Abstract
Urinary tract infections affect 150 million people worldwide, yet the diagnosis of this common infection is not straightforward. Misdiagnoses and incorrect prescriptions are frequent in the treatment of urinary tract infections; this also contributes to the increase in antibiotic resistance among pathogens. Present diagnostic practices take 2–3 days for pathogen identification and antibiotic susceptibility testing. New technologies are urgently needed for improved patient care as well as to promote antibiotic stewardship. An ideal new diagnostic technology will test clinical urine samples directly and identify the pathogen and determine its antibiotic susceptibilities within a few hours such that the patient can be prescribed the appropriate antibiotic treatment the same day. Screening tools, such as flow cytometers and new dipstick assays, can help with rapidly identifying negative samples and improving workflow and reducing costs. Several groups have made progress in optimizing mass spectrometry methods for direct urine processing, and there are also new multiplex PCR panels that are specific for UTI pathogens and antibiotic resistance. We also discuss several emerging technologies – microfluidics, biosensors, real-time microscopy systems, and sequence-based diagnostics – that show huge potential in delivering rapid results.
Introduction
Urinary tract infections (UTIs) caused 10.5 million hospital visits in the USA in 2007, of which 2.2 million were emergency department visits [1, 2]. Total healthcare costs for hospitalizations due to UTIs in 2011 in the USA were $2.8 billion [3]. At least 50% of all women suffer from a UTI at least once in their lifetime, and 20–30% of women with UTIs will suffer from recurring episodes [4]. UTIs account for 10–14% of all infections in the elderly and are also the second most common bacterial infection (after ear infections) affecting children [5, 6]. Around 7–9% of children will have had a UTI by age 19 [7]. About 25% of hospitalized patients are catheterized, which puts them at the risk of acquiring a catheter-associated UTI (CAUTI). CAUTIs account for 40% of healthcare-associated infections [8]. When complicated by factors such as urinary stones, long-term catheters, and urinary tract surgeries, UTIs carry the risk of urosepsis, which can have a mortality rate of 20–40% [9].
Despite UTIs being so common, they present differently across various age groups, making their diagnosis difficult [10]. For women with uncomplicated UTIs, urinary frequency, urgency, dysuria, and the absence of vaginal discharge are strongly diagnostic of UTIs. For such women, empirical antibiotic therapy is a valid choice. Symptoms for older women are unreliable since urinary frequency and urgency may be present in the absence of infection [10]. Diagnosis is challenging in children since symptoms may be inconsistent or vague, and children often cannot describe their symptoms well. Young children and infants, in particular, need not have the classic symptoms of urinary frequency, urgency, or dysuria [6].
Methods used to diagnose UTIs
Figure 1 shows the traditional and emerging methods used in the laboratory diagnosis of UTIs. Urine samples for analysis are most commonly obtained as “clean catch” mid-stream voided urine samples. The nitrite and leucocyte esterase tests are the two standard dipstick analyses used for urinalysis [11]. The nitrite test is positive for those bacteria that can reduce nitrate to nitrite and detects bacteria at concentrations >105 cfu/mL, and the leucocyte esterase test detects an enzyme produced by leucocytes whose level increases in the urine during infection. Positive nitrite and esterase tests can indicate UTIs, especially in the presence of diagnostic symptoms. However, it is not recommended to treat a positive nitrite or positive esterase test alone as an indication of UTI. The main drawback of these tests lies in their poor negative predictive values. They cannot be used to rule out infections because Gram-positive pathogens such as Enterococci and Staphylococcus do not produce nitrites, and leucocyte esterase levels may not be high enough to be detected early in an infection. Therefore, these dipstick tests are no longer employed at point-of-care; rather, they are used in the clinical laboratory along with other tests [11, 12].
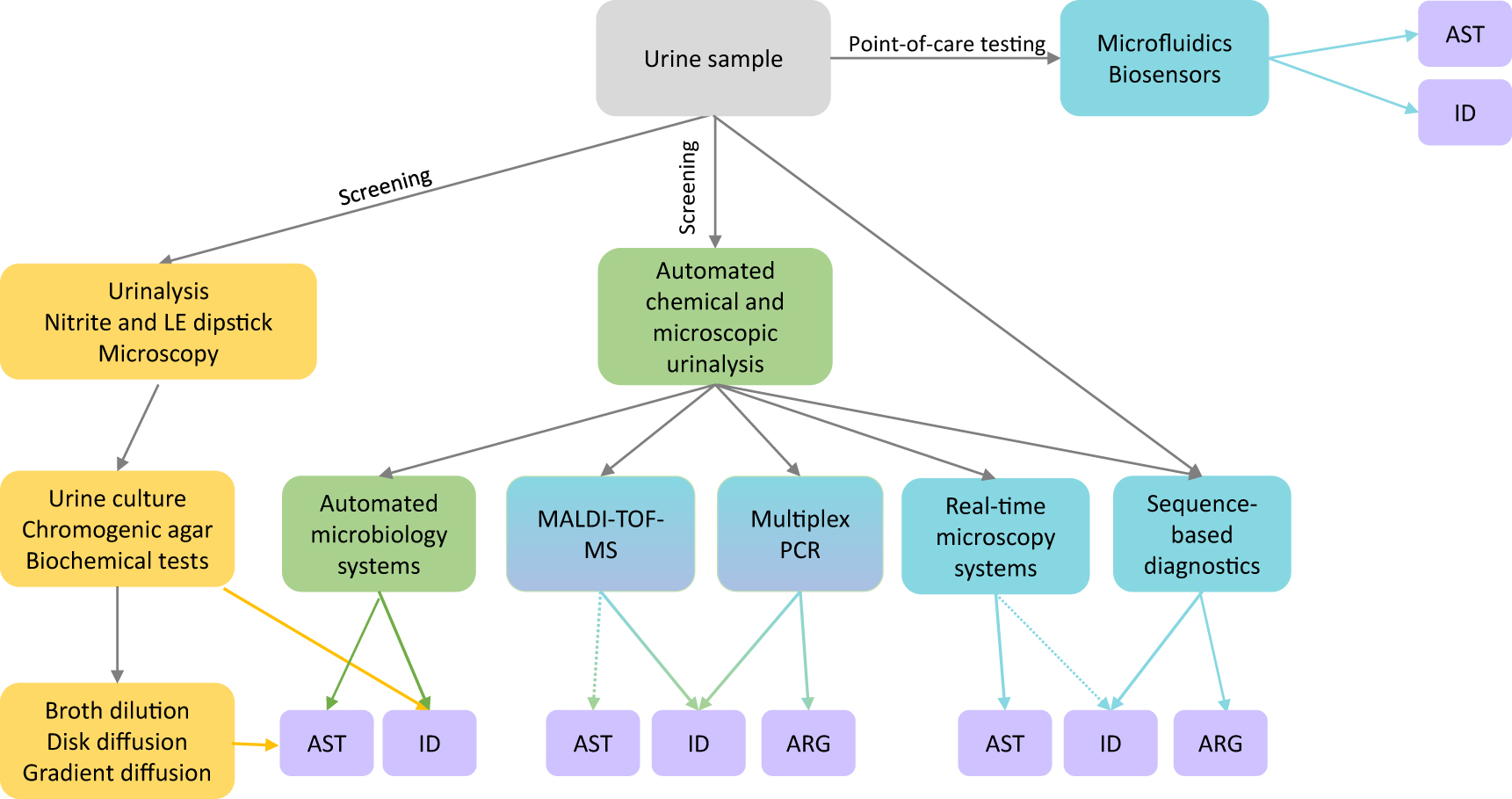
Laboratory methods for diagnosis of UTIs.
Traditionally (yellow boxes), lab diagnosis begins with urinalysis by dipstick tests and microscopy; positive samples may be sent for urine culture where the pathogen is identified (ID) by chromogenic agar or biochemical tests. Antibiotic susceptibility testing (AST) is determined following culture by broth dilution, disk diffusion, or gradient diffusion. In green boxes are represented currently available new automated technologies – automated urinalysis systems for high-throughput screening and automated microbiology systems that can deliver both pathogen ID and AST more rapidly than traditional systems. Methods are in progress (blue-green boxes) for directly using urine samples for pathogen ID (and potentially AST) by MALDI-TOF. Multiplex PCR methods are also commercially available for uropathogen identification and detection of antibiotic resistance genes (ARG). Emerging technologies (blue boxes), such as microfluidics and biosensor-based methods, have the potential for point-of-care testing. Real-time microscopy systems also are emerging as a tool for rapidly determining AST and possibly, ID. Urine samples may be directly used in metagenomic sequencing to determine bacterial species and to detect antibiotic resistance genes in the whole sample. Dotted arrows represent in progress.
Clinical labs may also perform microscopic analysis of the urine to detect white blood cells or bacteria. Pyuria and bacteriuria are helpful for UTI diagnosis when symptoms are present. In the absence of symptoms, bacteriuria may be a result of contamination or asymptomatic bacteriuria. Asymptomatic bacteriuria is present in 5% of healthy premenopausal women, 2–10% of pregnant women, and in 10–15% of older women [10]. It can occur in school children, patients with catheters, institutionalized elderly patients, and patients with complications such as diabetes or spinal cord injuries. Treatment for asymptomatic bacteriuria is recommended only for pregnant women [13].
Urine culture is considered the most diagnostic test for UTIs
The most common cause of UTIs across all age groups is Escherichia coli. Other common uropathogens are the Enterobacteriaceae family, Proteus, Klebsiella, Staphylococcus saprophyticus and Enterococcus sp. Group B Streptococcus is frequently isolated from pregnant women [10]. Common pathogens are easily identified with the use of chromogenic agar during urine culture. For the identification of other species, automated systems are often used, such as MALDI-TOF mass spectrometry. Under the current laboratory practices, sample collection to pathogen identification can take as long as 18–30 h [12].
Bacterial counts for the dominant species at above 105 cfu/mL were traditionally considered diagnostic; however, this value results in a large number of false negatives [10]. Studies have shown that 30–50% of women with symptomatic UTIs can have bacterial counts as low as 102 cfu/mL. Most laboratories use 104 cfu/mL as the diagnostic threshold [10]. However, high bacterial counts do not necessarily indicate infection. Treatment of asymptomatic bacteriuria contributes to future resistant infections, and therefore the European Association of Urology (EAU) does not recommend urine cultures in the absence of symptoms for uncomplicated UTIs. For pyelonephritis, catheter-associated UTIs, and complicated UTIs, urine culture and antibiotic susceptibility testing are recommended because diverse species can cause infections under these situations [14]. Traditional guidelines for interpretation of urine culture dictate that cultures with more than one dominant species are considered contaminated. However, many UTIs are polymicrobial, especially UTIs affecting the elderly population, CAUTIs, and complicated UTIs [15].
Before we discuss new tools that deliver rapid results, we briefly highlight an improved urine culture method. The expanded quantitative urine culture (EQUC) offers improved pathogen identification as compared to standard urine culture. Standard urine culture is effective with common uropathogens such as E. coli and other Gram-negative bacilli but misses many Gram-positive bacteria and other anaerobic, fastidious, or slow-growing bacteria. The EQUC method resulted from simple modifications to the standard method (plating volumes, growth medium, and incubation conditions). When 65 urine samples (41 from patients with overactive bladder and 24 from healthy controls) were cultured, standard culture missed 92% of the isolates that were obtained by EQUC [16]. In another study comparing the two culture methods, standard culture missed 67% of the uropathogens that were detected by EQUC in urine samples from 150 women [17]. Since the EQUC uses a longer incubation time than standard culture, increasing the time to result, the authors recommend using the EQUC as a supplemental tool for patients with UTI symptoms that give negative results with standard urine culture, and for patients with recurring UTIs [17].
Antibiotic resistance among uropathogens
The rise in antibiotic resistance among uropathogens is of growing concern and adds to the economic burden of healthcare. The CDC (Center for Disease and Control) 2019 report estimated that 2.8 million antibiotic-resistant infections and 35,000 deaths occur every year in the USA [18]. Antibiotic resistance increases mortality by 30–40%. The number of hospitalizations resulting from UTIs increased by 76% between 1998 and 2011 in the USA; this increase has been attributed to antibiotic resistance [3]. Resistance to fluoroquinolones such as ciprofloxacin has increased among the Enterobacteriaceae to such an extent that they are no longer recommended for empiric antibiotic therapy for UTIs [19]. Trimethoprim-sulfamethoxazole is recommended as the first line of antibiotics for UTIs only when the local resistance rate is <20% [20]. The prevalence of extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae has limited the available options for antibiotic therapies [21]. ESBL-encoding plasmids often also encode resistance to trimethoprim-sulfamethoxazole and fluoroquinolones [22]. In a systematic meta-analysis, ESBL-producing Enterobacteriaceae caused one out of seven pediatric UTIs and one out of 10 UTIs in renal transplant patients [23, 24]. The rates of isolation of ESBL-positive E. coli and Klebsiella pneumoniae increased between 2010 and 2014; the rates vary across countries [25]. Nitrofurantoin, one of the first antibiotics used for UTIs, is now prescribed as the first line of defense. Nitrofurantoin is effective against E. coli and Enterococci, but many uropathogens such as Proteus, Pseudomonas, Providencia, and Acinetobacter spp. are intrinsically resistant to nitrofurantoin [20]. Bacteria producing ESBLs are resistant to most antibiotics except carbapenems. Therefore, it is worrying that the CDC 2019 report listed carbapenem-resistant Acinetobacter and carbapenem-resistant Enterobacteriaceae as urgent threats [18].
The overuse of antibiotics, particularly the large-scale use of antibiotics in the poultry and cattle industry, is the primary reason for the increase in multi-drug resistance [18]. Inappropriate prescriptions contribute to the spread of resistance genes in the microbial community. A large study performed in the US showed that 30–50% of antibiotic prescriptions for outpatient visits in 2010–2011 were incorrect for either the indication, the dosage, or the duration [26]. Another study with data from more than 300 hospitals showed that antibiotic use among hospitalized patients could have been decreased by 40% [27]. Previous antibiotic therapy or prophylactic treatments or hospitalizations increase the risk of multi-drug resistant infections. Multi-drug resistance is higher among hospital-acquired UTIs than community-acquired UTIs and higher among elderly patients in nursing care homes and other institutes than those who are not [28]. Previous unnecessary treatments of asymptomatic bacteriuria, particularly during long-term catheterization, can result in later resistant infections.
Antibiotic treatment, once thought to be only beneficial, is now known to be detrimental to the natural human microbiota. Empiric broad-spectrum antibiotic therapy, which is routinely prescribed for UTIs, can have unintended consequences for the microbiome. Infections by Clostridioides difficile, causing life-threatening diarrhea, are a direct result of the disruption of the gut microbiome by antibiotic use; C. difficile infections are now the most common healthcare-associated infections and listed as one of five urgent threats by the CDC [18]. The CDC 2019 report stated that 223,900 people required hospital care for C. difficile in 2017 [18]. It is believed that the bladder and the urinary tract are sterile, but recent studies show that microbes can reside here and help in protecting the system from infection by external pathogens [29]. In 2011, IDSA (Infectious Disease Society of America) and European Society for Microbiology and Infectious Diseases (ESCMID) have issued guidelines stating that while choosing antibiotics and the duration of the therapy, the adverse effects of antibiotic therapy must be considered along with the efficacy of the chosen treatment [30].
Knowledge of antibiotic susceptibility of an infection, in addition to the identification of the pathogen, is essential to providing appropriate therapy. The standard method of determining Antibiotic susceptibility testing (AST) is through phenotypic bacterial growth assays in the presence of antibiotics, where the strain is classified as resistant, susceptible, or intermediate. Conventional manual methods of determining phenotypic AST include broth dilution, disk diffusion, and gradient diffusion, and involve manual sample preparation steps and additional time of 16–24 h from culture isolation (Figure 1). Newer high-throughput instruments that are now employed by several large laboratories enable automated sample preparation, increased sensitivity, and reduced time of 10–16 h. These commercially available and FDA-approved instruments include the Microscan Walkaway (Beckman Coulter), Phoenix Automated Microscopy System (Beckton-Dickinson), and the Vitek-2 (bioMerieux) (Table 1).
Technologies used in laboratory diagnosis of UTIs.
| Technology/method | Commercially availablea | FDA approved | Uses | Pros | Cons | |
|---|---|---|---|---|---|---|
| Dipstick assays | Nitrite and leucocyte esterase tests | Yes – several (Scanwell Health UTI has in-home test for use with smartphone) | Yes | Urinalysis, screening | Rapid tests, inexpensive | Poor specificity; negative tests do not rule out infection |
| Immunological test | RapidBac (Silver Lake Research Corporation) | Yes (for vet use) | Detects bacteriuria | Rapid, inexpensive test; high sensitivity for Gram-negative bacteria | Poor sensitivity for Gram-positive infections | |
| Automated urine analyzers | Flow cytometry | UF-1000i, UF-5000 (Sysmex) | Yes | Chemical and/or microscopic urine analysis; for screening purposes | High negative predictive value; can reduce workflow | No pathogen ID or AST information; screening utility depends on cutoffs employed and cutoffs need to be optimized for each laboratory; bacterial counts tend to be higher since they do not distinguish between live and dead cells |
| Light scattering | 216 Dx (Bacterioscan) | Yes | Can be developed for AST | |||
| Microscopy-based sediment analyzer | SediMAX (77 Electronika) | More reliable and less time-consuming than manual microscopic analysis | ||||
| Chemical urinalysis | Clinitek Novus (Siemens), Urisys 1100, Urisys 2400 (Roche Diagnostics) | Yes | High-throughput chemical analysis | |||
| Chemical and microscopic analysis | iQ workcell (Beckman Coulter); Cobas 6500 (Roche); UN series (Sysmex) | Yes | Chemical and microscopic automated analysis; can flag samples for urine culture | |||
| Urine culture | Standard culture | Chromogenic agar | ID | Gold standard for UTI diagnosis | Time-consuming; not all uropathogens are cultivated by standard culture | |
| Expanded quantitative urine culture | ID | Can detect slow-growing, anaerobic and fastidious pathogens | Time-consuming, takes longer than standard culture | |||
| Automated microbiology systems | Vitek-2 (bioMerieux), Microscan Walkaway (Beckman coulter), Phoenix (BD) | Yes | ID, AST | Automated, faster than traditional AST methods | Still takes 10–16 h for AST results | |
| MALDI-TOF MS | Vitek-MS (bioMerieux), MALDI Biotyper (Bruker) | Yes | ID | Rapid pathogen ID from bacterial culture; potential for direct urine testing | Currently not suitable for AST or for polymicrobial infections | |
| Multiplex PCR | Guidance UTI (Pathnostics); Step-2 (AusDiagnostics), real-time PCR solutions by Thermo Fisher | Nob | ID, ARG | Sensitive pathogen ID in a couple of hours; direct urine testing possible | Cannot give MIC information or phenotypic AST | |
| Emerging technologies | Microfluidics | ID, AST | Small, cost-effective, high-throughput, multiplex analysis and point-of-care; rapid AST in 0.5–6 h | Requires more validations and clinical trials with different pathogen/antibiotic combinations | ||
| Real-time microscopy | oCelloScope (Biosense Solutions) | No | AST | Can determine MIC values; provides rapid AST results in 0.5–4 h | Requires more validations and clinical trials with different pathogen/antibiotic combinations | |
| Biosensors | ID, AST | Small, cost-effective, amenable to point-of-care; rapid ID and AST results in less than 6 h | Requires more validations and clinical trials with different pathogen/antibiotic combinations | |||
| Sequence-based diagnostics | ID, ARG | Can diagnose polymicrobial infections; can provide ID for each pathogen and the antibiotic resistance genes encoded by each species | Currently more expensive and time-consuming than conventional methods; does not deliver phenotypic MIC results | |||
-
aNot an exhaustive list. bFDA approved multiplex PCR panels are available for other diseases such as respiratory, gastrointestinal, meningitis/encephalitis. ID, pathogen identification; AST, antibiotic susceptibility testing; ARG, antibiotic resistance gene.
New diagnostic technologies
A timely diagnosis of UTIs is essential to prevent inappropriate antibiotic treatments and to promote antibiotic stewardship, thereby. The pathogen and its antibiotic susceptibility should be identified within a few hours of sample collection, such that the correct therapy can be started within the same day that the patient visits the doctor. New diagnostic technologies have the potential to promote both patient care and management as well as antibiotic stewardship by delivering rapid results. Urine culture is the most time-consuming step in current laboratory diagnosis; therefore, new technologies need to be able to work directly on urine samples without the need for enrichment or bacterial isolation. New methods need to account for the possible polymicrobial nature of UTIs and allow AST for each dominant pathogen present in the urine sample. For any technology to be implemented in a clinical laboratory, it also needs to be easy to use and cost-effective.
Below we first discuss new screening tools, followed by rapid pathogen identification tools utilizing existing molecular platforms such as mass spectrometry and multiplex PCR. We then discuss emerging platforms for rapid antibiotic susceptibility testing using biosensors, microfluidics, real-time microscopy systems, and sequence-based diagnostics (Table 1).
Screening methods
The methods that we discuss in this section aim to improve laboratory workflow and reduce costs by screening urine samples to quickly identify and remove negative samples while not missing any positive samples.
Automated instruments such as flow cytometers, light scattering instruments, and urine sediment analyzers make useful screening tools with a high negative predictive value (Table 1). Flow cytometers analyze urine by sorting and counting cells, and they can distinguish between bacteria, yeast, white blood cells, red blood cells, and epithelial cells. Several studies have evaluated the FDA-approved Sysmex UF-1000i. The range of bacterial cell counts diagnostic for UTI varies from 103 to 105 cfu/mL of urine, and accordingly, different studies have used different cutoffs for the number of bacteria per µL of urine. The screening utility of flow cytometers depends on the cutoffs applied, and each laboratory must optimize the cutoffs based on the prevalence of UTI it encounters [31, 32]. In a recent study on 546 hospitalized children, 10 bacteria/µL was used as a threshold to achieve a negative predictive value of 100% [33]. Other studies, where UTI diagnosis was based on bacterial counts of >105 cfu/mL, achieved 96% sensitivity and 99.7% negative predictive value with a cutoff of 230 bacteria/µL [34], or 97% sensitivity and 94% specificity with cutoffs of 125 bacteria/µL and 40 leucocytes/µL [35]. Another study that screened urine samples from children found that using leucocyte counts (35 cells/µL cutoff) gave more reliable results than bacterial or red blood cell counts [36]. One study evaluated the newer Sysmex Uf-5000 instrument and found that bacterial cutoffs of 58/µL gave 99.4% sensitivity and a negative predictive value of 99.7% [37]. The new Sysmex UF-5000 also has better capabilities to distinguish between Gram-negative and Gram-positive bacteria. A recent study that evaluated this instrument with 1,430 urine samples found that it works better with Gram-negative bacteria [31]. Most studies agree that flow cytometry screening significantly reduces the number of samples for further processing [36, 38].
The Bacterioscan 216 Dx is an FDA-approved semi-automated system that uses light scattering profiles in the urine samples to detect bacteria in 3 h. When tested with urine samples from children, the 216 Dx had similar sensitivity but higher specificity than dipstick urinalysis. With >104 cfu/mL as UTI diagnostic cutoff, the 216 Dx had 92% sensitivity, 82.7% specificity, and a negative predictive value of 98.6% [39].
Automated urine analyzers such as the Clinitek Novus (Siemens) and Urisys series (Roche) provide high-throughput chemical urinalysis by assaying for protein, glucose, nitrites, and leucocyte esterase, among other parameters (Table 1). Automated urine sediment analyzers (e.g. SediMAX) can microscopically analyze the urine sediment for leucocytes, erythrocytes, epithelial cells, bacteria, and yeast. Chemical urine analyzers have also been integrated with microscopic analyzers or flow cytometers to provide complete automated urine analysis (e.g. Clinitek AUWi (Siemens), UN series (Sysmex), Cobas 6500 (Roche)), and these systems can flag samples for further analysis, such as urine culture (Table 1). Studies that have evaluated systems such as these agree that they are useful for screening negative samples, and that they provide more reliable results than manual urinalysis, particularly for labor-intensive microscopic analysis. However, for samples with suspected bacteriuria, manual microscopic analysis is also recommended to confirm pathology [40], [41], [42], [43].
Since the current nitrite and esterase dipstick assays are not reliable as point-of-care screening assays owing to their poor negative predictive value, the development of a sensitive and specific dipstick assay would greatly enable the rapid screening of samples. A new monoclonal antibody-based immunoassay, RapidBac (Silver Lake Research Corporation, Monrovia, CA), has potential as a point-of-care dipstick assay. RapidBac has been approved for veterinarian use and was recently evaluated for use on human urine samples [44]. When 966 human urine samples were tested using the RapidBac kits, the tests were 90% sensitive and 94% specific for Gram-negative bacteria in samples with >103 cfu/mL, when compared with results from standard urine culture. For samples with >105 cfu/mL, the sensitivity was 99%. In addition, the assay time was just 20 min [44].
Novel screening assays are being developed that need further clinical evaluation. For instance, an ATP-sensing paper was designed based on the ATP-driven bioluminescence reaction; it could detect microbial ATP in urine as a marker for UTI with bacterial counts of >105 cfu/mL. This ATP-sensing paper can be combined with smartphone detection for cost-effective screening [45].
Mass-spectrometry
MALDI-TOF (matrix-assisted laser desorption ionization-time of flight) mass spectrometers are increasingly used in clinical laboratories for unambiguous species-level identification of bacteria and yeast. Two FDA-cleared MALDI-TOF MS systems are available – the VITEK® MS system (bioMérieux Inc.) and the MALDI Biotyper® CA system (Bruker Daltonics Inc.). Although initial instrument costs are high, MALDI-TOF-MS offers savings on technical labor and reagent costs and delivers rapid and accurate results. The main disadvantage is that the method requires a pure culture of bacteria and is typically used with isolated bacterial colonies following standard urine culture. The challenge in using urine samples directly is that the urine matrix (pH, electrolyte concentration, cellular composition) varies not only from person to person but also from the same person at different times. Nevertheless, many groups have explored different urine processing steps such as differential centrifugation or dual filtration with promising results [46], [47], [48]. To save on workflow times, many studies have also examined the combination of screening tools followed by direct testing of urine samples with MALDI-TOF. The results from studies that prescreened samples with flow cytometry followed by MALDI-TOF ranged in accuracy from 74 to 94% when compared with conventional identification [46, 49, 50]. A combination of MALDI-TOF-MS and urinalysis (leucocyte esterase positive, nitrate positive, and bacterial counts of >500/µL) resulted in increased sensitivity and specificity for pathogen identification than by either method alone and provided results within 1 h [47].
MALDI-TOF continues to be limited by its inability to process polymicrobial urine samples and to provide AST. However, recent developments in the field have enabled the detection of carbapenemase-producing Enterobacteriaceae with MALDI-TOF by using imipenem as a marker [51, 52]. Other mass spectrometry methods also offer the potential for rapid diagnosis and AST. An LC-MS/MS-based method employed machine-learning algorithms to identify peptidic signatures for specific uropathogens and targeted proteomics to search the test urine samples for the peptidic signatures [53]. This method provided identification in less than 4 h directly from urine. Peptidic libraries were created for 15 of the most common uropathogens that cause about 84% of all UTIs [53]. It is conceivable to develop peptidic signatures for common antibiotic resistance mechanisms and get knowledge of antibiotic susceptibility as well. Some mass-spectrometry methods have been recently used to parse specific antibiotic resistance [54].
Multiplex PCR
Multiplex PCR offers a cost-effective and rapid approach to pathogen identification. Quantification of the pathogen DNA is necessary for diagnosing UTIs since it is necessary to distinguish between contamination and infection. Many studies have examined real-time PCR assays for specific pathogens, but only a few studies have looked at multiplex PCR panels for UTI pathogens [55], [56], [57], [58]. All such studies have found that multiplex PCR compares favorably with a standard urine culture. When samples are positive by culture but negative by PCR, it is usually because the specific pathogen was not represented in the panel [55, 57]. Multiplex PCR delivers substantial time savings over standard urine culture.
The most extensive panel to be investigated was a commercially available multiplex real-time PCR test (Guidance UTI Test, Pathnostics, Irvine, CA) that could detect 31 uropathogens [57]. This study retrospectively reviewed 582 elderly patients with UTI symptoms and compared results from multiplex PCR panels with that from urine culture. PCR was more sensitive, detecting pathogens in 56% of samples vs. 37% by urine culture. Where samples were positive by both methods, the results were 90% in agreement. PCR also detected more polymicrobial infections than urine culture (166 vs. 39 cases). Eight pathogens were only identified by PCR and not detected by standard culture. The Guidance UTI test also tests for antibiotic susceptibility, but these have not been evaluated yet.
Multiplex PCR panels may also be used for antibiotic susceptibility testing by representing common antibiotic resistance markers. One such commercially available multiplex tandem PCR panel that tests for common antibiotic resistance genes present in Enterobacteriaceae (AusDiagnostics, Sydney, Australia) was evaluated recently. The PCR panel delivered AST results in 3 h, and the study concluded that such panels could help in getting appropriate and immediate antibiotic therapy [59].
Microfluidics platforms for AST
Miniaturized microfluidic devices are very attractive options for emerging diagnostic tools for antibiotic susceptibility testing. Microfluidic platforms are cost-effective and amenable to automation, multiplexing, high-throughput processing, and single-cell analysis. Their portability confers great potential as point-of-care devices [60, 61]. Their small size enables them to use minimal volumes of samples and reagents. Small confined spaces within the microfluidic device allow the detection of bacterial cell division and growth in far less time than conventional phenotypic AST assays. Various groups have developed microfluidic platforms capable of direct testing on urine samples. Some platforms offer both pathogen identification (or classification) and AST [62], [63], [64], [65], while some only deliver AST [66, 67]. It can be argued that AST information is more relevant than pathogen identification for prescribing treatment.
The different platforms vary in their method of pathogen identification and detection of bacterial growth. Bacterial growth in the presence or absence of antibiotics may be measured by monitoring the change in the length of bacteria trapped in pneumatic microchannels [62], using fluorescent dyes like resazurin that indicate cell viability [66], dark-field imaging of agglutinated bacterial clusters [64], ATP-bioluminescence assay [65], and by nucleic acid amplification [63, 67, 68]. Pathogens could be identified by classifying bacteria trapped in pneumatic microchannels by size and shape [62], using pathogen-specific 16S rRNA probes [64], by high-resolution melt curve profiles [63], and using pathogen-specific capture antibodies [65]. Most of these platforms could deliver results within 3–6 h, with some platforms delivering in under 30 min. An ultra-fast digital loop-mediated isothermal amplification method (7 min) could determine AST of E. coli after exposure to antibiotics for just 15 min [67], and this method could be integrated into commercially available microfluidic chips [68]. Some platforms are also capable of detecting polymicrobial infections and reporting AST for each identified pathogen [62, 63]. Studies on these platforms, although performed with a limited number of clinical samples or limited to AST for only E. coli, show high sensitivities and specificities and good agreement of AST results with that of standard AST methods. For these methods to be developed further, they need to be tested extensively with a large number of clinical samples and with several pathogens and antibiotic combinations.
Real-time microscopy systems
Bright-field optical microscopic imaging systems can monitor bacterial growth and cell-division in real-time directly in urine in the presence or absence of antibiotics and, thus, deliver AST results much faster than standard phenotypic methods (usually under 3 h) [69], [70], [71], [72], [73], [74]. Many of these systems can also determine minimum inhibitory concentrations (MIC) of antibiotics, which are more useful for guiding treatment than binary resistant/susceptible classifications [69, 71, 73]. Some of the microscopy-based methods have also been integrated into microfluidic chips and offer single-cell analysis [69, 70]. Although the AST/MIC determined by microscopy-based methods agree well with the results from standard AST methods, most of the methods have been tested only on a few samples and a few strains and require more validation and development before implementation in a clinical laboratory. The oCelloScope (BioSense Solutions Aps, Denmark) is one such technology that uses time-lapse bright-field imaging of up to 96 bacteria-antibiotic combinations and can be used on urine samples and is commercially available [72, 75, 76].
The bacterial cells may be imaged while immobilized or while freely moving. Imaging of freely moving cells gives the advantage of reduced sample preparation. A new video microscopy-based method imaged freely moving bacteria in urine and applied deep machine learning algorithms to learn phenotypic features of bacteria such as bacterial motion, morphology, and cell division, and how these features changed in response to antibiotics [71]. Although training the algorithm was time-consuming, MICs could then be determined rapidly in less than 30 min. The method was validated with E. coli and its response to five antibiotics. If the models are trained on different pathogens, this method should also be able to identify pathogens in urine samples. Another method used light-scattering microscopy to image bacteria and monitor growth by tracking bacterial multiplication directly in urine samples [74] and could determine AST classification for isolates in 15 min and clinical urine samples in 90 min.
Some real-time microscopy systems use immobilized bacterial cells. In one such system, bacteria were immobilized in agarose within a high-throughput microfluidic chip, and single-cell morphological changes were imaged and analyzed by an automated algorithm. MICs were determined for isolates from 189 clinical samples representing five species (E. coli, Pseudomonas aeruginosa, K. pneumoniae, Staphylococcus aureus, and Enterococcus sp.) in 3–4 h [69]. In another microfluidic chip, single-cell imaging allowed the classification of E. coli as resistant or susceptible to nine antibiotics in less than 30 min. This method was validated on 50 clinical samples with ciprofloxacin-resistant or sensitive UPEC isolates and needs to be tested with other pathogens [70]. A third example of a rapid microscopy-based AST system does not use a microfluidic chip; rather, single bacterial cells were tethered to a glass surface, and the sub-micron motion of single cells was imaged. An automated software was used to analyze the images and correlate them to cell viability in response to antibiotics. This method could determine MICs for polymyxin B in 2 h from urine samples spiked with UPEC [73].
Biosensors
Biosensors are small and cost-effective, deliver rapid results, and have the potential to be integrated into point-of-care devices. Several biosensors – with electrochemical, mechanical, or optical transducers – have been explored for the detection of UTIs [77]. Inexpensive sensors can detect pathogens at very low concentrations in urine samples – a nanoplasmonic sensor detects E. coli as low as 102 cfu/mL [78], and an impedance sensor could detect a range of E. coli from 7 to 7 × 108 cfu/mL [79]. However, very few have been used for direct testing on urine samples, and very few can provide both pathogen identification as well as antibiotic susceptibility testing.
One such integrated electrochemical biosensor was recently developed. It used universal and pathogen-specific 16S rRNA probes to identify pathogens and determine MIC in less than 6 h (including the time for brief culturing of urine samples with and without antibiotics) [80]. The sensor was validated with ∼100 urine samples; the assay had 98.5% sensitivity, 96.6% specificity, 93.0% positive predictive value, and 99.3% negative predictive value. MIC was determined for ciprofloxacin and was 97.6% in agreement with standard methods. Although species identification is currently possible only for six pathogens, this sensor can be applied to polymicrobial samples and shows promise for more extensive development and testing [80].
A new study delivered molecular biosensors directly into viable bacterial cells using a technique called nanotube assisted microwave electroporation. This enabled high target concentration resulting in strong signals from single cells; combining this technique with a microfluidic device achieved pathogen identification within 30 min and AST in 3 h [81]. The method was demonstrated both on clinical isolates and patient samples.
Sequence-based diagnostics
Advances in next-generation sequencing have led to large-scale sequencing of microbial genomes. Whole-genome sequencing of clinical bacterial isolates or metagenomic sequencing of clinical samples is being considered as alternatives to standard clinical diagnostic practices. Sequencing-based diagnostics are highly sensitive and are able to identify pathogens at the species level. Next-generation sequencing can detect organisms that are difficult to cultivate, such as slow-growing, anaerobic, or fastidious organisms. Metagenomic sequencing of urine samples can generate a comprehensive picture of the bacterial, eukaryotic, and viral communities in the urinary tract [82]. However, the sensitivity of next-generation sequencing itself can be a limitation making it challenging to interpret sequencing results. The most dominant species seen by sequencing may not be the pathogen responsible for the infection [83]. Since voided urine samples are particularly prone to contamination from the surrounding genital areas, sequencing results must be interpreted with caution. To overcome these limitations, next-generation sequencing needs to be tested in well-designed clinical studies.
Only a handful of studies have compared the diagnosis of UTIs by sequencing-based methods with that of conventional methods. Next-generation sequencing of the PCR amplified 16S-23S rRNA regions was used as a diagnostic tool to identify causative pathogens in 60 urine samples. This method was found to be superior to just 16s rRNA sequencing in its ability to discriminate between closely related species and also superior to culture-dependent methods in its sensitivity [84]. MinIon nanopore sequencing technology, a faster and less expensive sequencing technology, was tested on 10 clinical urine samples and five artificially spiked urine samples. MinIon sequencing could correctly identify the pathogens and most of the acquired antibiotic resistance genes, but it did poorly at differentiating between allelic variants of resistance genes [85]. Although MinIon technology is more expensive than conventional diagnostics, it had a turnaround time of 4 h and, thus, shows promise for further development.
Shotgun DNA sequencing of urinary cell-free DNA, containing debris of genomes of dead cells (human and microbial), was recently explored for its diagnostic potential with urine samples from 141 kidney transplant recipients, some with bacterial UTI [86]. Pathogen identification results obtained by sequencing agreed with those from standard methods, and in addition, bacteria and viruses not detected by culture methods could be identified by sequencing (for example, a case of UTI caused by Haemophilus influenzae, a very unusual uropathogen, was identified). Additionally, cell-free DNA sequencing was able to give information on the bacterial population growth rates by examining skewed genome coverage (greater coverage of sequences surrounding the origin of replication indicates faster growth rate). They also observed good agreement with the detection of antibiotic resistance gene fragments and phenotypic AST results. Thus, a large body of information could be obtained from a single diagnostic assay [86].
Whole-genome sequences or metagenomic sequences can be mined for the presence of antibiotic resistance genes, and this data used to generate a resistance profile. Thus, sequencing-based diagnostics offer the potential of pathogen identification and AST in a single assay. As attractive as this method is, there are several hurdles to be overcome. Genotypic antibiotic susceptibility results from whole-genome sequencing are far more expensive and more time-consuming than standard diagnostic tests and phenotypic AST [87]. As sequencing technologies continue to improve, costs and turnaround times are predicted to decrease in the future. Robust bioinformatic tools are also being developed that enable rapid and accurate analysis of sequencing data [88]. The primary challenge is to predict phenotype from genotype because the mere presence or absence of antibiotic resistance genes does not always predict susceptibility. Many factors that determine the expression of genes (promoter that drives expression, genomic context, presence of regulatory factors), or the environmental context of the pathogen (for example, being in a biofilm naturally increases resistance), contribute to antibiotic resistance phenotypes. Determining which antibiotics an infection is susceptible to is more relevant to prescribing treatment than knowing which antibiotics the infection is resistant to. Accurate prediction from genotype also requires knowledge of all genetic variations responsible for a resistance phenotype and requires that databases be updated as new genetic mechanisms are uncovered [87, 88].
Conclusions
By providing patients with a timely diagnosis and tailored antibiotic therapies, healthcare providers cannot only improve patient care but also contribute to antibiotic stewardship. As we have discussed, many new technologies and methods are being developed that aim to both identify the pathogens and determine their antibiotic susceptibilities very rapidly by directly testing clinical urine samples (Figure 1, Table 1). Screening tools such as the flow cytometers are already in use in clinical laboratories and greatly help to reduce the number of samples for downstream processing. Multiplex PCR panels are commercially available and show promising results. MALDI-TOF mass spectrometers are also routinely used in clinical laboratories, but with new developments in urine pre-processing, urine samples can be directly applied for MALDI-TOF-MS. Clinicians and healthcare providers need to be aware of the emerging microfluidics, biosensors, and real-time microscopy platforms that have demonstrated very rapid turnaround times. Further development of these technologies, with validation on a more significant number of samples and with a higher number of pathogen-antibiotic combinations, will pave the way for tools that are ready to be implemented in the clinical laboratory.
Acknowledgments
All persons who have made substantial contributions to the work reported in the manuscript (e.g., technical help, writing and editing assistance, general support), but who do not meet the criteria for authorship, are named in the Acknowledgements and have given us their written permission to be named. If we have not included an Acknowledgements, then that indicates that we have not received substantial contributions from non-authors.
-
Research funding: Not applicable.
-
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and approved its submission. All persons who meet authorship criteria are listed as authors, and all authors certify that they have participated sufficiently in the work to take public responsibility for the content, including participation in the concept, design, analysis, writing, or revision of the manuscript. Furthermore, each author certifies that this material or similar material has not been and will not be submitted to or published in any other publication before its appearance in Journal ‘Journal of Laboratory Medicine’.
-
Competing interests: Author Mohammed Harris is a part-time consultant for Locus Science LLC.
-
Informed consent: Not applicable.
-
Ethical approval: Not applicable.
References
1. Harding, GK, Ronald, AR. The management of urinary infections: what have we learned in the past decade? Int J Antimicrob Agents 1994;4:83–8. https://doi.org/10.1016/0924-8579(94)90038-8.Search in Google Scholar
2. Schappert, SM, Rechtsteiner, EA. Ambulatory medical care utilization estimates for 2007. Vital Health Stat 13 2011;169:1–38.Search in Google Scholar
3. Simmering, JE, Tang, F, Cavanaugh, JE, Polgreen, LA, Polgreen, PM. The increase in hospitalizations for urinary tract infections and the associated costs in the United States, 1998–2011. Open Forum Infect Dis 2017;4:ofw281. https://doi.org/10.1093/ofid/ofw281.Search in Google Scholar
4. Foxman, B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am J Med 2002;113(1A Suppl):5S–13S. https://doi.org/10.1016/s0002-9343(02)01054-9.Search in Google Scholar
5. Ruben, FL, Dearwater, SR, Norden, CW, Kuller, LH, Gartner, K, Shalley, A, et al.. Clinical infections in the noninstitutionalized geriatric age group: methods utilized and incidence of infections. The Pittsburgh Good Health Study. Am J Epidemiol 1995;141:145–57. https://doi.org/10.1093/oxfordjournals.aje.a117402.Search in Google Scholar PubMed
6. Becknell, B, Schober, M, Korbel, L, Spencer, JD. The diagnosis, evaluation and treatment of acute and recurrent pediatric urinary tract infections. Expert Rev Anti Infect Ther 2015;13:81–90. https://doi.org/10.1586/14787210.2015.986097.Search in Google Scholar PubMed PubMed Central
7. Shaikh, N, Morone, NE, Bost, JE, Farrell, MH. Prevalence of urinary tract infection in childhood: a meta-analysis. Pediatr Infect Dis J 2008;27:302–8. https://doi.org/10.1097/inf.0b013e31815e4122.Search in Google Scholar PubMed
8. Haley, RW, Culver, DH, White, JW, Morgan, WM, Emori, TG. The nationwide nosocomial infection rate. A new need for vital statistics. Am J Epidemiol 1985;121:159–67. https://doi.org/10.1093/oxfordjournals.aje.a113988.Search in Google Scholar PubMed
9. Wagenlehner, FME, Lichtenstern, C, Rolfes, C, Mayer, K, Uhle, F, Weidner, W, et al.. Diagnosis and management for urosepsis. Int J Urol 2013;20:963–70. https://doi.org/10.1111/iju.12200.Search in Google Scholar PubMed
10. Chu, CM, Lowder, JL. Diagnosis and treatment of urinary tract infections across age groups. Am J Obstet Gynecol 2018;219:40–51. https://doi.org/10.1016/j.ajog.2017.12.231.Search in Google Scholar PubMed
11. Kumar, S, Dave, A, Wolf, B, Lerma, EV. Urinary tract infections. Dis Mon 2015;61:45–59. https://doi.org/10.1016/j.disamonth.2014.12.002.Search in Google Scholar PubMed
12. Davenport, M, Mach, KE, Shortliffe, LMD, Banaei, N, Wang, T-H, Liao, JC. New and developing diagnostic technologies for urinary tract infections. Nat Rev Urol 2017;14:296–310. https://doi.org/10.1038/nrurol.2017.20.Search in Google Scholar PubMed PubMed Central
13. Nicolle, LE, Gupta, K, Bradley, SF, Colgan, R, DeMuri, GP, Drekonja, D, et al.. Clinical practice guideline for the management of asymptomatic bacteriuria: 2019 update by the Infectious Diseases Society of America. Clin Infect Dis 2019;68:1611–5. https://doi.org/10.1093/cid/ciz021.Search in Google Scholar PubMed
14. Bonkat, G, Pickard, R, Bartoletti, R, Cai, T, Bruyere, F, Geerlings, SE, et al.. EAU guidelines on urological infections; 2018. Available from: https://uroweb.org/guideline/urological-infections/ [Accessed 28 Aug 2019].Search in Google Scholar
15. Kline, KA, Lewis, AL. Gram-positive uropathogens, polymicrobial urinary tract infection, and the emerging microbiota of the urinary tract. Microbiol Spectr 2016;4. https://doi.org/10.1128/microbiolspec.UTI-0012-2012.Search in Google Scholar PubMed PubMed Central
16. Hilt, EE, McKinley, K, Pearce, MM, Rosenfeld, AB, Zilliox, MJ, Mueller, ER, et al.. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol 2014;52:871–6. https://doi.org/10.1128/jcm.02876-13.Search in Google Scholar PubMed PubMed Central
17. Price, TK, Dune, T, Hilt, EE, Thomas-White, KJ, Kliethermes, S, Brincat, C, et al.. The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol 2016;54:1216–22. https://doi.org/10.1128/jcm.00044-16.Search in Google Scholar
18. CDC. Antibiotic resistance threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2019. Available from: http://www.cdc.gov/DrugResistance/Biggest-Threats.html.Search in Google Scholar
19. Chen, YH, Ko, WC, Hsueh, PR. The role of fluoroquinolones in the management of urinary tract infections in areas with high rates of fluoroquinolone-resistant uropathogens. Eur J Clin Microbiol Infect Dis 2012;31:1699–704. https://doi.org/10.1007/s10096-011-1457-x.Search in Google Scholar PubMed
20. Concia, E, Bragantini, D, Mazzaferri, F. Clinical evaluation of guidelines and therapeutic approaches in multi drug-resistant urinary tract infections. J Chemother 2017;29:19–28. https://doi.org/10.1080/1120009x.2017.1380397.Search in Google Scholar
21. Boyd, LB, Atmar, RL, Randall, GL, Hamill, RJ, Steffen, D, Zechiedrich, L. Increased fluoroquinolone resistance with time in Escherichia coli from >17,000 patients at a large county hospital as a function of culture site, age, sex, and location. BMC Infect Dis 2008;8:4. https://doi.org/10.1186/1471-2334-8-4.Search in Google Scholar PubMed PubMed Central
22. Paterson, DL, Bonomo, RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev 2005;18:657–86. https://doi.org/10.1128/cmr.18.4.657-686.2005.Search in Google Scholar
23. Flokas, ME, Detsis, M, Alevizakos, M, Mylonakis, E. Prevalence of ESBL-producing Enterobacteriaceae in paediatric urinary tract infections: a systematic review and meta-analysis. J Infect 2016;73:547–57. https://doi.org/10.1016/j.jinf.2016.07.014.Search in Google Scholar PubMed
24. Alevizakos, M, Nasioudis, D, Mylonakis, E. Urinary tract infections caused by ESBL-producing Enterobacteriaceae in renal transplant recipients: a systematic review and meta-analysis. Transpl Infect Dis 2017;19. https://doi.org/10.1111/tid.12759.Search in Google Scholar PubMed
25. Mazzariol, A, Bazaj, A, Cornaglia, G. Multi-drug-resistant Gram-negative bacteria causing urinary tract infections: a review. J Chemother 2017;29:2–9. https://doi.org/10.1080/1120009x.2017.1380395.Search in Google Scholar PubMed
26. Fleming-Dutra, KE, Hersh, AL, Shapiro, DJ, Bartoces, M, Enns, EA, File, TM, et al.. Prevalence of inappropriate antibiotic prescriptions among US ambulatory care visits, 2010–2011. J Am Med Assoc 2016;315:1864–73. https://doi.org/10.1001/jama.2016.4151.Search in Google Scholar PubMed
27. Fridkin, S, Baggs, J, Fagan, R, Magill, S, Pollack, LA, Malpiedi, P, et al.. Vital signs: improving antibiotic use among hospitalized patients. MMWR Morb Mortal Wkly Rep 2014;63:194–200.Search in Google Scholar
28. López Romo, A, Quirós, R. Appropriate use of antibiotics: an unmet need. Ther Adv Urol 2019;11:1756287219832174.10.1177/1756287219832174Search in Google Scholar PubMed PubMed Central
29. Brubaker, L, Wolfe, A. The urinary microbiota: a paradigm shift for bladder disorders? Curr Opin Obstet Gynecol 2016;28:407–12. https://doi.org/10.1097/gco.0000000000000298.Search in Google Scholar PubMed PubMed Central
30. Gupta, K, Hooton, TM, Naber, KG, Wullt, B, Colgan, R, Miller, LG, et al.. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis 2011;52:e103–20. https://doi.org/10.1093/cid/ciq257.Search in Google Scholar PubMed
31. Kim, SY, Park, Y, Kim, H, Kim, J, Koo, SH, Kwon, GC. Rapid screening of urinary tract infection and discrimination of Gram-positive and Gram-negative bacteria by automated flow cytometric analysis using Sysmex UF-5000. J Clin Microbiol 2018;56. https://doi.org/10.1128/JCM.02004-17.Search in Google Scholar PubMed PubMed Central
32. Mejuto, P, Luengo, M, Díaz-Gigante, J. Automated flow cytometry: an alternative to urine culture in a routine clinical microbiology laboratory? Int J Microbiol 2017;2017:8532736. https://doi.org/10.1155/2017/8532736.Search in Google Scholar PubMed PubMed Central
33. Conkar, S, Mir, S. Urine flow cytometry in the diagnosis of urinary tract infection. Indian J Pediatr 2018;85:995–9. https://doi.org/10.1007/s12098-018-2689-x.Search in Google Scholar PubMed
34. Broeren, MAC, Bahçeci, S, Vader, HL, Arents, NLA. Screening for urinary tract infection with the Sysmex UF-1000i urine flow cytometer. J Clin Microbiol 2011;49:1025–9. https://doi.org/10.1128/jcm.01669-10.Search in Google Scholar
35. Manoni, F, Fornasiero, L, Ercolin, M, Tinello, A, Ferrian, M, Hoffer, P, et al.. Cutoff values for bacteria and leukocytes for urine flow cytometer Sysmex UF-1000i in urinary tract infections. Diagn Microbiol Infect Dis 2009;65:103–7. https://doi.org/10.1016/j.diagmicrobio.2009.06.003.Search in Google Scholar PubMed
36. Duong, HP, Wissing, KM, Tram, N, Mascart, G, Lepage, P, Ismaili, K. Accuracy of automated flow cytometry-based leukocyte counts to rule out urinary tract infection in febrile children: a prospective cross-sectional study. J Clin Microbiol 2016;54:2975–81. https://doi.org/10.1128/jcm.01382-16.Search in Google Scholar
37. De Rosa, R, Grosso, S, Lorenzi, G, Bruschetta, G, Camporese, A. Evaluation of the new Sysmex UF-5000 fluorescence flow cytometry analyser for ruling out bacterial urinary tract infection and for prediction of Gram negative bacteria in urine cultures. Clin Chim Acta 2018;484:171–8. https://doi.org/10.1016/j.cca.2018.05.047.Search in Google Scholar PubMed
38. García-Coca, M, Gadea, I, Esteban, J. Relationship between conventional culture and flow cytometry for the diagnosis of urinary tract infection. J Microbiol Methods 2017;137:14–8.10.1016/j.mimet.2017.03.010Search in Google Scholar PubMed
39. Hassan, F, Bushnell, H, Taggart, C, Gibbs, C, Hiraki, S, Formanek, A, et al.. Evaluation of bacterioscan 216dx in comparison to urinalysis as a screening tool for diagnosis of urinary tract infections in children. J Clin Microbiol 2019;57. https://doi.org/10.1128/JCM.00571-19.Search in Google Scholar PubMed PubMed Central
40. Bakan, E, Bayraktutan, Z, Baygutalp, NK, Gul, MA, Umudum, FZ, Bakan, N. Evaluation of the analytical performances of Cobas 6500 and Sysmex UN series automated urinalysis systems with manual microscopic particle counting. Biochem Med 2018;28:020712. https://doi.org/10.11613/BM.2018.020712.Search in Google Scholar PubMed PubMed Central
41. Foudraine, DE, Bauer, MP, Russcher, A, Kusters, E, Cobbaert, CM, van der Beek, MT, et al.. Use of automated urine microscopy analysis in clinical diagnosis of urinary tract infection: defining an optimal diagnostic score in an academic medical center population. J Clin Microbiol 2018;56. https://doi.org/10.1128/JCM.02030-17.Search in Google Scholar PubMed PubMed Central
42. van Delft, S, Goedhart, A, Spigt, M, van Pinxteren, B, de Wit, N, Hopstaken, R. Prospective, observational study comparing automated and visual point-of-care urinalysis in general practice. BMJ Open 2016;6:e011230. https://doi.org/10.1136/bmjopen-2016-011230.Search in Google Scholar PubMed PubMed Central
43. Gonzalez, M, Razzano, D, Ebid, A, Schubert, FD. Clinical and antibiotic management of urinary tract infections pre- and postimplementation of the CLINITEK AUWi system from Siemens to screen out negative urine samples submitted for culture: a retrospective cohort study. Lab Med 2017;49:18–24. https://doi.org/10.1093/labmed/lmx057.Search in Google Scholar PubMed
44. Stapleton, AE, Cox, ME, DiNello, RK, Geisberg, M, Abbott, A, Roberts, PL, et al.. Performance of a new rapid immunoassay test kit for point-of-care diagnosis of significant bacteriuria. J Clin Microbiol 2015;53:2805–9. https://doi.org/10.1128/jcm.00353-15.Search in Google Scholar PubMed PubMed Central
45. Calabretta, MM, Álvarez-Diduk, R, Michelini, E, Roda, A, Merkoçi, A. Nano-lantern on paper for smartphone-based ATP detection. Biosens Bioelectron 2019:111902. https://doi.org/10.1016/j.bios.2019.111902.Search in Google Scholar PubMed
46. Zboromyrska, Y, Rubio, E, Alejo, I, Vergara, A, Mons, A, Campo, I, et al.. Development of a new protocol for rapid bacterial identification and susceptibility testing directly from urine samples. Clin Microbiol Infect 2016;22:561.e1–6. https://doi.org/10.1016/j.cmi.2016.01.025.Search in Google Scholar PubMed
47. Huang, B, Zhang, L, Zhang, W, Liao, K, Zhang, S, Zhang, Z, et al.. Direct detection and identification of bacterial pathogens from urine with optimized specimen processing and enhanced testing algorithm. J Clin Microbiol 2017;55:1488–95. https://doi.org/10.1128/jcm.02549-16.Search in Google Scholar
48. Kitagawa, K, Shigemura, K, Onuma, K-I, Nishida, M, Fujiwara, M, Kobayashi, S, et al.. Improved bacterial identification directly from urine samples with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Clin Lab Anal 2018;32. https://doi.org/10.1002/jcla.22301.Search in Google Scholar PubMed PubMed Central
49. Wang, XH, Zhang, G, Fan, YY, Yang, X, Sui, WJ, Lu, XX. Direct identification of bacteria causing urinary tract infections by combining matrix-assisted laser desorption ionization-time of flight mass spectrometry with UF-1000i urine flow cytometry. J Microbiol Methods 2013;92:231–5. https://doi.org/10.1016/j.mimet.2012.12.016.Search in Google Scholar PubMed
50. Íñigo, M, Coello, A, Fernández-Rivas, G, Rivaya, B, Hidalgo, J, Quesada, MD, et al.. Direct identification of urinary tract pathogens from urine samples, combining urine screening methods and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 2016;54:988–93.10.1128/JCM.02832-15Search in Google Scholar PubMed PubMed Central
51. Oviaño, M, Ramírez, CDLL, Barbeyto, LP, Bou, G. Rapid direct detection of carbapenemase-producing Enterobacteriaceae in clinical urine samples by MALDI-TOF MS analysis. J Antimicrob Chemother 2017;72:1350–4.10.1093/jac/dkw579Search in Google Scholar PubMed
52. Dortet, L, Tandé, D, de Briel, D, Bernabeu, S, Lasserre, C, Gregorowicz, G, et al.. MALDI-TOF for the rapid detection of carbapenemase-producing Enterobacteriaceae: comparison of the commercialized MBT STAR®-Carba IVD Kit with two in-house MALDI-TOF techniques and the RAPIDEC® CARBA NP. J Antimicrob Chemother 2018;73:2352–9. https://doi.org/10.1093/jac/dky209.Search in Google Scholar PubMed
53. Roux-Dalvai, F, Gotti, C, Leclercq, M, Hélie, M-C, Boissinot, M, Arrey, TN, et al.. Fast and accurate bacterial species identification in urine specimens using LC-MS/MS mass spectrometry and machine learning. Mol Cell Proteomics 2019;18:2492–505. https://doi.org/10.1074/mcp.tir119.001559.Search in Google Scholar PubMed PubMed Central
54. Fondrie, WE, Liang, T, Oyler, BL, Leung, LM, Ernst, RK, Strickland, DK, et al.. Pathogen identification direct from polymicrobial specimens using membrane glycolipids. Sci Rep 2018;8:15857. https://doi.org/10.1038/s41598-018-33681-8.Search in Google Scholar PubMed PubMed Central
55. Lehmann, LE, Hauser, S, Malinka, T, Klaschik, S, Stüber, F, Book, M. Real-time polymerase chain-reaction detection of pathogens is feasible to supplement the diagnostic sequence for urinary tract infections. BJU Int 2010;106:114–20. https://doi.org/10.1111/j.1464-410X.2009.09017.x.Search in Google Scholar PubMed
56. Lehmann, LE, Hauser, S, Malinka, T, Klaschik, S, Weber, SU, Schewe, J-C, et al.. Rapid qualitative urinary tract infection pathogen identification by SeptiFast real-time PCR. PLoS One 2011;6:e17146. https://doi.org/10.1371/journal.pone.0017146.Search in Google Scholar PubMed PubMed Central
57. Wojno, KJ, Baunoch, D, Luke, N, Opel, M, Korman, H, Kelly, C, et al.. Multiplex pcr based urinary tract infection (uti) analysis compared to traditional urine culture in identifying significant pathogens in symptomatic patients. Urology 2019;136:199–26. https://doi.org/10.1016/j.urology.2019.10.018.Search in Google Scholar PubMed
58. van der Zee, A, Roorda, L, Bosman, G, Ossewaarde, JM. Molecular diagnosis of urinary tract infections by semi-quantitative detection of uropathogens in a routine clinical hospital setting. PLoS One 2016;11:e0150755. https://doi.org/10.1371/journal.pone.0150755.Search in Google Scholar PubMed PubMed Central
59. Schmidt, K, Stanley, KK, Hale, R, Smith, L, Wain, J, O’Grady, J, et al.. Evaluation of multiplex tandem PCR (MT-PCR) assays for the detection of bacterial resistance genes among Enterobacteriaceae in clinical urines. J Antimicrob Chemother 2019;74:349–56. https://doi.org/10.1093/jac/dky419.Search in Google Scholar PubMed
60. Li, Y, Yang, X, Zhao, W. Emerging microtechnologies and automated systems for rapid bacterial identification and antibiotic susceptibility testing. SLAS Technol 2017;22:585–608. https://doi.org/10.1177/2472630317727519.Search in Google Scholar PubMed PubMed Central
61. Khan, ZA, Siddiqui, MF, Park, S. Current and emerging methods of antibiotic susceptibility testing. Diagnostics 2019;9. https://doi.org/10.3390/diagnostics9020049.Search in Google Scholar PubMed PubMed Central
62. Li, H, Torab, P, Mach, KE, Surrette, C, England, MR, Craft, DW, et al.. Adaptable microfluidic system for single-cell pathogen classification and antimicrobial susceptibility testing. Proc Natl Acad Sci USA 2019;116:10270–9. https://doi.org/10.1073/pnas.1819569116.Search in Google Scholar PubMed PubMed Central
63. Athamanolap, P, Hsieh, K, O’Keefe, CM, Zhang, Y, Yang, S, Wang, T-H. Nanoarray digital PCR with high-resolution melt enables broad bacteria identification and pheno-molecular antimicrobial susceptibility test. Anal Chem 2019;91:12784–92. https://doi.org/10.1021/acs.analchem.9b02344.Search in Google Scholar PubMed PubMed Central
64. Wu, T-F, Chen, Y-C, Wang, W-C, Fang, Y-C, Fukuoka, S, Pride, DT, et al.. A rapid and low-cost pathogen detection platform by using a molecular agglutination assay. ACS Cent Sci 2018;4:1485–94. https://doi.org/10.1021/acscentsci.8b00447.Search in Google Scholar PubMed PubMed Central
65. Dong, T, Zhao, X. Rapid identification and susceptibility testing of uropathogenic microbes via immunosorbent ATP-bioluminescence assay on a microfluidic simulator for antibiotic therapy. Anal Chem 2015;87:2410–8. https://doi.org/10.1021/ac504428t.Search in Google Scholar PubMed
66. Avesar, J, Rosenfeld, D, Truman-Rosentsvit, M, Ben-Arye, T, Geffen, Y, Bercovici, M, et al.. Rapid phenotypic antimicrobial susceptibility testing using nanoliter arrays. Proc Natl Acad Sci USA 2017;114:E5787–95. https://doi.org/10.1073/pnas.1703736114.Search in Google Scholar PubMed PubMed Central
67. Schoepp, NG, Schlappi, TS, Curtis, MS, Butkovich, SS, Miller, S, Humphries, RM, et al.. Rapid pathogen-specific phenotypic antibiotic susceptibility testing using digital LAMP quantification in clinical samples. Sci Transl Med 2017;9. https://doi.org/10.1126/scitranslmed.aal3693.Search in Google Scholar PubMed PubMed Central
68. Rolando, JC, Jue, E, Schoepp, NG, Ismagilov, RF. Real-time, digital LAMP with commercial microfluidic chips reveals the interplay of efficiency, speed, and background amplification as a function of reaction temperature and time. Anal Chem 2019;91:1034–42. https://doi.org/10.1021/acs.analchem.8b04324.Search in Google Scholar PubMed PubMed Central
69. Choi, J, Yoo, J, Lee, M, Kim, E-G, Lee, JS, Lee, S, et al.. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci Transl Med 2014;6:267ra174. https://doi.org/10.1126/scitranslmed.3009650.Search in Google Scholar PubMed
70. Baltekin, Ö, Boucharin, A, Tano, E, Andersson, DI, Elf, J. Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. Proc Natl Acad Sci USA 2017;114:9170–5. https://doi.org/10.1073/pnas.1708558114.Search in Google Scholar PubMed PubMed Central
71. Yu, H, Jing, W, Iriya, R, Yang, Y, Syal, K, Mo, M, et al.. Phenotypic antimicrobial susceptibility testing with deep learning video microscopy. Anal Chem 2018;90:6314–22. https://doi.org/10.1021/acs.analchem.8b01128.Search in Google Scholar PubMed
72. Fredborg, M, Rosenvinge, FS, Spillum, E, Kroghsbo, S, Wang, M, Sondergaard, TE. Rapid antimicrobial susceptibility testing of clinical isolates by digital time-lapse microscopy. Eur J Clin Microbiol Infect Dis 2015;34:2385–94. https://doi.org/10.1007/s10096-015-2492-9.Search in Google Scholar PubMed PubMed Central
73. Syal, K, Shen, S, Yang, Y, Wang, S, Haydel, SE, Tao, N. Rapid antibiotic susceptibility testing of uropathogenic E. coli by tracking submicron scale motion of single bacterial cells. ACS Sens 2017;2:1231–9. https://doi.org/10.1021/acssensors.7b00392.Search in Google Scholar PubMed
74. Mo, M, Yang, Y, Zhang, F, Jing, W, Iriya, R, Popovich, J, et al.. Rapid antimicrobial susceptibility testing of patient urine samples using large volume free-solution light scattering microscopy. Anal Chem 2019;91:10164–71. https://doi.org/10.1021/acs.analchem.9b02174.Search in Google Scholar PubMed PubMed Central
75. Canali, C, Spillum, E, Valvik, M, Agersnap, N, Olesen, T. Real-time digital bright field technology for rapid antibiotic susceptibility testing. Methods Mol Biol 2018;1736:75–84. https://doi.org/10.1007/978-1-4939-7638-6_7.Search in Google Scholar PubMed
76. Fredborg, M, Andersen, KR, Jørgensen, E, Droce, A, Olesen, T, Jensen, BB, et al.. Real-time optical antimicrobial susceptibility testing. J Clin Microbiol 2013;51:2047–53. https://doi.org/10.1128/jcm.00440-13.Search in Google Scholar
77. Sin, MLY, Mach, KE, Wong, PK, Liao, JC. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev Mol Diagn 2014;14:225–44. https://doi.org/10.1586/14737159.2014.888313.Search in Google Scholar PubMed PubMed Central
78. Gomez-Cruz, J, Nair, S, Manjarrez-Hernandez, A, Gavilanes-Parra, S, Ascanio, G, Escobedo, C. Cost-effective flow-through nanohole array-based biosensing platform for the label-free detection of uropathogenic E. coli in real time. Biosens Bioelectron 2018;106:105–10. https://doi.org/10.1016/j.bios.2018.01.055.Search in Google Scholar PubMed
79. Settu, K, Chen, C-J, Liu, J-T, Chen, C-L, Tsai, J-Z. Impedimetric method for measuring ultra-low E. coli concentrations in human urine. Biosens Bioelectron 2015;66:244–50. https://doi.org/10.1016/j.bios.2014.11.027.Search in Google Scholar PubMed
80. Altobelli, E, Mohan, R, Mach, KE, Sin, MLY, Anikst, V, Buscarini, M, et al.. Integrated biosensor assay for rapid uropathogen identification and phenotypic antimicrobial susceptibility testing. Eur Urol Focus 2017;3:293–9. https://doi.org/10.1016/j.euf.2015.12.010.Search in Google Scholar PubMed PubMed Central
81. Gao, J, Li, H, Torab, P, Mach, KE, Craft, DW, Thomas, NJ, et al.. Nanotube assisted microwave electroporation for single cell pathogen identification and antimicrobial susceptibility testing. Nanomedicine 2019;17:246–53. https://doi.org/10.1016/j.nano.2019.01.015.Search in Google Scholar PubMed PubMed Central
82. Moustafa, A, Li, W, Singh, H, Moncera, KJ, Torralba, MG, Yu, Y, et al.. Microbial metagenome of urinary tract infection. Sci Rep 2018;8:4333. https://doi.org/10.1038/s41598-018-22660-8.Search in Google Scholar PubMed PubMed Central
83. Mouraviev, V, McDonald, M. An implementation of next generation sequencing for prevention and diagnosis of urinary tract infection in urology. Can J Urol 2018;25:9349–56.Search in Google Scholar
84. Sabat, AJ, van Zanten, E, Akkerboom, V, Wisselink, G, van Slochteren, K, de Boer, RF, et al.. Targeted next-generation sequencing of the 16S-23S rRNA region for culture-independent bacterial identification – increased discrimination of closely related species. Sci Rep 2017;7:3434. https://doi.org/10.1038/s41598-017-03458-6.Search in Google Scholar PubMed PubMed Central
85. Schmidt, K, Mwaigwisya, S, Crossman, LC, Doumith, M, Munroe, D, Pires, C, et al.. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother 2017;72:104–14. https://doi.org/10.1093/jac/dkw397.Search in Google Scholar PubMed
86. Burnham, P, Dadhania, D, Heyang, M, Chen, F, Westblade, LF, Suthanthiran, M, et al.. Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat Commun 2018;9:2412. https://doi.org/10.1038/s41467-018-04745-0.Search in Google Scholar PubMed PubMed Central
87. Ellington, MJ, Ekelund, O, Aarestrup, FM, Canton, R, Doumith, M, Giske, C, et al.. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect 2017;23:2–22. https://doi.org/10.1016/j.cmi.2016.11.012.Search in Google Scholar PubMed
88. Boolchandani, M, D’Souza, AW, Dantas, G. Sequencing-based methods and resources to study antimicrobial resistance. Nat Rev Genet 2019;20:356–70. https://doi.org/10.1038/s41576-019-0108-4.Search in Google Scholar PubMed PubMed Central
© 2021 Mohammed Harris and Tracy Fasolino, published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Obituary
- In Memoriam of Beniam Ghebremedhin
- Review
- New and emerging technologies for the diagnosis of urinary tract infections
- Original Articles
- Blood indices and circulating tumor cells for predicting metastasis of hepatocellular carcinoma after liver transplantation
- Comparison of APACHE II scores and mortality with CRP/albumin, neutrophil/lymphocyte and thrombocyte/lymphocyte ratios in patients admitted to internal medicine and anesthesia reanimation intensive care unit
- The correlation between the prevalence of gestational diabetes mellitus and maternal age in Southern China
- The investigation of serum protein profiles in anal fistula for potential biomarkers
- Evaluation of antioxidant and antiinflammatory activity of ethanolic extracts of Polygonum senticosum in lipopolysaccharide-induced RAW 264.7 macrophages
- Prevalence of SARS-CoV-2 antibodies in hospital employees, Central Germany
- Fast forward evolution in real time: the rapid spread of SARS-CoV-2 variant of concern lineage B.1.1.7 in Saxony-Anhalt over a period of 5 months
- Letters to the Editor
- It did not stop there: rapid substitution of circulating SARS-CoV-2 variant of concern B.1.1.7 (Alpha) by variant of concern B.1.617.2 (Delta) and further evolution of different Delta sublineages in Southern Saxony-Anhalt in late summer 2021
- In reply to a review article “Review of potentials and limitations of indirect approaches for estimating reference limits/intervals of quantitative procedures in laboratory medicine”
- Reply to the letter of Katayev and Fleming
Articles in the same Issue
- Frontmatter
- Obituary
- In Memoriam of Beniam Ghebremedhin
- Review
- New and emerging technologies for the diagnosis of urinary tract infections
- Original Articles
- Blood indices and circulating tumor cells for predicting metastasis of hepatocellular carcinoma after liver transplantation
- Comparison of APACHE II scores and mortality with CRP/albumin, neutrophil/lymphocyte and thrombocyte/lymphocyte ratios in patients admitted to internal medicine and anesthesia reanimation intensive care unit
- The correlation between the prevalence of gestational diabetes mellitus and maternal age in Southern China
- The investigation of serum protein profiles in anal fistula for potential biomarkers
- Evaluation of antioxidant and antiinflammatory activity of ethanolic extracts of Polygonum senticosum in lipopolysaccharide-induced RAW 264.7 macrophages
- Prevalence of SARS-CoV-2 antibodies in hospital employees, Central Germany
- Fast forward evolution in real time: the rapid spread of SARS-CoV-2 variant of concern lineage B.1.1.7 in Saxony-Anhalt over a period of 5 months
- Letters to the Editor
- It did not stop there: rapid substitution of circulating SARS-CoV-2 variant of concern B.1.1.7 (Alpha) by variant of concern B.1.617.2 (Delta) and further evolution of different Delta sublineages in Southern Saxony-Anhalt in late summer 2021
- In reply to a review article “Review of potentials and limitations of indirect approaches for estimating reference limits/intervals of quantitative procedures in laboratory medicine”
- Reply to the letter of Katayev and Fleming

