Abstract
In recent years, the field of causal inference from observational data has emerged rapidly. The literature has focused on (conditional) average causal effect estimation. When (remaining) variability of individual causal effects (ICEs) is considerable, average effects may be uninformative for an individual. The fundamental problem of causal inference precludes estimating the joint distribution of potential outcomes without making assumptions. In this work, we show that the ICE distribution is identifiable under (conditional) independence of the individual effect and the potential outcome under no exposure, in addition to the common assumptions of consistency, positivity, and conditional exchangeability. Moreover, we present a family of flexible latent variable models that can be used to study individual effect modification and estimate the ICE distribution from cross-sectional data. How such latent variable models can be applied and validated in practice is illustrated in a case study on the effect of hepatic steatosis on a clinical precursor to heart failure. Under the assumptions presented, we estimate that 20.6% (95% Bayesian credible interval: 8.9%, 33.6%) of the population has a harmful effect greater than twice the average causal effect.
1 Introduction
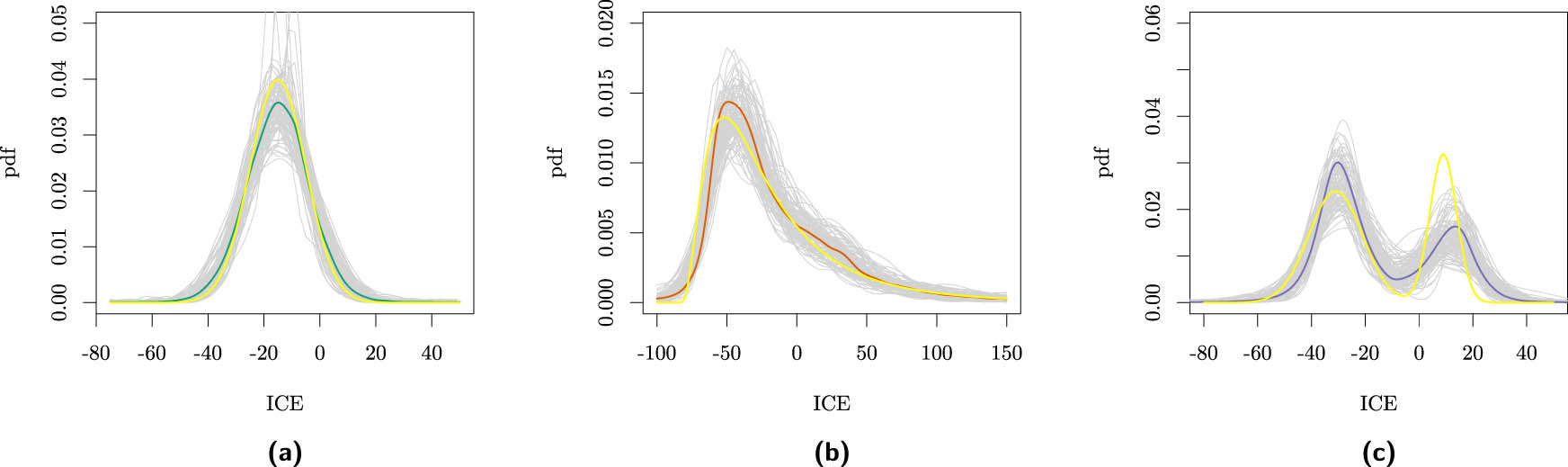
The main result of an epidemiological study is often summarized by an average treatment effect (ATE). Consequently, the ATE might (subconsciously) be interpreted as the causal effect for each individual, while individuals may react differently to exposures. These individual causal effects (ICEs) can be highly variable and may even have opposite signs [1,2]. The effect distribution within a (sub)population might be skewed, high in variability, or multimodal. Examples of ICE distributions for which the ATE is equal to

Examples of effect distributions where the (conditional) ATE equals
Nowadays, the variability in ICEs is typically studied by estimating conditional average treatment effects (CATEs), which equals the mean of the conditional ICE distribution given certain observed features. Standard causal inference methods can typically include covariates to account for such effect modification, e.g., strata-specific marginal structural models [5], and many machine learning (ML) algorithms have been proposed for flexible CATE estimation (see Caron et al. [6] for a detailed review). In this ML literature, a CATE based on many observed features is sometimes considered equivalent to the ICE [7]. In the ideal case of almost homogeneous conditional causal effects, such as for a subpopulation with the effect distribution shown in Figure 1(a), the CATE can be used to make treatment decisions for an individual. However, the CATE may not be informative enough when given the levels of the measured features the ICEs among the subpopulation are still highly variable, as shown in Figure 1(b) and (c). Despite accounting for many features of the patient, other and unknown features might modify the exposure effect so that in the subpopulation, the exposure is expected to harm some, while others benefit (e.g., Figure 1(d)). Despite the increasing number of covariates collected in epidemiological studies, significant effect modifiers may remain unmeasured so that the personalized CATE deviates from the personal ICE [8]. Therefore, (remaining) effect heterogeneity should be taken into account for decision-making [9–13].
In recent work, we have explained how CATE estimation using ML methods may be extended to additionally estimate the remaining conditional variance of the ICE assuming conditional independence of the individual effect and the potential outcome under no exposure, a causal assumption [8]. The conditional ICE distribution is only identified by this conditional variance and the CATE when assuming (conditionally) Gaussian-distributed effects. Under the conditional independence assumption, the Gaussianity of the effect is not a causal assumption because it could be verified with sufficient observed data. In this work, we shift focus from the (conditional) ICE variance to the (conditional) ICE distribution without assuming the shape of the effect distribution. We will explain under what assumptions the (conditional) ICE distribution becomes identifiable so that latent variable models may be used to estimate the distribution. We will present a semi-parametric linear mixed model (LMM) to estimate the (conditional) ICE distribution when the assumptions apply and discuss the complications in estimation due to the heterogeneity of the effect of confounders on the outcome.
First, we will introduce our notation and framework to formalize individual effect heterogeneity in Section 2. In Section 3, we will review the literature related to ICE and illustrate how and under which assumptions the ICE distribution can be identified from an RCT and observational data, respectively. Subsequently, the semi-parametric causal mixed models and possible methods to fit these models are presented in Section 4. In Section 5, the Framingham heart study (FHS) is considered to illustrate how the reasoning presented in this work could be used in practice. The modeling considerations are discussed in detail. Finally, we reflect on the work’s importance, limitations, and future research in Section 6.
2 Notation and framework
Probability distributions of factual and counterfactual outcomes are defined in the potential outcome framework [14,15]. Let
We will consider only two exposure levels,
Throughout this work, we will assume causal consistency, implying that potential outcomes are independent of the assigned exposure levels of other individuals (no interference) and that there are no different versions of the exposure levels [19].
Assumption 1
(Causal consistency)
This definition of causal consistency is also referred to as the stable unit treatment value assumption [20, Assumption 1.1]. Furthermore, we will assume that for all levels of measured features
Assumption 2
(Positivity)
Next, we will introduce a structural model and random variables necessary to formalize the relevant causal relations and joint distribution of potential outcomes. The variability in exposure assignment
Note that
There exists confounding when
Assumption 3
(Conditional exchangeability)
As discussed in Section 1, causal effect heterogeneity is often addressed by studying the CATEs in subpopulations defined by measured features
The CATE equals the mean of the conditional ICE distribution,
Assumption 4
(Conditional independent effect deviation)
For the parameterization of the cause–effect relations in terms of SCM (1), this is equivalent to assuming
Realize that Assumption 4 is violated when there exists a feature that, despite conditioning on
3 Identifiability of the ICE distribution
As a result of the fundamental problem of causal inference, studying properties of the conditional ICE distribution other than the CATE (which equals the mean) received less attention in the literature. For binary outcomes
Besides the non-parametric approaches, non-verifiable assumptions on the joint distribution of the potential outcomes
In this section, we discuss the non-identifiability of the ICE as a result of the fundamental problem in more detail and explain why Assumption 4 can result in identifiability. To build intuition, we begin by showing how the fundamental problem of causal inference results in the non-identifiability of the conditional variance.
3.1 Conditional variance
In the absence of remaining effect heterogeneity in subpopulations defined by levels of
Proposition 1
If Assumptions
1–3
apply, and
The result of Proposition 1 concerns observational quantities and can thus be verified when sufficient data are available without making additional assumptions. In that case, it is also possible that the equality in conditional variances does not hold, which by the contrapositive of Proposition 1 is equivalent to the existence of remaining effect heterogeneity. In particular, if the conditional variance among treated individuals is larger than non-treated individuals, then the conditional ICE distribution is non-degenerate, as shown in Proposition 2.
Proposition 2
then
For an exposure that would reduce the heterogeneity in the outcomes, i.e.,
is not identifiable without making additional assumptions.[2] The converse of Propositions 1 and 2 is false since, in terms of SCM (1),
If the conditional covariance,
3.2 Conditional ICE distribution
Understanding the conditional variance is a first step beyond the CATEs when interested in the conditional ICE distribution. We continue by expressing the ICE distribution in terms of the observed outcome distribution given
The conditional distribution of
Lemma 1
If Assumptions
1–3
apply, and the cause–effect relations are parameterized in terms of SCM (1), then
where
Inference on the ICE distribution thus requires inference on the conditional distribution of
Proposition 3
Let the random variable
In words, if Proposition 3 applies,
4 Causal mixed models
We continue by focusing on fitting the unknown
where
Note that
The distribution of
where the distributions
4.1 Methods to fit the random effects model
LMMs are often used in epidemiological studies, and particularly, the fitting of LMMs with (independent) Gaussian random effects (
When interested in the ICE distribution, the distribution of the random effect should be well specified and thus not restricted to be Gaussian. Verbeke et al. [47] suggested modeling the random effect as a mixture of normals with an unknown number of components. This model can be fitted using an expectation–maximization algorithm [47], and an alternative estimation procedure has been proposed by Proust and Jacqmin-Gadda [48]. To study features of between-individual variation, Zhang and Davidian [49] proposed to fit an LMM by approximating the density of the random effect by a semi-non-parametric representation. For both approaches, an optimal tuning parameter is often selected based on information criteria [48,49].
The Gaussian mixture distribution for the random effect can also be fitted in a Bayesian framework [50]. As the ICE distribution is unknown, information criteria can be used to set the optimal number of components [51, Chapter 22]. A one-step approach using a uniform prior distribution on the number of components of the mixture distribution of the random effect has been proposed to avoid model selection [52]. Model selection is also not necessary when one does fix the number of components to a “large” constant
Finally, the random effect distribution can be modeled non-parametrically. For example, a Bayesian non-parametric fit of a hierarchical model can be obtained using a Dirichlet process prior [54] or a truncated Dirichlet process prior [55], respectively, for the distribution of the random effect. Despite the existence of all methods, precise estimation of the distribution of a latent variable remains challenging and is sensitive to the proposed model.
4.2 Example of ICE distribution estimation
To demonstrate the use of a causal mixed model, we will next consider a simple example with data simulated for 1,000 individuals using equation (6) with

Estimated
We will model the ICE distribution with a Gaussian mixture that we fit using the Bayesian method with an upper bound for the number of components (discussed in Section 4.1) since this one-step approach can be easily implemented in both SAS and R. Moreover, since we are interested in estimating the entire distribution of the ICE, a Bayesian method has the advantage that (pointwise) uncertainty quantification of the ICE density can be directly obtained. On the contrary, when using a frequentist approach, the uncertainty of the density should be derived from the uncertainty in the model parameters.
We model the observed data as
where
and the weakly informative prior
are used. When (8) is well specified and Assumptions 1–4 apply, by Proposition 3,
The mean values of the posterior means and the coverage of the Bayesian credible sets are presented in Table 1 for
Actual values of
| Actual value | Average posterior means | Coverage credible sets | |||||||
|---|---|---|---|---|---|---|---|---|---|
| (a) | (b) | (c) | (a) |
|
(c) | (a) |
|
(c) | |
|
|
0.07 | 0.27 | 0.39 | 0.08 | 0.27 | 0.36 | 0.95 | 0.94 | 0.90 |
|
|
|
|
|
|
|
|
0.90 | 0.94 | 0.90 |
|
|
|
|
|
|
|
|
0.92 | 0.89 | 0.91 |
|
|
|
|
|
|
|
|
0.95 | 0.89 | 0.90 |
|
|
|
2.86 | 7.41 |
|
4.14 | 6.83 | 0.96 | 0.93 | 0.88 |
|
|
1.44 | 89.60 | 14.75 | 3.42 | 90.50 | 15.90 | 0.97 | 0.89 | 0.86 |
For these characteristics, the average of the posterior means, each estimated on 1,000 individuals, and the coverage of the 95% Bayesian credible sets, based on the 100 simulations, are presented.
In practice, the model itself should be validated after verifying the proper convergence of the Markov chains. The causal Assumptions 1, 3, and 4 cannot be verified, but the associational model can be validated using the observational data, as we will elaborate on in Section 5. It is essential to realize that when Assumption 4 is violated, the ICE distribution is not identified by the

Estimated
4.3 Confounding-effect heterogeneity
As stated in equation (6), the distribution of
For causal inference from observational data, it is often necessary to adjust for features so that Assumption 3 applies. If the interest is in the ATE, we in principle only need to take into account the confounder’s effect on the outcome’s mean, i.e., we need to adjust for
However, if one is interested in the ICE distribution, then it is necessary to consider the effect of the confounder on the entire distribution of the outcome, i.e., adjust for
Otherwise, a difference in (the shape of the) distribution between the exposed and unexposed individuals can be caused by the non-exchangeable confounders. Then, the estimated distribution of

Estimated
In the case of confounding-effect heterogeneity, the distribution of
where
Similarly, the
5 Case study: the FHS
In this section, we consider heterogeneity in the effect of non-alcoholic fatty liver disease on cardiac structure and function, as studied by Chiu et al. [58] in the FHS population. We will work with a subset of the FHS third-generation and offspring cohorts
The sample standard deviation of the LVFP equals 2.27 for individuals exposed to hepatic steatosis and only 1.74 for those who were not. This difference might be the result of causal effect heterogeneity. As the Bayesian analysis discussed in Section 4.2 is computationally intensive, we started by fitting a traditional Gaussian LMM to investigate which candidate confounders affected the estimated mean or variance of the effect of hepatic steatosis on LVFP. Confounders accounted for in the original study were age, sex, smoking, alcohol use, diabetes, systolic blood pressure (SBP), use of antihypertensive medication (HRX), use of lipid-lowering medication, total cholesterol, high-density lipoprotein cholesterol, triglycerides, and fasting glucose. The relevant confounders were age, sex, diabetes, SBP, and HRX (details can be found in Section E of Appendix).
In this case study, our focus is on the population ICE distribution; thus, we do not consider modifiers that are no confounders. We have fitted the model
where
The model was fitted using the Bayesian method discussed in Section 4.2. The initial values for the parameters in the Markov chain were based on the final Gaussian LMM used to select the confounders and can be found in Table A6 in Appendix. Per chain, we have used 100,000 burn-in iterations, followed by 500,000 MCMC iterations. We have used a thinning rate of 100 to save computer memory space. In total, 5 chains, each contained 5,000 (thinned) MCMC iterations that were saved. Each chain was split into two pieces, and we investigated the convergence of the
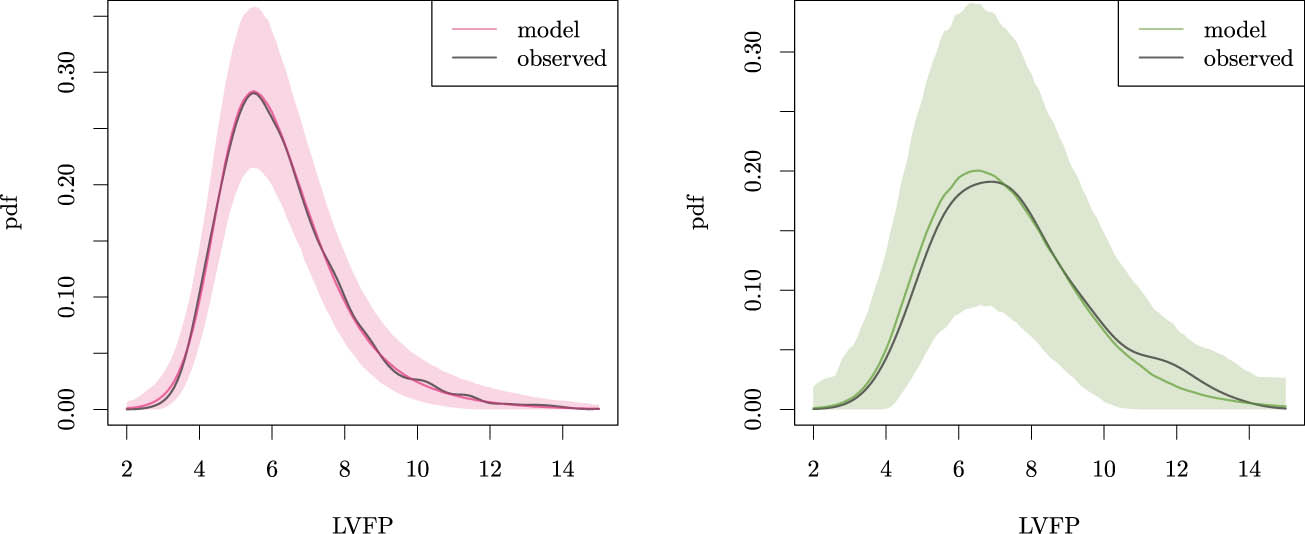
We should ensure that the model accurately fits the observed data to draw inferences on the ICE distribution. To validate the model, we study the posterior predictive distribution of the LVFP for both the exposed and unexposed individuals (

Posterior predictive distribution of the LVFP for model (13), with corresponding 95% BCIs and the kernel density of the observed data for unexposed (left) and exposed individuals (right).
As shown in Table 2, the standard deviation of the LVFP is higher for individuals who are either older, female, have diabetes, have higher blood pressure, or use HRX for both exposure groups. In Section 4.3, we have explained how confounding-effect heterogeneity can affect the distribution of
Sample standard deviation of the LVFP in sub-samples partitioned by the (dichotomized) confounders and the exposure
| Age
|
Sex | Diabetes |
|
HRX | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | ||
| Hepatic steatosis | 0 | 1.4 | 1.9 | 1.5 | 1.9 | 1.7 | 2.0 | 1.5 | 1.9 | 1.6 | 2.1 |
| 1 | 1.7 | 2.6 | 1.8 | 2.6 | 2.2 | 2.5 | 2.0 | 2.3 | 1.9 | 2.7 | |
Thus, model (13) does appropriately describe the observed conditional distributions. The ICE distribution is only identifiable in the absence of other confounders and when the dependence of

Posterior distribution of the effect of hepatic steatosis on the LVFP using model (13) (solid line) and pointwise 95% BCIs. Moreover, distributions obtained when either adjusting for confounding-effect heterogeneity, using a Gaussian residual or a Gaussian LMM, respectively, are presented for comparison (Section 5.1).
5.1 Sensitivity analysis
We performed a sensitivity analysis by extending model (13) with confounder-specific residual variances to demonstrate how one could try to adjust for confounding-effect heterogeneity. We have fitted the model
where again
The posterior predictive checks in the different exposure and confounder strata can be found in Section F.2 of Appendix. The minor differences between the posterior predictive distribution and the observed distribution presented in Section F.1 of Appendix became even smaller. The ATE has a posterior mean of 0.38 (95% BCI: 0.19, 0.56), and the posterior mean of the
We want to emphasize that all latent variables must be modeled with a distribution that is flexible enough to fit the observed outcomes accurately. For comparison, we have also presented the estimated ICE distribution when modeling the residual with a Gaussian distribution without adjusting for confounding-effect heterogeneity (ATE equals 0.42 (0.17, 0.69)) and

Distribution of the number of
6 Discussion
Methods for causal inference have rapidly evolved over the past decades. As a result, making causal claims from observational studies became more common, relying on expert knowledge to back up the untestable identifiability assumption of conditional exchangeability. However, such methods focus on learning (conditional) ATEs rather than effect distributions. Therefore, the results are only informative at the level of an individual when (remaining) effect heterogeneity is low. In this work, we have presented an identifiability assumption that suffices to move from the (conditional) ATE to quantification of the (conditional) distribution of causal effects.
In the case of effect heterogeneity, exposure (or absence thereof) increases variability in the outcome as individuals respond differently. When the common causal Assumptions 1–3 apply and additionally (conditional) independence of the ICE and the potential outcome under no exposure (Assumption 4) can be assumed, inference on the ICE distribution can be drawn from cross-sectional data. The joint distribution of potential outcomes should be linked to the law of observations, which can be learned from the data. Then, deviations from the (C)ATE could be quantified. In case of serious deviations, the shape of the (conditional) ICE distribution can inform about the remaining heterogeneity of the exposure effect. It may illustrate that there is still a severe lack of understanding of the exposure effect as the presence of unmeasured modifiers is considerable. The distribution of the unmeasured modifiers may differ across populations, so estimated ICE distributions could be helpful in the understanding of differences in causal effects [62].
In contrast to the well-established models to estimate CATEs (e.g., [63] and [64]), the focus of the methods presented in this article is inference on the distribution that quantifies remaining effect heterogeneity. In particular, in settings where limited features are available, the CATEs will not be accurate proxies of the actual ICEs. In the examples presented in this article, we have used simple linear models (models (8), (13), and (14)) to estimate the ICE distribution for illustration. However, the causal theory derived in Section 3 applies to more general models of the form presented in (7). A misspecified mean model will affect the fit of the random exposure effect. The conditional distributions of the data-generating distribution should thus be validated, e.g., by Bayesian posterior predictive checking as illustrated in the case study. If the posterior distribution for unexposed individuals is off, a more flexible mean model (involving the expected effects of modifiers and confounders) may be necessary. For cases where rich data are available, it will be promising to investigate how flexible ML methods can estimate the conditional means in our mixed model [8,65,66]. For the example and case study, we used a specific Bayesian approach to fit the random-effects model, as it inherently quantifies the uncertainty of the ICE density estimate and simplifies the process by using a one-step approach. However, the reasoning presented in this article does not rely on the estimation method, and we have listed Bayesian and non-Bayesian alternatives in Section 4.1. What method can be used for a specific application will depend on the available resources, e.g., the Bayesian methods can result in lengthy computation times depending on the complexity of the associational model (7) and the size of the data.
As mentioned earlier, the fit of the associational model for
Acknowledgements
We sincerely thank Michelle Long and Alison Pedley for sharing the SAS script to reproduce the sub-sample of the Framingham Heart Study as used in Section 5. The authors thank the associate editor, and three anonymous reviewers for their valuable comments.
-
Funding information: The authors state no funding involved.
-
Author contributions: RAJP conceptualized the study, developed the methodology, implemented the simulation code, conducted the case study, and drafted the original manuscript. ERvdH supervised the project and contributed to the review and editing of the manuscript. Both authors read and approved the final manuscript.
-
Conflict of interest: The authors state no conflict of interest.
-
Data availability statement: The SAS and R codes used for simulation and analysis of the example presented in Sections 4.2 and 4.3 and for the analysis in Section 5 can be found at https://github.com/RAJP93/ICE-distribution.
Appendix A Proof of Proposition 1
If
By Assumption 2,
B Proof of Proposition 2
By Assumptions 1–3,
If
C Proof of Lemma 1
The conditional ICE distribution,
where
As
Since
Finally, by causal consistency and positivity,
D Proof of Proposition 3
By definition of SCM (1),
where
E Confounder selection case study
As the Bayesian analysis was computationally intensive, we started by fitting a Gaussian linear mixed model (LMM) to investigate which candidate confounders did change the mean or variance of the effect of hepatic steatosis on the LVFP. More precisely, we started by fitting
where
Then, we started to remove single components of
Change in the estimate of exposure effect (
| Remove fixed effect | – | Age | Sex | Smoke | DPW | DIAB | SBP | HRX | LRX | CHOL | HDL | TRIG | GLU |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
0.425 | 0.387 | 0.467 | 0.408 | 0.410 | 0.429 | 0.569 | 0.470 | 0.424 | 0.424 | 0.412 | 0.427 | 0.433 |
| Relative change |
|
0.098 |
|
|
0.008 | 0.337 | 0.104 |
|
|
|
0.004 | 0.018 | |
|
|
0.424 | 0.383 | 0.466 | 0.407 | 0.409 | 0.428 | 0.568 | 0.471 | 0.422 | 0.411 | 0.427 | 0.432 | |
| Relative change |
|
0.098 |
|
|
0.008 | 0.338 | 0.111 |
|
|
0.006 | 0.018 | ||
|
|
0.422 | 0.379 | 0.464 | 0.405 | 0.406 | 0.426 | 0.565 | 0.471 | 0.420 | 0.414 | 0.430 | ||
| Relative change |
|
0.099 |
|
|
0.009 | 0.337 | 0.115 |
|
|
0.017 | |||
|
|
0.420 | 0.362 | 0.415 | 0.414 | 0.421 | 0.427 | 0.558 | 0.473 | 0.397 | 0.427 | |||
| Relative change |
|
|
|
0.002 | 0.016 | 0.330 | 0.126 |
|
0.017 | ||||
|
|
0.421 | 0.361 | 0.413 | 0.415 | 0.427 | 0.558 | 0.470 | 0.401 | 0.430 | ||||
| Relative change |
|
|
|
0.016 | 0.326 | 0.118 |
|
0.022 | |||||
|
|
0.415 | 0.361 | 0.406 | 0.422 | 0.550 | 0.468 | 0.402 | 0.424 | |||||
| Relative change |
|
|
0.016 | 0.326 | 0.128 |
|
0.023 | ||||||
|
|
0.422 | 0.368 | 0.414 | 0.556 | 0.482 | 0.407 | 0.462 | ||||||
| Relative change |
|
|
0.319 | 0.143 |
|
0.096 | |||||||
|
|
0.414 | 0.350 | 0.547 | 0.473 | 0.368 | 0.449 | |||||||
| Relative change |
|
0.321 | 0.142 |
|
0.084 |
In each step of the procedure, the covariate with the least impact is removed.
At this stage age, SBP, HRX, TRIG, and GLU affect
where
Gaussian LMM parameter estimates ignoring confounder effect heterogeneity
| Fixed effects | Variance components | ||
|---|---|---|---|
| Intercept | 6.330 | ||
| Hepatic steatosis | 0.414 | Hepatic steatosis | 1.805 |
| age | 0.484 | ||
| Hepatic steatosis*age | 0.003 | ||
| SBP | 0.238 | ||
| Hepatic steatosis*SBP | 0.242 | ||
| HRX | 0.392 | ||
| Hepatic steatosis*HRX | 0.071 | ||
| TRIG | 0.034 | ||
| Hepatic steatosis*TRIG |
|
||
| GLU | 0.006 | ||
| Hepatic steatosis*GLU | 0.146 | ||
| Residual | 2.538 | ||
Gaussian LMM parameter estimates for the model after first selection procedure accounting for confounder effect heterogeneity
| Fixed effects | Variance components | ||
|---|---|---|---|
| Intercept | 6.335 | ||
| Hepatic steatosis | 0.408 | Hepatic steatosis | 0.940 |
| age | 0.476 | (Age > 49) | 1.069 |
| Hepatic steatosis*age | 0.003 | ||
| SBP | 0.248 | (SBP > 120) | 0.503 |
| Hepatic steatosis*SBP | 0.215 | ||
| HRX | 0.402 | HRX | 1.489 |
| Hepatic steatosis*HRX | 0.090 | ||
| TRIG | 0.027 | (TRIG > 98) | 0.051 |
| Hepatic steatosis*TRIG |
|
||
| GLU |
|
(GLU < 96) | 0.194 |
| Hepatic steatosis*GLUCOSE | 0.138 | ||
| Residual | 1.441 | ||
Subsequently, we again added one of the other candidate confounders to the model and investigated how the variance of
Change in the estimate of the variance of the random effect of fatty liver (variance of
| Add random effect | – | Sex | Smoke | (
|
DIAB | (CHOL > 189) | (HDL > 52) | 1-LRX |
|---|---|---|---|---|---|---|---|---|
| Variance
|
0.940 | 0.712 | 0.841 | 0.945 | 0.873 | 0.958 | 0.945 | 0.979 |
| Relative change | 0.243 | 0.105 | 0.005 |
|
0.019 | 0.006 | 0.042 | |
| Variance
|
0.712 | 0.689 | 0.731 | 0.589 | 0.759 | 0.719 | 0.743 | |
| Relative change |
|
0.027 |
|
0.066 | 0.010 | 0.044 | ||
| Variance
|
0.589 | 0.552 | 0.635 | 0.636 | 0.593 | 0.623 | ||
| Relative change |
|
0.078 | 0.080 | 0.007 | 0.057 |
In each step of the procedure, the covariate with the most impact is added.
As a final step, we check whether we can remove any of the candidate confounders from the model without significantly changing the variance of
Change in the estimated mean and the variance of the random effect of fatty liver after removing candidate confounders from the LMM
| Remove | – | Age | Sex | DIAB | SBP | HRX | TRIG | GLU |
|---|---|---|---|---|---|---|---|---|
| Variance
|
0.589 | 0.423 | 0.830 | 0.712 | 0.886 | 0.956 | 0.612 | 0.595 |
|
|
0.378 | 0.309 | 0.392 | 0.393 | 0.521 | 0.459 | 0.390 | 0.387 |
| Variance
|
0.595 | 0.465 | 0.824 | 0.745 | 0.886 | 0.972 | 0.627 | |
|
|
0.387 | 0.326 | 0.395 | 0.432 | 0.537 | 0.467 | 0.405 | |
| Variance
|
0.627 | 0.525 | 0.813 | 0.776 | 0.934 | 0.997 | ||
|
|
0.405 | 0.334 | 0.372 | 0.458 | 0.567 | 0.471 |
Final LMM parameter estimates accounting for age, sex, diabetes, SBP, and HRX use
| Fixed effects | Variance components | ||
|---|---|---|---|
| Intercept | 5.882 | ||
| Hepatic steatosis | 0.319 | Hepatic steatosis | 0.627 |
| Age | 0.366 | (age > 49) | 0.706 |
| Hepatic steatosis*age | 0.007 | ||
| SBP | 0.311 | (SBP > 120) | 0.724 |
| Hepatic steatosis*SBP | 0.174 | ||
| HRX | 0.418 | HRX | 1.370 |
| Hepatic steatosis*HRX | 0.057 | ||
| SEX | 0.650 | SEX | 0.927 |
| Hepatic steatosis*SEX | 0.118 | ||
| DIAB | 0.471 | DIAB | 0.775 |
| Hepatic steatosis*DIAB | 0.197 | ||
| Residual | 1.063 | ||
With this procedure, we have selected age, sex, DIAB, SBP, and HRX as confounders. The parameter estimates of the final Gaussian LMM are presented in Table A6 and were used as initial values in the Bayesian procedure of the case study.
F Supplementary figures
Figures A1, A2, A3.
Kernel density estimates of the

Trace plots of simulated ICEs for two unexposed individuals while fitting model (13).

Posterior predictive distribution of the LVFP for model (13), with corresponding 95% BCIs and the kernel density of the observed data for individuals that are older than 49 years (right) or not (left).
F.1 Posterior predictive distribution per confounder strata
Figures A4, Figure A5, Figure A6, Figure A7, A8.

Posterior predictive distribution of the LVFP for model (13), with corresponding 95% BCIs and the kernel density of the observed data for males (left) and females (right).

Posterior predictive distribution of the LVFP for model (13), with corresponding 95% BCIs and the kernel density of the observed data for individuals with (right) and without (left) diabetes.

Posterior predictive distribution of the LVFP for model (13), with corresponding 95% BCIs and the kernel density of the observed data for individuals with an SBP above 120 mmHg (right) or not (left).

Posterior predictive distribution of the LVFP for model (13), with corresponding 95% BCIs and the kernel density of the observed data for individuals with (right) and without (left) HRX use.

Posterior predictive distribution of the LVFP for model (14), with corresponding 95% BCIs and the kernel density of the observed data for unexposed (left) and exposed individuals (right).
F.2 Model adjusting for confounding-effect heterogeneity
Figures A9, Figure A10, Figure A11, Figure A12, Figure A13, A14.

Posterior predictive distribution of the LVFP for model (14), with corresponding 95% BCIs and the kernel density of the observed data for individuals that are older than 49 years (right) or not (left).

Posterior predictive distribution of the LVFP for model (14), with corresponding 95% BCIs and the kernel density of the observed data for males (left) and females (right).
Posterior predictive distribution of the LVFP for model (14), with corresponding 95% BCIs and the kernel density of the observed data for individuals with (right) and without (left) diabetes.

Posterior predictive distribution of the LVFP for model (14), with corresponding 95% BCIs and the kernel density of the observed data for individuals with an SBP above 120 mmHg (right) or not (left).

Posterior predictive distribution of the LVFP for model with (14), with corresponding 95% BCIs and the kernel density of the observed data for individuals with (right) and without (left) HRX use.

Posterior predictive distribution of the LVFP for model (13) but with Gaussian-distributed
F.3 Model with Gaussian residual
F.4 Gaussian LMM

Posterior predictive distribution of the LVFP for model (13) but with Gaussian-distributed
References
[1] Hand DJ. On comparing two treatments. Am Stat. 1992;46(3):190–2. 10.1080/00031305.1992.10475881. Suche in Google Scholar
[2] Greenland S, Fay MP, Brittain EH, Shih JH, Follmann DA, Gabriel EE. On causal inferences for personalized medicine: How hidden causal assumptions led to erroneous causal claims about the D-value. Am Stat. 2019;74(3):243–8. 10.1080/00031305.2019.1575771. Suche in Google Scholar PubMed PubMed Central
[3] Holland PW. Statistics and causal inference. J Am Stat Assoc. 1986;81(396):945–60. 10.1080/01621459.1986.10478354. Suche in Google Scholar
[4] Kennedy EH, Balakrishnan S, Wasserman LA. Semiparametric counterfactual density estimation. Biometrika. 2023;110(4):875–96. 10.1093/biomet/asad017. Suche in Google Scholar
[5] Robins JM, Hernán MA, Brumback B Marginal structural models and causal inference in epidemiology. Epidemiology. 2000;11(5):550–60. 10.1097/00001648-200009000-00011. Suche in Google Scholar PubMed
[6] Caron A, Baio G, Manolopoulou I. Estimating individual treatment effects using non-parametric regression models: A review. J R Stat Soc Ser A (Stat Soc). 2022;185(3):1115–49. 10.1111/rssa.12824. Suche in Google Scholar
[7] Lu M, Sadiq S, Feaster DJ, Ishwaran H. Estimating individual treatment effect in observational data using random forest methods. J Comput Graph Stat. 2018;27(1):209–19. 10.1080/10618600.2017.1356325. Suche in Google Scholar PubMed PubMed Central
[8] Post RAJ, Petkovic M, van den Heuvel IL, van den Heuvel ER. Flexible machine learning estimation of conditional average treatment effects: a blessing and a curse. Epidemiology. 2024;35(1):32–40. 10.1097/EDE.0000000000001684. Suche in Google Scholar PubMed
[9] Kallus N. What’s the harm? Sharp bounds on the fraction negatively affected by treatment. 2022. https://arxiv.org/abs/2205.10327. Suche in Google Scholar
[10] Ben-Michael E, Imai K, Jiang Z. Policy learning with asymmetric counterfactual utilities. J Am Stat Assoc. 2024;119(548):3045–58. 10.1080/01621459.2023.2300507. Suche in Google Scholar
[11] Li H, Zheng C, Cao Y, Geng Z, Liu Y, Wu P. Trustworthy policy learning under the counterfactual no-harm criterion. In: Proceedings of the 40th International Conference on, volume 202 of Proceedings of Research, pp. 20575–98. PMLR. July 2023. https://proceedings.mlr.press/v202/li23ay.html. Suche in Google Scholar
[12] Mueller S, Pearl J. Personalized decision making - a conceptual introduction. J Causal Infer. 2023;11(1):20220050. 10.1515/jci-2022-0050. Suche in Google Scholar
[13] Sarvet AL, Stensrud MJ. Perspective on ‘Harm’ in personalized medicine. Am J Epidemiol. 2023;kwad162. 10.1093/aje/kwad162. Suche in Google Scholar PubMed
[14] Neyman J. On the application of probability theory to agricultural experiments. essay on principles. Stat Sci. 1923;5(4):465–72. 10.1214/ss/1177012031. Suche in Google Scholar
[15] Rubin DB. Estimating causal effects of treatments in randomized and nonrandomized studies. J Educat Psychol. 1974;66(5):688–701. 10.1037/h0037350. Suche in Google Scholar
[16] Robins J, Greenland S. The probability of causation under a stochastic model for individual risk. Biometrics. 1989;45(4):1125–38. 10.2307/2531765. Suche in Google Scholar
[17] VanderWeele TJ, Robins JM. Stochastic counterfactuals and stochastic sufficient causes. Stat Sin. 2012;22(1):379–92. 10.5705/ss.2008.186. Suche in Google Scholar PubMed PubMed Central
[18] Hernán MA, Robins JM. Causal inference: what if. 1st edition. Boca Raton: Chapman and Hall/CRC, Boca Raton, Florida; 2020. https://www.hsph.harvard.edu/miguel-hernan/causal-inference-book/. Suche in Google Scholar
[19] Cole SR, Frangakis CE. The consistency statement in causal inference: a definition or an assumption? Epidemiology. 2009;20(1):1115–49. 10.1097/EDE.0b013e31818ef366. Suche in Google Scholar PubMed
[20] Imbens GW, Rubin DB. Causal inference for statistics, social, and biomedical sciences: an introduction. 1st edition. Cambridge: Cambridge University Press; 2015. ISBN 9781139025751. 10.1017/CBO9781139025751. Suche in Google Scholar
[21] Pearl J. Causality: models, reasoning, and inference. 2nd edition. Cambridge: Cambridge University Press; 2009. ISBN 9780511803161. 10.1017/CBO9780511803161. Suche in Google Scholar
[22] Peters J, Janzing D, Schölkopf B. Elements of causal inference: foundations and learning algorithms. Cambridge: The MIT Press; 1st edition. 2018. ISBN 9780262037310. 10.1080/00949655.2018.1505197. Suche in Google Scholar
[23] Munafò MR, Tilling K, Taylor AE, Evans DM, Smith GD. Collider scope: when selection bias can substantially influence observed associations. Int J Epidemiol. 2017;47(1):226–35. 10.1093/ije/dyx206. Suche in Google Scholar PubMed PubMed Central
[24] Pearl J. Three counterfactual interpretations and their identification. Synthese. 1999;121(1):93–149. 10.1023/A:1005233831499. Suche in Google Scholar
[25] Lu Z, Geng Z, Li W, Zhu S, Jia J. Evaluating causes of effects by posterior effects of causes. Biometrika. 2022;110(2):449–65. 10.1093/biomet/asac038. Suche in Google Scholar
[26] Zhang C, Geng Z, Li W, Ding P. Identifying and bounding the probability of necessity for causes of effects with ordinal outcomes. 2024. https://arxiv.org/abs/2411.01234. Suche in Google Scholar
[27] Tian J, Pearl J. Probabilities of causation: Bounds and identification. An Math Artif Intel. 2000;28(1):287–313. 10.1023/A:1018912507879. Suche in Google Scholar
[28] Li A, Pearl J. Probabilities of causation with nonbinary treatment and effect. 2022. 10.48550/arXiv.2208.09568. Suche in Google Scholar
[29] Huang EJ, Fang EX, Hanley DF, Rosenblum M. Inequality in treatment benefits: Can we determine if a new treatment benefits the many or the few? Biostatistics. 2016;18(2):308–24. 10.1093/biostatistics/kxw049. Suche in Google Scholar PubMed PubMed Central
[30] Huang EJ, Fang EX, Hanley DF, Rosenblum M. Constructing a confidence interval for the fraction who benefit from treatment, using randomized trial data. Biometrics. 2019;75(4):1228–39. 10.1111/biom.13101. Suche in Google Scholar PubMed PubMed Central
[31] Gadbury GL, Iyer HK, Albert JM. Individual treatment effects in randomized trials with binary outcomes. J Stat Plan Infer2004;121(2):163–74. 10.1016/S0378-3758(03)00115-0. Suche in Google Scholar
[32] Zhang Z, Wang C, Nie L, Soon G. Assessing the heterogeneity of treatment effects via potential outcomes of individual patients. J R Stat Soc Ser C (Appl Stat). 2013;62(5):687–704. 10.1111/rssc.12012. Suche in Google Scholar PubMed PubMed Central
[33] Wu P, Ding P, Geng Z, Liu Y. Quantifying individual risk for binary outcome. 2024. https://arxiv.org/abs/2402.10537. Suche in Google Scholar
[34] Su Y, Li X. Treatment effect quantiles in stratified randomized experiments and matched observational studies. Biometrika. 2023;111(1):235–54. 10.1093/biomet/asad030. Suche in Google Scholar
[35] Lei L, Candès EJ. Conformal inference of counterfactuals and individual treatment effects. J R Stat Soc Ser B (Stat Meth). 2021;83(5):911–38. 10.1111/rssb.12445. Suche in Google Scholar
[36] Jin Y, Ren Z, Candès EJ. Sensitivity analysis of individual treatment effects: A robust conformal inference approach. Proc Nat Acad Sci. 2023;120(6):e2214889120. 10.1073/pnas.2214889120. Suche in Google Scholar PubMed PubMed Central
[37] Yin M, Shi C, Wang Y, Blei DM. Conformal sensitivity analysis for individual treatment effects. J Am Stat Assoc. 2022;119:1–14. 10.1080/01621459.2022.2102503. Suche in Google Scholar
[38] Chernozhukov V, Wúthrich K, Zhu Y. Toward personalized inference on individual treatment effects. Proc Nat Acad Sci. 2023;120(7):e2300458120. 10.1073/pnas.2300458120. Suche in Google Scholar PubMed PubMed Central
[39] Chernozhukov V, Wüthrich K, Zhu Y. Distributional conformal prediction. Proc Nat Acad Sci, 2021;118(48):e2107794118. 10.1073/pnas.2107794118. Suche in Google Scholar PubMed PubMed Central
[40] Yin Y, Liu L, Geng Z. Assessing the treatment effect heterogeneity with a latent variable. Stat Sin. 2018;28(1):115–35. 10.5705/ss.202016.0150Suche in Google Scholar
[41] Laubender RP, Mansmann U, Lauseker M. Estimating the distribution of heterogeneous treatment effects from treatment responses and from a predictive biomarker in a parallel-group rct: A structural model approach. Biometric J. 2020;62(3):697–711. 10.1002/bimj.201800370. Suche in Google Scholar PubMed
[42] Shahn Z, Madigan D. Latent class mixture models of treatment effect heterogeneity. Bayesian Anal. 2017;12(3):831–54. 10.1214/16-BA1022. Suche in Google Scholar
[43] Meister A. Density deconvolution. Berlin, Heidelberg: Springer Berlin Heidelberg; 2009. p. 5–105. ISBN 978-3-540-87557-4. 10.1007/978-3-540-87557-4_2. Suche in Google Scholar
[44] Hansen BB. The prognostic analogue of the propensity score. Biometrika. 2008;95(2):481–8. 10.1093/biomet/asn004. Suche in Google Scholar
[45] Browne WJ, Draper D. A comparison of Bayesian and likelihood-based methods for fitting multilevel models. Bayesian Anal. 2006;1(3):473–514. 10.1214/06-BA117. Suche in Google Scholar
[46] McCulloch CE, Neuhaus JM. Misspecifying the shape of a random effects distribution: why getting it wrong may not matter. Stat Sci. 2011;26(3):388–402. 10.1214/11-STS361. Suche in Google Scholar
[47] Verbeke G, Lesaffre E. A linear mixed-effects model with heterogeneity in the random-effects population. J Am Stat Assoc. 1996;91(433):217–21. 10.2307/2291398. Suche in Google Scholar
[48] Proust C, Jacqmin-Gadda H. Estimation of linear mixed models with a mixture of distribution for the random effects. Comput Meth Programs Biomed. 2005;78(2):165–73. 10.1016/j.cmpb.2004.12.004. Suche in Google Scholar PubMed PubMed Central
[49] Zhang D, Davidian M. Linear mixed models with flexible distributions of random effects for longitudinal data. Biometrics. 2001;57(3):795–802. 10.1111/j.0006-341X.2001.00795.x. Suche in Google Scholar
[50] Kleinman KP, Ibrahim JG. A semiparametric bayesian approach to the random effects model. Biometrics. 1998;54(3):921. 10.2307/2533846. Suche in Google Scholar
[51] Gelman A, Carlin JB, Stern HS, Dunson DB, Vehtari A, Rubin DB. Bayesian data analysis. New York: CRC Press; third edition. 2021. ISBN 9781439898208. Suche in Google Scholar
[52] Ho RKW, Hu I. Flexible modelling of random effects in linear mixed models - A Bayesian approach. Comput Stat Data Anal. 2008;52(3):1347–61. 10.1016/j.csda.2007.09.005. Suche in Google Scholar
[53] Rousseau J, Mengersen K. Asymptotic behaviour of the posterior distribution in overfitted mixture models. J R Stat Soc Ser B (Methodol). 2011;73(5):689–710. 10.1111/j.1467-9868.2011.00781.x. Suche in Google Scholar
[54] Dunson DB. Bayesian nonparametric hierarchical modeling. Biomet J. 2009;51(2):273–84. 10.1002/bimj.200800183. Suche in Google Scholar PubMed
[55] Ohlssen DI, Sharples LD, Spiegelhalter DJ. Flexible random-effects models using bayesian semi-parametric models: applications to institutional comparisons. Stat Med. 2007;26(9):2088–112. 10.1002/sim.2666. Suche in Google Scholar PubMed
[56] Gelman A. Prior distributions for variance parameters in hierarchical models (comment on article by Browne and Draper). Bayesian Anal. 2006;1(3):515–34. 10.1214/06-BA117A. Suche in Google Scholar
[57] Bonvini M, Kennedy EH. Sensitivity analysis via the proportion of unmeasured confounding. J Am Stat Assoc. 2021;117(539):1–11. 10.1080/01621459.2020.1864382. Suche in Google Scholar
[58] Chiu LS, Pedley A, Massaro JM, Benjamin EJ, Mitchell GF, McManus DD, et al. The association of non-alcoholic fatty liver disease and cardiac structure and function-?framingham heart study. Liver Int. 2020;40(10):2445–54. 10.1111/liv.14600. Suche in Google Scholar PubMed PubMed Central
[59] Ghidey W, Lesaffre E, Verbeke G. A comparison of methods for estimating the random effects distribution of a linear mixed model. Stat Meth Med Res. 2010;19(6):575–600. 10.1177/0962280208091686. Suche in Google Scholar PubMed
[60] Rubio FJ, Steel MFJ. Flexible linear mixed models with improper priors for longitudinal and survival data. Electron J Stat. 2018;12(1):572–98. 10.1214/18-EJS1401. Suche in Google Scholar
[61] Vehtari A, Gelman A, Simpson D, Carpenter B, Bürkner PC. Rank-normalization, folding, and localization: An improved R for assessing convergence of MCMC (with discussion). Bayesian Anal. 2021;16(2):667–718. 10.1214/20-BA1221. Suche in Google Scholar
[62] Seamans MJ, Hong H, Ackerman B, Schmid I, Stuart EA. Generalizability of subgroup effects. Epidemiology. 2021;32(3):389–92. 10.1097/EDE.0000000000001329. Suche in Google Scholar PubMed PubMed Central
[63] Hahn RP, Murray JS, Carvalho CM. Bayesian regression tree models for causal inference: Regularization, confounding, and heterogeneous effects (with discussion). Bayesian Anal 2020;15(3):965–1056. 10.1214/19-BA1195. Suche in Google Scholar
[64] Wager S, Athey S. Estimation and inference of heterogeneous treatment effects using random forests. J Am Stat Assoc. 2018;113(523):1228–42. 10.1080/01621459.2017.1319839. Suche in Google Scholar
[65] Hajjem A, Bellavance F, Larocque D. Mixed-effects random forest for clustered data. J Stat Comput Simulat. 2014;84(6):1313–28. 10.1080/00949655.2012.741599. Suche in Google Scholar
[66] Pellagatti M, Masci C, Ieva F, Paganoni AM. Generalized mixed-effects random forest: A flexible approach to predict university student dropout. Stat Anal Data Min ASA Data Sci J. 2021;14(3):241–57. 10.1002/sam.11505. Suche in Google Scholar
© 2025 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Research Articles
- Decision making, symmetry and structure: Justifying causal interventions
- Targeted maximum likelihood based estimation for longitudinal mediation analysis
- Optimal precision of coarse structural nested mean models to estimate the effect of initiating ART in early and acute HIV infection
- Targeting mediating mechanisms of social disparities with an interventional effects framework, applied to the gender pay gap in Western Germany
- Role of placebo samples in observational studies
- Combining observational and experimental data for causal inference considering data privacy
- Recovery and inference of causal effects with sequential adjustment for confounding and attrition
- Conservative inference for counterfactuals
- Treatment effect estimation with observational network data using machine learning
- Causal structure learning in directed, possibly cyclic, graphical models
- Mediated probabilities of causation
- Beyond conditional averages: Estimating the individual causal effect distribution
- Matching estimators of causal effects in clustered observational studies
- Ancestor regression in structural vector autoregressive models
- Single proxy synthetic control
- Bounds on the fixed effects estimand in the presence of heterogeneous assignment propensities
- Minimax rates and adaptivity in combining experimental and observational data
- Highly adaptive Lasso for estimation of heterogeneous treatment effects and treatment recommendation
- A clarification on the links between potential outcomes and do-interventions
- Valid causal inference with unobserved confounding in high-dimensional settings
- Spillover detection for donor selection in synthetic control models
- Causal additive models with smooth backfitting
- Experiment-selector cross-validated targeted maximum likelihood estimator for hybrid RCT-external data studies
- Applying the Causal Roadmap to longitudinal national registry data in Denmark: A case study of second-line diabetes medication and dementia
- Orthogonal prediction of counterfactual outcomes
- Variable importance for causal forests: breaking down the heterogeneity of treatment effects
- Multivariate zero-inflated causal model for regional mobility restriction effects on consumer spending
- Rate doubly robust estimation for weighted average treatment effects
- Adding covariates to bounds: what is the question?
- Review Article
- The necessity of construct and external validity for deductive causal inference
Artikel in diesem Heft
- Research Articles
- Decision making, symmetry and structure: Justifying causal interventions
- Targeted maximum likelihood based estimation for longitudinal mediation analysis
- Optimal precision of coarse structural nested mean models to estimate the effect of initiating ART in early and acute HIV infection
- Targeting mediating mechanisms of social disparities with an interventional effects framework, applied to the gender pay gap in Western Germany
- Role of placebo samples in observational studies
- Combining observational and experimental data for causal inference considering data privacy
- Recovery and inference of causal effects with sequential adjustment for confounding and attrition
- Conservative inference for counterfactuals
- Treatment effect estimation with observational network data using machine learning
- Causal structure learning in directed, possibly cyclic, graphical models
- Mediated probabilities of causation
- Beyond conditional averages: Estimating the individual causal effect distribution
- Matching estimators of causal effects in clustered observational studies
- Ancestor regression in structural vector autoregressive models
- Single proxy synthetic control
- Bounds on the fixed effects estimand in the presence of heterogeneous assignment propensities
- Minimax rates and adaptivity in combining experimental and observational data
- Highly adaptive Lasso for estimation of heterogeneous treatment effects and treatment recommendation
- A clarification on the links between potential outcomes and do-interventions
- Valid causal inference with unobserved confounding in high-dimensional settings
- Spillover detection for donor selection in synthetic control models
- Causal additive models with smooth backfitting
- Experiment-selector cross-validated targeted maximum likelihood estimator for hybrid RCT-external data studies
- Applying the Causal Roadmap to longitudinal national registry data in Denmark: A case study of second-line diabetes medication and dementia
- Orthogonal prediction of counterfactual outcomes
- Variable importance for causal forests: breaking down the heterogeneity of treatment effects
- Multivariate zero-inflated causal model for regional mobility restriction effects on consumer spending
- Rate doubly robust estimation for weighted average treatment effects
- Adding covariates to bounds: what is the question?
- Review Article
- The necessity of construct and external validity for deductive causal inference

