHg and/or DBP≥80 mmHg [24], or as SBP ≥140 mmHG and/or DBP ≥90 mmHg [28]. The studies on T2DM defined DM as fasting blood glucose (FBG) ≥126 mg/dL [38], 41], HbA1c ≥6.5 % [32], 35], 38], self-reported physician diagnosis or T2DM treatment [38], and 2-h post glucose ≥200 mg/dl [40], with homeostasis model assessment of insulin resistance (HOMA2-IR) computed using fasting glucose and insulin values [41]. GDM was defined at 24–28 weeks of pregnancy as FBG ≥5.1 mmol/L, or 1 h plasma glucose (PG) ≥10 mmol/L and 2hPG ≥8.5 mmol/L after 75 g oral glucose tolerance test (OGTT) [37]. Carotid atherosclerosis was defined based on the intima-media thickness (>1 mm), plaque score (≥1), and maximum level of stenosis of the ECCA (≥50 %) [26], 27], 30]. CVD, CHD and stroke diagnoses were based on a validated verbal autopsy procedure [34], and obesity was defined as those with body mass index (BMI) values of ≥30 kg/m2 Finally, metabolic syndrome cases were those with three or more of the following identified FPG ≥110 mg/dL, triglycerides ≥150 mg/dL, HDL ≤40 mg/dL for men or ≤50 mg/dL for women, SBP ≥130 mmHg, DBP ≥85 mmHg, and waist girth ≥90 cm for men ≥80 cm for women [34]. Studies focused predominantly on non-fatal outcomes, with only one of the studies focusing on fatal events (Table 1) [34].
Environmental arsenic exposure
Environmental arsenic exposure was measured in water [26], [27], [28], [29], [30], [31], [32], [33], [34], [40], spot morning [24], 41] or spot [25], [33], [34], [35], [36], [37], [38] urine (adjusted [24], 25], 33], 34], 38], 41] or not adjusted for creatinine [35], [36], [37]), blood [31], 36], and blood plasma [39]. Most studies performed elemental analyses, with some performing speciation analyses (Table 1) [25], [33], [34], [35], [37], 38], 41]. In the SNP-arsenic-cardiometabolic disease analyses, most studies focused on total arsenic (tAs) or inorganic arsenic (iAs) [24], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [[38], [39], [40], [41] with only two studies including analyses exploring the interactions between SNP and arsenic metabolism markers: Primary methylation index [PMI] (defined by MMA/iAs) [25], Secondary Methylation Index (SMI) (defined by DMA/MMA) [25], iAs% [37], MMA% [25], 37], or DMA% [25], 37]. Studies reported predominantly on point arsenic exposure [24], [25], [26], [27], [[30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41] or cumulative arsenic exposure (CAE) [26], 28], 29], and for two studies it was assumed that multiple arsenic measurements were performed [28], 29].
Chemical analyses were performed using inductively couple plasma mass spectrometry (ICP-MS) [32], 36], 38], 39], high-performance liquid chromatography coupled to ICP-MS (HPLC-ICP-MS) [25], 33], 34], 37], 41], high-resolution ICP-MS (HR-ICP-MS) [31], anion exchange liquid chromatography coupled to ICP-MS (AEC-ICP-MS) [38], hydride generation atomic absorption spectroscopy AAS (HG-AAS) [26], 27], 30], 32], 40], graphite furnace AAS (GF-AAS) [24], 31], 33], 34], liquid chromatography-atomic fluorescence spectrometry (LC-AFS) [35] or results from previous reports (without detail on the chemical analyses performed) [28], 29].
From the 18 included studies, some were conducted in areas with high arsenic exposure (i.e., at least one category or the mean was reported or estimated to be >100 μg/L arsenic in water, or the study area was reported to be an arsenic-rich or arsenic hyperendemic area) [28], [29], [30], [31], [32], [33], [34], [36], 38], 39] and others in areas with low-moderate arsenic exposure (i.e., at least one category was reported or estimated to be <100 μg/L arsenic in water, or the study area was reported to have low or moderate environmental arsenic exposure) [24], [25], [26], [27], [[30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41]. Several studies required participants to have a minimum length of residence or unchanged life circumstances to be included in the study (i.e., living in the study area for ≥1 year [24], 36], ≥2 years [40], ≥5 years [31], 33], 34] or ≥6 years [37], or residing in the study area for ≥5 days in the week [29]), whilst other studies followed participants over time or assumed exposure was constant (Table 1).
Genes and single nucleotide polymorphisms
Across studies, 676 unique SNPs in 148 genes were explored. supplementary Table 5 provides an overview of all studied SNPs including alleles and the consequence of the genetic variation. All studies used a hypothesis-driven candidate gene approach selecting candidate genes based on i) their involvement in arsenic metabolism [25], 27], [29], [30], [31], [33], 34], 37], 40], 41], ii) their role in oxidative stress or endogenous defence against reactive oxygen species (ROS) [24], 28], iii) their role related to T2DM metabolic pathways [32], 35], 38], iv), their role in the development of atherosclerosis [26], 29], 30], or v) their role in the endothelin system relevant to obesity [39], vi) if genes have been reported to modify associations between arsenic and CVD in previous epidemiological studies [31], 34], vii) genes where arsenic exposure has been associated with gene products known as risk factors or predictors for CVD [31], 34] or impaired vascular development [36], and ix) if genes have been related to arsenic exposure in animal, in vitro, or epidemiologic studies, and are known to play a key role in CVD risk [33].
Genotyping was performed using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) [24], [25], [26], [27], [28], [30], the TaqMan SNP Genotyping Assay [25], 29], 40], the Illumina GoldenGate asay [31], 33], 34], the oligo-ligation-assay SNPlex [38], 39], the Illumina Cardio-Metabo DNA Analysis BeadChip (MetaboChip) [41], the Sequenome MassARRAY iPLEX [32], a multiplex PCR [35], a high throughput-SNP approach (3-rounds of multiplex PCR with NGS) [37], or using the Improved Multiple Ligase Detection Reaction (iMLDR) method (Table 1) [36].
Quality assessment of the individual studies
Most of the included studies [24], [25], [26], [27], [28], [29], [30], [32], 36], 37], 40] have sample sizes of less than 1000 participants or had a relatively low frequency of the variant allele, which greatly limits the power to detect modest genetic effect sizes. All studies are candidate gene-environment studies, which increases the likelihood of false positive results. Overall, studies varied between the potential for bias, confounding and representativeness of the study population (supplementary Tables 6–8). Within the studies based on cohorts, there was a lack of clarity on the adequacy of the follow-up of participants as well as on the independence of loss to follow-up by exposure group.
In the interaction analyses, most studies adjusted for at least age [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], gender or sex [24], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [[38], [39], [40], [41], smoking status [27], 29], [31], [32], [33], [34], [35], [36], [38], 41], BMI [27], [31], [32], [33], [34], [35], [36], [37], [41], and educational attainment [31], 33], 34], [36], [37], [38], [41], whilst some further adjusted for hypertension or blood pressure [27], [33], [34], [35], [40], diabetes status or blood glucose [27], 31], 33], 34], 36], 41], alcohol use [27], 35], 36], 38], total cholesterol [27], 36], low-density lipoprotein cholesterol (LDL) [28], 36], high-density lipoprotein cholesterol (HDL) [36], triglycerides [28], 36], waist circumference [41], glomerular filtration rate (GFR) [41], C-reactive protein (CRP) [36], ethnicity [37], place of residence [38], betel nut chewing [25], fish consumption [38], obesity [40], renal disease history [24], skin lesions [32], serum folate [37], urinary cotinine levels [38], homocysteine [37], vitamin B2 [41], B6 [41] or B12 [37], change in creatinine between visits [34], urinary lead levels [24], arsenobetaine [37], 38], (change in) urinary total arsenic [33], 37], AS3MT polymorphism [37], 41], and SOD2 polymorphism [24].
Hypertension and blood pressure
The four studies exploring SNPs in arsenic-related hypertension or blood pressure, explored the following candidate genes: APOE, [31] AS3MT, [31] CAT, [28] CBS, [31] CCM3, [36] CYBA, [28], 31] GSTM1, [31] GSTO1, [31] GSTP1, [31]GSTT1, [31] HMOX1, [31] ICAM1, [31] IL6, [31] MTHFR, [31] NOS3, [28], 31] OGG1, [24] PNP, [31] S1PR1, [31] SOD2, [24], 28], 31] TNF, [31] and VCAM1 [31]. See supplementary Table 5 for abbreviations, alleles and consequences, and Table 2 for individual study results). Compared to participants with low urinary arsenic exposure (<8 μg/g creatinine) who were SOD2 or OGG1 reference allele homozygotes, those that were SOD2 (rs4880-C) or OGG1 (rs1052133-G) allele homozygotes or heterozygotes did not have a significantly higher odds of hypertension at low urinary arsenic levels. However, people with higher urinary arsenic levels (≥8 μg/g creatinine) and the reference SOD2 [OR 3.1, 95%CI: 1.2–7.9] or OGG1 homozygous alleles [OR 3.2, 95%CI: 1.4–7.4] had a significantly higher hypertension odds relative to the low urinary arsenic reference, which was even higher in participants who carried the SOD2 (rs4880-C) [OR 4.2, 95%CI: 1.7–10.3, p<0.01] or OGG1 (rs1052133-G) [OR 3.4, 95 % CI:1.1–10.7, p<0.05] alleles [24]. Findings from Hsueh et al. 2007 on CAE further supported these results for the SOD2 rs4880-C allele: hypertension odds for participants with high water CAE (≥10.5 mg/l-year) carrying the SNP was significantly higher (OR 9.0, 95 % CI: 2.3–35.0, p<0.05) compared to SOD2 reference allele homozygotes and low CAE (<10.5 mg/l-yr), whilst the higher hypertension odds for participants with the reference alleles and high CAE was lower at OR 5.7 (95%CI: 1.6–20.9, p<0.05). Furthermore, both NOS3 and CYBA variants were suggested to play a role in susceptibility to arsenic-related hypertension, but not CAT (Table 2 for details) [28]. Evidence from the HEALS study on 235 SNPs in 18 genes [31] found that 44 SNPs interacted with water arsenic for one or more blood pressure outcomes, including SNPs in the aforementioned genes SOD2 (analyses did not include rs4880 or another strong proxy for rs4880 [24], 28]), NOS3 (of which one of the same SNPs as Hsueh et al. [rs1799983 [28]]) and CYBA (analyses did not include rs4673 or another strong proxy [28]) [31]. However, after FDR adjustment, the only variant that had a statistically significant interaction (Q<0.05) with water arsenic and annual pulse pressure was CYBA (rs3794624-A) in recessive genetic models (β123 2.10 mmHg, 95 % CI: 1.01–3.20) (Table 2) [31]. Finally, one study on variants in CCM3 identified interactions between rs9818496, rs3804610 and rs6784267 and arsenic exposure with higher systolic blood pressure (Table 2) [36].
Diabetes mellitus type 2 and gestational diabetes mellitus
Studies on SNPs in arsenic-related T2DM included 128 unique candidate genes (supplementary Table 5) [32], 35], 38], 40], 41]. Two studies that solely focused on different SNPs in the key enzyme catalysing the transfer of methyl groups to arsenic, AS3MT, indicated that carriers of two SNPs (rs10748835, rs11191439) were more likely to develop arsenic-related DM (greater than additive with water arsenic ≥52 ppb and rs11191439-T alleles, approximately multiplicative with water arsenic ≥52 ppb and rs17881215-C alleles) compared to those carrying reference homozygous alleles [40], and that the association between iAs% and DMA% with HOMA2-IR varied depending on the AS3MT (rs12768205) genetic variant (Table 2) [41]. A follow-up of a cancer case-control study found statistically significant interactions (after correction for multiple comparisons) between CDC123 (rs1051055), NOTCH2 (rs699780), and TCF2 (rs2688) – and a nominally significant interaction in THADA (rs1549723) with arsenic-related T2DM when using piecewise linear regression in the interaction model [32]. However, when assessing the subset of participants exposed to water arsenic <148 μg/L, only NOTCH2 (rs699780) remained significant [32]. Interestingly, a non-monotonic dose-response association was found in participants with the NOTCH2 rs699780-C heterozygous or homozygous alleles – whilst the dose-response association in the reference allele homozygotes was approximately linear [32]. Furthermore, a study on 354 SNPs found no significant interactions with total urinary arsenic and the included SNPs after correction for multiple testing. However, some nominally significant interactions were observed: IL8RA (rs1008563 [p-value 0.004], rs1008562 [p-value 0.01]), TXN (rs4135168 [p-value 0.004]), NR3C2 (rs13117325 [p-value 0.007], (rs2137335) [p-value 0.01]), COX5A (rs1133322 [p-value 0.01]) and GCLC (rs11415624 [p-value 0.01]) (Table 2) [38].
One cross-sectional study on the combined effects of N6AMT1 and AS3MT SNPs and arsenic exposure on GDM odds found no statistically significant combined effects between AS3MT SNPs and arsenic metabolism markers [37]. Odds of GDM were greater among those carrying the N6AMT1 rs1997605-G/ rs100367-G homozygous alleles and lower urinary iAs%, lower MMA% and greater DMA% compared to those carrying the reference homozygous and heterozygous alleles. However, additive statistically significant interactions were only found between N6AMT1 rs1997605/rs1003671 risk genotypes and lower iAs% or higher DMA% (Table 2) [37].
Carotid atherosclerosis and carotid intima-media thickness
Four studies explored genetic susceptibility to arsenic in relation to carotid atherosclerosis [26], 27], 30] or CIMT [33] in genes encoding well-known proteins involved in arsenic metabolism (AS3MT, [27], 33] PNP [27], 33] and members of the glutathione S-transferase (GST) superfamily of enzymes [GSTM1, [30], 33] GSTO1, [27], 33] GSTO2, [27] GSTP1, [30], 33] GSTT1, [30], 33]), oxidative stress (HMOX1, [33] NOS3, [33] SOD2, [33] CYBA [33]), or inflammation and endothelial function (APOE, [33] TNF, [33] IL6, [33] ICAM1, [33] S1PR1, [33] VCAM1 [33]), as well as a tumour-suppressive gene TP53 [30] (which may play a role in vascular apoptosis), and the combined effect of variants in APOE and MCP-1 [26].
While none were statistically significant after adjusting for multiple testing, there were 9 SNPs in APOE, AS3MT, PNP and TNF that had a nominally statistically significant interaction with water arsenic in CIMT (Table 2), with joint presence of the risk genotype and arsenic exposure having a much greater difference in CIMT in comparison to the risk genotype or high arsenic exposure alone [33]. In genotype and haplotype analyses, participants with water arsenic exposure >50 μg/L had a significant or nominally significant interaction for carotid atherosclerosis risk with the PNP A-T haplotype (listed rs1049564, rs1130650) [OR 2.08; 95%CI, 1.32–3.30, P int 0.006], the AS3MT rs11191439 genotypic variant (OR 2.63, 95%CI: 1.00–6.92, P int 0.075), and the GST A-G-G haplotype (listed rs662, rs705379, rs7493) [OR 1.62; 95 % CI, 1.05–2.50, P int 0.045] compared to a reference of arsenic exposure <50 μg/L and the reference haplotype/genotype [27]. Those who had both risk genotypes of APOE and MCP-1 and had either high water arsenic exposure (>10 μg/L) or high cumulative arsenic exposure (>0.22 mg/L-year), likewise, had higher odds compared to participants who carried reference genotypes and had low arsenic exposure, with ORs of 10.3 (1.0–102.5) and 15.7 (1.7–141.2) respectively [26]. Further analyses indicate that participants with the GSTP1 rs1695 or TP53 rs1042522 exposed to water arsenic >50 μg/L had significantly higher odds of carotid atherosclerosis, with ORs of 6.0 (95%CI: 3.3–10.7) and 3.1 (95%CI: 1.7–5.7) [highly significant synergistic effect] respectively, compared to participants with the reference alleles and water arsenic ≤50 μg/L – whilst participants below ≤50 μg/L arsenic exposure had no significant difference in risk and those at >50 μg/L arsenic exposure with reference alleles had lower ORs of 2.7 (95%CI: 1.5–4.5) for GSTP1 and 1.4 (95%CI: 0.7–3.0) for TP53 respectively (Table 2) [30].
Metabolic syndrome
One study focused on metabolic syndrome and the GSTO1 rs4925 SNP [25], which was the only study to explore SNP-arsenic-disease interactions utilising arsenic metabolism markers PMI, SMI, MMA% and DMA% (other studies only explored tAs or iAs). The study findings suggest that participants with lower PMI and the GSTO1 rs4925-D homozygous plus heterozygous alleles had significantly higher odds of MetS (OR 4.00, 95%CI: 1.65–9.71, p=0.002) compared to those with higher PMI and reference homozygous alleles – with those exposed to lower PMI and carrying reference alleles having lower MetS odds (OR 2.03, 95%CI: 1.03–4.01, p=0.04) [25]. Lower MMA%, and higher SMI or DMA% were also associated with MetS – with some suggestive evidence that the association may be modified by GSTO1 genotype (Table 2) [25].
Obesity
One study focused on arsenic-related obesity and genetic variation in genes of the Endothelin (EDN) system (EDN1, EDN2, EDN3, EDNRA, EDNRB), selected as i) endothelin is able to regulate adiponectin levels (thus may impact obesity), and ii) EDN receptors have been reported as potential factors mediating arsenic-related adipocyte dysfunction [39]. Two SNPs in the EDNRB gene (rs3759475, rs5351) had lower odds of obesity in participants carrying the EDNRB variant alleles compared to the reference homozygous alleles at high blood arsenic levels (≥50 nmol/L), with ORs of 0.51 (95 % CI: 0.32–0.82) and 0.53 (95%CI: 0.33–0.85) for those carrying rs5351-A and rs3759475-C homozygous plus heterozygous alleles respectively [39]. However, the associations were not statistically significant at medium (20–50 nmol/L) or low (≤20 nmol/L) arsenic levels (Table 2) [39].
Cardiovascular disease and ECG abnormalities
The two studies with a focus on arsenic with CVD (including CHD, stroke) or ECG abnormalities (in line with MI or ischemia, conduction defect, arrhythmia, atrial enlargement or ventricular hypertrophy, prolonged ventricular repolarization) included the following candidate genes: APOE, [34] AS3MT, [29], 34] CBS, [34] CYBA, [34] GSTM1, [34] GSTO1, [29], 34] GSTO2 [29] GSTP1, [34] GSTT1, [34] HMOX1, [34] ICAM1, [34] IL6, [34] MTHFR, [34] NOS3, [34] PNP, [34] PON1, [29] PON2 [29], S1PR1, [34] SOD2, [34] TNF, [34] and VCAM1. [34] A constructed PON R-C-S haplotype [PON1 (rs662), PON1 (rs705379), PON2 (rs7493)] had synergistic interaction effects with cumulative arsenic exposure and ECG abnormalities. Illustratively, those with the R-C-S haplotype and cumulative arsenic exposure >14.7 ppm-yr had a statistically significant (p=0.014) higher odds (OR 19.19, 95%CI: 1.86–197.76) when compared to those without the R-C-S haplotype exposed to ≤14.7 ppm-yr cumulative arsenic, whilst those with the R-C-S haplotype and ≤14.7 ppm-yr cumulative arsenic exposure and those without the haplotype and >14.7 ppm-yr cumulative arsenic exposure did not have any statistically significant higher odds of ECG abnormality (OR 1.57, 95%CI: 0.19–13.00; and 4.27, 95%CI: 0.83–22.08 respectively) [29]. Furthermore, a case-cohort study nested in HEALS found only two (out of 170) SNPs had statistically significant multiplicative interactions for water arsenic related CVD. Participants with the ICAM (rs281432) and VCAM1 (rs3176867) variant alleles and ≥ 45 μg/L water arsenic exposure had adjusted hazard ratios (aHRs) of 2.98 (95%CI: 1.87–4.77) and 2.13 (95%CI: 1.37–3.31) respectively compared with those carrying the reference ICAM and VCAM1 alleles and <45 μg/L water arsenic exposure. These aHRs were higher than participants with ≥45 μg/L not carrying the variants, who had aHRs of 1.67 (95%CI: 1.14–2.43) and 1.30 (95%CI: 0.83–2.05) for ICAM and VCAM1 respectively (Table 2) [34].
Overall results
Table 3 summarises all the SNPs which were found to have statistically significant interactions with arsenic exposure in relation to cardiometabolic outcomes. Other studies exploring the same SNP but reporting a non-significant result (for the same or a different cardiometabolic outcome), were also listed to provide appropriate context. For each gene (product) information is provided on the biological roles and processes the gene (product) is thought to be involved in (Figure 2, Table 3).
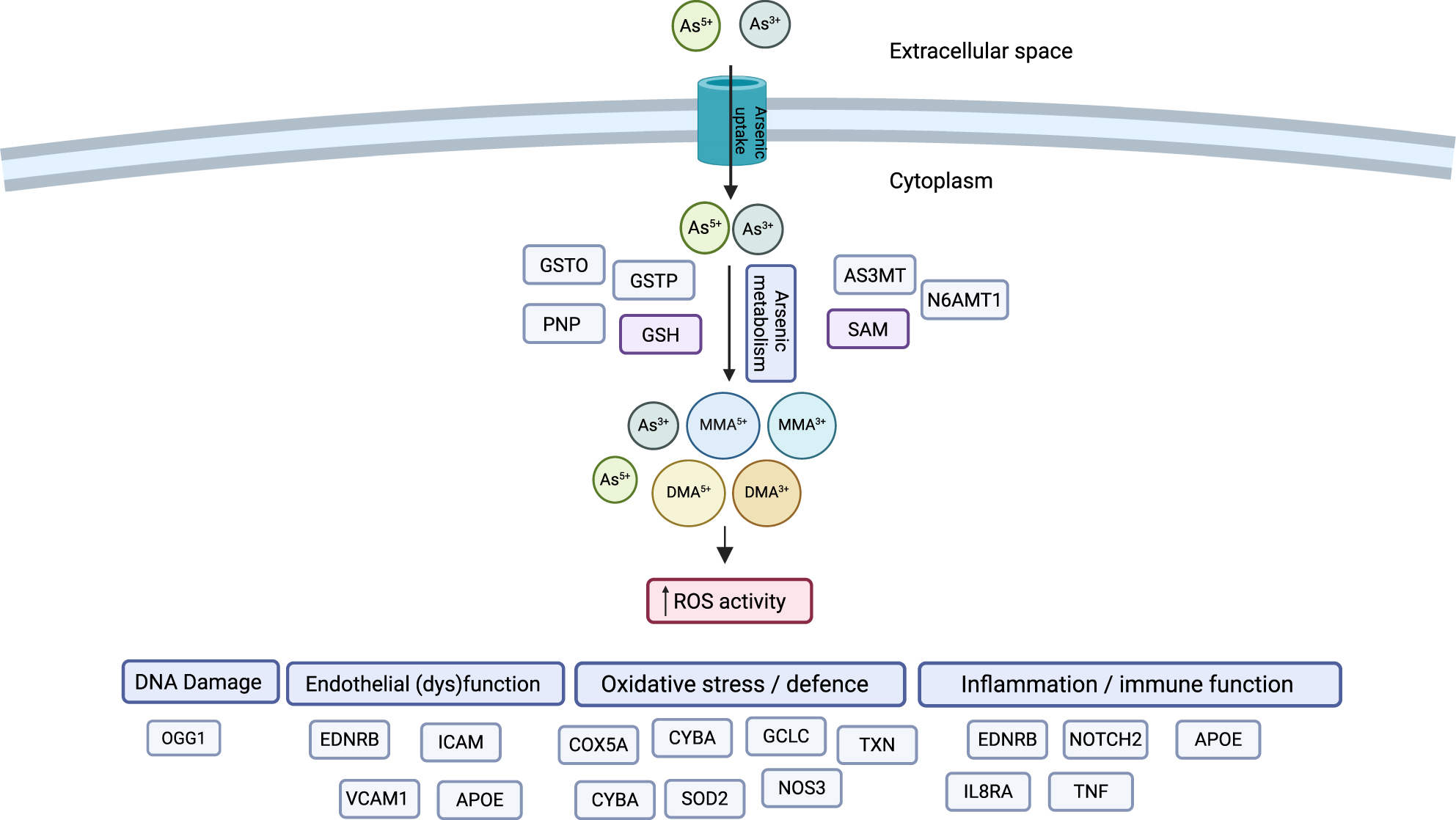
Genes and molecular pathways reported to interact with arsenic exposure to affect risk of arsenic-related adverse cardiometabolic outcomes. After ingestion, iAs (predominantly iAs5+ in drinking water) is absorbed in the gastro-intestinal tract and undergoes a series of reduction-oxidation and methylation steps to generate monomethylated arsenic (MMA) and, subsequently, dimethylated arsenic (DMA) species, whereby DMA5+ is the human biomethylation end-product. Biological mechanisms underlying adverse cardiometabolic outcomes have largely been hypothesised to be related to an increase in reactive oxygen species (ROS) activity. Different molecular pathways (arsenic metabolism, DNA damage, endothelial (dys)function, oxidative stress / defence, inflammation / immune function) are in dark blue and genes for which SNPs were identified to interact with arsenic exposure on risk adverse arsenic-related cardiometabolic outcomes are in light blue/grey (Table 3).
Discussion
To our knowledge, this is the first systematic review synthesising the evidence on genetic susceptibility to adverse arsenic-related cardiometabolic outcomes (i.e., genetic interactions with arsenic exposure). Across 676 SNPs in 148 genes assessed in 18 candidate gene studies, 40 SNPs in 24 genes, 4 haplotypes and combined SNPs in MCP-1/APOE, were indicative of SNP-arsenic interactions associated with adverse cardiometabolic outcomes. Polymorphisms that were reported to significantly interact with arsenic in relation to cardiometabolic outcomes or to result in higher/lower susceptibility to arsenic, were predominantly affecting genes involved in arsenic metabolism (AS3MT, GSTO1, GSTP1, N6AMT1, PNP), oxidative stress or defence (COX5A, CYBA, GCLC, NOS3, SOD2, TXN), DNA damage repair (OGG1), endothelial (dys)function, inflammation or immune function (APOE, EDNRB, NOTCH2, IL8RA, ICAM, MCP-1, TNF, VCAM1), tumour suppressor activity (TP53), or have been previously implicated in influencing the cardiovascular related physiological response (CCM3), diabetes mellitus (CDC123, COX5A, ILRA8 NR3C2, TCF2), or atherosclerosis (PON, TP53). Importantly, most SNPs were only investigated in a single study and only one SNP (SOD2 rs4880) had been tested for replication with the same cardiometabolic outcome (hypertension), albeit different arsenic exposure metrics (urinary arsenic [24], water CAE [28]). Overall, there is a lack of high-quality studies exploring the interactions between genetic variations and environmental arsenic exposure in relation to cardiometabolic diseases, as well as a shortage of replicated findings across existing studies.
In 2012 the first genome-wide association study (GWAS) on arsenic-related toxicity phenotypes was performed in the HEALS study [43], identifying two genetic variants in AS3MT (rs9527, rs11191527) that had independent associations with arsenic metabolism (appearing to impact DMA production [44]), of which one (rs9527) was also shown to interact with arsenic to influence incident skin lesion risk [43]. A more recent HEALS GWAS reported suggestive evidence for an additive interaction with arsenic for a protein-altering variant of forminotransferase cyclodeaminase [FTCD] (rs61735836) – a SNP located in a gene important for histidine catabolism which generates one-carbon units [9]. These studies provide evidence for the relevance of arsenic metabolism in arsenic toxicity phenotypes, as well as the involvement of the one-carbon/folate cycle in arsenic metabolism and toxicity [9], 43], 45]. However, the candidate genes studies identified in this review reported only nominally significant SNP-arsenic interactions for AS3MT (rs9527) and for SNPs in other genes involved in arsenic metabolism in relation to CVD and DBP (which did not remain significant after multiple comparisons adjustment) [31], 34], no significant arsenic-SNP interaction for AS3MT (rs11191527) with CVD [34], and did not include FTCD (rs61735836) in any study. Nevertheless, the current finding that multiple SNPs in A3SMT interact with arsenic exposure in relation to cardiometabolic outcomes [33], 40], 41], coupled with existing epidemiological and experimental evidence on the role of arsenic metabolism in disease progression [46], 47], suggests that genetic variants in the arsenic metabolism pathway likely modulate arsenic’s effect on cardiometabolic disease. Involvement of the exact genetic variants influencing arsenic metabolism need to be further confirmed in studies with substantially greater power to detect interaction effects relevant to cardiometabolic health.
Within this study, several genes related to oxidative stress have been proposed to be involved in the differential susceptibility to arsenic-related cardiometabolic disease. For example, in two studies CYBA, which encodes a subunit of the NADPH oxidase involved in ROS generation, was found to have a significant interaction with arsenic exposure in relation to hypertension and blood pressure [28], 31]. Interestingly, in vitro studies have indicated that NADPH oxidase activity and subcellular localisation can also be influenced by arsenic [48], 49]. Furthermore, two other studies identified potential interaction effects between SOD2, a superoxide dismutase, and arsenic in relation to hypertension [24], 28] – suggesting SOD2 may limit mitochondrial defence capacity against ROS. Considering that arsenic is able to induce the generation of ROS, and ROS has been implicated in atherosclerosis and CVD development [50], it seems biologically plausible that genetic variants affecting oxidative stress may influence the effect of arsenic on cardiometabolic outcomes [34].
Importantly, all gene-arsenic studies identified, focused on candidate gene SNPs that were selected a priori, which should be evaluated with caution [51], [52], [53]. By selecting genes a priori, the roles of other (unknown) SNPs potentially relevant for arsenic susceptibility cannot be included. Notably, in well-powered GWAS analyses of complex phenotypes, robust, replicable, signals were often different from genes hypothesised to be related to an outcome in candidate gene studies [54], [55], [56]. Thus, candidate genes previously hypothesised to be implicated in a disease have often shown limited evidence for association in well-powered large GWAS. In absence of additional supporting evidence, this may suggest that candidate genes hypothesised to influence arsenic susceptibility in relation to cardiometabolic disease may not be “truly” associated with arsenic-related cardiometabolic disease [51], 52]. Furthermore, the gene-arsenic candidate gene studies described in this review are likely substantially underpowered to observe “true” associations of the small effect sizes in complex traits or diseases (candidate gene-environment studies [cGxE] studies often assume larger effect sizes than appropriate when basing the effect size on the environmental exposure of the effect) [52]. Whilst this may seem to be in contrast with the statistically significant findings reported in this review, candidate gene studies are simultaneously inherently prone to false positives as they focus on a small number of variants out of millions of possibilities. Given this low prior probability, strong statistical evidence is required – including the use of appropriate multiple testing thresholds and replication of findings across different studies [51], 52], 57]. Yet, across the included studies there was a complete lack of replication of observed SNP-arsenic interactions for the same cardiometabolic outcome, and multiple testing adjustments were often only made for all SNPs included in the study instead of all plausible genetic variants. Studies that did explore the same SNP but on different outcomes or arsenic species did not consistently report results in the same direction. Thirdly, cGxE studies are often marred by statistical inaccuracies that most cGxE studies do not account for: i) transformations of the arsenic exposure scale can alter evidence of multiplicative interaction terms [51], ii) covariate-gene and covariate-arsenic terms are often not included in models, preventing the proper adjustment of the impact of covariates on interaction terms [51], and iii) multiple comparisons may make erroneous inferences more likely which needs to properly adjusted for [51], 52].
Limitations in the ascertainment of arsenic exposure can also influence uncertainty levels when assessing the arsenic-SNP interactions. For example; i) the use of environmental media may not accurately reflect “true” arsenic exposure due to the exclusion of other exposure routes, ii) studies using elemental analysis instead of speciation analysis may not accurately account for arsenic species considered non-toxic (e.g., arsenobetaine), iii) studies that measured arsenic at one time-point without taking into consideration changes in exposure may have an inaccurate perception of “true” chronic exposure, or iv) studies that measured arsenic at a community, county or state level or estimated arsenic based on a geographical location may not accurately reflect individual exposure. Such potential exposure misclassifications could result in the underestimation of the assessed associations if the misclassification was non-differential by the outcome of interest.
This systematic review has various limitations. First, the available evidence was limited in quality and replication, complicating robust information synthesis and preventing the performance of meta-synthesis. The lack of GWAS of SNP-arsenic interactions on cardiometabolic outcomes has prevented robust discovery. Secondly, most studies were conducted in Taiwan and Bangladesh. This may influence the generalisability of results across different study populations as different populations may have different genetic ancestry, and demographic histories including arsenic exposure. Likewise, the lack of ethnically diverse populations may risk missing genetic variations that increase arsenic tolerance. For example, some Andean populations that have been exposed to arsenic over the past 7000 years are reported to carry genetic variations that increase arsenic metabolism efficiency and reduce susceptibility [58], 59]. Thirdly, whilst this systematic review was not able to formally assess the influence of publication bias, the presence of publication bias (i.e., lack of publication of negative or null results, including unpublished results in publications that were published) is likely increasing the number of false-positive gene-arsenic findings [51], 52]. Despite this, this systematic review has several strengths, including a broad definition of cardiometabolic outcomes, a detailed comprehensive search strategy to gather available evidence, and no restriction in terms of language or publication date.
Conclusions
There is a lack of high-quality studies examining the interplay between genetic variations and environmental arsenic exposure in relation to cardiometabolic diseases, along with a lack of replicated results across current research. Variants proposed to contribute to differential arsenic susceptibility were found in genes related to arsenic metabolism, oxidative stress and defense, endothelial function, DNA repair, tumor suppression, inflammation or immune response, or were previously linked to cardiometabolic diseases. Whilst the involvement of many of these pathways appear biologically plausible, further research in large prospective cohort studies that use genome wide GxE analyses and are conducted in ethnographically diverse cohorts, combined with in vivo and in vitro studies that identify or confirm the functional impacts of identified SNPs, are needed to further understand the interplay between arsenic and genotypic variants in relation to cardiometabolic disease. Furthermore, considering the polygenic nature of cardiometabolic outcomes and the low statistical power in single marker GWAS, further research should consider the use of genome-wide composite measures (polygenic scores [PGSs]) in GxE analyses. [60].
Funding source: Gates Cambridge Trust
Award Identifier / Grant number: OP114
Funding source: BHF Cambridge Centre for Research Excellence
Award Identifier / Grant number: RE/24/130011
-
Research ethics: Not applicable.
-
Informed consent: Not applicable.
-
Author contributions: KRvD conceived the presented idea and developed the research protocol with support from AB and CW. KRvD conducted the literature searches with support from a medical librarian. KRvD conducted all title and abstract screening, full-text screening, data extraction, and quality assessment. JK, MJ and NK acted as second screeners, data extractors and quality assessors. KRvD drafted the initial manuscript, tables, and figures with support of all other co-authors. AB and CW provided critical feedback and guidance throughout the project. All authors reviewed the manuscript and contributed intellectually to the project.
-
Use of Large Language Models, AI and Machine Learning Tools: Not applicable.
-
Conflict of interest: The authors have no conflict of interest to declare.
-
Research funding: KRvD received funding from the Gates Cambridge Trust (OP114) for her PhD studies, and received funding for publication of this article from the Gates Foundation. KRvD is funded by the BHF Cambridge Centre for Research Excellence RE/24/130011.This work was supported by core funding from the British Heart Foundation (RG/18/13/33946: RG/F/23/110103), NIHR Cambridge Biomedical Research Centre (NIHR203312) [*], BHF Chair Award (CH/12/2/29428), and by Health Data Research UK, which is funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation and the Wellcome Trust. *The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care.
-
Data availability: All data is available in the publication and its supplement materials.
References
1. Moon, KA, Oberoi, S, Barchowsky, A, Chen, Y, Guallar, E, Nachman, KE, et al.. A dose-response meta-analysis of chronic arsenic exposure and incident cardiovascular disease. Int J Epidemiol 2017;46:1924–39. https://doi.org/10.1093/ije/dyx202.Suche in Google Scholar PubMed PubMed Central
2. Navas-Acien, A, Silbergeld, EK, Streeter, RA, Clark, JM, Burke, TA, Guallar, E. Arsenic exposure and type 2 diabetes: A systematic review of the experimental and epidemiologic evidence. Env Heal Perspect 2006;114:641–8. https://doi.org/10.1289/ehp.8551.Suche in Google Scholar PubMed PubMed Central
3. Abhyankar, LN, Jones, MR, Guallar, E, Navas-Acien, A. Arsenic exposure and hypertension: A systematic review. Environ Health Perspect 2012;120:494–500. https://doi.org/10.1289/ehp.1103988.Suche in Google Scholar PubMed PubMed Central
4. Minatel, BC, Sage, AP, Anderson, C, Hubaux, R, Marshall, EA, Lam, WL, et al.. Environmental arsenic exposure: From genetic susceptibility to pathogenesis. Environ Int 2018;112:183–97. https://doi.org/10.1016/j.envint.2017.12.017.Suche in Google Scholar PubMed
5. Pierce, BL, Kibriya, MG, Tong, L, Jasmine, F, Argos, M, Roy, S, et al.. Genome-wide association study identifies chromosome 10q24.32 variants associated with arsenic metabolism and toxicity phenotypes in Bangladesh. PLoS Genet 2012;8:e1002522. https://doi.org/10.1371/journal.pgen.1002522.Suche in Google Scholar PubMed PubMed Central
6. Gribble, MO, Voruganti, VS, Cole, SA, Haack, K, Balakrishnan, P, Laston, SL, et al.. Linkage analysis of urine arsenic species patterns in the strong heart family study. Toxicol Sci 2015;148:89–100. https://doi.org/10.1093/toxsci/kfv164.Suche in Google Scholar PubMed PubMed Central
7. Agusa, T, Fujihara, J, Takeshita, H, Iwata, H. Individual variations in inorganic arsenic metabolism associated with AS3MT genetic polymorphisms. Int J Mol Sci 2011;12:2351–82. https://doi.org/10.3390/ijms12042351.Suche in Google Scholar PubMed PubMed Central
8. Minatel, BC, Sage, AP, Anderson, C, Hubaux, R, Marshall, EA, Lam, WL, et al.. Environmental arsenic exposure: From genetic susceptibility to pathogenesis. Environ Int 2018;112:183–97. https://doi.org/10.1016/j.envint.2017.12.017.Suche in Google Scholar
9. Pierce, BL, Tong, L, Dean, S, Argos, M, Jasmine, F, Rakibuz-Zaman, M, et al.. A missense variant in FTCD is associated with arsenic metabolism and toxicity phenotypes in Bangladesh. Barsh GS. PLoS Genet 2019;15:e1007984. https://doi.org/10.1371/journal.pgen.1007984.Suche in Google Scholar PubMed PubMed Central
10. Hsu, L-I, Chen, W-P, Yang, T-Y, Chen, Y-H, Lo, W-C, Wang, Y-H, et al.. Genetic polymorphisms in glutathione S-transferase (GST) superfamily and risk of arsenic-induced urothelial carcinoma in residents of southwestern Taiwan. J Biomed Sci 2011;18. https://doi.org/10.1186/1423-0127-18-51.Suche in Google Scholar PubMed PubMed Central
11. De Chaudhuri, S, Ghosh, P, Sarma, N, Majumdar, P, Sau, TJ, Basu, S, et al.. Genetic variants associated with arsenic susceptibility: Study of purine nucleoside phosphorylase, arsenic (+3) methyltransferase, and glutathione S-transferase omega genes. Environ Health Perspect 2008;116:501–5. https://doi.org/10.1289/ehp.10581.Suche in Google Scholar PubMed PubMed Central
12. Ahsan, H, Chen, Y, Kibriya, MG, Islam, MN, Slavkovich, VN, Graziano, JH, et al.. Susceptibility to arsenic-induced hyperkeratosis and oxidative stress genes myeloperoxidase and catalase. Cancer Lett 2003;201:57–65. https://doi.org/10.1016/S0304-3835(03)00471-3.Suche in Google Scholar PubMed
13. Thirumaran, RK, Bermejo, JL, Rudnai, P, Gurzau, E, Koppova, K, Goessler, W, et al.. Single nucleotide polymorphisms in DNA repair genes and basal cell carcinoma of skin. Carcinogenesis 2006;27:1676–81. https://doi.org/10.1093/carcin/bgi381.Suche in Google Scholar PubMed
14. Page, MJ, McKenzie, JE, Bossuyt, PM, Boutron, I, Hoffmann, TC, Mulrow, CD, et al.. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021;372. https://doi.org/10.1136/BMJ.N71.Suche in Google Scholar PubMed PubMed Central
15. Moola, S, Munn, Z, Sears, K, Sfetcu, R, Currie, M, Lisy, K, et al.. Conducting systematic reviews of association (etiology): The Joanna Briggs Institute’s approach. Int J Evid Based Healthc 2015;13:163–9. https://doi.org/10.1097/XEB.0000000000000064.Suche in Google Scholar PubMed
16. Minatel, BC, Sage, AP, Anderson, C, Hubaux, R, Marshall, EA, Lam, WL, et al.. Environmental arsenic exposure: From genetic susceptibility to pathogenesis. Environ Int 2018;112:183–97. https://doi.org/10.1016/J.ENVINT.2017.12.017.Suche in Google Scholar
17. Whelton, PK, Carey, RM, Aronow, WS, Casey, DE, Collins, KJ, Himmelfarb, CD, et al.. ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ ASH/ASPC/NMA/PCNA guideline for the prevention, detection, evaluation, and management of high blood pressure in adults a report of the American College of Cardiology/American Heart Association Task Force on Clinical practice guidelines. Hypertension 2018;71:E13–115. https://doi.org/10.1161/HYP.0000000000000065/-/DC2.Suche in Google Scholar
18. Williams, B, Mancia, G, Spiering, W, Agabiti Rosei, E, Azizi, M, Burnier, M, et al.. ESC/ESH Guidelines for the management of arterial hypertensionThe Task Force for the management of arterial hypertension of the European Society of Cardiology (ESC) and the European Society of Hypertension (ESH). Eur Heart J 2018;39: 3021–104. https://doi.org/10.1093/EURHEARTJ/EHY339.Suche in Google Scholar
19. World Health Organization. Classification of diabetes mellitus 2019. 2019.Suche in Google Scholar
20. Association American Diabetes. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes–2021. Diabetes Care 2021;44:S15–33. https://doi.org/10.2337/DC21-S002.Suche in Google Scholar PubMed
21. Diagnosis in adults | Diagnosis | Diabetes - type 2 | CKS | NICE. [cited 17 Jul 2022]. Available: https://cks.nice.org.uk/topics/diabetes-type-2/diagnosis/diagnosis-in-adults/.Suche in Google Scholar
22. Wells, G, Shea, B, O’Connell, D, Peterson, J, Welch, V, Losos, M, et al.. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. OHRI. [cited 12 Aug 2021]. Available: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.Suche in Google Scholar
23. Sohani, ZN, Meyre, D, de Souza, RJ, Joseph, PG, Gandhi, M, Dennis, BB, et al.. Assessing the quality of published genetic association studies in meta-analyses: the quality of genetic studies (Q-Genie) tool. BMC Genet 2015;16. https://doi.org/10.1186/S12863-015-0211-2.Suche in Google Scholar PubMed PubMed Central
24. Chen, SC, Chen, CC, Kuo, CY, Huang, CH, Lin, CH, Lu, ZY, et al.. Elevated risk of hypertension induced by arsenic exposure in Taiwanese rural residents: possible effects of manganese superoxide dismutase (MnSOD) and 8-oxoguanine DNA glycosylase (OGG1) genes. Arch Toxicol 2012;86:869–78. https://doi.org/10.1007/S00204-011-0797-8.Suche in Google Scholar PubMed
25. Chen, JW, Wang, SL, Wang, YH, Sun, CW, Huang, YL, Chen, CJ, et al.. Arsenic methylation, GSTO1 polymorphisms, and metabolic syndrome in an arseniasis endemic area of southwestern Taiwan. Chemosphere 2012;88:432–8. https://doi.org/10.1016/J.CHEMOSPHERE.2012.02.059.Suche in Google Scholar
26. Hsieh, Y-C, Hsieh, F-I, Lien, L-M, Chou, Y-L, Chiou, H-Y, Chen, C-J. Risk of carotid atherosclerosis associated with genetic polymorphisms of apolipoprotein E and inflammatory genes among arsenic exposed residents in Taiwan. 2008;227:1–7 [cited 11 Jul 2023]. https://doi.org/10.1016/j.taap.2007.10.013.Suche in Google Scholar PubMed
27. Hsieh, YC, Lien, LM, Chung, WT, Hsieh, FI, Hsieh, PF, Wu, MM, et al.. Significantly increased risk of carotid atherosclerosis with arsenic exposure and polymorphisms in arsenic metabolism genes. Environ Res 2011;111:804–10. https://doi.org/10.1016/J.ENVRES.2011.05.003.Suche in Google Scholar PubMed
28. Hsueh, YM, Lin, PP, Chen, HW, Shiue, HS, Chung, CJ, Tsai, CT, et al.. Genetic polymorphisms of oxidative and antioxidant enzymes and arsenic-related hypertension. J Toxicol Environ Heal – Part A 2005;68:1471–84. https://doi.org/10.1080/15287390590967414.Suche in Google Scholar PubMed
29. Liao, YT, Li, WF, Chen, CJ, Prineas, RJ, Chen, WJ, Zhang, ZM, et al.. Synergistic effect of polymorphisms of paraoxonase gene cluster and arsenic exposure on electrocardiogram abnormality. Toxicol Appl Pharmacol 2009;239:178–83. https://doi.org/10.1016/J.TAAP.2008.12.017.Suche in Google Scholar
30. Wang, Y-H, Wu, M-M, Hong, C-T, Lien, L-M, Hsieh, Y-C, Tseng, H-P, et al.. Effects of arsenic exposure and genetic polymorphisms of p53, glutathione S-transferase M1, T1, and P1 on the risk of carotid atherosclerosis in Taiwan. Atherosclerosis 2007;192:305–12. https://doi.org/10.1016/j.atherosclerosis.2006.07.029.Suche in Google Scholar PubMed
31. Farzan, SF, Karagas, MR, Jiang, J, Wu, F, Liu, M, Newman, JD, et al.. Gene-arsenic interaction in longitudinal changes of blood pressure: Findings from the Health Effects of Arsenic Longitudinal Study (HEALS) in Bangladesh. Toxicol Appl Pharmacol 2015;288:95–105. https://doi.org/10.1016/j.taap.2015.07.017.Suche in Google Scholar PubMed PubMed Central
32. Pan, WC, Kile, ML, Seow, WJ, Lin, X, Quamruzzaman, Q, Rahman, M, et al.. Genetic susceptible locus in NOTCH2 interacts with arsenic in drinking water on risk of type 2 diabetes. PLoS One 2013;8:e70792. https://doi.org/10.1371/journal.pone.0070792.Suche in Google Scholar PubMed PubMed Central
33. Wu, F, Jasmine, F, Kibriya, MG, Liu, M, Cheng, X, Parvez, F, et al.. Interaction between arsenic exposure from drinking water and genetic susceptibility in carotid intima-media thickness in Bangladesh. Toxicol Appl Pharmacol 2014;276:195–203. https://doi.org/10.1016/j.taap.2014.02.014.Suche in Google Scholar PubMed PubMed Central
34. Wu, F, Jasmine, F, Kibriya, MG, Liu, M, Cheng, X, Parvez, F, et al.. Interaction between arsenic exposure from drinking water and genetic polymorphisms on cardiovascular disease in Bangladesh: A prospective case-cohort study. Environ Health Perspect 2015;123:451–7. https://doi.org/10.1289/EHP.1307883.Suche in Google Scholar PubMed PubMed Central
35. Fan, C, Zhan, Z, Zhang, X, Lou, Q, Guo, N, Su, M, et al.. Research for type 2 diabetes mellitus in endemic arsenism areas in central China: role of low level of arsenic exposure and KEAP1 rs11545829 polymorphism. Arch Toxicol 2022;96:1673–83. https://doi.org/10.1007/S00204-022-03279-1/TABLES/5.Suche in Google Scholar
36. Gao, Y, Zhao, Z, Yang, L, Liu, X, Xing, X, Zhang, H, et al.. Arsenic exposure assists ccm3 genetic polymorphism in elevating blood pressure. Oncotarget 2018;9:4915–23. https://doi.org/10.18632/oncotarget.23518.Suche in Google Scholar PubMed PubMed Central
37. Liang, X, Guo, G, Wang, Y, Wang, M, Chen, X, Zhang, J, et al.. Arsenic metabolism, N6AMT1 and AS3MT single nucleotide polymorphisms, and their interaction on gestational diabetes mellitus in Chinese pregnant women. Environ Res 2023;221:115331. https://doi.org/10.1016/J.ENVRES.2023.115331.Suche in Google Scholar PubMed
38. Grau-Perez, M, Navas-Acien, A, Galan-Chilet, I, Briongos-Figuero, LS, Morchon-Simon, D, Bermudez, JD, et al.. Arsenic exposure, diabetes-related genes and diabetes prevalence in a general population from Spain. Environ Pollut 2018;235:948–55. https://doi.org/10.1016/j.envpol.2018.01.008.Suche in Google Scholar PubMed PubMed Central
39. Martínez-Barquero, V, De Marco, G, Martínez-Hervas, S, Rentero, P, Galan-Chilet, I, Blesa, S, et al.. Polymorphisms in Endothelin System Genes, Arsenic Levels and Obesity Risk. PLoS One 2015;10:e0118471. https://doi.org/10.1371/JOURNAL.PONE.0118471.Suche in Google Scholar PubMed PubMed Central
40. Drobná, Z, Del Razo, LM, García-Vargas, GG, Sánchez-Peña, LC, Barrera-Hernández, A, Stýblo, M, et al.. Environmental exposure to arsenic, AS3MT polymorphism and prevalence of diabetes in Mexico. J Expo Sci Environ Epidemiol 2013;23:151–5. https://doi.org/10.1038/jes.2012.103.Suche in Google Scholar PubMed PubMed Central
41. Grau-Perez, M, Kuo, C-C, Gribble, MO, Balakrishnan, P, Spratlen, MJ, Vaidya, D, et al.. Association of low-moderate arsenic exposure and arsenic metabolism with incident diabetes and insulin resistance in the strong heart family study. Environ Health Perspect 2017;125. https://doi.org/10.1289/EHP2566.Suche in Google Scholar PubMed PubMed Central
42. Wu, HY, Chen, KP, Tseng, WP, Hsu, CL. Epidemiologic studies on blackfoot disease. I. Prevalence and incidence of the disease by age, sex, occupation and geographical distribution. Mem Coll Med Natl Taiwan Univ 1961;7:33–50.Suche in Google Scholar
43. Pierce, BL, Kibriya, MG, Tong, L, Jasmine, F, Argos, M, Roy, S, et al.. Genome-Wide Association Study Identifies Chromosome 10q24.32 Variants Associated with Arsenic Metabolism and Toxicity Phenotypes in Bangladesh. PLOS Genet 2012;8. https://doi.org/10.1371/journal.pgen.1002522.Suche in Google Scholar PubMed PubMed Central
44. Jansen, RJ, Argos, M, Tong, L, Li, J, Rakibuz-Zaman, M, Islam, MT, et al.. Determinants and consequences of arsenic metabolism efficiency among 4,794 individuals: Demographics, lifestyle, genetics, and toxicity. Cancer Epidemiol Biomarkers Prev 2016;25:381–90. https://doi.org/10.1158/1055-9965.EPI-15-0718.Suche in Google Scholar PubMed PubMed Central
45. Pierce, BL, Tong, L, Argos, M, Gao, J, Jasmine, F, Roy, S, et al.. Arsenic metabolism efficiency has a causal role in arsenic toxicity: Mendelian randomization and gene-environment interaction. Int J Epidemiol 2013;42:1862–72. https://doi.org/10.1093/ije/dyt182.Suche in Google Scholar PubMed PubMed Central
46. El-Ghiaty, MA, El-Kadi, AOS. The Duality of Arsenic Metabolism: Impact on Human Health. Annu Rev Pharmacol Toxicol 2023;63:341–58. https://doi.org/10.1146/ANNUREV-PHARMTOX-051921-020936.Suche in Google Scholar PubMed
47. Wāng, Y, Ma, L, Wang, C, Gao, T, Han, Y, Xu, DX. Cardiovascular adverse effects and mechanistic insights of arsenic exposure: a review. Environ Chem Lett 2024;22:1437–72. https://doi.org/10.1007/S10311-023-01677-0.Suche in Google Scholar
48. Qian, Y, Ke, JL, Chen, Y, Flynn, DC, Castranova, V, Shi, X. Cdc42 regulates arsenic-induced NADPH oxidase activation and cell migration through actin filament reorganization. J Biol Chem 2005;280:3875–84. https://doi.org/10.1074/jbc.M403788200.Suche in Google Scholar PubMed
49. Smith, KR, Klei, LR, Barchowsky, A. Arsenite stimulates plasma membrane NADPH oxidase in vascular endothelial cells. Am J Physiol Lung Cell Mol Physiol 2001;280. https://doi.org/10.1152/AJPLUNG.2001.280.3.L442.Suche in Google Scholar
50. Stea, F, Bianchi, F, Cori, L, Sicari, R. Cardiovascular effects of arsenic: Clinical and epidemiological findings. Environ Sci Pollut Res 2014;21:244–51. https://doi.org/10.1007/S11356-013-2113-Z/FIGURES/2.Suche in Google Scholar
51. Border, R, Keller, MC. Commentary: Fundamental problems with candidate gene-by-environment interaction studies – reflections on Moore et Thoemmes (2016). J Child Psychol Psychiatry 2017;58:328. https://doi.org/10.1111/JCPP.12669.Suche in Google Scholar
52. Dick, DM, Agrawal, A, Keller, MC, Adkins, A, Aliev, F, Monroe, S, et al.. Candidate Gene-Environment Interaction Research: Reflections and Recommendations. Perspect Psychol Sci 2015;10:37. https://doi.org/10.1177/1745691614556682.Suche in Google Scholar PubMed PubMed Central
53. Duncan, LE, Ostacher, M, Ballon, J. How genome-wide association studies (GWAS) made traditional candidate gene studies obsolete. Neuropsychopharmacol 2019;44:1518–23. https://doi.org/10.1038/s41386-019-0389-5.Suche in Google Scholar PubMed PubMed Central
54. Ripke, S, O’Dushlaine, C, Chambert, K, Moran, JL, Kähler, AK, Akterin, S, et al.. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 2013;45:1150–9. https://doi.org/10.1038/ng.2742.Suche in Google Scholar PubMed PubMed Central
55. Bosker, FJ, Hartman, CA, Nolte, IM, Prins, BP, Terpstra, P, Posthuma, D, et al.. Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol Psychiatry 2011;16:516–32. https://doi.org/10.1038/MP.2010.38.Suche in Google Scholar
56. Auer, PL, Stitziel, NO. Genetic association studies in cardiovascular diseases: do we have enough power? Trends Cardiovasc Med 2017;27:397. https://doi.org/10.1016/J.TCM.2017.03.005.Suche in Google Scholar PubMed PubMed Central
57. Duncan, LE, Keller, MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. Am J Psychiatry 2011;168:1041–9. https://doi.org/10.1176/APPI.AJP.2011.11020191.Suche in Google Scholar PubMed PubMed Central
58. Apata, M, Arriaza, B, Llop, E, Moraga, M. Human adaptation to arsenic in Andean populations of the Atacama Desert. Am J Phys Anthropol 2017;163:192–9. https://doi.org/10.1002/AJPA.23193.Suche in Google Scholar PubMed
59. Schlebusch, CM, Lewis, CM, Vahter, M, Engström, K, Tito, RY, Obregón-Tito, AJ, et al.. Possible positive selection for an arsenic-protective haplotype in humans. Environ Health Perspect 2013;121:53–8. https://doi.org/10.1289/EHP.1205504.Suche in Google Scholar
60. Lin, WY, Huang, CC, Liu, YL, Tsai, SJ, Kuo, PH. Polygenic approaches to detect gene–environment interactions when external information is unavailable. Brief Bioinform 2019;20:2236–52. https://doi.org/10.1093/BIB/BBY086.Suche in Google Scholar
Supplementary Material
This article contains supplementary material (https://doi.org/10.1515/reveh-2025-0041).
© 2025 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.


