Abstract
During their biogenesis, the ribosomal subunits undergo numerous structural and compositional changes to achieve their final architecture. RNA helicases are a key driving force of such remodelling events but deciphering their particular functions has long been challenging due to lack of knowledge of their molecular functions and RNA substrates. Advances in the biochemical characterisation of RNA helicase activities together with new insights into RNA helicase binding sites on pre-ribosomes and structural snapshots of pre-ribosomal complexes containing RNA helicases now open the door to a deeper understanding of precisely how different RNA helicases contribute to ribosomal subunit maturation.
1 Introduction
RNA-dependent nucleoside-triphosphatases (NTPases) are key regulators of RNA metabolism through their functions in RNA/ribonucleoprotein (RNP) complex remodelling. A sub-group of NTPases, often referred to as RNA helicases, are characterized by the presence of tandem RecA-like domains and conserved sequence motifs (Bourgeois et al. 2016). The cellular process requiring the largest number of RNA helicases is the biogenesis of ribosomal subunits where, in S. cerevisiae (yeast), approximately 20 RNA helicases are involved (Martin et al. 2013; Mitterer and Pertschy 2022). Assembly of the four ribosomal RNAs (rRNAs) and 79 ribosomal proteins (RPs) to form the mature small (40S) and large (60S) ribosomal subunits (SSU and LSU, respectively) is a precisely orchestrated process involving the coordinated action of more than 200 trans-acting assembly factors (AFs) (Bohnsack and Bohnsack 2019; Klinge and Woolford 2019). These include structural components that mediate protein—protein and protein—RNA interactions and diverse types of enzymes. The portfolio of AFs also includes small nucleolar (sno)RNPs, which install specific rRNA modifications or chaperone rRNA folding via basepairing interactions of their snoRNA components with the pre-rRNA. In yeast, nascent pre-ribosomal particles are formed co-transcriptionally and undergo a hierarchical series of remodelling and maturation events including processing, modification and folding of the (pre-)rRNAs, and recruitment and positioning of the RPs.
The RNA helicases involved in ribosome biogenesis were initially identified based on their enrichment in the nucleolus, the initiation site of ribosomal subunit assembly, their physical association with pre-ribosomal particles and/or the fact that their depletion/deletion affects pre-rRNA processing. However, attaining mechanistic understanding of the precise molecular functions of these helicases is a major challenge. A key aspect of elucidating the molecular functions of RNA helicases is to determine their sites of action on pre-ribosomal particles and identify their substrates. The growing understanding that the molecular actions of RNA helicases extend beyond unwinding RNA duplexes to include roles in duplex annealing and displacement of RNA-binding proteins (RBPs) raises the further question of how each of the helicases involved in ribosome biogenesis exert their function on a mechanistic level. Knowledge on these topics is particularly important for understanding the bona fide roles of RNA helicases involved in ribosome biogenesis as it allows to differentiate direct and indirect effects of helicase action. In this article, we will describe the biochemical activities of RNA helicases involved in yeast ribosome biogenesis and review the current knowledge on their binding sites on pre-ribosomal particles and pre-rRNA substrates. We will also discuss the topic of indirect consequences of RNA helicase action on the yeast ribosome assembly pathway and highlight evidence supporting direct remodelling of pre-ribosomal subunits by RNA helicases.
2 Different modes of action of RNA helicases in ribosome biogenesis
Ribosome assembly is best characterized in the yeast model system where 19 RNA helicases are known to be involved (Martin et al. 2013; Mitterer and Pertschy 2022). These helicases belong to the DEAH/RHA (Prp43, Dhr1, Dhr2), Ski2-like (Mtr4) and DExD box (Dbp2, Dbp3, Dbp4, Dbp6, Dbp7, Dbp8, Dbp9, Dpb10, Drs1, Fal1, Has1, Mak5, Rok1, Rrp3, Spb4) families.
Prp43 has served as a prototype for structural and biochemical analyses of DEAH/RHA helicases, and mechanistic views of RNA duplex unwinding by this type of enzyme are based on these findings (reviewed in K. E. Bohnsack et al. 2022a). In line with biochemical data, crystal structures of Prp43 revealed that it does not display RNA sequence specificity as it predominately contacts the RNA backbone (He et al. 2010; Tauchert et al. 2016, 2017; Walbott et al. 2010). Binding of DEAH/RHA helicases to single-stranded regions of their substrate RNAs induced formation of a “closed” conformation in which the two RecA-like domains are positioned such that ATP hydrolysis is triggered. This, in turn, causes further conformational rearrangements of the RecA-like domains with the more flexible RecA2 domain opening towards to the 5′ end of the RNA such that an additional nucleotide is accommodated in the active site. Continuous ATP hydrolysis-coupled transitions between the closed and open states thus allow the protein to translocate in a 3′-5′ direction relative to the single stranded RNA substrate (Figure 1A, upper panel) (Hamann et al. 2019). Ultimately, release of the RNA substrate occurs mainly by rearrangements of the C-terminal domains that lead to opening of the RNA-binding tunnel formed by the two RecA-like domains. Translocation of DEAH/RHA helicases has the potential to displace an associated RNA strand leading to duplex unwinding as described for DNA helicases. However, within the context of an RNP where the helicases are held in a fixed position, translocation activity can allow DEAH/RHA helicases to function as molecular winches, disrupting RNA duplexes and displacing RBPs at a distance (Figure 1A, lower panel) (Semlow et al. 2016). In the context of ribosome assembly, Prp43 is activated by two cofactor proteins belonging to the G-patch family, Prx1 (Gno1) and Sqs1 (Pfa1) (M. T. Bohnsack et al. 2009; Chen et al. 2014; Lebaron et al. 2009; Pertschy et al. 2009; Robert-Paganin et al. 2017). These cofactor proteins enhance Prp43 activity by tethering the RecA-like domains in a favourable conformation, thus stimulating the transitions between the open and closed states of the RecA-like domains, accelerating ATP hydrolysis and translocation of the enzyme on the substrate (Bohnsack et al. 2021; Enders et al. 2022). Of the other DEAH/RNA helicases involved in yeast ribosome biogenesis, Dhr2 remains uncharacterized, but in vitro unwinding activity of Dhr1 has been demonstrated on a physiologically relevant RNA duplex, implying that it functions similarly to Prp43 on a molecular level (Lin et al. 2022; Zhu et al. 2016).
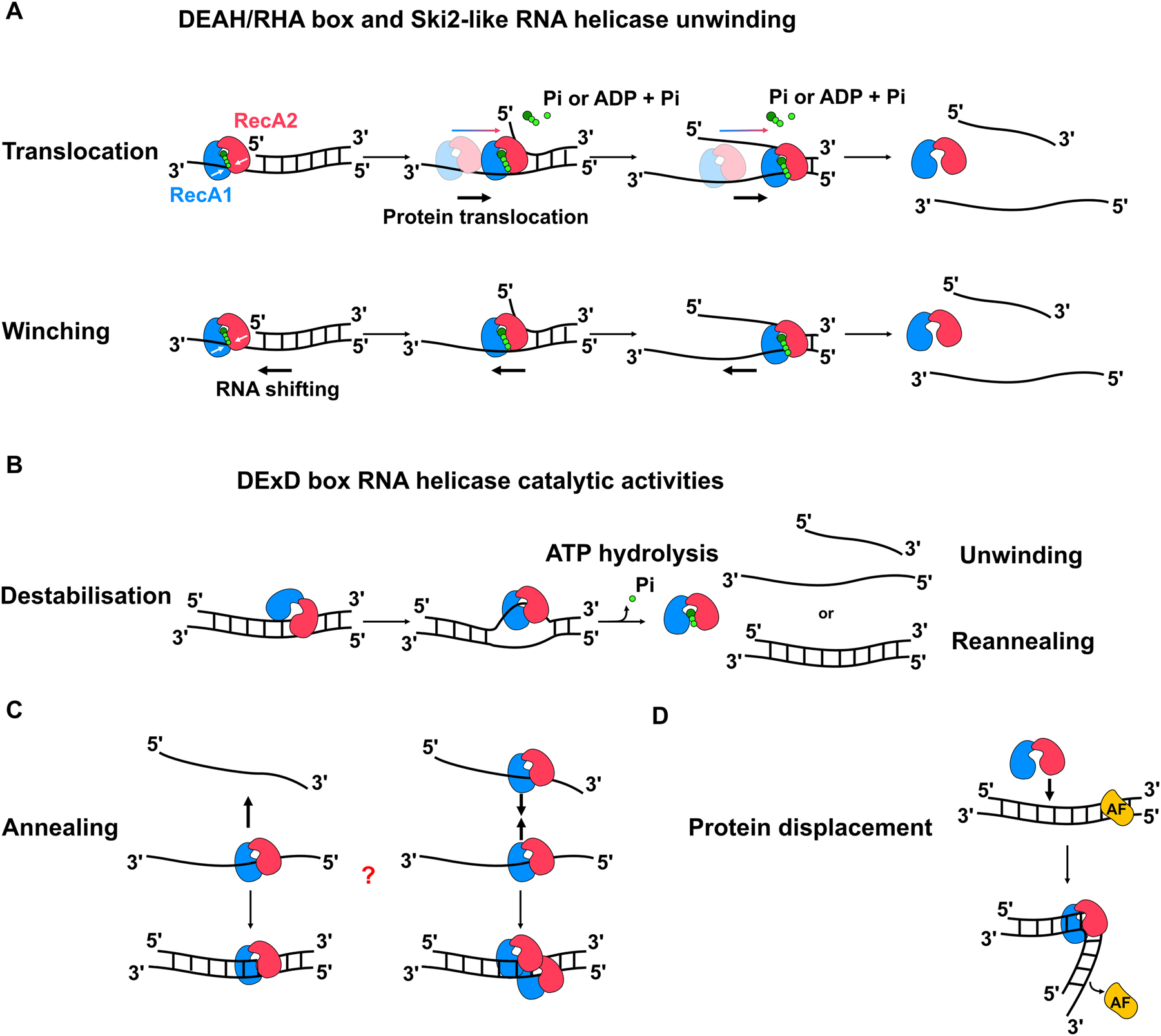
Molecular functions of RNA helicases. Schematic views of different modes of action of different types of RNA helicases are depicted. The two RecA-like domains of the helicase core are shown in blue and red, ATP is depicted as green circles and RNA strands are shown as black lines. (A) RNA duplex unwinding by strand displacement (upper) and winching (lower) by DEAH/RHA and Ski2-like helicases. (B) Canonical model of local strand unwinding by DExD box proteins. (C) Alternative models of “RNA annealing” by RNA duplex formation (left) and RNA strand bridging by helicase multimerization (right). (D) Displacement of an RNA-binding protein (RBP) from a duplex upon RNA helicase binding.
The Ski2-like RNA helicase Mtr4, which associates with the RNA processing and decay machinery, the RNA exosome, has been extensively characterized in vitro (Hoof et al. 2000). Mtr4 contains a four-domain helix bundle, called the ‘Ratchet helix’, which forms a ring-like structure that accommodates RNA passage (Figure 2B) (Johnson and Jackson 2013). This helix is important for Mtr4 affinity for polyadenylated substrates. Two anti-parallel coiled-coil domains extend over the helicase core and terminate in a globular region. These two domains are bent into an arch-like structure forming the ‘Arch domain’ (Figure 2B). Both the ratchet and arch domains are necessary for Mtr4 unwinding activity (Taylor et al. 2014). Similar to DEAH/RHA RNA helicases, Mtr4 displays an ATP-dependent, 3′—5′ polarity unwinding activity in vitro (Figure 1A, upper) (Jia et al. 2012; Wang et al. 2008). This mode of action is well suited to the role of Mtr4 in resolving RNA structural elements to allow channelling into the barrel of the RNA exosome.
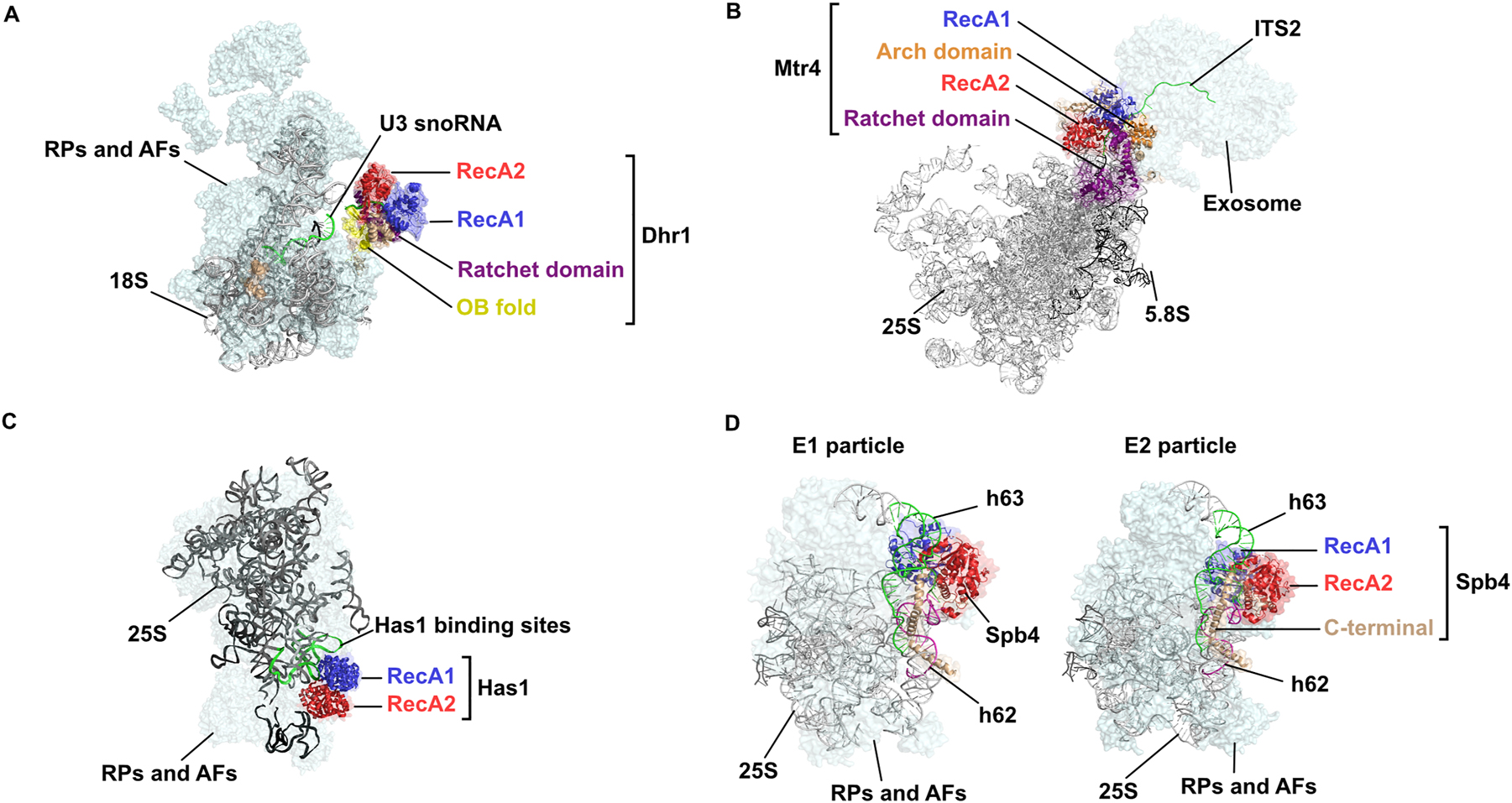
Structural views of RNA helicases in pre-ribosomal complexes. Cryo-EM structures of selected pre-ribosomal particles containing RNA helicases are shown: (A) Dhr1 (PDB: 6ZQG), (B) Mtr4 (PDB: 6FT6_6FSZ), (C) Has1 (PDB: 6C0F), (D) Sbp4 (PDB: E1: 7R72; E2: 7NAD). In each panel, the RNA helicase RecA1 and RecA2 domains are shown in blue and red, respectively, and other helicase regions: Ratchet domain in purple, OB fold in yellow, Arch domain in orange and other helicase regions/C-terminal extensions in beige. All presented in cartoon mode with a transparent surface. rRNAs are depicted in cartoon mode in grey/black and the rRNA regions/snoRNAs contacted by the RNA helicases are shown in green (A – U3 snoRNA; B – internal transcribed spacer 2 (ITS2); C – domain I; D – helix (h) 63) or pink (D – helix (h) 62). Selected ribosomal proteins (RPs) and assembly factors (AFs) present in the particles are shown in pale cyan in surface view.
In contrast to DEAH/RHA helicases, the classical model of RNA duplex unwinding by DExD box proteins involves non-processive, non-directional local strand unwinding (Figure 1B) (Linder and Jankowsky 2011). DExD RNA helicases lacking ATP or substrate adopt an open, inactive conformation. Cooperative binding of ATP and recruitment to an RNA substrate induce formation of a closed state in which the conserved sequence motifs of the two RecA-like domains, ATP and the RNA substrate are positioned in a geometry incompatible with RNA duplex structure. The conformational changes arising from ATP and RNA binding therefore lead to displacement of one RNA strand in a process termed local strand unwinding. Typically, this rearrangement of the catalytic site also induces ATP hydrolysis, which in turn leads to dissociation of the DExD box protein from the remaining RNA strand. Such activity is well suited to resolving short stretches of basepairing between RNA strands and several DExD box proteins involved in yeast ribosome biogenesis (Dbp3, Dbp4, Rok1 and Rrp3) have been shown capable of unwinding short (10 bp) RNA duplexes in vitro (Garcia et al. 2012). Despite the fact that the majority of RNA helicases involved in yeast ribosome biogenesis are DExD box proteins, it is not immediately intuitive to understand how such limited activity can be leveraged to remodel such large RNP complexes.
This conundrum may in part be resolved by the growing appreciation that the term “RNA helicase” has the potential to be misleading, as molecular functions other than unwinding are attributed to DExD box proteins; it has emerged that some DExD box proteins may act as RNA strand annealers and/or function by displacing RBPs (Figure 1C, D) (Karbstein 2022). The ability of DExD box proteins to displace RBPs is well established (Figure 1D) (Jankowsky et al. 2001) and has also been observed, for example, for the SSU RNA helicase Rok1 (Khoshnevis et al. 2016; Young et al. 2013). An RNA annealing function can involve promoting formation of RNA duplexes (Figure 1C, left), but the term has also been used in the context of proteins that bridge interactions between RNA strands (Figure 1C, right). RNA unwinding and strand annealing activities are not mutually exclusive and several DExD box proteins, including Rok1, have been reported to display both activities in vitro (Garcia et al. 2012; Halls et al. 2007; Yang and Jankowsky 2005; Young et al. 2013). Such flexibility in molecular function raises the possibility that DExD box proteins could independently chaperone RNA intricate re-folding events without the need for other factors. It is likely that balance between unwinding and annealing functions depends on ATP and ADP concentrations as well as the current arrangement of the RNA substrate. Of the DExD box proteins involved in yeast ribosome biogenesis, Rok1, Dbp6 and, to a lesser extent, Dbp2 have been demonstrated to display annealing activity in vitro (Khoshnevis et al. 2016; Khreiss et al. 2023; Ma et al. 2013; Young et al. 2013), but the mechanistic basis of RNA annealing by these proteins remains unknown.
3 Identification of RNA helicase pre-rRNA binding sites and targets
A key frontier in understanding RNA helicases functions during ribosome assembly on the mechanistic level has been the identification of the targets of their remodelling activities. In several cases, genetic approaches have provided important clues toward RNA helicase substrates by revealing the effects of catalytically inactive mutants on specific steps of subunit assembly and inter-dependent relationships with other AFs, including specific snoRNAs (Jaafar et al. 2021; Sardana et al. 2015). Alongside, the development of techniques for physically mapping the binding sites of RBPs on substrate RNAs and momentous advances in structural biology of macromolecular complexes have opened the door to elucidating RNA helicase binding sites on pre-ribosomal particles and pre-rRNA targets.
An approach extensively employed to determine pre-rRNA binding sites of AFs, including various RNA helicases, is the UV-crosslinking and analysis of cDNA (CRAC) approach (Bohnsack et al. 2012; Granneman et al. 2009). This method involves deep sequencing of a cDNA library generated by reverse transcription of trimmed RNA fragments covalently crosslinked to a target protein. The CRAC approach has so far been successfully utilized to identify pre-rRNA crosslinking sites of Rok1, Has1, Prp43, Dbp6, Dbp7, Mak5, Mtr4, Sbp4 and Dbp10 (Aquino et al. 2021a; Bohnsack et al. 2009; Brüning et al. 2018; Gnanasundram et al. 2019; Khreiss et al. 2023; Manikas et al. 2016; Martin et al. 2014). In some cases, individual binding sites were identified (e.g. Mak5), whereas in others, multiple crosslinking sites were revealed. In the case of e.g. Dbp7, Spb4 and Dbp10, the crosslinking sites spatially cluster suggesting that they reflect different crosslinks of RNA helicase molecules bound at one defined locus. In other cases where multiple distinct crosslinking sites were observed, it is possible that, especially for DEAH/RHA RNA helicases (e.g. Prp43), the multiple crosslinking sites reflect snapshots or stalling points of the helicase as it translocates relative to its pre-rRNA substrate. The transcriptome-wide nature of the CRAC approach has also revealed crosslinking sites of several RNA helicases (Dhr1, Rok1, Has1, Prp43 and Dbp7) on specific snoRNAs. Mapping RNA helicase crosslinking sites on snoRNAs has often revealed sequences known to basepair with rRNAs, suggesting functional relevance. The complementary crosslinking, ligation and sequencing of hybrids (CLASH) approach, which ligates two RNA fragments bound by the same protein to generate chimeric sequencing reads (Kudla et al. 2011; Travis et al. 2014), has provided experimental verification that Rok1, Has1 and Prp43 not only contact pre-rRNA and snoRNA sequences with the potential to basepair, but that the RNA helicase contacts these sequences when they are actually basepaired (Aquino et al. 2021b; Brüning et al. 2018; Martin et al. 2014). Such data support RNA helicase functions in modulating snoRNA-pre-rRNA interactions, but a limitation of the CRAC approach is that the RNA crosslinking sites identified cannot be attributed to specific regions of the protein. In the case of RNA helicases involved in ribosome biogenesis, it therefore remains unknown whether the sites identified represent remodelling targets of the helicase catalytic site or if they rather reflect binding platforms of the helicase.
Alongside such protein—RNA crosslinking-based studies, a recent protein—protein crosslinking analysis of pre-60S particles provided insights into the context of several RNA helicases (Sailer et al. 2022). Mapping of the obtained crosslinking sites on available (pre-)ribosome structures allows the orientation of RNA helicases to be estimated. For example, crosslinking between the RecA1 domain of Dbp9 and the ribosomal protein uL6 suggests proximity of the helicase active site and the eukaryotic expansion segment (ES)27 in domain II of the 25S rRNA.
Cryo-EM analyses of pre-ribosomal particles has recently allowed visualization of a handful of RNA helicases together with their substrates (Figure 2). Such structural snapshots not only reveal the architecture of RNA helicase—pre-rRNA interactions, but also give temporal insights, by revealing the composition of the pre-ribosomal particle at the time that the RNA helicase is in the identified position. So far, only four RNA helicases have been captured on pre-ribosomal particles subjected to cryo-EM: Dhr1, Mtr4, Has1 and Sbp4 (Figure 2; Cheng et al. 2020; Cruz et al. 2022; Kater et al. 2017; Olsen and Johnson 2021; Sanghai et al. 2018). The minimal number of RNA helicases thus far visualized on pre-ribosomal particles likely reflects the facts that (i) the interactions RNA helicases make with pre-ribosomal subunits can be transient and potentially weak, and (ii) many RNA helicases function during the early stages of ribosomal subunit assembly, when the particles are very flexible and therefore challenging for structural analyses. Notably, none of the pre-ribosomal particles visualized by cryo-EM thus far contain RNA modification-guiding snoRNPs. Dedicated strategies targeting pre-ribosomal particles associated with particular RNA helicases, as was recently done for Spb4 (Cruz et al. 2022), will likely be necessary to obtain structural views of further RNA helicases in situ.
The first visualizations of RNA helicases engaged on pre-rRNA substrates were obtained for DEAH and Ski2-like helicases that possess translocase activity (Figure 2A, B). It is possible that the long residence times of such helicases on their substrates increase the chance of capturing them in an active state. A series of SSU processomes undergoing maturation revealed how the DEAH/RHA helicase Dhr1 engages its U3 snoRNA—18S rRNA target duplex (Cheng et al. 2020). Initially, Dhr1 binds in an open conformation and substrate interaction is sterically inhibited. Structural rearrangements then alter Dhr1 position and topology such that the helicase engages its snoRNA—pre-rRNA target in an active closed conformation (Figure 2A). Together with biochemical data showing that Dhr1 can efficiently unwind RNA duplexes in vitro (Lin et al. 2022), the structural data support a model in which Dhr1 actively resolves the snoRNA—pre-rRNA duplex to promote expulsion of the U3 snoRNA, SSU processome disassembly and the transition to a pre-40S particle. Similarly, the Ski2-like helicase Mtr4, which displays robust helicase activity in vitro, has been visualized in an active conformation on pre-rRNAs in the context of its role as an adaptor and unwindase for the RNA exosome (Taylor et al. 2014). In the SSU processome, the RNA exosome degrades a fragment of the 5′ external transcribed spacer and the pathway of this pre-rRNA can be traced from the core of the particle, through the Mtr4 active site, where RNA secondary structures are resolved, and into the barrel of the exosome to the ribonuclease sites (Du et al. 2020). Likewise, structural evidence supports the role of Mtr4 in dissolving structured regions in the internal transcribed spacer region 2 to allow processing of the 3′ end of the 5.8S rRNA during late pre-60S biogenesis (Figure 2B) (Schuller et al. 2018).
Defining the precise pre-ribosomal targets of DExD box proteins is especially interesting due to their diverse biochemical functions, but has thus far proved challenging. Has1 contributes to the biogenesis of both ribosomal subunits (Gnanasundram et al. 2019). The lack of the helicase causes accumulation of the U14 snoRNA on pre-SSU particles. An active role in promoting U14 dissociation from pre-ribosomes is supported by CRAC and CLASH data that demonstrate crosslinking of Has1 to the U14 snoRNA and its cognate 18S rRNA interaction sites while they are basepaired (Brüning et al. 2018; Liang and Fournier 2006). However, as Has1 helicase activity has not yet been demonstrated, it remains unknown if it is able to trigger disassembly of such structures or whether U14 release is a consequence of other remodelling events. While Has1 has not been visualized on pre-40S complexes, structures showing Has1 associated with helix (h)16 of the 25S rRNA (domain I) of pre-60S particles are available. In the context of pre-LSU particles, Has1 is required for the recruitment of several ribosomal proteins to domain I (Dembowski et al. 2013). Although the ATPase activity of Has1 is required for maturation of the LSU pre-rRNAs, the structural snapshots captured show Has1 in a non-active conformation (Figure 2C) (Sanghai et al. 2018; Zhou et al. 2019). ATP is not resolved/present in the binding pocket and while the h16 rRNA threads through the two RecA-like domains, the contacts are not consistent with those observed for other helicases in active conformations. Therefore, the precise target of Has1 remodelling activity still remains elusive. Recent structural analysis of pre-60S particles purified via Sbp4 elegantly illustrate how local strand unwinding by a DExD box protein can initiate substantial remodelling of RNA structures within a large RNP like a pre-60S particle (Figure 2D) (Cruz et al. 2022). Crosslinking data and structural analyses reveal Spb4 binding to h62 and h63 at the base of ES27 of the 25S rRNA (Brüning et al. 2018; Cruz et al. 2022). In the earlier particles analysed (Figure 2D, left), five nucleotides of h63 are accommodated in the Spb4 active site with contact to the ribose backbone, consistent with those of active DExD box helicases, defining this region as the direct target of Sbp4 activity. Intriguingly, as the pre-60S particles mature, h63 and h62 are unwound and h62 is arranged in an alternative conformation (h62alt; Figure 2D, right). This post-remodelling state is stabilized by the long C-terminal extension of Spb4; one region of the C-terminal extension associates in a sequence-specific manner with nucleotides adjacent to those in the helicase active site, while another wraps around h62alt. These interactions effectively extend the bubble of local strand unwinding to approximately 35 nucleotides, thus encompassing h62. Formation of h62alt is an important stepping stone in the formation of the h61-h64 junction that precedes assembly of the h61, h62, h64 three-way junction (Cruz et al. 2022).
For Dhr1, Mtr4 and Sbp4, the combination of biochemical, structural and functional data provides clear evidence of direct pre-rRNA remodelling by helicases during ribosome assembly and highlight the diverse molecular mechanism by which these proteins can act. These examples serve as excellent templates upon which to further explore the functions of other RNA helicases in ribosome assembly.
4 Differentiating direct effects of RNA helicase action during ribosome assembly and indirect consequences
The combination of elucidating the biochemical properties of RNA helicases and determining their RNA target sites is necessary to truly understand their cellular functions. In the context of a complex RNP biogenesis pathway like ribosomal subunit assembly, lack of a particular helicase will cause both direct and indirect effects that can be difficult to differentiate. While direct effects reflect the precise RNA structural elements remodeled by the RNA helicase, indirect effects can arise in several different ways. For example, RNA helicase-mediated remodelling of a specific pre-rRNA structure can lead to structural changes at distant sites within the same particle. Alternatively, indirect effects can be temporal with the helicase action having indirect consequences on downstream steps in the assembly pathway. Differentiating direct and indirect effects of helicase action is further compounded by the fact that these enzymes, especially the DExD box proteins, often likely fulfil both catalytic and non-catalytic roles, i.e. as well as utilizing their remodelling activities, their presence in pre-ribosomal particles and the protein-protein interactions they make are often also important.
The rearrangement of h61-h64 of the 25S rRNA by Sbp4 and the Mtr4-mediated resolution of structural elements of the 5′ ETS and ITS2 for RNA exosome-mediated processing or decay described above exemplify clearly direct effects of RNA helicase action (Cruz et al. 2022; Olsen and Johnson 2021). Several RNA helicases have been functionally linked to modulating snoRNA dynamics on pre-ribosomes, because specific snoRNAs accumulate on pre-ribosomes in the absence of the helicase (see for example: Bohnsack et al. 2008; Koš and Tollervey 2005; Liang and Fournier 2006; Sardana et al. 2015). In the case of Dhr1, a direct role in unwinding U3 snoRNA-18S rRNA basepairing is confirmed by structural evidence (Cheng et al. 2020; Sardana et al. 2015). Prp43 is implicated in releasing a subset of snoRNPs from their pre-60S binding sites (Bohnsack et al. 2009). During spliceosome disassembly, Prp43 acts as a winch, disrupting RNA structures and displacing RBPs remotely (Semlow et al. 2016). It is possible that Prp43 uses a similar mode of action during pre-60S biogenesis, but the finding that the helicase crosslinks to the affected snoRNAs and their cognate pre-rRNA sequences while they are basepaired rather favours the model that Prp43 acts as a translocating unwindase. For DExD box proteins, the situation is more complex. For example, the U14 snoRNA accumulates on pre-40S particles when either Dbp4 or Has1 is lacking (Koš and Tollervey 2005; Liang and Fournier 2006). Although this was initially expected to reflect functional redundancy, the identification of Has1 RNA substrates rather suggest a role of Has1 in disrupting U14 snoRNA—18S rRNA basepairing (Brüning et al. 2018). Dbp4 may instead perform a remodelling event upstream that, when not fulfilled, impairs Has1 action and/or U14 release (Soltanieh et al. 2015). This scenario has been observed for another DExD box protein, Rok1, which in an ATP-bound form stabilizes association of the AF Rrp5 with pre-40S particles (Khoshnevis et al. 2016). ATP hydrolysis triggers dissociation of Rrp5, thus promoting release of snR30. Notably, Has1 has also been linked to release of Rrp5 via displacement of the AF Rrp36, thus also contributing indirectly to release of snR30 (Liu et al. 2021). The DExD box protein Dbp7 crosslinks to domain V/VI of the 25S rRNA as well as a number of snoRNAs with basepairing sites in this region (Aquino 2021a; Bohnsack et al. 2022b; Jaafar et al. 2021). In the absence of Dbp7, many of these snoRNAs accumulate on pre-60S particles, but it is unlikely that Dbp7 directly unwinds multiple stretches of snoRNA—pre-rRNA basepairing. Rather an individual snoRNA—pre-rRNA duplex or a pre-rRNA structure within domain V represents the direct Dbp7 substrate. A recently described example of indirect consequences of an unknown remodelling event mediated by a DExD box protein is the finding that lack of Dbp3 causes widespread reduction of 2′-O-methylation across the 25S rRNA (Aquino et al. 2021b). Although Dbp3 can unwind short RNA duplexes in vitro (Garcia et al. 2012), in this case, lack of knowledge of the in vivo substrate RNAs impedes deeper mechanistic understanding of the role of this helicase.
5 Conclusions and outlook
Deciphering the precise functions of RNA helicases during ribosomal subunit assembly has proved to be a demanding endeavour. However, concerted efforts in biochemistry, structural biology and functional analyses have recently provided the first break-through examples where RNA helicase functions can be precisely and specifically attributed. This sets a precedent that will no doubt lead to further insights into RNA helicase functions during ribosome assembly. However, the best characterized RNA helicases so far are those working on relatively well-defined, stable particles (SSU processome and late pre-60S) that are accessible for structural analyses. Achieving a similar level of understanding for RNA helicases operating in highly dynamic early pre-60S particles or nascent SSU processomes will likely require further innovation and technological developments. RNA helicases have long been considered driving forces for ribosomal subunit assembly, but discovery of their precise molecular functions will allow their general roles within the pathway to be better understood. It is likely to emerge that the remodelling events catalysed by RNA helicases not only allow progression to the next maturation step but also act as key checkpoints allowing dynamic regulation and quality control.
Funding source: Deutsche Forschungsgemeinschaft
Award Identifier / Grant number: SFB860 A12
Funding source: Deutsche Forschungsgemeinschaft
Award Identifier / Grant number: Unassigned
-
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Research funding: This work was funded by the Deutsche Forschungsgemeinschaft (SFB860 A12). We would like to thank Jimena Davila Gallesio for comments on the manuscript.
-
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
Aquino, G.R.R., Hackert, P., Krogh, N., Pan, K.T., Jaafar, M., Henras, A.K., Nielsen, H., Urlaub, H., Bohnsack, K.E., and Bohnsack, M.T. (2021a). The RNA helicase Dbp7 promotes domain V/VI compaction and stabilization of inter-domain interactions during early 60S assembly. Nat. Commun. 12: 6152, https://doi.org/10.1038/s41467-021-26208-9.Search in Google Scholar PubMed PubMed Central
Aquino, G.R.R., Krogh, N., Hackert, P., Martin, R., Gallesio, J.D., van Nues, R.W., Schneider, C., Watkins, N.J., Nielsen, H., Bohnsack, K.E., et al.. (2021b). RNA helicase-mediated regulation of snoRNP dynamics on pre-ribosomes and rRNA 2′- O -methylation. Nucleic Acids Res. 49: 4066–4084, https://doi.org/10.1093/nar/gkab159.Search in Google Scholar PubMed PubMed Central
Bohnsack, K.E. and Bohnsack, M.T. (2019). Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J. 38: e100278, https://doi.org/10.15252/embj.2018100278.Search in Google Scholar PubMed PubMed Central
Bohnsack, K.E., Ficner, R., Bohnsack, M.T., and Jonas, S. (2021). Regulation of DEAH-box RNA helicases by G-patch proteins. Biol. Chem. 402: 561–579, https://doi.org/10.1515/hsz-2020-0338.Search in Google Scholar PubMed
Bohnsack, K.E., Henras, A.K., Nielsen, H., and Bohnsack, M.T. (2022a). Making ends meet: a universal driver of large ribosomal subunit biogenesis. Trends Biochem. Sci. 48: 213–215 https://doi.org/10.1016/j.tibs.2022.09.003.Search in Google Scholar PubMed
Bohnsack, K.E., Kanwal, N., and Bohnsack, M.T. (2022b). Prp43/DHX15 exemplify RNA helicase multifunctionality in the gene expression network. Nucleic Acids Res. 50: 9012–9022, https://doi.org/10.1093/nar/gkac687.Search in Google Scholar PubMed PubMed Central
Bohnsack, M.T., Kos, M., and Tollervey, D. (2008). Quantitative analysis of snoRNA association with pre-ribosomes and release of snR30 by Rok1 helicase. EMBO Rep. 9: 1230–1236, https://doi.org/10.1038/embor.2008.184.Search in Google Scholar PubMed PubMed Central
Bohnsack, M.T., Martin, R., Granneman, S., Ruprecht, M., Schleiff, E., and Tollervey, D. (2009). Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol. Cell 36: 583–592, https://doi.org/10.1016/j.molcel.2009.09.039.Search in Google Scholar PubMed PubMed Central
Bohnsack, M.T., Tollervey, D., and Granneman, S. (2012). Identification of RNA helicase target sites by UV cross-linking and analysis of cDNA. In: Methods in enzymology. Elsevier, Amsterdam, pp. 275–288.10.1016/B978-0-12-396546-2.00013-9Search in Google Scholar PubMed
Bourgeois, C.F., Mortreux, F., and Auboeuf, D. (2016). The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. Rev. Mol. Cell Biol. 17: 426–438, https://doi.org/10.1038/nrm.2016.50.Search in Google Scholar PubMed
Brüning, L., Hackert, P., Martin, R., Gallesio, J.D., Aquino, G.R.R., Urlaub, H., Sloan, K.E., and Bohnsack, M.T. (2018). RNA helicases mediate structural transitions and compositional changes in pre-ribosomal complexes. Nat. Commun. 9: 5383, https://doi.org/10.1038/s41467-018-07783-w.Search in Google Scholar PubMed PubMed Central
Chen, Y.-L., Capeyrou, R., Humbert, O., Mouffok, S., Kadri, Y.A., Lebaron, S., Henras, A.K., and Henry, Y. (2014). The telomerase inhibitor Gno1p/PINX1 activates the helicase Prp43p during ribosome biogenesis. Nucleic Acids Res. 42: 7330–7345, https://doi.org/10.1093/nar/gku357.Search in Google Scholar PubMed PubMed Central
Cheng, J., Lau, B., La Venuta, G., Ameismeier, M., Berninghausen, O., Hurt, E., and Beckmann, R. (2020). 90S pre-ribosome transformation into the primordial 40S subunit. Science 369: 1470–1476, https://doi.org/10.1126/science.abb4119.Search in Google Scholar PubMed
Cruz, V.E., Sekulski, K., Peddada, N., Sailer, C., Balasubramanian, S., Weirich, C.S., Stengel, F., and Erzberger, J.P. (2022). Sequence-specific remodeling of a topologically complex RNP substrate by Spb4. Nat. Struct. Mol. Biol. 29: 1228–1238, https://doi.org/10.1038/s41594-022-00874-9.Search in Google Scholar PubMed
Dembowski, J.A., Kuo, B., and Woolford, J.L. (2013). Has1 regulates consecutive maturation and processing steps for assembly of 60S ribosomal subunits. Nucleic Acids Res. 41: 7889–7904, https://doi.org/10.1093/nar/gkt545.Search in Google Scholar PubMed PubMed Central
Du, Y., An, W., Zhu, X., Sun, Q., Qi, J., and Ye, K. (2020). Cryo-EM structure of 90S small ribosomal subunit precursors in transition states. Science 369: 1477–1481, https://doi.org/10.1126/science.aba9690.Search in Google Scholar PubMed
Enders, M., Ficner, R., and Adio, S. (2022). Regulation of the DEAH/RHA helicase Prp43 by the G-patch factor Pfa1. Proc. Natl. Acad. Sci. U.S.A. 119: e2203567119, https://doi.org/10.1073/pnas.2203567119.Search in Google Scholar PubMed PubMed Central
Garcia, I., Albring, M.J., and Uhlenbeck, O.C. (2012). Duplex destabilization by four ribosomal DEAD-box proteins. Biochemistry 51: 10109–10118, https://doi.org/10.1021/bi301172s.Search in Google Scholar PubMed
Gnanasundram, S.V., Kos-Braun, I.C., and Koš, M. (2019). At least two molecules of the RNA helicase Has1 are simultaneously present in pre-ribosomes during ribosome biogenesis. Nucleic Acids Res. 47: 10852–10864, https://doi.org/10.1093/nar/gkz767.Search in Google Scholar PubMed PubMed Central
Granneman, S., Kudla, G., Petfalski, E., and Tollervey, D. (2009). Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. U.S.A. 106: 9613–9618, https://doi.org/10.1073/pnas.0901997106.Search in Google Scholar PubMed PubMed Central
Halls, C., Mohr, S., Del, Campo, M., Yang, Q., Jankowsky, E., and Lambowitz, A.M. (2007). Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and-independent mechanisms, and general RNA chaperone activity. J. Mol. Biol. 365: 835–855, https://doi.org/10.1016/j.jmb.2006.09.083.Search in Google Scholar PubMed PubMed Central
Hamann, F., Enders, M., and Ficner, R. (2019). Structural basis for RNA translocation by DEAH-box ATPases. Nucleic Acids Res. 47: 4349–4362, https://doi.org/10.1093/nar/gkz150.Search in Google Scholar PubMed PubMed Central
He, Y., Andersen, G.R., and Nielsen, K.H. (2010). Structural basis for the function of DEAH helicases. EMBO Rep. 11: 180–186, https://doi.org/10.1038/embor.2010.11.Search in Google Scholar PubMed PubMed Central
Jaafar, M., Contreras, J., Dominique, C., Martín-Villanueva, S., Capeyrou, R., Vitali, P., Rodríguez-Galán, O., Velasco, C., Humbert, O., Watkins, N.J., et al.. (2021). Association of snR190 snoRNA chaperone with early pre-60S particles is regulated by the RNA helicase Dbp7 in yeast. Nat. Commun. 12: 6153, https://doi.org/10.1038/s41467-021-26207-w.Search in Google Scholar PubMed PubMed Central
Jankowsky, E., Gross, C.H., Shuman, S., and Pyle, A.M. (2001). Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 291: 121–125, https://doi.org/10.1126/science.291.5501.121.Search in Google Scholar PubMed
Jia, H., Wang, X., Anderson, J.T., and Jankowsky, E. (2012). RNA unwinding by the Trf4/Air2/Mtr4 polyadenylation (TRAMP) complex. Proc. Natl. Acad. Sci. U.S.A. 109: 7292–7297, https://doi.org/10.1073/pnas.1201085109.Search in Google Scholar PubMed PubMed Central
Johnson, S.J. and Jackson, R.N. (2013). Ski2-like RNA helicase structures: common themes and complex assemblies. RNA Biol. 10: 33–43, https://doi.org/10.4161/rna.22101.Search in Google Scholar PubMed PubMed Central
Karbstein, K. (2022). Attacking a DEAD problem: the role of DEAD-box ATPases in ribosome assembly and beyond. In: Methods in enzymology. Elsevier, Amsterdam, pp. 19–38.10.1016/bs.mie.2022.03.033Search in Google Scholar PubMed PubMed Central
Kater, L., Thoms, M., Barrio-Garcia, C., Cheng, J., Ismail, S., Ahmed, Y.L., Bange, G., Kressler, D., Berninghausen, O., Sinning, I., et al.. (2017). Visualizing the assembly pathway of nucleolar pre-60S ribosomes. Cell 171: 1599–1610.10.1016/j.cell.2017.11.039Search in Google Scholar PubMed PubMed Central
Khoshnevis, S., Askenasy, I., Johnson, M.C., Dattolo, M.D., Young-Erdos, C.L., Stroupe, M.E., and Karbstein, K. (2016). The DEAD-box protein Rok1 orchestrates 40S and 60S ribosome assembly by promoting the release of Rrp5 from pre-40S ribosomes to allow for 60S maturation. PLoS Biol. 14: e1002480, https://doi.org/10.1371/journal.pbio.1002480.Search in Google Scholar PubMed PubMed Central
Khreiss, A., Capeyrou, R., Lebaron, S., Albert, B., Bohnsack, K.E., Bohnsack, M.T., Henry, Y., Henras, A.K., and Humbert, O. (2023). The DEAD-box protein Dbp6 is an ATPase and RNA annealase interacting with the peptidyl transferase center (PTC) of the ribosome. Nucleic Acids Res. 51: 744–764, https://doi.org/10.1093/nar/gkac1196.Search in Google Scholar PubMed PubMed Central
Klinge, S. and Woolford, J.L. (2019). Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 20: 116–131, https://doi.org/10.1038/s41580-018-0078-y.Search in Google Scholar PubMed PubMed Central
Koš, M. and Tollervey, D. (2005). The putative RNA helicase Dbp4p is required for release of the U14 snoRNA from preribosomes in Saccharomyces cerevisiae. Mol. Cell 20: 53–64, https://doi.org/10.1016/j.molcel.2005.08.022.Search in Google Scholar PubMed
Kudla, G., Granneman, S., Hahn, D., Beggs, J.D., and Tollervey, D. (2011). Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc. Natl. Acad. Sci. U.S.A. 108: 10010–10015, https://doi.org/10.1073/pnas.1017386108.Search in Google Scholar PubMed PubMed Central
Lebaron, S., Papin, C., Capeyrou, R., Chen, Y.-L., Froment, C., Monsarrat, B., Caizergues-Ferrer, M., Grigoriev, M., and Henry, Y. (2009). The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J. 28: 3808–3819, https://doi.org/10.1038/emboj.2009.335.Search in Google Scholar PubMed PubMed Central
Liang, X. and Fournier, M.J. (2006). The helicase Has1p is required for snoRNA release from pre-rRNA. Mol. Cell. Biol. 26: 7437–7450, https://doi.org/10.1128/mcb.00664-06.Search in Google Scholar PubMed PubMed Central
Lin, R., Correll, C.C., and Johnson, A.W. (2022). In vitro characterization of Dhr1 from Saccharomyces cerevisiae. In: Methods in enzymology. Elsevier, Amsterdam, pp. 77–101.10.1016/bs.mie.2022.03.058Search in Google Scholar PubMed
Linder, P. and Jankowsky, E. (2011). From unwinding to clamping — the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12: 505–516, https://doi.org/10.1038/nrm3154.Search in Google Scholar PubMed
Liu, X., Huang, H., and Karbstein, K. (2021). Blocking a dead-end assembly pathway creates a point of regulation for the DEAD-box ATPase Has1 and prevents platform misassembly. BioRxiv. https://doi.org/10.1101/2021.09.06.459192.Search in Google Scholar
Ma, W.K., Cloutier, S.C., and Tran, E.J. (2013). The DEAD-box protein Dbp2 functions with the RNA-binding protein Yra1 to promote mRNP assembly. J. Mol. Biol. 425: 3824–3838, https://doi.org/10.1016/j.jmb.2013.05.016.Search in Google Scholar PubMed PubMed Central
Manikas, R.-G., Thomson, E., Thoms, M., and Hurt, E. (2016). The K+-dependent GTPase Nug1 is implicated in the association of the helicase Dbp10 to the immature peptidyl transferase centre during ribosome maturation. Nucleic Acids Res. 44: 1800–1812, https://doi.org/10.1093/nar/gkw045.Search in Google Scholar PubMed PubMed Central
Martin, R., Hackert, P., Ruprecht, M., Simm, S., Brüning, L., Mirus, O., Sloan, K.E., Kudla, G., Schleiff, E., and Bohnsack, M.T. (2014). A pre-ribosomal RNA interaction network involving snoRNAs and the Rok1 helicase. RNA 20: 1173–1182, https://doi.org/10.1261/rna.044669.114.Search in Google Scholar PubMed PubMed Central
Martin, R., Straub, A.U., Doebele, C., and Bohnsack, M.T. (2013). DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol. 10: 4–18, https://doi.org/10.4161/rna.21879.Search in Google Scholar PubMed PubMed Central
Mitterer, V. and Pertschy, B. (2022). RNA folding and functions of RNA helicases in ribosome biogenesis. RNA Biol. 19: 781–810, https://doi.org/10.1080/15476286.2022.2079890.Search in Google Scholar PubMed PubMed Central
Olsen, K.J. and Johnson, S.J. (2021). Mtr4 RNA helicase structures and interactions. Biol. Chem. 402: 605–616, https://doi.org/10.1515/hsz-2020-0329.Search in Google Scholar PubMed PubMed Central
Pertschy, B., Schneider, C., Gnädig, M., Schäfer, T., Tollervey, D., and Hurt, E. (2009). RNA helicase Prp43 and its Co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J. Biol. Chem. 284: 35079–35091, https://doi.org/10.1074/jbc.m109.040774.Search in Google Scholar
Robert-Paganin, J., Halladjian, M., Blaud, M., Lebaron, S., Delbos, L., Chardon, F., Capeyrou, R., Humbert, O., Henry, Y., Henras, A.K., et al.. (2017). Functional link between DEAH/RHA helicase Prp43 activation and ATP base binding. Nucleic Acids Res. 45: 1539–1552, https://doi.org/10.1093/nar/gkw1233.Search in Google Scholar PubMed PubMed Central
Sailer, C., Jansen, J., Sekulski, K., Cruz, V.E., Erzberger, J.P., and Stengel, F. (2022). A comprehensive landscape of 60S ribosome biogenesis factors. Cell Rep. 38: 110353, https://doi.org/10.1016/j.celrep.2022.110353.Search in Google Scholar PubMed PubMed Central
Sanghai, Z.A., Miller, L., Molloy, K.R., Barandun, J., Hunziker, M., Chaker-Margot, M., Wang, J., Chait, B.T., and Klinge, S. (2018). Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature 556: 126–129, https://doi.org/10.1038/nature26156.Search in Google Scholar PubMed PubMed Central
Sardana, R., Liu, X., Granneman, S., Zhu, J., Gill, M., Papoulas, O., Marcotte, E.M., Tollervey, D., Correll, C.C., and Johnson, A.W. (2015). The DEAH-box helicase Dhr1 dissociates U3 from the pre-rRNA to promote formation of the central pseudoknot. PLoS Biol. 13: e1002083, https://doi.org/10.1371/journal.pbio.1002083.Search in Google Scholar PubMed PubMed Central
Schuller, J.M., Falk, S., Fromm, L., Hurt, E., and Conti, E. (2018). Structure of the nuclear exosome captured on a maturing preribosome. Science 360: 219–222, https://doi.org/10.1126/science.aar5428.Search in Google Scholar PubMed
Semlow, D.R., Blanco, M.R., Walter, N.G., and Staley, J.P. (2016). Spliceosomal DEAH-box ATPases remodel pre-mRNA to activate alternative splice sites. Cell 164: 985–998, https://doi.org/10.1016/j.cell.2016.01.025.Search in Google Scholar PubMed PubMed Central
Soltanieh, S., Osheim, Y.N., Spasov, K., Trahan, C., Beyer, A.L., and Dragon, F. (2015). DEAD-box RNA helicase Dbp4 is required for small-subunit processome formation and function. Mol. Cell. Biol. 35: 816–830, https://doi.org/10.1128/mcb.01348-14.Search in Google Scholar PubMed PubMed Central
Tauchert, M.J., Fourmann, J.-B., Christian, H., Lührmann, R., and Ficner, R. (2016). Structural and functional analysis of the RNA helicase Prp43 from the thermophilic eukaryote Chaetomium thermophilum. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 72: 112–120, https://doi.org/10.1107/s2053230x15024498.Search in Google Scholar
Tauchert, M.J., Fourmann, J.-B., Lührmann, R., and Ficner, R. (2017). Structural insights into the mechanism of the DEAH-box RNA helicase Prp43. eLife 6: e21510, https://doi.org/10.7554/eLife.21510.Search in Google Scholar PubMed PubMed Central
Taylor, L.L., Jackson, R.N., Rexhepaj, M., Klauer King, A., Lott, L.K., van Hoof, A., and Johnson, S.J. (2014). The Mtr4 ratchet helix and arch domain both function to promote RNA unwinding. Nucleic Acids Res. 42: 13861–13872, https://doi.org/10.1093/nar/gku1208.Search in Google Scholar PubMed PubMed Central
Travis, A.J., Moody, J., Helwak, A., Tollervey, D., and Kudla, G. (2014). Hyb: a bioinformatics pipeline for the analysis of CLASH (crosslinking, ligation and sequencing of hybrids) data. Methods 65: 263–273, https://doi.org/10.1016/j.ymeth.2013.10.015.Search in Google Scholar PubMed PubMed Central
van, Hoof, A., Lennertz, P., and Parker, R. (2000). Yeast exosome mutants accumulate 3-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20: 441–452, https://doi.org/10.1128/mcb.20.2.441-452.2000.Search in Google Scholar
Walbott, H., Mouffok, S., Capeyrou, R., Lebaron, S., Humbert, O., van Tilbeurgh, H., Henry, Y., and Leulliot, N. (2010). Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 29: 2194–2204, https://doi.org/10.1038/emboj.2010.102.Search in Google Scholar PubMed PubMed Central
Wang, X., Jia, H., Jankowsky, E., and Anderson, J.T. (2008). Degradation of hypomodified tRNAiMet in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA 14: 107–116, https://doi.org/10.1261/rna.808608.Search in Google Scholar PubMed PubMed Central
Yang, Q. and Jankowsky, E. (2005). ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry 44: 13591–13601, https://doi.org/10.1021/bi0508946.Search in Google Scholar PubMed
Young, C.L., Khoshnevis, S., and Karbstein, K. (2013). Cofactor-dependent specificity of a DEAD-box protein. Proc. Natl. Acad. Sci. U.S.A. 110: E2668–E2676, https://doi.org/10.1073/pnas.1302577110.Search in Google Scholar PubMed PubMed Central
Zhou, D., Zhu, X., Zheng, S., Tan, D., Dong, M.-Q., and Ye, K. (2019). Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate. Protein Cell 10: 120–130, https://doi.org/10.1007/s13238-018-0526-7.Search in Google Scholar PubMed PubMed Central
Zhu, J., Liu, X., Anjos, M., Correll, C.C., and Johnson, A.W. (2016). Utp14 recruits and activates the RNA helicase Dhr1 to undock U3 snoRNA from the preribosome. Mol. Cell. Biol. 36: 965–978, https://doi.org/10.1128/mcb.00773-15.Search in Google Scholar
© 2023 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Highlight: Integrative Structural Biology of Dynamic Macromolecular Assemblies
- Highlight: integrative structural biology of dynamic macromolecular assemblies
- Bayesian methods in integrative structure modeling
- The many faces of ribosome translocation along the mRNA: reading frame maintenance, ribosome frameshifting and translational bypassing
- Translation termination in human mitochondria – substrate specificity of mitochondrial release factors
- Molecular functions of RNA helicases during ribosomal subunit assembly
- Interaction of nucleoporins with nuclear transport receptors: a structural perspective
- Protein transport along the presequence pathway
- Autophagic and non-autophagic functions of the Saccharomyces cerevisiae PROPPINs Atg18, Atg21 and Hsv2
- Influence of phosphorylation on intermediate filaments
- Mediator structure and function in transcription initiation
- Structure and phase separation of the C-terminal domain of RNA polymerase II
- The DEAD-box RNA helicase Dbp5 is a key protein that couples multiple steps in gene expression
- Structure and function of spliceosomal DEAH-box ATPases
- Molecular simulations of DEAH-box helicases reveal control of domain flexibility by ligands: RNA, ATP, ADP, and G-patch proteins
Articles in the same Issue
- Frontmatter
- Highlight: Integrative Structural Biology of Dynamic Macromolecular Assemblies
- Highlight: integrative structural biology of dynamic macromolecular assemblies
- Bayesian methods in integrative structure modeling
- The many faces of ribosome translocation along the mRNA: reading frame maintenance, ribosome frameshifting and translational bypassing
- Translation termination in human mitochondria – substrate specificity of mitochondrial release factors
- Molecular functions of RNA helicases during ribosomal subunit assembly
- Interaction of nucleoporins with nuclear transport receptors: a structural perspective
- Protein transport along the presequence pathway
- Autophagic and non-autophagic functions of the Saccharomyces cerevisiae PROPPINs Atg18, Atg21 and Hsv2
- Influence of phosphorylation on intermediate filaments
- Mediator structure and function in transcription initiation
- Structure and phase separation of the C-terminal domain of RNA polymerase II
- The DEAD-box RNA helicase Dbp5 is a key protein that couples multiple steps in gene expression
- Structure and function of spliceosomal DEAH-box ATPases
- Molecular simulations of DEAH-box helicases reveal control of domain flexibility by ligands: RNA, ATP, ADP, and G-patch proteins

