Constructing a prognostic model for colon cancer patients on basis of coagulation genes enriched in cancer-associated fibroblasts to guide personalized immunotherapy
-
Rui Gao
and Xiaoxiang Chen
Abstract
Objectives
Colon cancer is a global health challenge. This research is designed to build a prognostic model that can personalize the guidance of immunotherapy among colon cancer patients.
Methods
Coagulation-associated prognostic genes which were subsequently integrated into a Least Absolute Shrinkage and Selection Operator algorithm for constructing a prognostic model were identified with the univariate Cox analyses. The potential of coagulation-related risk score (CRRS) in prognosis and immunotherapy outcomes was rigorously assessed. Finally, the cellular origin of genes in the CRRS model was explored with single-cell RNA-seq data, and the biological functions of core genes were further confirmed by cell function experiments.
Results
Our findings showed the CRRS model usefully classified patients into high-risk and low-risk groups. High-risk patients exhibited worse total survival. A nomogram was subsequently devised, enabling quantitative survival prediction by incorporating CRRS, age, sex, and TNM stage. Moreover, the CRRS model predicted the extent of cancer-associated fibroblasts (CAFs) infiltration. The analysis further indicated diminished immune responsiveness in high-risk patients, and single-cell data analysis pinpointed TIMP1+ CAF as a potential contributor to cancer progression.
Conclusions
The CRRS model can be adopted as a prognostic device for colon cancer patients and low-risk patients are more suitable for treatment with immune checkpoint inhibitors. TIMP1 secreted by CAF can promote the malignant progression of colon cancer.
Introduction
As information published by the International Agency for Research on Cancer, it is estimated that there will be more than 1.9 million novel cases and 904,000 deaths from colorectal cancer in 2022 [1]. The incidence of colorectal cancer is currently highest in the developed states. However, as developing countries develop rapidly, and people improve their living standards, it is predicted that novel cases of CRC will grow to 2–5 million by 2035 [2]. Consequently, CRC has emerged as a significant menace to humanity’s well-being and public health.
Presently, the primary approach for treating CRC centers around comprehensive treatment predominantly involving surgical resection [3]. Nevertheless, surgical intervention cannot eliminate metastatic colorectal cancer (mCRC), which is accompanied by a meager 5-year survival rate of around 14 % [4]. Recent years have witnessed a rapid advancement in CRC immunotherapy, with two programmed cell death 1 (PD-1) – blocking antibodies gaining approval from the Food and Drug Administration (FDA) for treating mCRC patients with mismatch repair defect and microsatellite instability [5]. Regrettably, only a minority of patients are benefited by this treatment [6]. Therefore, a comprehensive understanding of the factors contributing to treatment failure in CRC is imperative to enhance patient outcomes.
According to an analysis derived from a prospective observational study, thrombosis and infection stand out as the primary causes of non-cancer-related mortality in cancer patients [7]. Venous thromboembolism represents the most prevalent complication stemming from coagulation dysfunction and can often serve as a prominent feature of tumors [8]. The components of the tumor microenvironment (TME) associated with hemostasis collectively constitute what is known as the tumor coagulome. Research has established that the coagulome and TME interact in ways that either foster or hinder tumor progression and can even impact the effectiveness of immunotherapy by reshaping the TME [9]. Elements, such as the coagulation cascade, platelet activation, and neutrophil extracellular traps, are implicated in cancer-associated thrombosis and contribute to the creation of an immunosuppressive TME. Anticoagulants have shown potential in augmenting the effect of immune checkpoint inhibitors (ICIs), and the FDA has sanctioned the use of ICIs in combination with anticoagulants for treating various malignant tumors [10]. These findings underscore the potential significance of coagulation in the TME and the escape mechanisms employed by tumors from the immune system. Nonetheless, there remains a dearth of knowledge regarding the impact of coagulation and its immunomodulatory effect on the prognosis of colon cancer patients.
It endeavors to build a prognostic model known as the Coagulation-Related Risk Score (CRRS) in colon cancer patients on basis of a compilation of genes related to coagulation. The genes involved in CRRS were analyzed to find the key factors affecting the prognosis and treatment of patients.
Materials and methods
Data collection of bulk transcriptomes
The clinical data and RNA-seq data originated from the TCGA dataset. The TCGA-COAD dataset included 476 tumor tissues and 41 normal tissues. Additionally, to validate our model’s accuracy, we obtained expression data from GSE39582 [11] and corresponding platform annotation files from the Gene Expression Omnibus database, comprising 585 tumor samples. In the analysis of colon cancer patient prognosis, individuals with incomplete clinical data were excluded, leading to 452 patients in the TCGA-COAD dataset and 578 patients in the GSE39582 dataset. Detailed clinicopathological characteristics are offered in Table S1 of the Supplementary materials. The immunohistochemical pictures used in this study were provided by the online database The Human Protein Atlas (HPA) (https://www.proteinatlas.org/ENSG00000102265-TIMP1).
Construction and validation of coagulation-related signatures
The coagulation pathway data were offered by the KEGG database (https://www.genome.jp/kegg/), encompassing hsa04610 and hsa04611 [12], as well as HALLMARK_COAGULATION from the MsigDB database (https://www.gsea-msigdb.org/gsea/msigdb). There were 290 coagulation-associated genes (CRGs) included. CRGs related to prognosis were recognized with univariate Cox regression analysis. Later, a prognostic model on basis of LASSO regression was built with the R package “glmnet.” The coagulation-related risk score (CRRS) was derived from the model and adopted to fall patients into high- and low-risk groups. The production of Kaplan–Meier survival curves were made with the “gsurvplot” function in the R software package “survminer”. The TCGA-COAD cohort acted as the training set for internal validation, while the GSE39582 cohort was employed for external validation. The accuracy and predictive capability of CRRS were assessed via receiver operating characteristic curves (ROCs) using the “survivalROC” package. This study used 4.3.0 R software.
Building and evaluation of nomogram for colon patients
The “Survival” and “RMS” packages were employed to build a predictive nomogram for colon patients, utilizing variables from the TCGA-COAD training set. The prognostic efficiency of the predictive model was gauged with Harrell’s Concordance Index.
Functional enrichment analysis of DEGs
The “limma” R package was adopted to analyze differentially expressed genes between high- and low-risk groups. Gene set enrichment analysis on basis of DEGs and their logFCs was made with the R package “clusterProfiler” [13]. The consequent enrichment maps were visualized by applying R software. Additionally, hidden variations in pathway activity in every sample were assessed with GSVA, analyzing enrichment scores for pathways across all samples with the “GSVA” package in R [14].
Exploration of the tumor immune microenvironment
The calculation of immune scores and estimate scores for colon patients was made with the “estimate” R package [15]. We obtained the TCGA pan-cancer immune cell infiltration file (“infiltration_estimation_for_TCGA.csv”) from the TIMER 2.0 database (http://timer.cistrome.org/), enabling correlation analysis between immune cell infiltration and risk values.
Estimation of CAFs infiltration
Four methods were adopted to forecast the abundance of CAFs, such as the Estimation of Ratio of Immune and Cancer Cells (ERIC) algorithm [16], the xCell algorithm [17], the Microenvironmental Cell Population Counter (MCPcounter) algorithm [18], and the Tumor Immune Dysfunction and Rejection algorithm [19]. Spearman correlations were then analyzed between CRRS and stromal scores, and different CAFs infiltration level scores (EPIC, xCell, MCPcounter, and TIDE), validating the correlation between the model and CAFs infiltration.
Role of CRRS in predicting immunotherapy
To predict immunotherapy outcomes using immune checkpoint blockers (ICBs), we computed predictive scores such as TIDE scores and Immunophenotype scores (IPS). The gene expression profile of the TCGA-COAD cohort was uploaded to the Cancer Immunome Atlas, and IPS for each patient was downloaded [20]. The TIDE score, a new way to evaluate the efficacy of ICBs, is available on the TIDE website [19].
TIDE through two main mechanisms simulation of tumor immune escape, in cytotoxic T lymphocytes (CTL) to induce T cell dysfunction in high levels of tumor, as well as the prevention of T cells in the low level of CTL tumor infiltrating [19]. Tumor Inflammatory Signature includes genes related to antigen presentation, chemokine expression, cytotoxic activity, and adaptive immune resistance and has been retrospectively studied in clinical experiments [21]. We compared the prognostic power of CRRS, TIDE, and TIS by analyzing ROC curves for the three models.
Discussion single-cell sequencing data
The single-cell dataset GSE166555 [22] originated from the Tumor Immune Single-cell Hub (TISCH) database (http://tisch.comp-genomics.org/home/), encompassing 13 tumor tissues and 12 corresponding para-cancer tissues. Quality control procedures involved filtering and normalizing the original gene expression matrix using the “Seurat” R software package [23]. Subsequently, data dimensions were reduced using the Unified Move Approximation and Projection (UMAP) method, and the results were clustered and visualized using T-distributed Random Neighborhood Embedding (t-SNE) projection. Cells were annotated using standard cell surface markers.
Cell culture
We bought HCT116 and DLD1 cells from American Types Culture Collection, no mycoplasma infection. HCT116 cells were cultured in DMEM medium completely (KGL1206, KeyGEN BioTECH), DLD1 cell culture in the 1640 complete medium (KGL1501, KeyGEN BioTECH). All the complete media contained 10 % fetal bovine serum (FBS), 80 U/mL penicillin and 0.08 mg/mL streptomycin. Cells were cultured in an incubator at 37 °C containing 5 % CO2.
Cell counting kit-8 (CCK-8) assay
Cell viability was estimated applying CCK-8 (KGA9305, KeyGEN BioTECH, China) based on producer’s guidance. Transfected and untransfected HCT116 and DLD1 cells were inoculated in 96-well plates with a density of 1×104/well. Then CCK-8 reagent was added at different times (0 h, 24 h, 48 h, 72 h) to measure the OD value under the wavelength of 450 nm (Infinite 200 PRO, Tecan, Switzerland) and evaluate cell survival rate.
Cell transfection
The cloning of TIMP1 cDNAs into Lentiviral pLVX plasmid (SyngenTech, Beijing, China) was made, next to verification by sequencing. Then, Lipofectamine 3000 (L3000015, Invitrogen, USA) was adopted to transfect the pLVX-TIMP1 plasmids into HCT116 and DLD1 cells. CRC cells overexpressing TIMP1 were selected after 7 days of treatment with 10 μg/mL purinomycin (ST551, Beyotime, China).
Colony formation assay
The seeding of 1,000 cells per well into 6-well plates was made. The medium was changed every three days, next to 2-week incubation. With 4 % paraformaldehyde fixed colonies for 10 min, 15 min and then use 0.1 % crystal violet stain (C0121 Beyotime, China), in the end, rinsed in running water clean, air-dried under send pictures on the board.
Transwell assay
Transwell chamber was adopted to make the migration and invasion assays. For migration assay, the seeding of transfected and untransfected cells into the upper chamber (3472, Corning, USA) with serum-free medium (KeyGEN BioTECH, China) and the bottom of the chamber containing medium containing 20 % serum was made. For the invasion assay, Matrigel (356234, Corning, USA) was used to coat the chamber, and the following steps were like the migration assay. After 48 h incubation, the fixture of cells was made with 4 % paraformaldehyde, next to staining with 0.1 % crystal violet, and photographing.
Western blot
Proteins of colon cancer cells with and without TIMP1 overexpression were extracted using RIPA lysis buffer (P0013B, Beyotime, China). Equivalent amounts of proteins were isolated by sodium dodecyl sulfo-polyacrylamide gel electrophoresis in a 12 % gel, next to a PVDF membrane. Blocking was performed with 5 % skim milk powder (P0216, Beyotime, China) for 2 h, followed by incubation of membranes with TIMP1 antibody (1:1,500, 16644-1-AP, Proteintech, USA) overnight at 4 °C, next to incubation with secondary antibody (BL003A, Biosharp, China) the following day after three washes with TBST (G0001-2L, Servicebio, China). Resistance to GAPDH antibodies (1:10000, 10494-1-AP, Proteintech, USA) as a control. Finally, the stripes were visualized using a chemiluminescence instrument (Tanon 5200, China).
Statistical analysis
R software was adopted to make all statistical analyses. Results were of statistical significance in case of p-values of below 0.05 (two-sided). Kaplan–Meier analysis assessed and compared survival between high- and low-risk groups. Univariate Cox regression analysis screened genes related to prognosis, and independent risk elements influencing colon patients were identified after factors with p values <0.05 underwent multivariate Cox regression analysis. Cell counts were performed using ImageJ (1.54g), and the corresponding statistical plots were counted and plotted by GraphPad Prism 8. T-test or Wilcoxon rank sum test was adopted for comparing the two groups. Correlation analysis of continuous variables was made with Spearman’s test.
Results
Construction of coagulation-related signatures for colon cancer prognosis prediction
We initiated our analysis with a univariate Cox regression assessment to identify CRGs within the TCGA-COAD cohort. This scrutiny led to the selection of 23 prognostic genes exhibiting p-values <0.05 (Figure 1A). Later, LASSO Cox regression analysis was made on these 23 genes to construct a multi-gene prognosis model that minimized overfitting risk. The optimal value of λ yielded the inclusion of eight genes in our model (Figure 1B). The CRRS was then computed using the formula: CRRS=(0.447229484560575×PLCB1 expression)+(0.362128614897294×RASGRP2 expression)+(0.0457263309301581×PLCG2 expression)+(0.350092455079463×SERPINA5 expression)+(−0.443300975035417×F2RL2 expression)+(0.283241161078688×DUSP14 expression)+(−0.167780688007362×MMP10 expression)+(0.225325634783633×TIMP1 expression). With the median value as a limit, the classification of patients into high-risk and low-risk groups was made. Figure 1C shows risk curves, survival status, and heatmap of individual gene expression.
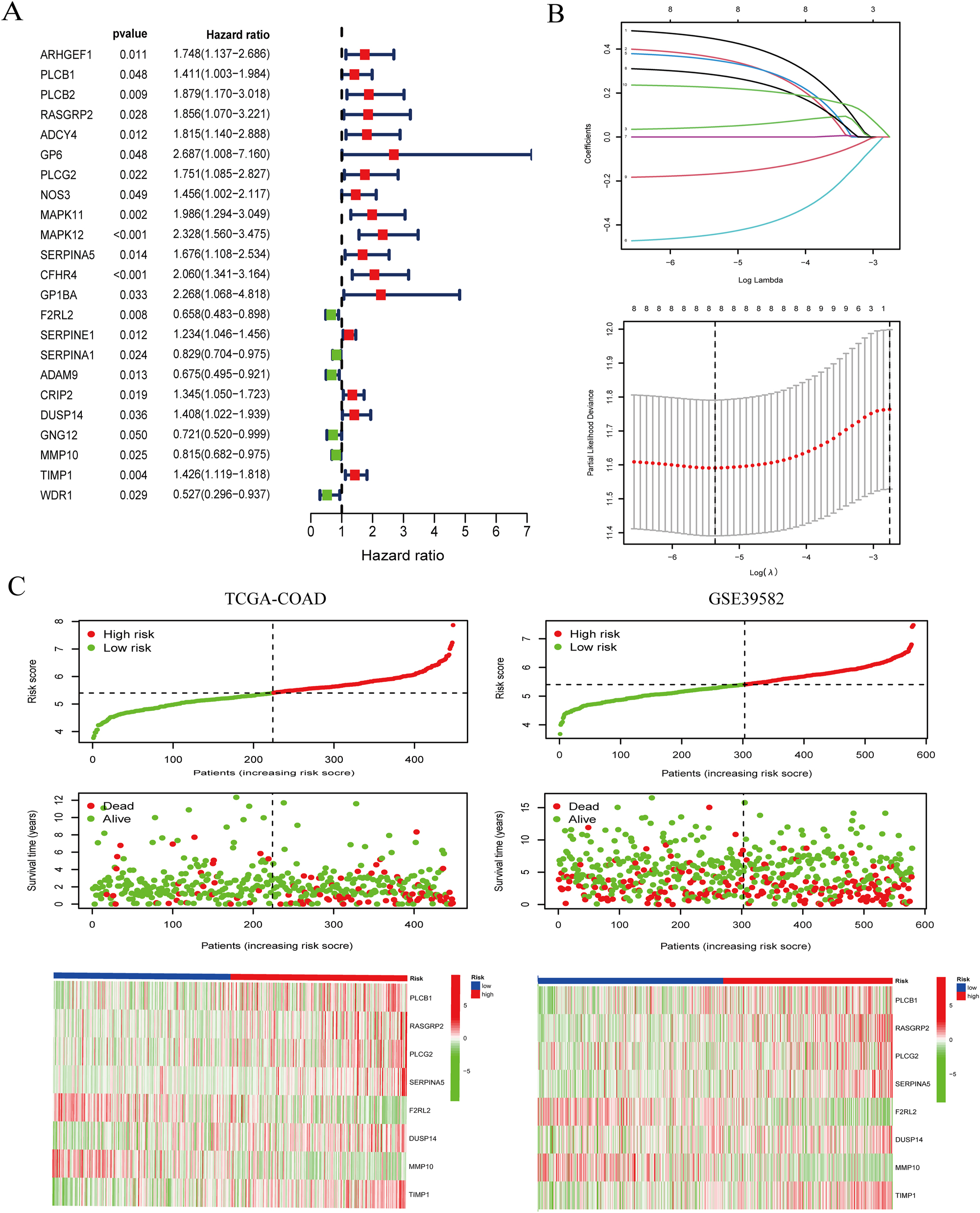
Construction of coagulation-related signatures for colon cancer prognosis prediction. (A) The forest plots of prognostic genes with p <0.05 were recognized by univariate Cox regression analysis. (B) The core genes were screened with LASSO Cox regression analysis. (C) The allocation of the risk score for every patient (top panel), survival situation of patients (middle panel), heatmaps for eight-gene signature between the high-risk group and low-risk group (bottom panel).
Validation of the model in training and verification sets
We proceeded to assess the prognostic capacity of our model in both the TCGA-COAD training set and the verification set (GSE39582). Survival analysis comparing low-risk and high-risk patients in the TCGA-COAD training set revealed greatly reduced overall survival (OS) among colon cancer patients in the high-risk group, a trend consistent with the validation set (Figure 2A and B). Receiver operating characteristic curves were adopted to gauge the sensitivity and specificity of our coagulation-related signatures in predicting colon cancer patient prognosis (Figure 2C and D). According to the outcomes, the areas under the curve for 1, 3, and 5-year OS predictions in the TCGA-COAD training set were 0.708, 0.713, and 0.701, while in the verification set, these AUCs were 0.575, 0.586, and 0.570. In summary, our 8-gene risk model related to coagulation effectively distinguishes high-risk from low-risk patients and demonstrates robust prognostic capabilities.

Verification of the model in training and verification sets. (A and B) Kaplan–Meier survival curve of OS between high-risk scores and low-risk scores in the TCGA training cohort and GSE39582 verification cohort. (C and D) The ROC curves at 1-, 3- and 5-year in the training and verification cohorts.
Construction of nomogram for colon cancer patients
To ascertain if CRRS qualifies as an independent clinical prognostic factor for colon cancer patients, univariate and multivariate Cox regression analyses were made. Age, sex, stage, TNM stage, and CRRS were evaluated as covariates, revealing that CRRS emerged as an independent predicting agent of colon cancer patient prognosis (Figure 3A). To improve the prediction of colon cancer patient survival, a nomogram amalgamating CRRS and clinical determinants were crafted based on the TCGA-COAD cohort (Figure 3B). After assigning scores to each prognostic parameter for patients, we calculated the cumulative survival rate, with higher total scores indicating a worse prognosis. Calibration plots portrayed 1-, 3-, and 5-year survival forecasts for colon cancer patients in the training set (Figure 3C). Furthermore, CRRS demonstrated significant associations with tumor stage and TNM stage, as confirmed by correlation analysis in both the training and verification sets (Figure 3D and E).
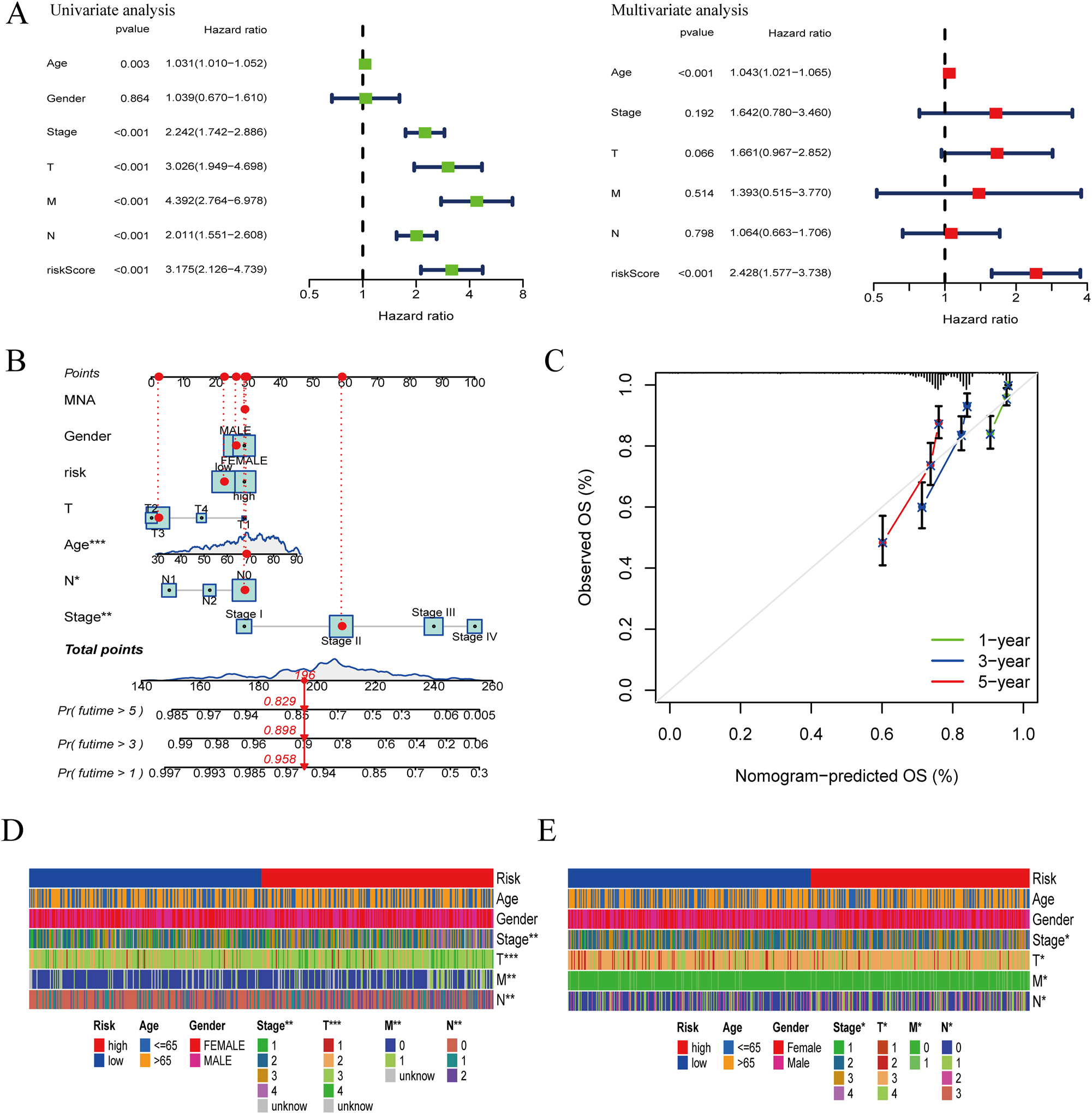
Construction of nomogram for colon cancer patients. (A) Univariate analysis and multivariate analysis including CRRS and clinical elements. (B) Nomogram integrating risk scores and clinical factors. (C) Calibration of the nomogram at 1-, 3- and 5-year survival rate in the training cohort. (D) Heat map of risk scores and clinical relevance in the TCGA-COAD set. (E) Heat map of risk scores and clinical relevance in the GSE39582 set (*p<0.05; **p<0.01; ***p<0.001).
Enrichment analysis of risk score-related functions
To delve deeper into the underlying mechanisms associated with CRRS, we scrutinized enriched KEGG gene sets within the high-risk and low-risk groups. As displayed in Figure 4A, genes in the high-risk group showed enrichment in pathways such as cell adhesion molecules (cams), dilated cardiomyopathy, ECM receptor interaction, focal adhesion, and systemic lupus erythematosus. Conversely, the enrichment of genes in the low-risk group was made in pathways like aminoacyl tRNA biosynthesis, butanoate metabolism, citrate cycle (TCA cycle), peroxisome, and retinol metabolism (Figure 4B). Additionally, GSVA analysis highlighted significant correlations between CRRS and pathways including the TGF-β signaling pathway, MAPK signaling pathway, and calcium signaling pathway (Figure 4C).

Enrichment analysis of risk score-related functions. (A) Enrichment pathways in the KEGG dataset of the high-risk group. (B) Enrichment pathways in the KEGG dataset of low-risk group. (C) Heat map of GSVA analysis CRRS and signaling pathway correlation (*p<0.05; **p<0.01; ***p<0.001).
Tumor microenvironment analysis in high- and low-risk groups
A contrast of the tumor microenvironment (TME) features between high- and low-risk groups was made. The ESTIMATE algorithm provided stromal marks, immune marks, and estimate marks to assess cancer-related stroma, immune cell infiltration, and tumor purity, respectively. The results indicated that the high-risk group exhibited greatly higher stromal marks, immune marks, and estimate marks by comparing with the low-risk group (Figure 5A). To ensure reliability, we also analyzed the correlation between immune cells and CRRS applying various algorithms, including xCell, TIMER, QUANTISEQ, MCPcounter, EPIC, CIBERSORT-ABS, and CIBERSORT. The findings demonstrated a strong positive correlation between CRRS and CAFs predicted by the MCPcounter and EPIC algorithms (Figure 5B and C). Spearman correlation analysis confirmed the robustness of the CRRS model as a CAFs infiltration predictor. CRRS was positively correlated with stromal scores and CAFs infiltration scores estimated by four algorithms (xCell, MCPcounter, EPIC, and TIDE) in both the TCGA-COAD and GSE39582 cohorts (Figure 5D). These results collectively affirmed the reliability of the risk score as an index of CAFs infiltration. Furthermore, the MCPcounter and EPIC algorithms were employed to score CAFs in high- and low-risk patients from the TCGA-COAD and GSE39582 cohorts, underscoring stronger CAFs infiltration in the high-risk group (Figure 5E and F).

Tumor microenvironment analysis in high- and low-risk groups. (A) Stromal mark, immune mark and estimate mark between high- and low-risk groups in TCGA-COAD and GSE39582. (B) Bubble chart of correlation between immune cells and CRRS. (C) Scatterplot of MCPcounter and EPIC algorithms to assess the correlation between CAFs infiltration and CRRS. (D) According to Spearman’s correlation analysis, CRRS was positively related to stromal scores and multiple estimates of CAFs infiltration in the TCGA-COAD and GSE39582 cohorts. (E) EPIC algorithm and MCPcounter algorithm score high- and low-risk groups in the TCGA-COAD for CAFs. (F) EPIC algorithm and MCPcounter algorithm score high- and low-risk groups in the GSE39582 for CAFs (*p<0.05; ***p<0.001).
Comparison of immunotherapy in high- and low-risk groups
Immunotherapy has gained prominence as a cornerstone in the treatment of different solid cancers, such as colon cancer. Notably, the FDA accelerated the approval of pembrolizumab and nivolumab, two programmed cell death 1 (PD1) blocking antibodies, for mCRC patients with mismatch repair flaws and high microsatellite unsteadiness (dMMR-MSI-H), demonstrating promising efficacy [24]. To investigate whether CRRS correlates significantly with immunotherapy outcomes, we employed the TIDE method. In the TCGA-COAD cohort, a higher percentage of high-risk patients (67 %) displayed unresponsiveness to treatment compared to low-risk patients (43 %). A similar tendency was discovered in the GSE39582 cohort, where a greater number of high-risk patients (60 %) did not respond to treatment as opposed to low-risk patients (51 %). High-risk patients also exhibited higher TIDE scores (Figure 6A), collectively indicating a greater potential for immune evasion. Additionally, we evaluated the likelihood of patients benefiting from immune checkpoint inhibitors (ICIs) by analyzing Immunophenotype scores (IPS) in high- and low-risk groups within the TCGA-COAD cohort. The low-risk group appeared to possess a more immunogenic phenotype (Figure 6B). Furthermore, we assessed the association between risk scores and immune checkpoints in both the TCGA-COAD and GSE39582 cohorts, revealing positive associations between risk marks and the majority of immune checkpoints (Figure 6C). While TIDE and TIS have been employed to predict patient responses to immunotherapy, the area under the ROC curve of CRRS exceeded that of TIDE and TIS, signifying superior predictive value and establishing CRRS as a promising biomarker for immune-related prognosis (Figure 6D).

Comparison of immunotherapy in high- and low-risk groups. (A) TIDE scoring and percentage of responders and non-responders in the high- and low-risk groups in the TCGA-COAD and GSE39582. (B) IPS scores for high- and low-risk groups in the TCGA-COAD. (C) Heatmap of CRRS and immune checkpoint correlation for the TCGA-COAD and GSE39582 cohorts. (D) ROC analysis of CRRS, TIDE, and TIS on OS at 1-, 2-, and 3-year follow-up in the TCGA-COAD cohort (*p<0.05; **p<0.01; ***p<0.001).
Identification of TIMP1+ CAF as a key prognostic component through scRNA transcriptomic analysis
Given the close connection between CAFs and CRRS within the immune microenvironment, our focus turned to express model genes in CAF cells applying scRNA transcriptome analysis. After dimensionality reduction and cell clustering of GSE166555 single-cell data, 33 distinct cell clusters emerged, subsequently annotated into 13 cell types, including fibroblasts (Figure 7A–C). Importantly, the HALLMARK_COAGULATION gene set was predominantly expressed in fibroblasts (Figure 7D). Further correlation analysis between model gene expression and CAFs infiltration using the EPIC and MCPcounter algorithms identified PLCG2, RASGRP2, and TIMP1 as genes positively correlated with CAFs infiltration (Figure 7E). Notably, TIMP1 is abundantly expressed in fibroblasts (Figure 7F), and was notably overexpressed in CAFs of tumor tissues compared to normal fibroblasts of normal tissues (p<0.001; Figure 7G). Additionally, analysis of TIMP1 protein expression in relation to CAFs using IHC images from the HPA online database [25] further confirmed TIMP1 as a specific gene marker for CAFs, showing strong staining in the stromal regions of colon cancer tissues and no expression in normal tissues (Figure 7H).

Identification of TIMP1+ CAF as a key prognostic component through scRNA transcriptomic analysis. (A and B) UMAP plot showing clusters of cells defined according to marker genes. (C) Bubble plot showing expression of labeled genes in all cell clusters. Dot size suggests the percentage of expressed genes and color suggests the level of expression intensity. (D) UMAP plot showing coagulation gene pathways enriched in fibroblast clusters. (E) Scatterplot of correlation between model gene expression and degree of CAFs infiltration. (F) The expression of RASGRP2, PLCG2 and TIMP1 on all cells. (G) Expression of TIMP1 in fibroblasts between normal and tumor tissues. (H) Protein expression of TIMP1 in normal colon tissue (left) and colon cancer specimens (right) from the Human Protein Atlas database. Scale bar=200 μm.
TIMP1 promotes the proliferation and metastasis of CRC cells
To research the role of TIMP1 in CRC, we upregulated TIMP1 in HCT116 and DLD1 cells (Figure 8A) and found that TIMP1 overexpression promoted CRC cell proliferation (Figure 8B), colony formation (Figure 8C), migration and invasion (Figure 8D). Collectively, these data showed that TIMP1 serves as an oncogene in CRC.

TIMP1 drives the propagation and metastasis of CRC cells. (A) TIMP1 overexpression efficiency was determined by Western blot at the protein level in HCT116 and DLD1 cells. (B) CCK8 reagent was used to detect the propagation of HCT116 cells and DLD1 cells after TIMP1 overexpression. (C) The changes in the propagation ability of HCT116 cells and DLD1 cells were explored with colony formation assay after TIMP1 overexpression. (D) Transwell assay investigated the changes of migration and invasion ability of HCT116 cells and DLD1 cells after TIMP1 overexpression, scale bar=100 μm (*p<0.05; **p<0.01; ***p<0.001).
Discussion
It has been disclosed that the coagulation system can be activated by tumor cells, resulting in a hypercoagulable state in cancer patients [26]. Venous thromboembolism (VTE), a manifestation of the hypercoagulable state in cancer patients, serves as one of the early clinical signs of underlying malignancies, contributing significantly to cancer-related morbidity and mortality. Among malignancies, breast, colon, and lung cancers exhibit the strongest associations with thrombosis [27]. Notably, data from a prospective study involving 2,477 treated Japanese patients with colorectal cancer highlighted that VTE incidence rises with advancing cancer stages, and the presence of VTE at the time of cancer diagnosis exacerbates patient mortality [28]. Based on the close relationship between thrombosis and patient prognosis, it suggests the possibility of coagulation pathway as a therapeutic target for colon cancer, few research has delved into the correlation of coagulation pathways with colon cancer prognosis and immunotherapy outcomes. Consequently, our study aimed to elucidate the impact of coagulation pathways in colon cancer on patient prognosis, tumor immune evasion, and therapeutic responses.
In this investigation, our first goal was to compile a comprehensive list of CRGs. To achieve this, we selected gene sets from two pathways, hsa04610 (complement and coagulation cascade) and hsa0461 (platelet activation), found in the KEGG pathway database [12], along with the HALLMARK_COAGULATION gene set from the MsigDB database. These selections collectively formed our set of CRGs. Our initial step involved a univariate Cox analysis applying the TCGA-COAD dataset to identify 23 coagulation genes with significant prognostic associations. Subsequently, we built a CRRS on the basis of eight CRGs through LASSO regression to predict patient outcomes. This model effectively distinguished colon cancer patients with pronounced coagulation characteristics and poorer prognoses. The eight genes included in CRRS were PLCB1, RASGRP2, PLCG2, SERPINA5, F2RL2, DUSP14, MMP10, and TIMP1. CRRS revealed PLCB1, RASGRP2, PLCG2, SERPINA5, DUSP14, and TIMP1 as risk factors, while F2RL2 and MMP10 were protective factors. Numerous studies have already established CRGs’ associations with malignant tumor progression and adverse patient outcomes. For example, PLCB1, a member of the phospholipase family, has been linked to promoting cholangiocarcinoma (CCA) proliferation and metastasis, as well as resistance to gemcitabine/cisplatin chemotherapy [29]. RASGRP2 has been demonstrated to drive malignant progression in lung adenocarcinoma (LUAD) through mitochondria-dependent apoptosis, resulting in poorer patient prognosis [30]. PLCG2 has been strongly related to metastasis and reduced survival in small-cell lung cancer (SCLC) [31]. SERPINA5, a serine protease inhibitor, has been shown to facilitate gastric cancer (GC) progression [32]. Dual-specificity phosphatase 14 (DUSP14) has been implicated as an inducer of the epithelial–mesenchymal transition (EMT) process, promoting pancreatic cancer proliferation, migration, and invasion [33]. TIMP1 is an inhibitor of matrix metalloproteinases, has been linked to lymph node metastasis, distant metastasis, vascular infiltration, AJCC staging, and dysregulation of signaling pathways, contributing to tumor proliferation, metastasis, and resistance to apoptosis [34]. Increased TIMP-1 expression has also been related to CAFs accumulation in prostate/colon cancer tissues, exerting a pro-tumorigenic effect [35] and resistance to 5-fluorouracil in colon cancer [36].
Colon cancer patients in the TCGA-COAD training set and GSE39582 verification set were categorized into high-risk and low-risk groups on basis of risk marks. According to the survival curve, the high-risk group showed greatly shorter survival time than the low-risk group. Univariate and multivariate Cox regression analysis were adopted to verify that CRRS affected the prognosis of patients as an independent factor. CRRS were significantly associated with T (tumor), N (lymph node metastasis), M (distant metastasis), and stage in colon cancer patients. Tumor grade and stage are also one of the key factors affecting cancer-related thrombosis [37]. GSEA and GSVA enrichment analyses were then made to dig into the reasons for the diversities between the high- and low-risk groups. According to the outcomes, a relationship between the major enriched pathways was discovered in the high-risk group and the extracellular matrix. Subsequently, we found that patients in the high- and low-risk groups showed various immune status. Despite immune active patients in the high-risk group, they were not sensitive to immunotherapy and had a worse prognosis. In view of this abnormal phenomenon, we further analyzed the immune microenvironment and found that CRRS had the highest correlation with CAFs, and the degree of CAFs infiltration was positively related to the risk score. The high-risk group displayed greatly larger infiltration of CAFs than the low-risk group. As a significant ingredient of the tumor microenvironment, CAFs exert a great effect on tumor promotion and drug resistance [38]. CAFs can secret different cytokines, development elements, chemokines, exosomes, and other effector molecules to generate an immunosuppressive microenvironment, so that tumor cells escape the surveillance of the immune system [39]. We then adopted single-cell data to classify and annotate the cells and found that clotting-related genes were significantly enriched in CAFs, which further proved that CRRS was significantly correlated with CAFs. Pape et al. have displayed that CAFs exert a key effect on driving angiogenesis and hence cancer invasion [40]. Subsequently, we further understood the association between the eight genes in the model and CAFs and found that TIMP1 was significantly enriched in CAFs.
TIMP1 is a secreted protein, TIMP1 secreted by CAFs that drives the migration of colon cancer cells [41]. We through CCK8, clone formation and transwell experiments, confirmed that when the high TIMP1 expression, CRC cell proliferation, migration, and invasion ability were improved significantly. TIMP1+ CAF may be an adverse factor leading to poor prognosis in colon cancer patients with immune escape. Although we validated the value of our model in forecasting the prognosis and immunotherapy of colon cancer patients through bioinformatics and in vitro experiments, as well as the promoting effect of TIMP1-positive CAF on the malignant progression of colon cancer, further validation of our model in clinical samples is required in the future.
Conclusions
To sum up, the findings reveal that individuals categorized as high-risk not only face a dismal prognosis but also exhibit resistance to immunotherapy. scRNA-seq offers precise insights into distinct cell types and their distinct characteristics under varying biological conditions. Analysis through scRNA-seq unveiled that CAFs are the origin of coagulation in colon cancer, and one of the model genes TIMP1, demonstrated heightened expression in a specific subset of CAFs.
Award Identifier / Grant number: ZKX21042
Funding source: The research project of Jiangsu Health Development Research Center
Award Identifier / Grant number: JSHD2022057
Funding source: Jiangsu Provincial Medical Key Discipline Cultivation Unit
Award Identifier / Grant number: JSDW202239
Funding source: Natural Science Foundation of Jiangsu Province
Award Identifier / Grant number: BK20210031
Acknowledgments
Thank all authors for their contributions to this study.
-
Research ethics: The local Institutional Review Board deemed the study exempt from review.
-
Informed consent: Informed consent was obtained from all individuals included in this study.
-
Author contributions: All authors read and approved the final manuscript. RG and QZ contributed to the conception and design. SH and JQ extracted the data from the databases. QX contributed to the data analysis and interpretation. RG drafted the manuscript. HS and XC designed the idea of the study and examined and YP revised the manuscript.
-
Competing interests: All authors state no conflict of interest.
-
Research funding: This study was supported by grants from the Natural Science Foundation of Jiangsu Province (BK20210031), Key Project of Science and Technology Development of Nanjing Medicine (ZKX21042), Jiangsu Provincial Medical Key Discipline Cultivation Unit (JSDW202239), The research project of Jiangsu Health Development Research Center (JSHD2022057).
-
Data availability: The datasets displayed herein can be discovered in online repositories. These can be discovered in the GEO database (https://www.ncbi.nlm.nih.gov/geo), The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov) and the Tumor Immune Single-cell Hub (TISCH) database (http://tisch.comp-genomics.org/home/). The article/Supplementary material includes the original contributions displayed herein. Further search can be directed to the corresponding author.
References
1. Bray, F, Laversanne, M, Sung, H, Ferlay, J, Siegel, RL, Soerjomataram, I, et al.. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024;74:229–63. https://doi.org/10.3322/caac.21834.Search in Google Scholar PubMed
2. Dekker, E, Tanis, PJ, Vleugels, JLA, Kasi, PM, Wallace, MB. Colorectal cancer. Lancet 2019;394:1467–80. https://doi.org/10.1016/s0140-6736(19)32319-0.Search in Google Scholar PubMed
3. Miller, KD, Nogueira, L, Devasia, T, Mariotto, AB, Yabroff, KR, Jemal, A, et al.. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin 2022;72:409–36. https://doi.org/10.3322/caac.21731.Search in Google Scholar PubMed
4. Shin, AE, Giancotti, FG, Rustgi, AK. Metastatic colorectal cancer: mechanisms and emerging therapeutics. Trends Pharmacol Sci 2023;44:222–36. https://doi.org/10.1016/j.tips.2023.01.003.Search in Google Scholar PubMed PubMed Central
5. Fan, A, Wang, B, Wang, X, Nie, Y, Fan, D, Zhao, X, et al.. Immunotherapy in colorectal cancer: current achievements and future perspective. Int J Biol Sci 2021;17:3837–49. https://doi.org/10.7150/ijbs.64077.Search in Google Scholar PubMed PubMed Central
6. Shan, J, Han, D, Shen, C, Lei, Q, Zhang, Y. Mechanism and strategies of immunotherapy resistance in colorectal cancer. Front Immunol 2022;13:1016646. https://doi.org/10.3389/fimmu.2022.1016646.Search in Google Scholar PubMed PubMed Central
7. Khorana, AA, Francis, CW, Culakova, E, Kuderer, NM, Lyman, GH. Thromboembolism is a leading cause of death in cancer patients receiving outpatient chemotherapy. J Thromb Haemost 2007;5:632–4. https://doi.org/10.1111/j.1538-7836.2007.02374.x.Search in Google Scholar PubMed
8. Gheshmy, A, Carrier, M. Venous thromboembolism and occult cancer: impact on clinical practice. Thromb Res 2016;140:S8–11. https://doi.org/10.1016/s0049-3848(16)30091-3.Search in Google Scholar
9. Galmiche, A, Rak, J, Roumenina, LT, Saidak, Z. Coagulome and the tumor microenvironment: an actionable interplay. Trends Cancer 2022;8:369–83. https://doi.org/10.1016/j.trecan.2021.12.008.Search in Google Scholar PubMed
10. Shafqat, A, Omer, MH, Ahmed, EN, Mushtaq, A, Ijaz, E, Ahmed, Z, et al.. Reprogramming the immunosuppressive tumor microenvironment: exploiting angiogenesis and thrombosis to enhance immunotherapy. Front Immunol 2023;14:1200941. https://doi.org/10.3389/fimmu.2023.1200941.Search in Google Scholar PubMed PubMed Central
11. Marisa, L, de Reyniès, A, Duval, A, Selves, J, Gaub, MP, Vescovo, L, et al.. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 2013;10:e1001453. https://doi.org/10.1371/journal.pmed.1001453.Search in Google Scholar PubMed PubMed Central
12. He, Q, Yang, J, Jin, Y. Immune infiltration and clinical significance analyses of the coagulation-related genes in hepatocellular carcinoma. Brief Bioinform 2022;23:bbac291. https://doi.org/10.1093/bib/bbac291.Search in Google Scholar PubMed
13. Subramanian, A, Tamayo, P, Mootha, VK, Mukherjee, S, Ebert, BL, Gillette, MA, et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. https://doi.org/10.1073/pnas.0506580102.Search in Google Scholar PubMed PubMed Central
14. Hänzelmann, S, Castelo, R, Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. https://doi.org/10.1186/1471-2105-14-7.Search in Google Scholar PubMed PubMed Central
15. Yoshihara, K, Shahmoradgoli, M, Martínez, E, Vegesna, R, Kim, H, Torres-Garcia, W, et al.. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. https://doi.org/10.1038/ncomms3612.Search in Google Scholar PubMed PubMed Central
16. Racle, J, de Jonge, K, Baumgaertner, P, Speiser, DE, Gfeller, D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife 2017;6:e26476. https://doi.org/10.7554/elife.26476.Search in Google Scholar PubMed PubMed Central
17. Aran, D, Hu, Z, Butte, AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. https://doi.org/10.1186/s13059-017-1349-1.Search in Google Scholar PubMed PubMed Central
18. Becht, E, Giraldo, NA, Lacroix, L, Buttard, B, Elarouci, N, Petitprez, F, et al.. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016;17:218. https://doi.org/10.1186/s13059-016-1070-5.Search in Google Scholar PubMed PubMed Central
19. Jiang, P, Gu, S, Pan, D, Fu, J, Sahu, A, Hu, X, et al.. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 2018;24:1550–8. https://doi.org/10.1038/s41591-018-0136-1.Search in Google Scholar PubMed PubMed Central
20. Charoentong, P, Finotello, F, Angelova, M, Mayer, C, Efremova, M, Rieder, D, et al.. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 2017;18:248–62. https://doi.org/10.1016/j.celrep.2016.12.019.Search in Google Scholar PubMed
21. Ayers, M, Lunceford, J, Nebozhyn, M, Murphy, E, Loboda, A, Kaufman, DR, et al.. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. https://doi.org/10.1172/jci91190.Search in Google Scholar PubMed PubMed Central
22. Uhlitz, F, Bischoff, P, Peidli, S, Sieber, A, Trinks, A, Lüthen, M, et al.. Mitogen-activated protein kinase activity drives cell trajectories in colorectal cancer. EMBO Mol Med 2021;13:e14123. https://doi.org/10.15252/emmm.202114123.Search in Google Scholar PubMed PubMed Central
23. Butler, A, Hoffman, P, Smibert, P, Papalexi, E, Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. https://doi.org/10.1038/nbt.4096.Search in Google Scholar PubMed PubMed Central
24. Ganesh, K, Stadler, ZK, Cercek, A, Mendelsohn, RB, Shia, J, Segal, NH, et al.. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol 2019;16:361–75. https://doi.org/10.1038/s41575-019-0126-x.Search in Google Scholar PubMed PubMed Central
25. Pontén, F, Schwenk, JM, Asplund, A, Edqvist, PH. The Human Protein Atlas as a proteomic resource for biomarker discovery. J Intern Med 2011;270:428–46. https://doi.org/10.1111/j.1365-2796.2011.02427.x.Search in Google Scholar PubMed
26. Falanga, A, Schieppati, F, Russo, L. Pathophysiology 1. Mechanisms of thrombosis in cancer patients. Cancer Treat Res 2019;179:11–36. https://doi.org/10.1007/978-3-030-20315-3_2.Search in Google Scholar PubMed
27. Lee, AY, Levine, MN. Venous thromboembolism and cancer: risks and outcomes. Circulation 2003;107:I17–21. https://doi.org/10.1161/01.cir.0000078466.72504.ac.Search in Google Scholar
28. Ikeda, M, Uetake, H, Yoshino, T, Hata, T, Oba, MS, Takita, A, et al.. Incidence and risk factors for venous thromboembolism, bleeding, and death in colorectal cancer (Cancer-VTE Registry). Cancer Sci 2022;113:3901–11. https://doi.org/10.1111/cas.15527.Search in Google Scholar PubMed PubMed Central
29. Liang, S, Guo, H, Ma, K, Li, X, Wu, D, Wang, Y, et al.. A PLCB1-PI3K-AKT signaling axis activates EMT to promote cholangiocarcinoma progression. Cancer Res 2021;81:5889–903. https://doi.org/10.1158/0008-5472.can-21-1538.Search in Google Scholar
30. Liu, Y, Ouyang, Y, Feng, Z, Jiang, Z, Ma, J, Zhou, X, et al.. RASGRP2 is a potential immune-related biomarker and regulates mitochondrial-dependent apoptosis in lung adenocarcinoma. Front Immunol 2023;14:1100231. https://doi.org/10.3389/fimmu.2023.1100231.Search in Google Scholar PubMed PubMed Central
31. Chan, JM, Quintanal-Villalonga, Á, Gao, VR, Xie, Y, Allaj, V, Chaudhary, O, et al.. Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell 2021;39:1479–96.e18. https://doi.org/10.1016/j.ccell.2021.09.008.Search in Google Scholar PubMed PubMed Central
32. Fan, M, Xiong, X, Han, L, Zhang, L, Gao, S, Liu, L, et al.. SERPINA5 promotes tumour cell proliferation by modulating the PI3K/AKT/mTOR signalling pathway in gastric cancer. J Cell Mol Med 2022;26:4837–46. https://doi.org/10.1111/jcmm.17514.Search in Google Scholar PubMed PubMed Central
33. Wei, Y, Wang, G, Wang, C, Zhou, Y, Zhang, J, Xu, K. Upregulation of DUSP14 affects proliferation, invasion and metastasis, potentially via epithelial-mesenchymal transition and is associated with poor prognosis in pancreatic cancer. Cancer Manag Res 2020;12:2097–108. https://doi.org/10.2147/cmar.s240040.Search in Google Scholar
34. Song, G, Xu, S, Zhang, H, Wang, Y, Xiao, C, Jiang, T, et al.. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J Exp Clin Cancer Res 2016;35:148. https://doi.org/10.1186/s13046-016-0427-7.Search in Google Scholar PubMed PubMed Central
35. Gong, Y, Scott, E, Lu, R, Xu, Y, Oh, WK, Yu, Q. TIMP-1 promotes accumulation of cancer associated fibroblasts and cancer progression. PLoS One 2013;8:e77366. https://doi.org/10.1371/journal.pone.0077366.Search in Google Scholar PubMed PubMed Central
36. Liu, T, Xia, R, Li, C, Chen, X, Cai, X, Li, W. mRNA expression level of CDH2, LEP, POSTN, TIMP1 and VEGFC modulates 5-fluorouracil resistance in colon cancer cells. Exp Ther Med 2021;22:1023. https://doi.org/10.3892/etm.2021.10455.Search in Google Scholar PubMed PubMed Central
37. Falanga, A, Marchetti, M, Vignoli, A. Coagulation and cancer: biological and clinical aspects. J Thromb Haemost 2013;11:223–33. https://doi.org/10.1111/jth.12075.Search in Google Scholar PubMed
38. Li, Z, Sun, C, Qin, Z. Metabolic reprogramming of cancer-associated fibroblasts and its effect on cancer cell reprogramming. Theranostics 2021;11:8322–36. https://doi.org/10.7150/thno.62378.Search in Google Scholar PubMed PubMed Central
39. Mao, X, Xu, J, Wang, W, Liang, C, Hua, J, Liu, J, et al.. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer 2021;20:131. https://doi.org/10.1186/s12943-021-01428-1.Search in Google Scholar PubMed PubMed Central
40. Pape, J, Magdeldin, T, Stamati, K, Nyga, A, Loizidou, M, Emberton, M, et al.. Cancer-associated fibroblasts mediate cancer progression and remodel the tumouroid stroma. Br J Cancer 2020;123:1178–90. https://doi.org/10.1038/s41416-020-0973-9.Search in Google Scholar PubMed PubMed Central
41. Nakai, N, Hara, M, Takahashi, H, Shiga, K, Hirokawa, T, Maeda, Y, et al.. Cancer cell-induced tissue inhibitor of metalloproteinase-1 secretion by cancer-associated fibroblasts promotes cancer cell migration. Oncol Rep 2022;47:112. https://doi.org/10.3892/or.2022.8323.Search in Google Scholar PubMed
Supplementary Material
This article contains supplementary material (https://doi.org/10.1515/oncologie-2024-0142).
© 2024 the author(s), published by De Gruyter on behalf of Tech Science Press (TSP)
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Review Articles
- Autophagy and radiotherapy in esophageal cancer: modulating treatment sensitivity and overcoming challenges
- Functions of CAFs in microenvironment of non-small cell lung cancer: based on updated hallmarks of cancer
- Cisplatin-induced pyroptosis: a double-edged sword in cancer treatment
- Radiotherapy directed to inferior vena cava tumor thrombus among patients with renal cell carcinoma: an illustrative case and review of the literature
- Research Articles
- A lipid metabolism-related gene model reveals the prognosis and immune microenvironment of cutaneous melanoma
- Boanmycin induces apoptosis and overcomes venetoclax resistance in acute myeloid leukemia
- Identification of GNB1 as a downstream effector of the circRNA-0133711/miR-145-5p axis involved in breast cancer proliferation and metastasis
- Clinical characteristics and prognosis of lung metastases from unknown primary cancer sites
- Integrating bulk-RNA and single-cell analysis reveals heterogeneous expression of cuproptosis-related sorafenib-resistant genes in hepatocellular carcinoma
- Exploring the mechanism of genistein in treating hepatocellular carcinoma through network pharmacology and molecular docking
- Investigating the potential mechanisms of Litsea cubeba essential oil for anti-melanoma through experimental validation, network pharmacology, and molecular docking analysis
- Exploring the anti-lung cancer mechanism of Ganoderma lucidum and its relationship with the level of immune cell infiltration based on network pharmacology and molecular docking
- Constructing a prognostic model for colon cancer patients on basis of coagulation genes enriched in cancer-associated fibroblasts to guide personalized immunotherapy
- Rapid Communication
- Evaluation of HER2 immunohistochemistry-positive and immunohistochemistry-negative FISH amplification breast cancers using next-generation sequencing
- Short Commentary
- Unveiling the unexplored: shedding light on a novel aspect of colorectal carcinoma
Articles in the same Issue
- Frontmatter
- Review Articles
- Autophagy and radiotherapy in esophageal cancer: modulating treatment sensitivity and overcoming challenges
- Functions of CAFs in microenvironment of non-small cell lung cancer: based on updated hallmarks of cancer
- Cisplatin-induced pyroptosis: a double-edged sword in cancer treatment
- Radiotherapy directed to inferior vena cava tumor thrombus among patients with renal cell carcinoma: an illustrative case and review of the literature
- Research Articles
- A lipid metabolism-related gene model reveals the prognosis and immune microenvironment of cutaneous melanoma
- Boanmycin induces apoptosis and overcomes venetoclax resistance in acute myeloid leukemia
- Identification of GNB1 as a downstream effector of the circRNA-0133711/miR-145-5p axis involved in breast cancer proliferation and metastasis
- Clinical characteristics and prognosis of lung metastases from unknown primary cancer sites
- Integrating bulk-RNA and single-cell analysis reveals heterogeneous expression of cuproptosis-related sorafenib-resistant genes in hepatocellular carcinoma
- Exploring the mechanism of genistein in treating hepatocellular carcinoma through network pharmacology and molecular docking
- Investigating the potential mechanisms of Litsea cubeba essential oil for anti-melanoma through experimental validation, network pharmacology, and molecular docking analysis
- Exploring the anti-lung cancer mechanism of Ganoderma lucidum and its relationship with the level of immune cell infiltration based on network pharmacology and molecular docking
- Constructing a prognostic model for colon cancer patients on basis of coagulation genes enriched in cancer-associated fibroblasts to guide personalized immunotherapy
- Rapid Communication
- Evaluation of HER2 immunohistochemistry-positive and immunohistochemistry-negative FISH amplification breast cancers using next-generation sequencing
- Short Commentary
- Unveiling the unexplored: shedding light on a novel aspect of colorectal carcinoma

