Abstract
Contact sites, areas where two organelles are held in close proximity through the action of molecular tethers, enable non-vesicular communication between compartments. Mitochondria have been center stage in the contact site field since the discovery of the first contact between mitochondria and the endoplasmic reticulum (ER) over 60 years ago. However, only now, in the last decade, has there been a burst of discoveries regarding contact site biology in general and mitochondrial contacts specifically. The number and types of characterized contacts increased dramatically, new molecular mechanisms enabling contact formation were discovered, additional unexpected functions for contacts were shown, and their roles in cellular and organismal physiology were emphasized. Here, we focus on mitochondria as we highlight the most recent developments, future goals and unresolved questions in the field.
Introduction
The central role of organelles as insulators of biochemical reactions also gives rise to their two biggest functional complexities – first, the need to communicate and collaborate with other organelles to coordinate cellular function. Second the requirement to transfer metabolites and signals through a membrane. This necessity has caused the evolution of multiple ways of organelle communication and material transfer that are faster and more regulated than simple diffusion. One such way is by vesicular traffic that targets large amounts of cargo and membranes to specific organelles. However, vesicular traffic is not enabled for all organelles, gives pulsatile input and requires machinery and lipid recycling. An additional mode of communication is by creation of areas of close proximity between organelles named contact sites.
Contact sites (in short contacts), are specialized zones where one organelle is positioned in close proximity (sometimes as close as 10 nm) to another organelle without membrane fusion occurring. Contacts are mediated by molecular tethering forces created through protein-protein or protein-lipid interactions. Most contacts described to date have more than one tether hence depletion of a single tether does not always result in alteration of the contact between two organelles. Besides tethering molecules, contacts also harbor functional resident proteins and are dependent on regulators that can influence the contact function, duration and abundance in response to internal or external stimuli (Eisenberg-Bord et al. 2016). Recently it is also becoming appreciated that between the same two organelles, different types of contacts may occur that are geographically distinct, have unique regulation and a specific set of resident proteins.
In the 1950s, the first contact site was described between mitochondria and the endoplasmic reticulum (ER) (Bernhard et al. 1952; Bernhard and Rouiller 1956). It then took approximately 40 years until the first of the functions of this contact were elucidated (Rizzuto et al. 1998; Vance 1990) marking a change in the field with the realization that contacts have cellular implications. As the contact site field started to grow, it was believed that the central player is the ER, with multiple contact sites relying on it. In contrast, mitochondria were believed to only have the one contact, with the ER (Elbaz and Schuldiner 2011). Recently it is becoming clear that this original view was inaccurate and that mitochondria, just like the ER, form contact sites with almost all other organelles (Kakimoto et al. 2018; Shai et al. 2018; Valm et al. 2017). This puts mitochondria center stage again in the contact site field. This may be no surprise since mitochondria are not part of the main vesicle traffic routes of the secretory pathway, making contact sites the central means for them to communicate and exchange molecules.
For the past decade the development of new techniques has helped overcome many of the experimental challenges in the field, opening the way to the identification of new mitochondrial contacts, tethers, functions and regulators and leading to exponential growth of the field (Jing et al. 2019; Scorrano et al. 2019; Shai et al. 2018; Valm et al. 2017). Indeed, to date, contacts have already been shown to influence multiple aspects of mitochondrial function and structure (Eisenberg-Bord and Schuldiner 2017a; Gatta and Levine 2017; Lackner 2019; Prinz et al. 2020). Moreover, due to their central role, disturbed mitochondrial contact sites are emerging as important players in a spectrum of human diseases (López-Crisosto et al. 2015; Liu and Zhu 2017; Paillusson et al. 2016).
In this review we will focus on recent studies in mammalian cells and in the yeast Saccharomyces cerevisiae (from here on referred to as yeast) that left a mark in this decade of new concepts. By summarizing new innovations, we can appreciate the depth of knowledge already gained since the first studies of ER-mitochondria contact sites and the long road ahead to fully comprehend the functions of mitochondrial contacts and their interplay in the cell.
Newly discovered cellular functions for mitochondrial contact sites
Thirty years ago the first function of a contact site was discovered. It was shown that synthesis of phospholipids occurs on specialized ER subdomains that form contacts with mitochondria (termed Mitochondrial Associated Membranes or MAMs) and that this is important for lipid transfer between the two organelles (Vance 1990). Less than a decade later, direct calcium transfer was also shown to occur at the same contact site (Rizzuto et al. 1998). Since then, these two central functions of the ER-mitochondria contact have been studied in great detail (reviewed in Cockcroft and Raghu 2018; Marchi et al. 2014; Muallem et al. 2017; Lee and Min 2018). Since multiple reviews discuss these two functions we will not cover these topics.
Excitingly, in the last decade multiple new functions of contacts have been described emphasizing the fact that contacts have more widespread roles than previously thought. The role of ER-mitochondria contacts in regulating mitochondrial fission, for example, is an exciting new role that has also been recently extensively reviewed and hence we will not expand on it (Wu et al. 2018). Here, we will try to touch on some more recent studies that are adding to the diverse repertoire of mitochondrial contact site functions.
Transfer of acetyl-CoA derivatives
An interesting metabolic shuttle was recently demonstrated for the peroxisome-mitochondria (PerMit) contact in yeast. Using a split Venus contact site reporter, two tethering proteins – Fzo1 and Pex34 (for all acronyms of proteins spelled out please see Table 1), were discovered. Pex34 was shown to extend the PerMit when overexpressed, and play a role in the transfer of acetyl-CoA derivatives (citrate and acetyl-carnitine) to mitochondria during β-oxidation of fatty acids. The human homolog of Pex34, a Pex11 family member (PEX11β), was suggested to also play a role in the human PerMit contact (Kustatscher et al. 2019).
Full names (ordered alphabetically) for all abbreviations of proteins mentioned in the article.
| Protein abbreviation | Full name/gene name |
|---|---|
| ATG2A/2B/9A/11/36 | Autophagy related 2A/2B/9A/11/36 |
| ATL3 | Atlastin 3 |
| BAP31 | B-cell receptor-associated protein 31 |
| BCL2 | B-cell lymphoma 2 |
| BCL2L10 | B-cell lymphoma 2-like 10 |
| BECN1 | Beclin 1 |
| CLCC1 | Chloride channel CLIC-like protein 1 |
| CNX | Calnexin |
| Dnm1 | Dynamin-related 1 |
| DRP1 | Dynamin related protein 1 |
| Emc1-6 | ER membrane protein complex 1–6 |
| EMD | Emerin |
| Erg6 | Ergosterol biosynthesis 6 |
| FATE1 | Fetal and adult testis expressed 1 |
| FIS1 | Mitochondrial fission 1 |
| FUNDC1 | FUN14 domain-containing protein 1 |
| Fzo1 | Fuzzy onions homolog 1 |
| Gem1 | GTPase EF-hand protein of mitochondria 1 |
| GRAMD1a/c | GRAM domain-containing protein 1 a/c |
| GRP75 | Glucose-regulated protein 75 |
| Hac1 | Homologous to Atf/Creb1 |
| HSP70 | 70 kDa heat shock protein |
| IFN-ϒ | Interferon ϒ |
| INF2 | Inverted formin 2 |
| IP3 /IP3R | Inositol trisphosphate /Inositol trisphosphate receptor |
| IRBIT | IP3R binding protein released with inositol 1,4,5-trisphosphate |
| Ire1 | Inositol requiring 1 |
| IRE1α | Inositol-requiring enzyme 1α |
| Lam6 | Lipid transfer protein anchored at membrane contact sites 6 |
| MARCH5 (MITOL) | Membrane-associated ring finger (C3HC4) 5 |
| Mcp1/2 | Mdm10 complementing protein 1/2 |
| Mcr1 | Mitochondrial NADH-cytochrome b5 reductase 1 |
| Mdm 1/34/12/10/36 | Mitochondrial distribution and morphology 1/34/12/10/36 |
| MFN 1/2 | Mitofusin 1/2 |
| Mgm1 | Mitochondrial genome maintenance 1 |
| MIGA2 | Mitoguardin 2 |
| MIRO | Mitochondrial Rho |
| Mmm1 | Maintenance of mitochondrial morphology 1 |
| Mmr1 | Mitochondrial myo2p receptor-relate 1 |
| MOSPD2 | Motile sperm domain containing 2 |
| NFAT1 | Nuclear factor of activated T-cell 1 |
| Nrp3 | Nitrogen permease regulator 3 |
| Num1 | Nuclear migration 1 |
| ORP5/8 | Oxysterol-binding protein related-proteins 5/8 |
| PACS2 | Phosphofurin acidic cluster sorting protein 2 |
| PANX2 | Pannexin 2 |
| PDK4 | Pyruvate dehydrogenase kinase isozyme 4 |
| PDZD8 | PDZ domain containing 8 |
| PEX11β | Peroxisomal membrane protein 11β |
| Pex34/11 | Peroxin 34/11 |
| PIGBOS | PIGB opposite strand 1 |
| PINK1 | PTEN-induced kinase 1 |
| Ptc1 | Phosphatase type two C1 |
| PTPIP51 | Protein tyrosine phosphatase interacting protein 51 |
| RAB7 | Ras-related protein rab 7 |
| REEP1 | Receptor expression-enhancing protein 1 |
| RRBP1 | Ribosome-binding protein 1 |
| Sar1 | Secretion-associated, ras-related 1 |
| SPIREC1C | Protein spire homolog 1C |
| SYNJ2BP (OMP25) | Synaptojanin-2-binding protein (outer membrane protein 25 kDa) |
| TBC1D15 | TBC1 domain family member 15 |
| TG2 | Transglutaminase type 2 |
| TOM40/70/71/5 | Translocase of the outer mitochondrial membrane 40/70/71/5 kDa |
| TSPO | Translocator protein |
| VAP A/B | Vesicle-associated membrane protein-associated protein A/B |
| VDAC1 | Voltage-dependent anion-selective channel protein 1 |
| Vps13/1/39 | Vacuolar protein sorting 13/1/39 |
| VPS13A/C | Vacuolar protein sorting-associated protein 13A/C |
| Ypt7 | Yeast protein two 7 |
In contrast, Fzo1, which was shown to localize not only to the mitochondrial outer membrane (MOM), but also to peroxisomes (Shai et al. 2018) could also extend the PerMit when overexpressed but did not affect the transfer of acetyl-CoA derivatives. Interestingly, the human homolog, MFN2, has also been described as a tether in contact sites of mitochondria with the ER, lipid droplets (LD) and melanosomes (Boutant et al. 2017; Daniele et al. 2014; Naon et al. 2016).
Transfer of Coenzyme Q precursors
Coenzyme Q (CoQ) is an essential lipid for electron transfer in the mitochondrial respiratory chain. The enzymes that synthesize CoQ are located in the matrix side of the inner mitochondrial membrane (Awad et al. 2018), and most form a large complex known as the CoQ synthome (Tran and Clarke 2007). However, the major precursors of CoQ begin their synthesis in the ER thus requiring transfer between the two organelles.
The CoQ synthome is positioned in proximity to the ER-mitochondria contact site, both in yeast (Eisenberg-Bord et al. 2019; Subramanian et al. 2019) and in human cells (Subramanian et al. 2019) potentially to optimize such substrate flux.
In support of a role for the contact in CoQ biosynthesis, in yeast loss of the main tethering complex forming a contact between mitochondria and the ER (the ERMES complex (ER Mitochondria Encounter Structure)) composed of four subunits (Mmm1, Mdm12, Mdm10, Mdm34) (Kornmann et al. 2009), caused a disruption of the CoQ synthome (Eisenberg-Bord et al. 2019; Subramanian et al. 2019). ERMES disruption also lead to a decrease in both the production and the steady state levels of CoQ6 (the final derivative of the CoQ synthome in yeast) and its intermediates in isolated mitochondria (Eisenberg-Bord et al. 2019; Subramanian et al. 2019).
How is this coordination achieved? It was suggested that the protein Coq10 is in charge of positioning the CoQ synthome near ERMES since Coq10 is co-expressed with the ERMES subunit Mdm12 (Cherry et al. 2012). Additionally, deletion of the COQ10 gene reduced the co-localization of the CoQ synthome and the ERMES complex (Eisenberg-Bord et al. 2019).
Regardless of the exact molecular details, the new role of the ER-mitochondria contact site in CoQ biosynthesis may explain why loss of ERMES results in reduced respiratory capacity (Kornmann et al. 2009).
Inheritance
In yeast, a role for the mitochondria-ER-plasma membrane (PM) contact site (termed mitochondrial-ER cortex anchor (MECA)) in mitochondrial inheritance was shown. This role involves the Num1 protein (Klecker et al. 2013; Lackner et al. 2013). Num1 has a coiled coil domain that is able to directly bind mitochondria, promoting the formation of Num1 clusters. Num1 can also bind dynein, the main microtubule motor protein that positions the spindle between mother and bud. The function of the MECA is therefore to couple disruption in mitochondrial inheritance with a delay in dynein-mediated spindle positioning. This is suggested to safeguard mitosis of cells lacking properly inherited mitochondria (Kraft and Lackner 2017).
Precursor targeting
BAP31 is an ER membrane protein that is involved in several cellular pathways such as apoptosis, autophagy and signaling (Iwasawa et al. 2011; Namba 2019; Rosati et al. 2010). In human U2OS cells it was shown that BAP31 interacts with the central subunit of the TOM complex, TOM40 (Namba 2019). The complex of BAP31 together with TOM40 docks a precursor of the respiratory chain complex I (pre-NDUFS4). Interestingly, it was suggested that BAP31 supports the translocation of pre-NDUFS4 through TOM40 (De Rasmo et al. 2008; Namba 2019; Papa et al. 2012).
In yeast, the ER has also been shown to play a role in the translocation of inner mitochondrial membrane proteins through ER-SURF, a process that retrieves mitochondrial proteins from the ER surface and reroutes them to mitochondria (Hansen et al. 2018). ER-SURF was also suggested to occur through contact sites.
Apoptosis
Multiple manuscripts have uncovered a role for the ER-mitochondria contact site in apoptosis:
BAP31, mentioned above for its role in precursor targeting, has also been suggested to play a role in apoptosis (Namba 2019). Interestingly, the ER stress response inducer tunicamycin changes BAP31 location from the whole ER to the rough ER only. There, instead of TOM40, BAP31 interacts with the anti-apoptosis protein BCL2 which stimulates mitochondria-dependent apoptosis (Namba 2019). This suggests a mechanism to couple ER-mitochondria contact sites with apoptosis.
Another ER-mitochondria contact site protein that might play a role in apoptosis is the mammalian membrane channel protein PANX2 (Sosinsky et al. 2011). At endogenous levels, PANX2 is located in discrete patches in the contact. Functionally, overexpression of PANX2 displays a two-fold increase in the speed of Caspase3 activation and DNA fragmentation during apoptosis, when compared to control, suggesting that PANX2 might play a role in this process. The hypothesis is that PANX2 channels promote apoptosis by creating Ca2+ channels from the ER to mitochondria (Le Vasseur et al. 2019).
The mammalian protein, IRBIT, binds to the IP3 receptor (IP3R) (that also acts as a tether) and promotes ER-mitochondria contact sites, facilitating Ca2+ transfer and thus regulating apoptosis. IRBIT does so through interaction with the antiapoptotic protein BCL2L10, a BCL2 homolog (Bonneau et al. 2016). BCL2L10 interacts with IP3R via its BH4 domain. Both IRBIT and BCL2L10 interact within the contact to strengthen each other’s interaction with IP3R and form one complex with IP3R-VDAC1. Moreover, knockout (KO) of IRBIT reduced both the number and extent of ER-mitochondria contacts compared to control cells, suggesting that IRBIT participates in the formation or stabilization of these contacts. In normal conditions, the interaction between IRBIT and BCL2L10 functionally balances the amount of Ca2+ released through IP3R, probably by interference of IP3 binding to its receptor. However, under stress conditions that promote apoptosis, induced by staurosporine or tunicamycin, IRBIT is dephosphorylated and inhibits the interaction of BCL2L10 with IP3R thus preventing rescue from cell death. KO of IRBIT under these conditions greatly attenuated the release of Ca2+ from the ER indicating its important role in apoptosis (Bonneau et al. 2016).
A negative regulator of apoptosis in ER-mitochondria contacts is FATE1, a cancer-testis antigen. FATE1 regulates Ca2+– and drug-dependent apoptosis in cancer cells by modulating organelle distance (Doghman-Bouguerra et al. 2016). FATE1 expression in adrenocortical carcinoma (ACC) cells decreases ER–mitochondria contact and mitochondrial Ca2+ uptake, while its knockdown (KD) has an opposite effect. FATE1 also decreases sensitivity to mitochondrial Ca2+ –dependent proapoptotic stimuli and to the chemotherapeutic drug mitotane which is the current medical therapy in advanced ACC. Therefore, ER–mitochondria uncoupling activity of FATE1 is harnessed by cancer cells to escape apoptotic death and resist the action of chemotherapeutic drugs (Doghman-Bouguerra et al. 2016).
A contact site protein with both negative and positive roles in cell death is GRP75, an HSP70 molecular chaperone (Wadhwa et al. 1993) that is localized to ER-mitochondria contacts. In cancer cells and astrocytes, overexpression of GRP75 after exposure to cellular stress prevents cell death (Guo et al. 2012; Voloboueva et al. 2008). In contrast, overexpression of GRP75 in HT22 neuronal cells made them more susceptible to cell death resulting from oxidative glutamate toxicity. In the neuronal cells, higher amounts of GRP75 caused increased ER-mitochondria contact whereas, KO, KD or inhibition reduced contact formation. This reduction in contacts preserved mitochondrial function during glutamate oxidative stress by rescuing mitochondrial membrane potential, preserving mitochondrial morphology, attenuating the production of reactive oxygen species and maintaining mitochondrial respiratory capacity. Although KO or KD of GRP75 in HT22 cells made them less sensitive to glutamate oxidative stress, it failed to protect them from cell death induced by the ER stressors thapsigargin or brefeldin A or by rotenone that inhibits mitochondrial complex I. This suggest that ER-mitochondria contact sites, and GRP75 in particular, are important for cell death pathways and not directly affected by mitochondrial damage or ER stress. Altogether, GRP75 is an example of a tether protein that is located and functions at contacts, but can present the opposite behaviors to mitochondrial stress induction depending on cell type and cellular state (Honrath et al. 2017).
Heme homeostasis
Heme is an iron-containing cofactor and signaling molecule that facilitates multiple processes by regulating proteins in nearly every organelle in the cell. It is synthesized via eight highly conserved enzymes that reside partly in mitochondria and partly in the cytosol. Mature heme is formed inside the mitochondrial matrix and must be transported to the various organelles (Hanna et al. 2017; Piel et al. 2019). Originally it was thought that heme transporters export heme to the cytosol first, and from there it is shuttled to other organelles (Chiabrando et al. 2012). However, a fluorescent heme sensor (HS1) in live yeast cells, demonstrated that the traffic of heme from mitochondria to the nucleus is 25% higher than the trafficking of heme to the cytosol. This suggest a direct transfer of heme between the two organelles (Martinez-Guzman et al. 2019; see also below).
Gem1, a MOM GTPase, is known to negatively regulate the ERMES complex (Kornmann et al. 2011). Increase in ERMES expression as a result of deletion of GEM1 significantly elevated heme trafficking rate from mitochondria to the nucleus. However, in strains with deletion of proteins that are part of the ERMES complex, heme trafficking rates were unaffected. Moreover, deletion of MGM1, encoding a GTPase that promotes inner mitochondrial membrane fusion, did not change heme synthesis rate but decreased its nuclear transport (Westermann 2008). The alteration was restored after several hours, likely as a result of a compensatory mechanism that sensed the heme reduction in the nucleus (Martinez-Guzman et al. 2019).
Iron homeostasis
Iron-sulfur clusters (ISC) are heme precursors and essential co-factors produced in mitochondria. ISC biosynthesis is the essential function of mitochondria and hence it is highly regulated. Reduction in ISC biosynthesis results in increased cellular iron uptake (Stehling and Lill 2013). Similarly, in yeast, loss of ERMES also increases iron uptake and iron concentrations in the cell (3-4-fold compared to control) (Xue et al. 2017). A strong suppressor of ERMES loss is a point mutation in VPS13 (Lang et al. 2015). Indeed, this mutation also suppresses iron overload in the cell and attenuates the iron deficiency response (Xue et al. 2017). Like many other known ERMES functions, it is unclear if the complex participates directly in iron homeostasis or if this is a secondary effect to the disruption in mitochondrial morphology or respiration.
How does iron reach mitochondria? Iron uptake occurs in multiple cell types through endocytosis of the iron binding transferrin (Tf) molecule following binding to its receptor on the cell surface (Aisen et al. 2001). Iron is then released from Tf to the endosomal lumen due to the increased acidity (Dautry-Varsat et al. 1983). From the early endosome, iron is transferred to the mitochondrial matrix (Ponka 1997). It seems that in this aspect too contacts play a central role. Several Tf-containing endosomes located in the cell periphery were found in close proximity with mitochondria in epithelial cells. It was shown that indeed iron is transferred directly from the early Tf-endosome to mitochondria through a contact site that reduces endosome motility, but iron release was not required for the interaction itself (Das et al. 2016).
Regulation of the unfolded protein response (UPR)
Mitochondria were shown to play an important role in regulating ER stress (Malhotra and Kaufman 2011). During ER stress, iron uptake is increased and heme production is upregulated as heme is required for ER membrane proteins to synthesize more sterols and unsaturated lipids. In the absence of efficient heme biosynthesis, ER stress cannot be properly resolved (Cohen et al. 2017). In yeast upon ER stress induction, ERMES foci increase to double the number of ER-mitochondria contact sites (Kojima et al. 2019). Surprisingly, the abundance of ERMES foci was independent of the ER stress transducers, Ire1 or Hac1. This expansion in number seems to be an important aspect of regulating ER stress since deletion of ERMES subunits blocked the stress-dependent ER membrane expansion and increased susceptibility to stress. This suggests that there is a direct involvement of ER-mitochondria contact sites in modulating ER stress.
Recently, the human microprotein PIGBOS was shown to be a MOM resident protein, which interacts specifically with the ER protein CLCC1 and by this regulates the UPR (Chu et al. 2019). Thus, the PIGBOS-CLCC1 interaction has the potential to be a new tether in the ER-mitochondria contact site and a direct modulator of the above response. However, KO or overexpression of PIGBOS does not show a clear phenotype on the extent of the contact or the distance between the organelles. Since multiple independent tethering machineries act at the ER-mitochondria contact site, it may still be that loss of this specific tethering pair can be compensated by others and that the effect of the overexpression was limited by the levels of free CLCC1.
Another angle of mitochondrial involvement in ER stress comes from the finding that a mitochondrial protein can regulate IREα. IRE1 is a conserved ER sensor protein that recognizes unfolded proteins in the ER lumen and activates a transcriptional program to restore homeostasis (Walter and Ron 2011). In mammals, IRE1α functions can also promote apoptosis when ER stress is not resolved (Ghosh et al. 2014) and hence its levels must be tightly regulated. IRE1α was found to be a substrate of MITOL (MARCH5), a mitochondrial ubiquitin ligase (Takeda et al. 2019). The C-terminus of IRE1α can directly bind to the N-terminus of MITOL. In the absence of MITOL, IRE1α is not ubiquitinated, leading to reduced cell survival rates and high levels of cleaved Caspase3. Moreover, MITOL depletion increased IRE1α oligomerization suggesting that MITOL prevents apoptosis by capping IRE1α oligomerization. Both IRE1α and MITOL are enriched in MAMs. Silencing of PACS2 or MFN2, a tether of the ER-mitochondria contact, reduces IRE1α ubiquitylation, while overexpression of MITOL enhances ubiquitylation. Thus, the ubiquitylation of IRE1α may occur at contacts. This suggests that the ER-mitochondria contact regulates apoptosis not only through Ca2+ transfer but also through controlling IRE1α oligomerization by MITOL ubiquitylation (Takeda et al. 2019).
Bulk autophagy
VAPB is an integral ER protein that plays important roles in several ER contact sites from yeast to humans (Lev et al. 2008). Mammalian VAPB is also enriched in MAMs. VAPB tethers the ER to mitochondria by binding to the outer mitochondrial protein PTPIP51 (De Vos et al. 2012). Downregulation of VAPB or PTPIP51 stimulates autophagic flux, which can be compensated by an artificial tether that increases the ER-mitochondria contacts (Gomez-Suaga et al. 2017). From these results it seems like autophagy is not specifically regulated by the VAPB-PTPIP51 interaction but rather by the degree of association between the two organelles. It remains to be determined by which mechanism the degree of tethering between the organelles is sensed by the cell and regulated. One clue is that overexpression of VAPB-PTPIP51 resulted in an increase of IP3R-VDAC1 interactions and concomitant uptake of Ca2+ by mitochondria. Since chemical inhibition of Ca2+ uptake reversed the effects of the overexpression, it hints that Ca2+ flux is linked to the sensing or signaling (Gomez-Suaga et al. 2017).
Another key player in bulk autophagy is mammalian ATG2A/B that is required for lipid transfer during the phagophore expansion step. During autophagy, ATG2A translocates to the phagophore at the ER-mitochondria contact by binding TOM40 through a C-terminal domain. Additionally, ATG2A recruitment to the contact is dependent on TOM70. Once at the contact, ATG2A recruits ATG9A promoting phagophore growth by coupling vesicular and non-vesicular lipid transport into the expanding phagophore (Tang et al. 2019).
Another protein that inhibits autophagosome formation at ER-mitochondria contacts is the tumor suppressor, promyelocytic leukemia (PML). This role could explain why mutations in this protein provide an advantage to tumor cells that use autophagy as a cell survival strategy under stress conditions (Missiroli et al. 2016).
Pexophagy
Loss of the ERMES complex in yeast results in multiple phenotypes of peroxisomes including changes in peroxisomal number and size (Cohen et al. 2014; Esposito et al. 2019). One way by which the number of peroxisomes is regulated is by selective autophagy called pexophagy. Pexophagy acts to remove excessive or dysfunctional peroxisomes thus protecting cells from increased generation of harmful reactive oxygen species (Till et al. 2012). When pexophagy was induced by a combination of nitrogen starvation and use of oleic acid as a sole carbon source, the number of proximities between peroxisomes and ERMES foci increased to more than 50% of peroxisomes relative to only 20% on rich medium. Moreover, disruption of the ERMES tether reduced pexophagy. A similar effect was seen in a ∆pex11 background (Liu et al. 2018). Pex11 was suggested to be part of the PerMit tethering machinery by binding the subunit Mdm34 in ERMES (Mattiazzi Ušaj et al. 2015). In addition, the peroxisome fission proteins, Dnm1 and Vps1 that are required for progression of pexophagy (Mao et al. 2014) were recruited by Atg11 and Atg36 to ER-mitochondria contacts (Liu et al. 2018). This supports the notion that establishment of a three-way peroxisome-ER-mitochondria contact is required for efficient pexophagy.
Mitophagy
Much like pexophagy, mitophagy is essential for removal of damaged or excessive mitochondria, even under nutrient-rich conditions (in comparison to autophagy that is activated in response to starvation or stress) (Ding and Yin 2012). It is clear that multiple players take part in orchestrating mitophagy and that many of them reside in ER-mitochondria contacts.
In mammals, mitophagy can be mediated through PINK1, a mitochondrial kinase. Upon mitochondrial depolarization, PINK1 levels increase at the damaged mitochondrial surface, where it recruits Parkin, which then induces mitophagy (Choubey et al. 2014; Koyano et al. 2014; Michiorri et al. 2010; Scarffe et al. 2014). BECN1, a pro-autophagic protein that can interact with PINK1, is also known to recruit Parkin (Choubey et al. 2014). However, this recruitment seems to be cell type specific. In a neuronal cell line (SH-SY5Y), while endogenous PINK1 and BECN1 are both localized in the ER-mitochondria contact, Parkin recruitment to damaged mitochondria occurs in a BECN1 independent manner (Gelmetti et al. 2017). Nevertheless, in all cell lines tested, both Parkin and BECN1 require PINK1 for recruitment. Moreover, downregulation of BECN1 and PINK1 reduces the normal increase in ER-mitochondria contact site formation upon treatment with the mitochondrial uncoupler, CCCP, which is also known to stimulate mitophagy (Gelmetti et al. 2017).
Another protein that modulates mitophagy in mammalian cells is the mitochondrial protein FUNDC1. FUNDC1 accumulates at ER–mitochondria contact sites during hypoxia by associating with the ER membrane protein calnexin (CNX) (Wu et al. 2016). As mitophagy continues, FUNDC1/CNX interaction decreases and the exposed cytosolic loop of FUNDC1 binds DRP1 instead, promoting fission. Moreover, downregulation of FUNDC1, DRP1, or CNX prevents mitophagy in hypoxic conditions (Wu et al. 2016).
It may be that in different types of stress or in different cell types various modulators exist. For example, PACS2 accumulates at ER-mitochondria contact sites following an oxidized low-density lipoprotein (LDL) stimulation in human vascular smooth muscle cells and this accumulation increases contact extent. In the absence of PACS2, under similar stimulation, contacts do not expand, mitophagy is impaired, and apoptosis ensues (Moulis et al. 2019).
In summary, induction of mitophagy is clearly linked to ER-mitochondria contacts and the understanding that this platform is essential during normal cellular functions and stress may promote a better understanding of disease states where mitophagy is reduced.
Regulation of mitochondrial contact site extent and number
As new functions are added to the repertoire of mitochondrial contacts, it is also becoming clear that regulating them is essential. Indeed, the list of identified regulators is also growing. Contact regulators have tremendous influence on multiple aspects of contacts such as their formation, ablation, number, duration, stability and functions.
One of the first regulators discovered was the highly conserved yeast tail anchor protein Lam6 (GRAMD1a and GRAMD1c are the homologs in mammalian cells). Lam6 resides in several mitochondrial contact sites in yeast such as ER-mitochondria and vacuole-mitochondria (VaCuoLe and Mitochondria Patch; vCLAMP) (Elbaz-Alon et al. 2015; Gatta et al. 2015; Murley et al. 2015). Lam6 is embedded in the ER membrane through its transmembrane domain and binds opposing organelles through protein-protein interactions (Eisenberg-Bord et al. 2016; Elbaz-Alon et al. 2015; Gatta et al. 2015; Murley et al. 2015). Since it is found in more than one contact, it enables the cross-talk between contact sites and can regulate their expansions in physiological conditions.
Another example of a versatile tether in yeast, whose relative abundance at different contact sites can regulate their extent, is Vps13 (Bean et al. 2018). Vps13 was first connected to contact sites since a single point mutation was found to compensate for loss of ERMES complex functions (Lang et al. 2015). More recently, it was found to be a resident of several contact sites such as the vCLAMP, mitochondria–endosome and nucleus-vacuole junction (NVJ) (Lang et al. 2015, Park et al. 2016). Two additional proteins that enable the functional rescue of ERMES mutants when overexpressed are Mcp1 and Mcp2, which are both located on the MOM (Tan et al. 2013, Lang et al. 2015). Overexpression of Mcp1 re-localizes Vps13 to mitochondria suggesting that these two proteins work together. The N-terminus of Mcp1 is both necessary and sufficient for binding Vps13 and recruiting it to mitochondria. However, while a truncated version of Mcp1 succeeded in localizing Vps13 to mitochondria, it did not compensate for ERMES function, demonstrating an additional role of Mcp1 beyond recruitment of Vps13 (John Peter et al. 2017). Interestingly, expression of the vCLAMP tether Vps39 is essential for the Mcp1-Vps13 mediated rescue of ERMES (John Peter et al. 2017). It is not clear if Vps13-Mcp1 interaction creates a tether or is merely a physiological effector. Vps13 is highly conserved to humans where multiple family members have been shown to act in contacts and mutations in them cause genetic disorders (see below) (Kumar et al. 2018).
The yeast vCLAMP is also regulated. Phosphorylation of the vCLAMP protein Vps39 affects tethering under different carbon sources (Hönscher et al. 2014). Another regulator of vCLAMP in yeast is the evolutionary conserved multiprotein SEA complex (GATOR in mammals (Bar-Peled et al. 2013)) that regulates the TOR Complex 1 (Evans et al. 2011). Although the SEA complex is located at the vacuole membrane, it partially co-localizes with mitochondria (Ma et al. 2019). Moreover, about 20% of the proteins that interact with the SEA complex are mitochondrial. The deletion of any protein from the SEACIT sub-complex of the SEA complex (Algret et al. 2014) reduces vCLAMP formation in Vps39 overexpressed strains that normally induce and enlarge the contacts (González Montoro et al. 2018). In addition, a double deletion mutant of MDM34 (an ERMES component) and NRP3 (a SEACIT complex protein) has reduced fitness compared to single mutant strains (Ma et al. 2019).
In mammalian cells, a newly discovered contact regulator is TG2, a Ca2+-dependent post-translational modification enzyme that creates intra or inter molecular crosslinks between lysine and glutamine residues (Fesus and Piacentini 2002). TG2 interacts with proteins that are located in the ER and mitochondria, such as the above mentioned GRP75, in murine embryonic fibroblasts. Like GRP75, TG2 is enriched specifically in MAMs. Although the absence of TG2 increased the interaction between IP3R and GRP75, it decreased the number of contact sites observed between the ER and mitochondria, and elevated the distance between these organelles (D’Eletto et al. 2018). The ablation of TG2 also caused downregulation of several contact site proteins, among them the known tether MFN2, suggesting that the increase in IP3R-GRP75 interaction compensates for the disruption of ER-mitochondria contact sites. Another protein downregulated upon ablation of TG2 is TSPO, a MOM protein found also in the ER-mitochondria contact site. TSPO translocates cholesterol from the outer to the inner mitochondrial membrane (Batarseh and Papadopoulos 2010). Functionally, overexpression of TG2 increases Ca2+ trafficking between the ER and mitochondria (D’Eletto et al. 2018).
To date, most mitochondria contact site regulators were identified for ER-mitochondria contacts as this contact has attracted the most research. More research is required to uncover new regulators for other mitochondrial contact sites and tethers.
Newly-discovered mitochondrial contacts
For many years it was believed that all contacts formed in the cell involve the ER (Elbaz and Schuldiner 2011). The discovery of a mitochondrial contact site that does not contain the ER – the vCLAMP (Elbaz-Alon et al. 2014, Hönscher et al. 2014) opened the door for the possibility that more ER-independent contacts exist. Since then, more and more contacts are being discovered and characterized using multiple new tools and techniques that emerged and are improved over time. Currently it seems that all organelles, including mitochondria, can communicate through contact sites with almost all other organelles. A summary of all mitochondrial contact sites characterized to date in both yeast and mammalian cells and their identified tethers is shown in Figure 1. Below we will discuss the most recently uncovered mitochondrial contacts:
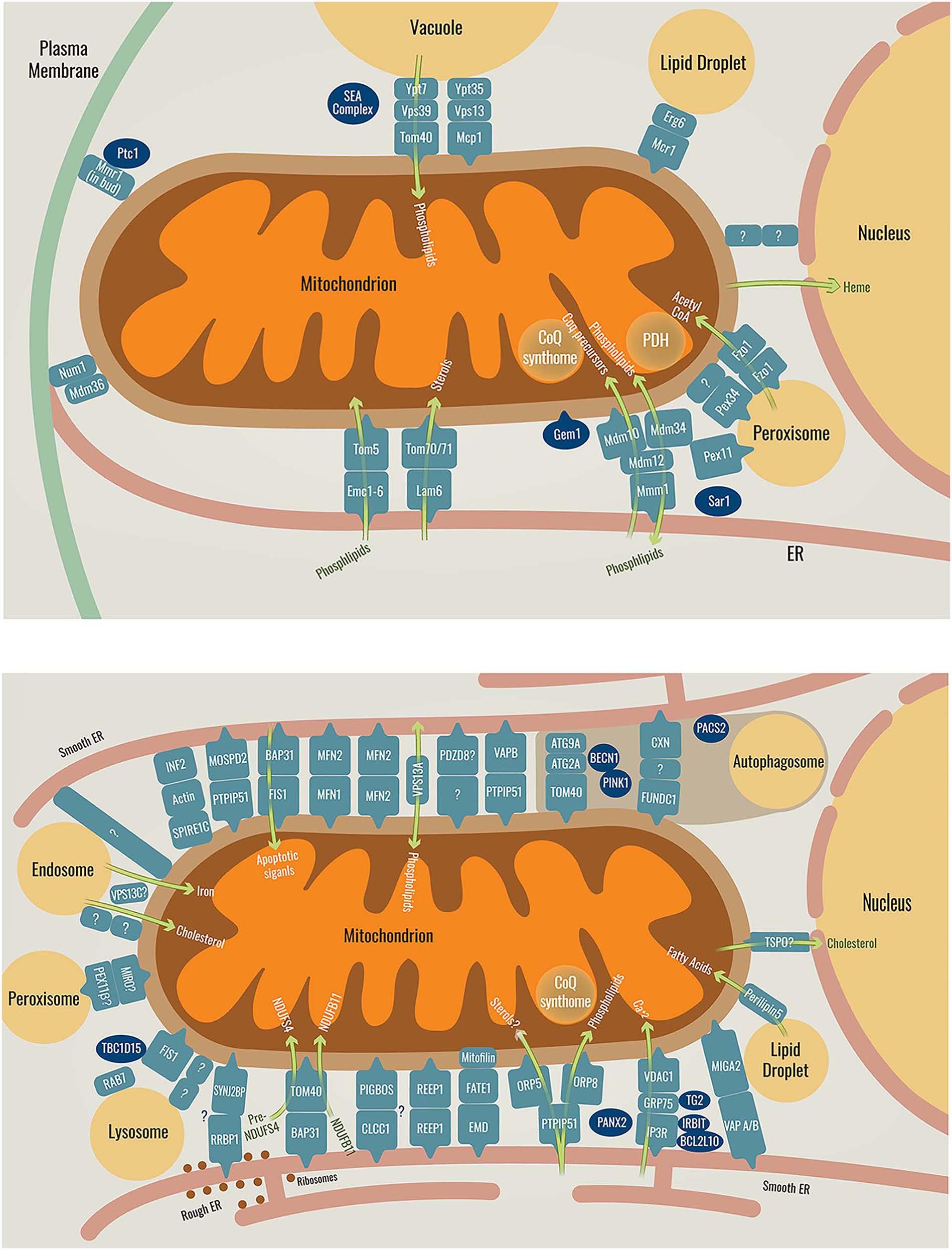
Schematic representation of the known mitochondrial contact sites and their resident tethers in yeast (top) or in mammals (bottom), as well as their functions. Tethers are shown in pale blue, regulators are shown in dark blue and the green arrows represent the function of the tether they pass through. The brown area in the mammalian cell contains the tethers and regulators responsible for autophagosome formation or inhibition. Proteins with a question mark are yet to be identified or fully proven. For more regulators in ER-mitochondria contacts see (Csordás et al. 2018). Full names of the depicted proteins can be seen in Table 1.
Mitochondria-nucleus contact in mammalian cells
A contact site between mitochondria and the nucleus was found in cell lines derived from breast cancer using electron microscopy. The contact, named Nucleus-Associated Mitochondria (NAM) (Desai et al. 2019), formed in response to staurosporine (STS), a drug that induces the mitochondrial retrograde stress response (Eisenberg-Bord and Schuldiner 2017b). TSPO, which was mentioned above as a resident protein of the ER-mitochondria contact, regulates the NAM as well. It is yet unclear if TSPO is a resident of both contacts or else that NAMs represent specialized cases of ER-mitochondria contacts on the outer nuclear membrane that is continuous with the ER.
In mouse embryonic fibroblasts, downregulation of TSPO increases the distance between the two organelles while its overexpression shows higher proximity of the MOM and the nuclear envelope. Overexpression of TSPO in these cells causes significant resistance to STS induced apoptosis and this is dependent on the ability of TSPO to bind cholesterol. After STS treatment, cholesterol is localized around the nucleus similarly to when TSPO is overexpressed (Desai et al. 2019). TSPO might be a NAM tether as many tethering molecules have lipid transfer capacity. More studies should be undertaken to further characterize this contact, its function and additional tethers and regulators.
Mitochondria-lysosome contact in mammalian cells
While it was shown already several years ago that mitochondria create a contact with the vacuole (the yeast lysosome), only recently the contact site between mitochondria and lysosomes was characterized in HeLa and HCT116 cells using electron microscopy (Wong et al. 2018). In live cells, mitochondria create a stable contact with several diverse sized vesicles simultaneously (Wong et al. 2018). The contact is regulated by TBC1D15, a GTPase-activating protein for RAB7. TBC1D15 is recruited to mitochondria by FIS1, stimulates RAB7 GTP hydrolysis and this untethers the mitochondria-lysosome contact. Expression of the constitutively active GTP-bound RAB7 mutant increases both contact formation and duration. A FIS1 mutation that prevents recruitment of TBC1D15 to mitochondria displayes the same phenotype (Wong et al. 2018).
In summary, contact sites of mitochondria with all organelles in both yeast and humans have now been described with the exception of a Golgi contact site, which will surely soon be found as well. An additional new avenue in contact site research is the capacity of membrane bound organelles to form contacts with membrane-less organelles (Gomes and Shorter 2019). Recently such contacts have been described for the ER (Ma and Mayr 2018, Lee et al. 2020) and LDs (Moldavski et al. 2015). It will be interesting to find if mitochondria can also have these kinds of contacts.
Three-way contacts
One of the most interesting recent discoveries in contact site biology is that contact sites can also mediate the physical interaction between more than two organelles. The phenomenon of contact formed between three organelles simultaneously or “three-way contacts” is changing our view that junctions between organelle membranes can be created only between two opposing membranes. For example, the ER protein Mdm1 interacts simultaneously with LDs and vacuoles to create a three-way contact in yeast (Hariri et al. 2019). Additional three-way contacts have been found in yeast between LDs, vacuoles and the nuclear membrane (Eisenberg-Bord et al. 2018) and between LDs, peroxisomes and the ER (Joshi et al. 2018).
Mitochondrial contact sites are central to cellular physiology and mediate multiple processes; hence it is no surprise that mitochondria too form three-way contacts. Indeed, mitochondria have been shown to form three-way contacts with peroxisomes and the ER in yeast (Cohen et al. 2014). In plant cells, mitochondria, peroxisomes and chloroplasts have been shown to form three-way contacts as well (Oikawa et al. 2019). A few examples of the molecular players involved in these types of contacts can be found below:
Mitochondria, ER, LDs
A three-way contact between LDs, mitochondria and the ER was identified in differentiating adipocytes (Freyre et al. 2019). In this contact, MIGA2, a MOM protein, acts as a physical tether by binding the LD surface through its amphipathic helix located at the C-terminus. In addition, MIGA2 contains a FFAT (double phenylalanine in an Acidic Tract) motif that is responsible for interacting with the ER proteins VAPA and VAPB. Removal of any one of these sequences causes dispersal of the three organelles in the cell. In differentiating adipocytes this tethering has been shown to be crucial for synthesis of triacylglycerols from non-lipid precursors such as glucose during de novo lipogenesis (Freyre et al. 2019).
Mitochondria, ER, PM
The MECA represents a contact between mitochondria, ER and the PM in yeast (Lackner et al. 2013, Klecker et al. 2013). Two important proteins for MECA establishment are Mdm36 on the MOM and Num1. Multidomain Num1 is a unique tether that interacts with membranes on both of its ends – the coiled coil domain at its N-terminus binds cardiolipin and phosphatidic acid on the mitochondrial membrane while the pleckstrin homology (PH) domain on its C-terminus specifically binds phosphatidylinositol 4,5-bisphosphate PI(4,5)P2 on the PM (Ping et al. 2016).
These findings support the notion that maintaining cellular homeostasis often requires signals and pathways to go through more than two organelles and that this can be facilitated by three-way contacts. Three-way contacts with mitochondria demonstrate a greater degree of organization in cellular space. This creates the potential to transfer metabolites even more efficiently, in complex pathways without being exposed to the cytosol where they may be consumed, degraded or modified. This may be important especially during differentiation or stress responses when the cell must react rapidly to changes. It will also be interesting to study how these three-way contacts are regulated and if they are dynamic.
Different contact sites between the same two organelles
While several tethering molecules and functions can be present, interspersed, in the same contact site, a newly emerging theme is the presence of spatially distinct contact sites forming between the same two organelles. Such contacts, although they connect the same two organelles, are in different places along the organelle membranes, perform non-overlapping functions and contain distinct tethers.
A recent example is the discovery of distinct domains of the ER-PM contact site in human cells, which are separated spatially and mediated by specialized tethers with different functions (Besprozvannaya et al. 2018).
The concept of spatially distinct contact sites with mitochondria is also starting to emerge. For example, in mammalian cells, a contact site of mitochondria specifically with the rough ER that differs from the contact with the smooth ER was shown (Giacomello and Pellegrini 2016). Proteomic mapping in human HEK293 cells revealed that the mitochondrial tail anchored protein SYNJ2BP (also known as OMP25) is a contact site protein (Hung et al. 2017). However, its overexpression in Cos7 cells increased the proximity specifically between the rough ER and mitochondria. The binding partner of SYNJ2BP on the ER membrane is the single-pass transmembrane protein RRBP1, which contains a PDZ-binding domain that may bind the PDZ domain of SYNJ2BP. Overexpression of SYNJ2BP in RRBP1 KO cells failed to increase the contact extent. Moreover, treatment with the translation inhibitor puromycin, which disassembles polysomes, reduced the extent of the overexpressed SYNJ2BP connection with endogenous RRBP1, without changing the abundance of either protein. Puromycin treatment also disrupted selectively the rough ER-mitochondria contacts (Hung et al. 2017). Hence, mitochondrial contacts with rough ER or smooth ER create distinct sub-domains with unique specialized functions and different tethering machinery (Hung et al. 2017, Giacomello and Pellegrini 2016).
Another recent example for different contact sites between the same organelles is the yeast vacuole-mitochondria contact. One contact is the vCLAMP whose tether is Vps39 that binds to the vacuole via the Rab7-like GTPase, Ypt7 and to mitochondria via Tom40 (González Montoro et al. 2018). Several mutations in the region necessary for Vps39 tethering to mitochondria resulted in reduction of Tom40 binding and association of the two organelles (González Montoro et al. 2018). The reduced tethering of Vps39 to Tom40 uncovered a second vacuole-mitochondria tethering pair – Vps13-Mcp1 (John Peter et al. 2017). In depth characterization of the localization and function of the Vps13-Mcp1 contact suggests that it is proximal to, but not overlapping with, that of the Ypt7-Vps39-Tom40 vCLAMP contact (González Montoro et al. 2018).
Hence, it is important that the field starts referring to contacts not only by the two organelles participating in them, but provide the exact tethers or functions studied or provide distinctive nomenclature.
Involvement of mitochondrial contact sites in disease
It is now clear that mitochondrial functions and homeostasis are dependent on formation of contact sites with other organelles. As mitochondria play a crucial role in many pathways, it is probably no wonder that damaging their capacity to communicate with other organelles plays a central role in diverse human diseases (López-Crisosto et al. 2015, Liu and Zhu 2017, Paillusson et al. 2016; for a summary see Figure 2).
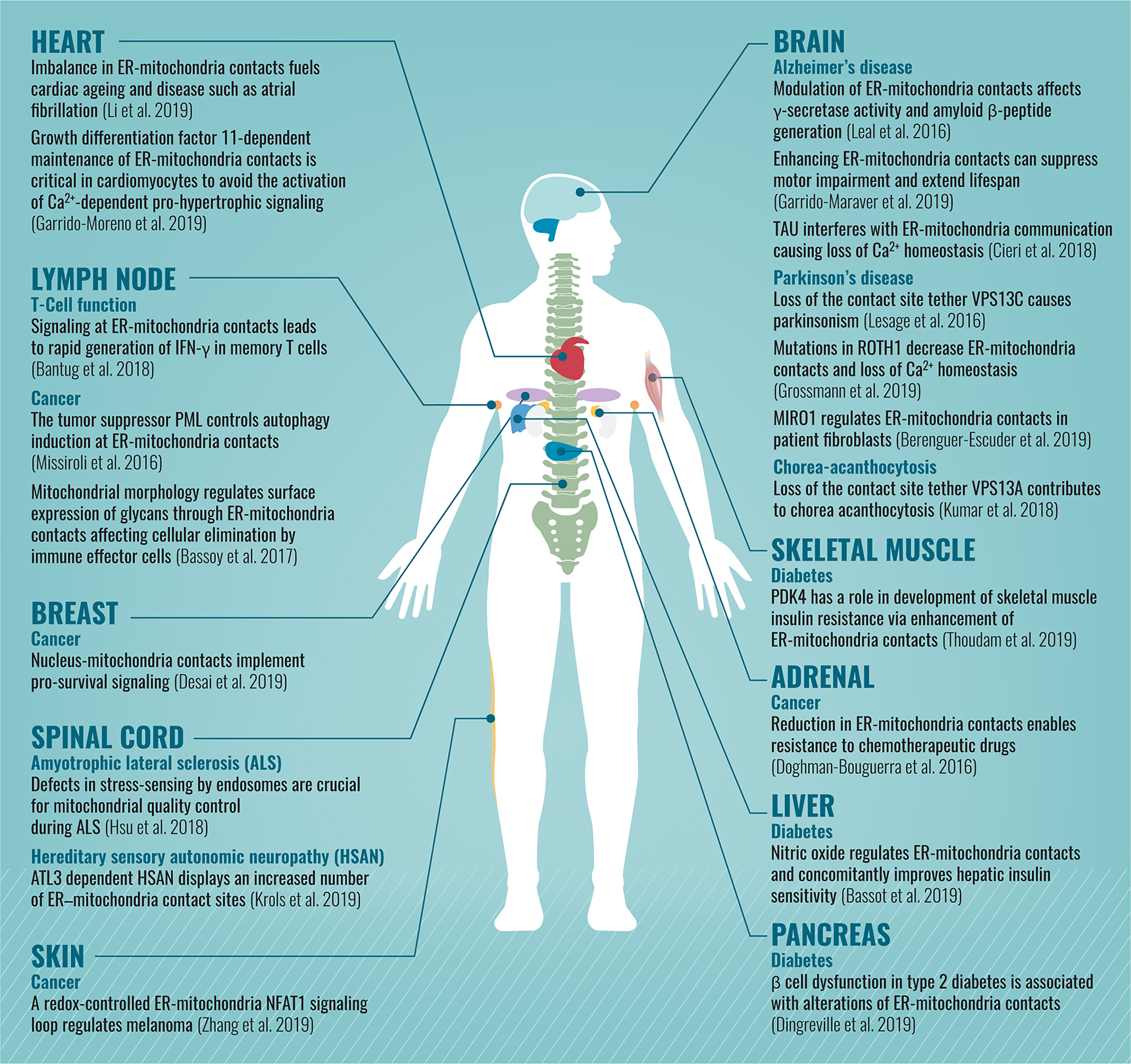
Schematic description of diseases (divided by the affected organs) that are associated with alterations in mitochondrial contact sites.
For many years, the molecules forming and regulating mitochondrial contact sites were unknown, making it impossible to find genetic diseases associated with their loss. In addition, since the phenotypes of mitochondrial contact site loss were unclear, the identification of the first diseases took time. In 2014 the first connection between a damaged mitochondrial contact site and a disease was made as the ER-mitochondria contact was implicated in Alzheimer’s disease progression (Area-Gomez et al. 2012).
Since most research in the contact site field has been focused on the ER-mitochondrial contact site, the majority of diseases displaying alterations in contact sites described to date are related to that between ER and mitochondria. However, the first examples for the involvement of other mitochondrial contact sites in diseases are starting to emerge. For example, Parkinson’s disease was found to be associated with a defect of the mitochondria-endosome contact (Lesage et al. 2016), and a connection between breast cancer and the mitochondria-nucleus contact site has been described (Desai et al. 2019).
With the rapid accumulation of knowledge on additional mitochondrial contact sites, we expect many more disease connections to emerge in the coming years.
It is of interest why defects in a fundamental and evolutionary conserved cellular process such as mitochondrial tethering to other organelles, ubiquitously present in most cell types, has cell-specific phenotypes. The most probable explanation is that the presence of multiple redundant tethering machineries ensures robustness in the face of mutations. In addition, it is possible that alternative contact sites can compensate for dysfunctional ones. The dependence of each tissue on specific tethers, the presence of back-up contacts and the requirement for mitochondrial function in each cell type and tissue may underlie the tissue-specificity of the diseases.
Perspective
Albert Einstein once said “As our circle of knowledge expands, so does the circumference of darkness surrounding it.” Similarly, as more and more groups flux into the emerging field of mitochondrial contact sites and new discoveries in the field are now a weekly phenomenon, so do new concepts requiring investigation arise.
Clearly the big challenge ahead is the realization that the various functions of mitochondrial contact sites and their regulation are cell type and condition dependent. Studies today focus mainly on one kind of cell line and establish their discoveries based on the organization of a specific cell network. This will have to be taken more into consideration as examples of various regulators, such as GRP75 or BECN1, that work in opposite ways in different cell lines, arise. Hence, to truly understand the complexity of mitochondrial contacts we must explore them in multiple environments, cell types and model systems. It will be interesting to explore which of the mechanisms described to date are universal and which are unique for specific conditions or cell types.
As we continue investigating the diverse mitochondrial contact sites, their functions and machineries, we understand more about the complexity of the cell network. The field has greatly relied on technological advances that enable new tools for visualizing contact sites and for analyzing their molecular composition. As these develop we will surely find new concepts in mitochondrial contact site research.
Funding source: Deutsche Forschungsgemeinschaft
Award Identifier / Grant number: SFB1190 (Gates and Contacts)
Funding source: H2020 European Research Council
Award Identifier / Grant number: CoG Peroxisystem (646604)
Acknowledgments
We would like to thank Michal Eisenberg-Bord, Dr. Inês Castro, Mira Rosenthal, Yotam David, Prof. Amnon Zung and Dr. Einat Zalckvar for helpful discussions and feedback on this paper. We would like to thank Hanna Vega for her graphical work for this manuscript. This work was supported by the DFG through an SFB1190 (Gates and Contacts) and by an ERC CoG Peroxisystem (646604). Maya Schuldiner is an Incumbent of the Dr. Gilbert Omenn and Martha Darling Professorial Chair in Molecular Genetics.
References
Aisen, P., Enns, C., and Wessling-Resnick, M. (2001). Chemistry and biology of eukaryotic iron metabolism. Int. J. Biochem. Cell Biol. 33: 940–959, https://doi.org/10.1016/S1357-2725(01)00063-2.Search in Google Scholar
Algret, R., Fernandez-Martinez, J., Shi, Y., Kim, S.J., Pellarin, R., Cimermancic, P., Cochet, E., Sali, A., Chait, B.T., Rout, M.P., et al. (2014). Molecular architecture and function of the SEA complex, a modulator of the TORC1 pathway. Mol. Cell Proteomics 13: 2855–2870, https://doi.org/10.1074/mcp.M114.039388.Search in Google Scholar PubMed PubMed Central
Area-Gomez, E., Del Carmen Lara Castillo, M., Tambini, M.D., Guardia-Laguarta, C., de Groof, A.J., Madra, M., Ikenouchi, J., Umeda, M., and Bird, T.D., Sturley, S.L., et al. (2012). Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 31: 4106–4123, https://doi.org/10.1038/emboj.2012.202.Search in Google Scholar PubMed PubMed Central
Awad, A.M., Bradley, M.C., Fernandez-Del-Rio, L., Nag, A., Tsui, H.S., and Clarke, C.F. (2018). Coenzyme Q10 deficiencies: pathways in yeast and humans. Essays Biochem. 62: 361–376, https://doi.org/10.1042/EBC20170106.Search in Google Scholar PubMed PubMed Central
Bantug, G.R., Fischer, M., Grahlert, J., Balmer, M.L., Unterstab, G., Develioglu, L., Steiner, R., Zhang, L., and Costa, A.S.H., Gubser, P.M., et al. (2018). Mitochondria-Endoplasmic reticulum contact sites Function as immunometabolic hubs that orchestrate the rapid recall response of memory CD8+ T cells. Immunity 48: 542–555 e546, https://doi.org/10.1016/j.immuni.2018.02.012.Search in Google Scholar PubMed PubMed Central
Bar-Peled, L., Chantranupong, L., Cherniack, A.D., Chen, W.W., Ottina, K.A., Grabiner, B.C., Spear, E.D., Carter, S.L., Meyerson, M., and Sabatini, D.M. (2013). A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340: 1100–1106, https://doi.org/10.1126/science.1232044.Search in Google Scholar PubMed PubMed Central
Bassot, A., Chauvin, M.A., Bendridi, N., Ji-Cao, J., Vial, G., Monnier, L., Bartosch, B., Alves, A., Cottet-Rousselle, C., Gouriou, Y., et al. (2019). Regulation of mitochondria-associated membranes (MAMs) by NO/sGC/PKG participates in the control of hepatic insulin response. Cells 8: 1319, https://doi.org/10.3390/cells8111319.Search in Google Scholar PubMed PubMed Central
Bassoy, E.Y., Kasahara, A., Chiusolo, V., Jacquemin, G., Boydell, E., Zamorano, S., Riccadonna, C., Pellegatta, S., Hulo, N., Dutoit, V., et al. (2017). ER-mitochondria contacts control surface glycan expression and sensitivity to killer lymphocytes in glioma stem-like cells. EMBO J. 36: 1493–1512, https://doi.org/10.15252/embj.201695429.Search in Google Scholar PubMed PubMed Central
Batarseh, A. and Papadopoulos, V. (2010). Regulation of translocator protein 18 kDa (TSPO) expression in health and disease states. Mol. Cell Endocrinol. 327: 1–12, https://doi.org/10.1016/j.mce.2010.06.013.Search in Google Scholar PubMed PubMed Central
Bean, B.D.M., Dziurdzik, S.K., Kolehmainen, K.L., Fowler, C.M.S., Kwong, W.K., Grad, L.I., Davey, M., Schluter, C., and Conibear, E. (2018). Competitive organelle-specific adaptors recruit Vps13 to membrane contact sites. J. Cell. Biol. 217: 3593–3607, https://doi.org/10.1083/jcb.201804111.Search in Google Scholar PubMed PubMed Central
Berenguer-Escuder, C., Grossmann, D., Massart, F., Antony, P., Burbulla, L.F., Glaab, E., Imhoff, S., Trinh, J., Seibler, P., Grunewald, A., et al. (2019). Variants in Miro1 cause alterations of ER-Mitochondria contact sites in fibroblasts from Parkinson’s Disease Patients. J. Clin. Med. 8: 2226, https://doi.org/10.3390/jcm8122226.Search in Google Scholar PubMed PubMed Central
Bernhard, W. and Rouiller, C. (1956). Close topographical relationship between mitochondria and ergastoplasm of liver cells in a definite phase of cellular activity. J. Biophys. Biochem. Cytol. 2: 73–78, https://doi.org/10.1083/jcb.2.4.73.Search in Google Scholar PubMed PubMed Central
Bernhard, W., Haguenau, F., Gautier, A., and Oberling, C. (1952). [Submicroscopical structure of cytoplasmic basophils in the liver, pancreas and salivary gland; study of ultrafine slices by electron microscope]. Z. Zellforsch Mikrosk Anat. 37: 281–300.10.1007/BF00343816Search in Google Scholar
Besprozvannaya, M., Dickson, E., Li, H., Ginburg, K.S., Bers, D.M., Auwerx, J., and Nunnari, J. (2018). GRAM domain proteins specialize functionally distinct ER-PM contact sites in human cells. Elife 7: e31019, https://doi.org/10.7554/eLife.31019.001.Search in Google Scholar
Bonneau, B., Ando, H., Kawaai, K., Hirose, M., Takahashi-Iwanaga, H., and Mikoshiba, K. (2016). IRBIT controls apoptosis by interacting with the Bcl-2 homolog, Bcl2l10, and by promoting ER-mitochondria contact. Elife 5: e19896, https://doi.org/10.7554/eLife.19896.001.Search in Google Scholar
Boutant, M., Kulkarni, S.S., Joffraud, M., Ratajczak, J., Valera-Alberni, M., Combe, R., Zorzano, A., and Canto, C. (2017). Mfn2 is critical for brown adipose tissue thermogenic function. EMBO J. 36: 1543–1558, https://doi.org/10.15252/embj.201694914.Search in Google Scholar PubMed PubMed Central
Cherry, J.M., Hong, E.L., Amundsen, C., Balakrishnan, R., Binkley, G., Chan, E.T., Christie, K.R., Costanzo, M.C., Dwight, S.S., Engel, S.R., et al. (2012). Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 40: D700–D705, https://doi.org/10.1093/nar/gkr1029.Search in Google Scholar PubMed PubMed Central
Chiabrando, D., Marro, S., Mercurio, S., Giorgi, C., Petrillo, S., Vinchi, F., Fiorito, V., Fagoonee, S., Camporeale, A., Turco, E., et al. (2012). The mitochondrial heme exporter FLVCR1b mediates erythroid differentiation. J. Clin. Invest. 122: 4569–4579.10.1172/JCI62422Search in Google Scholar PubMed PubMed Central
Choubey, V., Cagalinec, M., Liiv, J., Safiulina, D., Hickey, M.A., Kuum, M., Liiv, M., Anwar, T., Eskelinen, E.L., Kaasik, A. (2014). BECN1 is involved in the initiation of mitophagy: it facilitates PARK2 translocation to mitochondria. Autophagy 10: 1105–1119, https://doi.org/10.4161/auto.28615.Search in Google Scholar PubMed PubMed Central
Chu, Q., Martinez, T.F., Novak, S.W., Donaldson, C.J., Tan, D., Vaughan, J.M., Chang, T., Diedrich, J.K., Andrade, L., Kim, A., et al. (2019). Regulation of the ER stress response by a mitochondrial microprotein. Nat. Commun. 10: 4883, https://doi.org/10.1038/s41467-019-12816-z.Search in Google Scholar PubMed PubMed Central
Cieri, D., Vicario, M., Vallese, F., D’Orsi, B., Berto, P., Grinzato, A., Catoni, C., De Stefani, D., Rizzuto, R., Brini, M., Cali, T. (2018). Tau localises within mitochondrial sub-compartments and its caspase cleavage affects ER-mitochondria interactions and cellular Ca2+ handling. Biochim. Biophys. Acta Mol. Basis Dis. 1864: 3247–3256, https://doi.org/10.1016/j.bbadis.2018.07.011.Search in Google Scholar PubMed
Cockcroft, S. and Raghu, P. (2018). Phospholipid transport protein function at organelle contact sites. Curr. Opin. Cell. Biol. 53: 52–60, https://doi.org/10.1016/j.ceb.2018.04.011.Search in Google Scholar PubMed PubMed Central
Cohen, Y., Klug, Y.A., Dimitrov, L., Erez, Z., Chuartzman, S.G., Elinger, D., Yofe, I., Soliman, K., Gartner, J., Thoms, S., et al. (2014). Peroxisomes are juxtaposed to strategic sites on mitochondria. Mol. Biosyst. 10: 1742–1748, https://doi.org/10.1039/C4MB00001C.Search in Google Scholar PubMed
Cohen, N., Breker, M., Bakunts, A., Pesek, K., Chas, A., Argemi, J., Orsi, A., Gal, L., Chuartzman, S., Wigelman, Y., et al. (2017). Iron affects Ire1 clustering propensity and the amplitude of endoplasmic reticulum stress signaling. J. Cell. Sci. 130: 3222–3233, https://doi.org/10.1242/jcs.201715.Search in Google Scholar PubMed PubMed Central
Csordas, G., Weaver, D., and Hajnoczky, G. (2018). Endoplasmic reticulum-mitochondrial contactology: structure and signaling functions. Trends Cell. Biol. 28: 523–540, https://doi.org/10.1016/j.tcb.2018.02.009.Search in Google Scholar PubMed PubMed Central
Daniele, T., Hurbain, I., Vago, R., Casari, G., Raposo, G., Tacchetti, C., and Schiaffino, M.V. (2014). Mitochondria and melanosomes establish physical contacts modulated by Mfn2 and involved in organelle biogenesis. Curr. Biol. 24: 393–403, https://doi.org/10.1016/j.cub.2014.01.007.Search in Google Scholar PubMed
Das, A., Nag, S., Mason, A.B., and Barroso, M.M. (2016). Endosome-mitochondria interactions are modulated by iron release from transferrin. J. Cell. Biol. 214: 831–845, https://doi.org/10.1083/jcb.201602069.Search in Google Scholar PubMed PubMed Central
Dautry-Varsat, A., Ciechanover, A., and Lodish, H.F. (1983). pH and the recycling of transferrin during receptor-mediated endocytosis. Proc. Natl. Acad. Sci. USA 80: 2258–2262, https://doi.org/10.1073/pnas.80.8.2258.Search in Google Scholar PubMed PubMed Central
De Rasmo, D., Panelli, D., Sardanelli, A.M., and Papa, S. (2008). cAMP-dependent protein kinase regulates the mitochondrial import of the nuclear encoded NDUFS4 subunit of complex I. Cell Signal 20: 989–997, https://doi.org/10.1016/j.cellsig.2008.01.017.Search in Google Scholar PubMed
De Vos, K.J., Morotz, G.M., Stoica, R., Tudor, E.L., Lau, K.F., Ackerley, S., Warley, A., Shaw, C.E., and Miller, C.C. (2012). VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum. Mol. Genet. 21: 1299–1311, https://doi.org/10.1093/hmg/ddr559.Search in Google Scholar PubMed PubMed Central
D’Eletto, M., Rossin, F., Occhigrossi, L., Farrace, M.G., Faccenda, D., Desai, R., Marchi, S., Refolo, G., Falasca, L., Antonioli, M., et al. (2018). Transglutaminase Type 2 regulates ER-Mitochondria contact sites by interacting with GRP75. Cell. Rep. 25: 3573–3581.e4, https://doi.org/10.1016/j.celrep.2018.11.094.Search in Google Scholar PubMed
Desai, R., East, D.A., Hardy, L., Crosby, J., Rigon, M., Faccenda, D., Soledad-Alvarez, M., Singh, A., Mainenti, M., Kuhlman-Hussey, L., et al. (2019). Mitochondria form cholesterol tethered contact sites with the nucleus to regulate retrograde response. BioRxiv: 445411, https://doi.org/10.1101/445411.Search in Google Scholar
Ding, W.X. and Yin, X.M. (2012). Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol. Chem. 393: 547–564, https://doi.org/10.1515/hsz-2012-0119.Search in Google Scholar PubMed PubMed Central
Dingreville, F., Panthu, B., Thivolet, C., Ducreux, S., Gouriou, Y., Pesenti, S., Chauvin, M.A., Chikh, K., Errazuriz-Cerda, E., Van Coppenolle, F., et al. (2019). Differential effect of glucose on ER-Mitochondria Ca2+ exchange participates in insulin secretion and glucotoxicity-mediated dysfunction of beta-cells. Diabetes 68: 1778–1794, https://doi.org/10.2337/db18-1112.Search in Google Scholar PubMed
Doghman-Bouguerra, M., Granatiero, V., Sbiera, S., Sbiera, I., Lacas-Gervais, S., Brau, F., Fassnacht, M., Rizzuto, R., and Lalli, E. (2016). FATE1 antagonizes calcium- and drug-induced apoptosis by uncoupling ER and mitochondria. EMBO Rep. 17: 1264–1280, https://doi.org/10.15252/embr.201541504.Search in Google Scholar PubMed PubMed Central
Eisenberg-Bord, M. and Schuldiner, M. (2017a). Mitochatting – If only we could be a fly on the cell wall. Biochim. Biophys. Acta Mol. Cell. Res. 1864: 1469–1480, https://doi.org/10.1016/j.bbamcr.2017.04.012.Search in Google Scholar PubMed
Eisenberg-Bord, M. and Schuldiner, M. (2017b). Ground control to major TOM: mitochondria-nucleus communication. FEBS J. 284: 196–210, https://doi.org/10.1111/febs.13778.Search in Google Scholar PubMed
Eisenberg-Bord, M., Shai, N., Schuldiner, M., and Bohnert, M. (2016). A tether is a tether is a tether: tethering at membrane contact sites. Dev Cell 39: 395–409, https://doi.org/10.1016/j.devcel.2016.10.022.Search in Google Scholar PubMed
Eisenberg-Bord, M., Mari, M., Weill, U., Rosenfeld-Gur, E., Moldavski, O., Castro, I.G., Soni, K.G., Harpaz, N., and Levine, T.P., Futerman, A.H., et al. (2018). Identification of seipin-linked factors that act as determinants of a lipid droplet subpopulation. J. Cell. Biol. 217: 269–282, https://doi.org/10.1083/jcb.201704122.Search in Google Scholar PubMed PubMed Central
Eisenberg-Bord, M., Tsui, H.S., Antunes, D., Fernandez-Del-Rio, L., Bradley, M.C., Dunn, C.D., Nguyen, T.P.T., Rapaport, D., Clarke, C.F., and Schuldiner, M. (2019). The endoplasmic reticulum-mitochondria encounter structure complex coordinates coenzyme Q biosynthesis. Contact (Thousand Oaks) 2, https://doi.org/10.1177/2515256418825409.Search in Google Scholar PubMed PubMed Central
Elbaz, Y. and Schuldiner, M. (2011). Staying in touch: the molecular era of organelle contact sites. Trends Biochem. Sci. 36: 616–623, https://doi.org/10.1016/j.tibs.2011.08.004.Search in Google Scholar PubMed
Elbaz-Alon, Y., Rosenfeld-Gur, E., Shinder, V., Futerman, A.H., Geiger, T., and Schuldiner, M. (2014). A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell 30: 95–102, https://doi.org/10.1016/j.devcel.2014.06.007.Search in Google Scholar PubMed
Elbaz-Alon, Y., Eisenberg-Bord, M., Shinder, V., Stiller, S.B., Shimoni, E., Wiedemann, N., Geiger, T., and Schuldiner, M. (2015). Lam6 regulates the extent of contacts between organelles. Cell. Rep. 12: 7–14, https://doi.org/10.1016/j.celrep.2015.06.022.Search in Google Scholar PubMed PubMed Central
Esposito, M., Hermann-Le Denmat, S., and Delahodde, A. (2019). Contribution of ERMES subunits to mature peroxisome abundance. PLoS One 14: e0214287, https://doi.org/10.1371/journal.pone.0214287.Search in Google Scholar PubMed PubMed Central
Evans, D.S., Kapahi, P., Hsueh, W.C., and Kockel, L. (2011). TOR signaling never gets old: aging, longevity and TORC1 activity. Ageing Res. Rev. 10: 225–237, https://doi.org/10.1016/j.arr.2010.04.001.Search in Google Scholar PubMed PubMed Central
Fesus, L. and Piacentini, M. (2002). Transglutaminase 2: an enigmatic enzyme with diverse functions. Trends Biochem. Sci. 27: 534–539, https://doi.org/10.1016/S0968-0004(02)02182-5.Search in Google Scholar PubMed
Freyre, C.A.C., Rauher, P.C., Ejsing, C.S.,and Klemm, R.W. (2019). MIGA2 Links Mitochondria, the ER, and lipid droplets and promotes de novo lipogenesis in adipocytes. Mol. Cell 76: 811–825 e814, https://doi.org/10.1016/j.molcel.2019.09.011.Search in Google Scholar PubMed
Garrido-Maraver, J., Loh, S.H.Y., and Martins, L.M. (2020). Forcing contacts between mitochondria and the endoplasmic reticulum extends lifespan in a Drosophila model of Alzheimer’s disease. Biol. Open 9, https://doi.org/10.1242/bio.047530.Search in Google Scholar PubMed PubMed Central
Garrido-Moreno, V., Diaz-Vegas, A., Lopez-Crisosto, C., Troncoso, M.F., Navarro-Marquez, M., Garcia, L., Estrada, M., Cifuentes, M., and Lavandero, S. (2019). GDF-11 prevents cardiomyocyte hypertrophy by maintaining the sarcoplasmic reticulum-mitochondria communication. Pharmacol. Res. 146: 104273, https://doi.org/10.1016/j.phrs.2019.104273.Search in Google Scholar PubMed
Gatta, A.T. and Levine, T.P. (2017). Piecing together the patchwork of contact sites. Trends Cell. Biol. 27: 214–229, https://doi.org/10.1016/j.tcb.2016.08.010.Search in Google Scholar PubMed
Gatta, A.T., Wong, L.H., Sere, Y.Y., Calderon-Norena, D.M., Cockcroft, S., Menon, A.K., and Levine, T.P. (2015). A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. Elife 4: https://doi.org/10.7554/eLife.07253.001.Search in Google Scholar
Gelmetti, V., De Rosa, P., Torosantucci, L., Marini, E.S., Romagnoli, A., Di Rienzo, M., Arena, G., Vignone, D., Fimia, G.M., and Valente, E.M. (2017). PINK1 and BECN1 relocalize at mitochondria-associated membranes during mitophagy and promote ER-mitochondria tethering and autophagosome formation. Autophagy 13: 654–669, https://doi.org/10.1080/15548627.2016.1277309.Search in Google Scholar PubMed PubMed Central
Ghosh, R., Wang, L., Wang, E.S., Perera, B.G., Igbaria, A., Morita, S., Prado, K., Thamsen, M., and Caswell, D., Macias, H., et al. (2014). Allosteric inhibition of the IRE1alpha RNase preserves cell viability and function during endoplasmic reticulum stress. Cell 158: 534–548, https://doi.org/10.1016/j.cell.2014.07.002.Search in Google Scholar PubMed PubMed Central
Giacomello, M. and Pellegrini, L. (2016). The coming of age of the mitochondria-ER contact: a matter of thickness. Cell Death Differ. 23: 1417–1427, https://doi.org/10.1038/cdd.2016.52.Search in Google Scholar PubMed PubMed Central
Gomes, E. and Shorter, J. (2019). The molecular language of membraneless organelles. J. Biol. Chem. 294: 7115–7127, https://doi.org/10.1074/jbc.tm118.001192.Search in Google Scholar
Gomez-Suaga, P., Paillusson, S., Stoica, R., Noble, W., Hanger, D.P. and Miller, C.C.J. (2017). The ER-mitochondria tethering complex VAPB-PTPIP51 regulates autophagy. Curr. Biol. 27: 371–385, https://doi.org/10.1016/j.cub.2016.12.038.Search in Google Scholar PubMed PubMed Central
González Montoro, A., Auffarth, K., Honscher, C., Bohnert, M., Becker, T., Warscheid, B., Reggiori, F., van der Laan, M., Frohlich, F., and Ungermann, C. (2018). Vps39 interacts with Tom40 to establish one of two functionally distinct vacuole-mitochondria contact sites. Dev. Cell 45: 621–636 e627, https://doi.org/10.1016/j.devcel.2018.05.011.Search in Google Scholar PubMed
Grossmann, D., Berenguer-Escuder, C., Bellet, M.E., Scheibner, D., Bohler, J., Massart, F., Rapaport, D., Skupin, A., Fouquier d’Herouel, A., Sharma, M., et al. (2019). Mutations in RHOT1 disrupt endoplasmic reticulum-mitochondria contact sites interfering with calcium homeostasis and mitochondrial dynamics in Parkinson’s disease. Antioxid. Redox Signal 31: 1213–1234, https://doi.org/10.1089/ars.2018.7718.Search in Google Scholar PubMed PubMed Central
Guo, W., Yang, L., Li, H., Xie, Z., Liu, W., and Zuo, J. (2012). Glucose-regulated protein 75 overexpression attenuates ionizing radiation-mediated injury in PC12 cells by inducing the expression of topoisomerase IIalpha. Mol. Med. Rep. 6: 1423–1427, https://doi.org/10.3892/mmr.2012.1070.Search in Google Scholar PubMed
Hanna, D.A., Martinez-Guzman, O., and Reddi, A.R. (2017). Heme gazing: illuminating eukaryotic heme trafficking, dynamics, and signaling with fluorescent heme sensors. Biochemistry 56: 1815–1823, https://doi.org/10.1021/acs.biochem.7b00007.Search in Google Scholar PubMed PubMed Central
Hansen, K.G., Aviram, N., Laborenz, J., Bibi, C., Meyer, M., Spang, A., Schuldiner, M., and Herrmann, J.M. (2018). An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science 361: 1118–1122, https://doi.org/10.1126/science.aar8174.Search in Google Scholar PubMed
Hariri, H., Speer, N., Bowerman, J., Rogers, S., Fu, G., Reetz, E., Datta, S., Feathers, J.R., Ugrankar, R., Nicastro, D., and Henne, W.M. (2019). Mdm1 maintains endoplasmic reticulum homeostasis by spatially regulating lipid droplet biogenesis. J. Cell Biol. 218: 1319–1334, https://doi.org/10.1083/jcb.201808119.Search in Google Scholar PubMed PubMed Central
Honrath, B., Metz, I., Bendridi, N., Rieusset, J., Culmsee, C., and Dolga, A.M. (2017). Glucose-regulated protein 75 determines ER-mitochondrial coupling and sensitivity to oxidative stress in neuronal cells. Cell Death Discov. 3: 17076, https://doi.org/10.1038/cddiscovery.2017.76.Search in Google Scholar PubMed PubMed Central
Honscher, C., Mari, M., Auffarth, K., Bohnert, M., Griffith, J., Geerts, W., van der Laan, M., Cabrera, M., Reggiori, F., and Ungermann, C. (2014). Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell 30: 86–94, https://doi.org/10.1016/j.devcel.2014.06.006.Search in Google Scholar PubMed
Hsu, F., Spannl, S., Ferguson, C., Hyman, A.A., Parton, R.G., and Zerial, M. (2018). Rab5 and Alsin regulate stress-activated cytoprotective signaling on mitochondria. Elife 7: https://doi.org/10.7554/eLife.32282.001.Search in Google Scholar
Hung, V., Lam, S.S., Udeshi, N.D., Svinkina, T., Guzman, G., Mootha, V.K., Carr, S.A., and Ting, A.Y. (2017). Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife 6: https://doi.org/10.7554/eLife.24463.001.Search in Google Scholar
Iwasawa, R., Mahul-Mellier, A.L., Datler, C., Pazarentzos, E., and Grimm, S. (2011). Fis1 and Bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J. 30, 556–568, https://doi.org/10.1038/emboj.2010.346.Search in Google Scholar PubMed PubMed Central
Jing, J., Liu, G., Huang, Y., and Zhou, Y. (2019). A molecular toolbox for interrogation of membrane contact sites. J. Physiol., https://doi.org/10.1113/JP277761.Search in Google Scholar PubMed PubMed Central
John Peter, A.T., Herrmann, B., Antunes, D., Rapaport, D., Dimmer, K.S., and Kornmann, B. (2017). Vps13-Mcp1 interact at vacuole-mitochondria interfaces and bypass ER-mitochondria contact sites. J. Cell Biol. 216: 3219–3229, https://doi.org/10.1083/jcb.201610055.Search in Google Scholar PubMed PubMed Central
Joshi, A.S., Nebenfuehr, B., Choudhary, V., Satpute-Krishnan, P., Levine, T.P., Golden, A., and Prinz, W.A. (2018). Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat. Commun. 9: 2940, https://doi.org/10.1038/s41467-018-05277-3.Search in Google Scholar PubMed PubMed Central
Kakimoto, Y., Tashiro, S., Kojima, R., Morozumi, Y., Endo, T., and Tamura, Y. (2018). Visualizing multiple inter-organelle contact sites using the organelle-targeted split-GFP system. Sci. Rep. 8: 6175, https://doi.org/10.1038/s41598-018-24466-0.Search in Google Scholar PubMed PubMed Central
Klecker, T., Scholz, D., Fortsch, J., and Westermann, B. (2013). The yeast cell cortical protein Num1 integrates mitochondrial dynamics into cellular architecture. J. Cell Sci. 126, 2924–2930, https://doi.org/10.1242/jcs.126045.Search in Google Scholar PubMed
Kojima, R., Kakimoto, Y., Shinmyo, M., Kurokawa, K., Nakano, A., Endo, T., and Tamura, Y. (2019). A non-canonical unfolded protein response pathway and mitochondrial dynamics control the number of ER-mitochondria contact sites. BioRxiv: 684753, https://doi.org/10.1101/684753.Search in Google Scholar
Kornmann, B., Currie, E., Collins, S.R., Schuldiner, M., Nunnari, J., Weissman, J.S. and Walter, P. (2009). An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325: 477–481, https://doi.org/10.1126/science.1175088.Search in Google Scholar PubMed PubMed Central
Kornmann, B., Osman, C., and Walter, P. (2011). The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc. Natl. Acad. Sci. USA 108: 14151–14156, https://doi.org/10.1073/pnas.1111314108.Search in Google Scholar PubMed PubMed Central
Koyano, F., Okatsu, K., Kosako, H., Tamura, Y., Go, E., Kimura, M., Kimura, Y., Tsuchiya, H., and Yoshihara, H., Hirokawa, T., et al. (2014). Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510: 162–166, https://doi.org/10.1038/nature13392.Search in Google Scholar PubMed
Kraft, L.M. and Lackner, L.L. (2017). Mitochondria-driven assembly of a cortical anchor for mitochondria and dynein. J. Cell Biol. 216: 3061–3071, https://doi.org/10.1083/jcb.201702022.Search in Google Scholar PubMed PubMed Central
Krols, M., Asselbergh, B., De Rycke, R., De Winter, V., Seyer, A., Muller, F.J., Kurth, I., Bultynck, G., Timmerman, V., and Janssens, S. (2019). Sensory neuropathy-causing mutations in ATL3 affect ER-mitochondria contact sites and impair axonal mitochondrial distribution. Hum. Mol. Genet. 28: 615–627, https://doi.org/10.1093/hmg/ddy352.Search in Google Scholar PubMed PubMed Central
Kumar, N., Leonzino, M., Hancock-Cerutti, W., Horenkamp, F.A., Li, P., Lees, J.A., Wheeler, H., Reinisch, K.M., and De Camilli, P. (2018). VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217: 3625–3639, https://doi.org/10.1083/jcb.201807019.Search in Google Scholar PubMed PubMed Central
Kustatscher, G., Grabowski, P., Schrader, T.A., Passmore, J.B., Schrader, M., and Rappsilber, J. (2019). Co-regulation map of the human proteome enables identification of protein functions. Nat. Biotechnol. 37: 1361–1371, https://doi.org/10.1038/s41587-019-0298-5.Search in Google Scholar PubMed PubMed Central
Lackner, L.L., Ping, H., Graef, M., Murley, A., and Nunnari, J. (2013). Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc Natl Acad Sci USA 110: E458-E467, https://doi.org/10.1073/pnas.1215232110.Search in Google Scholar PubMed PubMed Central
Lackner, L.L. (2019). The expanding and unexpected functions of mitochondria contact sites. Trends Cell Biol. 29: 580–590, https://doi.org/10.1016/j.tcb.2019.02.009.Search in Google Scholar PubMed PubMed Central
Lang, A.B., John Peter, A.T., Walter, P., and Kornmann, B. (2015). ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol. 210: 883–890, https://doi.org/10.1083/jcb.201502105.Search in Google Scholar PubMed PubMed Central
Le Vasseur, M., Chen, V.C., Huang, K., Vogl, W.A., and Naus, C.C. (2019). Pannexin 2 localizes at ER-mitochondria contact sites. Cancers (Basel) 11, https://doi.org/10.3390/cancers11030343.Search in Google Scholar PubMed PubMed Central
Leal, N.S., Schreiner, B., Pinho, C.M., Filadi, R., Wiehager, B., Karlstrom, H., Pizzo, P., and Ankarcrona, M. (2016). Mitofusin-2 knockdown increases ER-mitochondria contact and decreases amyloid β-peptide production. J. Cell Mol. Med. 20: 1686–1695, https://doi.org/10.1111/jcmm.12863.Search in Google Scholar PubMed PubMed Central
Lee, S. and Min, K.T. (2018). The interface between ER and mitochondria: molecular compositions and functions. Mol. Cells 41: 1000–1007, https://doi.org/10.14348/molcells.2018.0438.Search in Google Scholar PubMed PubMed Central
Lee, J.E., Cathey, P.I., Wu, H., Parker, R., and Voeltz, G.K. (2020). Endoplasmic reticulum contact sites regulate the dynamics of membraneless organelles. Science 367, https://doi.org/10.1126/science.aay7108.Search in Google Scholar PubMed
Lesage, S., Drouet, V., Majounie, E., Deramecourt, V., Jacoupy, M., Nicolas, A., Cormier-Dequaire, F., Hassoun, S.M., Pujol, C., Ciura, S., et al. (2016). Loss of VPS13C function in autosomal-recessive Parkinsonism causes mitochondrial dysfunction and increases PINK1/Parkin-dependent mitophagy. Am. J. Hum. Genet. 98: 500–513, https://doi.org/10.1016/j.ajhg.2016.01.014.Search in Google Scholar PubMed PubMed Central
Lev, S., Ben Halevy, D., Peretti, D., and Dahan, N. (2008). The VAP protein family: from cellular functions to motor neuron disease. Trends Cell Biol. 18: 282–290, https://doi.org/10.1016/j.tcb.2008.03.006.Search in Google Scholar PubMed
Li, J., Zhang, D., Brundel, B., and Wiersma, M. (2019). Imbalance of ER and mitochondria interactions: prelude to cardiac ageing and disease? Cells 8: 1617, https://doi.org/10.3390/cells8121617.Search in Google Scholar PubMed PubMed Central
Liu, Y. and Zhu, X. (2017). Endoplasmic reticulum-mitochondria tethering in neurodegenerative diseases. Transl. Neurodegener. 6: 21, https://doi.org/10.1186/s40035-017-0092-6.Search in Google Scholar PubMed PubMed Central
Liu, X., Wen, X., and Klionsky, D.J. (2018). ER-mitochondria contacts are required for pexophagy in Saccharomyces cerevisiae. Contact (Thousand Oaks) 2, https://doi.org/10.1177/2515256418821584.Search in Google Scholar PubMed PubMed Central
Lopez-Crisosto, C., Bravo-Sagua, R., Rodriguez-Pena, M., Mera, C., Castro, P.F., Quest, A.F., Rothermel, B.A., Cifuentes, M., and Lavandero, S. (2015). ER-to-mitochondria miscommunication and metabolic diseases. Biochim. Biophys. Acta 1852: 2096–2105, https://doi.org/10.1016/j.bbadis.2015.07.011.Search in Google Scholar PubMed
Ma, W. and Mayr, C. (2018). A membraneless organelle associated with the endoplasmic reticulum enables 3’UTR-mediated protein-protein interactions. Cell 175: 1492–1506.e19, https://doi.org/10.1016/j.cell.2018.10.007.Search in Google Scholar PubMed PubMed Central
Ma, Y., Moors, A., Camougrand, N., and Dokudovskaya, S. (2019). The SEACIT complex is involved in the maintenance of vacuole-mitochondria contact sites and controls mitophagy. Cell. Mol. Life Sci. 76: 1623–1640, https://doi.org/10.1007/s00018-019-03015-6.Search in Google Scholar PubMed
Malhotra, J.D. and Kaufman, R.J. (2011). ER stress and its functional link to mitochondria: role in cell survival and death. Cold Spring Harb. Perspect. Biol. 3: a004424, https://doi.org/10.1101/cshperspect.a004424.Search in Google Scholar PubMed PubMed Central
Mao, K., Liu, X., Feng, Y., and Klionsky, D.J. (2014). The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy 10: 652–661, https://doi.org/10.4161/auto.27852.Search in Google Scholar PubMed PubMed Central
Marchi, S., Patergnani, S., and Pinton, P. (2014). The endoplasmic reticulum-mitochondria connection: one touch, multiple functions. Biochim. Biophys. Acta 1837: 461–469, https://doi.org/10.1016/j.bbabio.2013.10.015.Search in Google Scholar PubMed
Martinez-Guzman, O., M-Willoughby, M., Saini, A., V-Dietz, J., Bohovych, I., E-Medlock, A., Khalimonchuk, O., and R-Reddi, A. (2019). The heme biosynthetic enzyme, 5-aminolevulinic acid synthase (ALAS), and GTPases in control of mitochondrial dynamics and ER contact sites, regulate heme mobilization to the nucleus. bioRxiv: 734780, https://doi.org/10.1101/734780.Search in Google Scholar
Michiorri, S., Gelmetti, V., Giarda, E., Lombardi, F., Romano, F., Marongiu, R., Nerini-Molteni, S., Sale, P., Vago, R., Arena, G., et al. (2010). The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ. 17: 962–974, https://doi.org/10.1038/cdd.2009.200.Search in Google Scholar PubMed
Missiroli, S., Bonora, M., Patergnani, S., Poletti, F., Perrone, M., Gafa, R., Magri, E., Raimondi, A., Lanza, G., Tacchetti, C., et al. (2016). PML at mitochondria-associated membranes is critical for the repression of autophagy and cancer development. Cell Rep. 16: 2415–2427, https://doi.org/10.1016/j.celrep.2016.07.082.Search in Google Scholar PubMed PubMed Central
Moldavski, O., Amen, T., Levin-Zaidman, S., Eisenstein, M., Rogachev, I., Brandis, A., Kaganovich, D., and Schuldiner, M. (2015). Lipid droplets are essential for efficient clearance of cytosolic inclusion bodies. Dev. Cell 33: 603–610, https://doi.org/10.1016/j.devcel.2015.04.015.Search in Google Scholar PubMed
Moulis, M., Grousset, E., Faccini, J., Richetin, K., Thomas, G., and Vindis, C. (2019). The multifunctional sorting protein PACS-2 controls mitophagosome formation in human vascular smooth muscle cells through mitochondria-ER contact sites. Cells 8: 638, https://doi.org/10.3390/cells8060638.Search in Google Scholar PubMed PubMed Central
Muallem, S., Chung, W.Y., Jha, A., and Ahuja, M. (2017). Lipids at membrane contact sites: cell signaling and ion transport. EMBO Rep. 18: 1893–1904, https://doi.org/10.15252/embr.201744331.Search in Google Scholar PubMed PubMed Central
Murley, A., Sarsam, R.D., Toulmay, A., Yamada, J., Prinz, W.A., and Nunnari, J. (2015). Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J. Cell Biol. 209: 539–548, https://doi.org/10.1083/jcb.201502033.Search in Google Scholar PubMed PubMed Central
Namba, T. (2019). BAP31 regulates mitochondrial function via interaction with Tom40 within ER-mitochondria contact sites. Sci. Adv. 5: eaaw1386, https://doi.org/10.1126/sciadv.aaw1386.Search in Google Scholar PubMed PubMed Central
Naon, D., Zaninello, M., Giacomello, M., Varanita, T., Grespi, F., Lakshminaranayan, S., Serafini, A., Semenzato, M., Herkenne, S., Hernandez-Alvarez, M.I., et al. (2016). Critical reappraisal confirms that Mitofusin 2 is an endoplasmic reticulum-mitochondria tether. Proc. Natl. Acad. Sci. USA 113: 11249–11254, https://doi.org/10.1073/pnas.1606786113.Search in Google Scholar PubMed PubMed Central
Oikawa, K., Hayashi, M., Hayashi, Y., and Nishimura, M. (2019). Re-evaluation of physical interaction between plant peroxisomes and other organelles using live-cell imaging techniques. J. Integr. Plant Biol. 61, 836–852, https://doi.org/10.1111/jipb.12805.Search in Google Scholar PubMed
Paillusson, S., Stoica, R., Gomez-Suaga, P., Lau, D.H.W., Mueller, S., Miller, T., and Miller, C.C.J. (2016). There’s something wrong with my mam; the er-mitochondria axis and neurodegenerative diseases. Trends Neurosci. 39: 146–157, https://doi.org/10.1016/j.tins.2016.01.008.Search in Google Scholar PubMed PubMed Central
Papa, S., Rasmo, D.D., Technikova-Dobrova, Z., Panelli, D., Signorile, A., Scacco, S., Petruzzella, V., Papa, F., Palmisano, G., Gnoni, A., et al. (2012). Respiratory chain complex I, a main regulatory target of the cAMP/PKA pathway is defective in different human diseases. FEBS Lett. 586: 568–577, https://doi.org/10.1016/j.febslet.2011.09.019.Search in Google Scholar PubMed
Park, J.S., Thorsness, M.K., Policastro, R., McGoldrick, L.L., Hollingsworth, N.M., Thorsness, P.E. and Neiman, A.M. (2016). Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol. Biol. Cell 27: 2435–2449, https://doi.org/10.1091/mbc.E16-02-0112.Search in Google Scholar PubMed PubMed Central
Piel, R.B.3rd, Dailey, H.A.Jr. and Medlock, A.E. (2019). The mitochondrial heme metabolon: Insights into the complex(ity) of heme synthesis and distribution. Mol. Genet. Metab. 128: 198–203, https://doi.org/10.1016/j.ymgme.2019.01.006.Search in Google Scholar PubMed PubMed Central
Ping, H.A., Kraft, L.M., Chen, W., Nilles, A.E., and Lackner, L.L. (2016). Num1 anchors mitochondria to the plasma membrane via two domains with different lipid binding specificities. J Cell Biol. 213, 513–524, https://doi.org/10.1083/jcb.201511021.Search in Google Scholar PubMed PubMed Central
Ponka, P. (1997). Tissue-specific regulation of iron metabolism and heme synthesis: distinct control mechanisms in erythroid cells. Blood 89: 1–25, https://doi.org/10.1182/blood.v89.1.1.Search in Google Scholar
Prinz, W.A., Toulmay, A., and Balla, T. (2020). The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 21: 7–24, https://doi.org/10.1038/s41580-019-0180-9.Search in Google Scholar PubMed
Rizzuto, R., Pinton, P., Carrington, W., Fay, F.S., Fogarty, K.E., Lifshitz, L.M., Tuft, R.A., and Pozzan, T. (1998). Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280: 1763–1766, https://doi.org/10.1126/science.280.5370.1763.Search in Google Scholar PubMed
Rosati, E., Sabatini, R., Rampino, G., De Falco, F., Di Ianni, M., Falzetti, F., Fettucciari, K., Bartoli, A., Screpanti, I., and Marconi, P. (2010). Novel targets for endoplasmic reticulum stress-induced apoptosis in B-CLL. Blood 116: 2713–2723, https://doi.org/10.1182/blood-2010-03-275628.Search in Google Scholar PubMed
Scarffe, L.A., Stevens, D.A., Dawson, V.L., and Dawson, T.M. (2014). Parkin and PINK1: much more than mitophagy. Trends Neurosci. 37, 315–324, https://doi.org/10.1016/j.tins.2014.03.004.Search in Google Scholar PubMed PubMed Central
Scorrano, L., De Matteis, M.A., Emr, S., Giordano, F., Hajnoczky, G., Kornmann, B., Lackner, L.L., Levine, T.P., Pellegrini, L., Reinisch, K., et al. (2019). Coming together to define membrane contact sites. Nat. Commun. 10: 1287, https://doi.org/10.1038/s41467-019-09253-3.Search in Google Scholar PubMed PubMed Central
Shai, N., Yifrach, E., van Roermund, C.W.T., Cohen, N., Bibi, C., L, I.J., Cavellini, L., Meurisse, J., Schuster, R., Zada, L., et al. (2018). Systematic mapping of contact sites reveals tethers and a function for the peroxisome-mitochondria contact. Nat. Commun. 9: 1761, https://doi.org/10.1038/s41467-018-03957-8.Search in Google Scholar PubMed PubMed Central
Sosinsky, G.E., Boassa, D., Dermietzel, R., Duffy, H.S., Laird, D.W., MacVicar, B., Naus, C.C., Penuela, S., Scemes, E., Spray, D.C., et al. (2011). Pannexin channels are not gap junction hemichannels. Channels (Austin) 5, 193–197, https://doi.org/10.4161/chan.5.3.15765.Search in Google Scholar PubMed PubMed Central
Stehling, O. and Lill, R. (2013). The role of mitochondria in cellular iron-sulfur protein biogenesis: mechanisms, connected processes, and diseases. Cold Spring Harb. Perspect. Biol. 5: a011312, https://doi.org/10.1101/cshperspect.a011312.Search in Google Scholar PubMed PubMed Central
Subramanian, K., Jochem, A., Le Vasseur, M., Lewis, S., Paulson, B.R., Reddy, T.R., Russell, J.D., Coon, J.J., Pagliarini, D.J., and Nunnari, J. (2019). Coenzyme Q biosynthetic proteins assemble in a substrate-dependent manner into domains at ER-mitochondria contacts. J. Cell Biol. 218: 1353–1369, https://doi.org/10.1083/jcb.201808044.Search in Google Scholar PubMed PubMed Central
Takeda, K., Nagashima, S., Shiiba, I., Uda, A., Tokuyama, T., Ito, N., Fukuda, T., Matsushita, N., Ishido, S., Iwawaki, T., et al. (2019). MITOL prevents ER stress-induced apoptosis by IRE1α ubiquitylation at ER-mitochondria contact sites. EMBO J. 38: e100999, https://doi.org/10.15252/embj.2018100999.Search in Google Scholar PubMed PubMed Central
Tan, T., Ozbalci, C., Brugger, B., Rapaport, D., and Dimmer, K.S. (2013). Mcp1 and Mcp2, two novel proteins involved in mitochondrial lipid homeostasis. J. Cell Sci. 126: 3563–3574, https://doi.org/10.1242/jcs.121244.Search in Google Scholar PubMed
Tang, Z., Takahashi, Y., He, H., Hattori, T., Chen, C., Liang, X., Chen, H., Young, M.M. and Wang, H.G. (2019). TOM40 targets Atg2 to mitochondria-associated ER membranes for phagophore expansion. Cell Rep. 28, 1744-1757.e5, https://doi.org/10.1016/j.celrep.2019.07.036.Search in Google Scholar PubMed PubMed Central
Thoudam, T., Ha, C.M., Leem, J., Chanda, D., Park, J.S., Kim, H.J., Jeon, J.H., Choi, Y.K., Liangpunsakul, S., Huh, Y.H., et al. (2019). PDK4 augments ER-mitochondria contact to dampen skeletal muscle insulin signaling during obesity. Diabetes 68: 571–586, https://doi.org/10.2337/db18-0363.Search in Google Scholar PubMed PubMed Central
Till, A., Lakhani, R., Burnett, S.F., and Subramani, S. (2012). Pexophagy: the selective degradation of peroxisomes. Int. J. Cell Biol. 2012: 512721, https://doi.org/10.1155/2012/512721.Search in Google Scholar PubMed PubMed Central
Tran, U.C. and Clarke, C.F. (2007). Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 7: S62–S71, https://doi.org/10.1016/j.mito.2007.03.007.Search in Google Scholar PubMed PubMed Central
Usaj, M.M., Brloznik, M., Kaferle, P., Zitnik, M., Wolinski, H., Leitner, F., Kohlwein, S.D., Zupan, B., and Petrovic, U. (2015). Genome-wide localization study of yeast Pex11 identifies peroxisome-mitochondria interactions through the ERMES complex. J. Mol. Biol. 427: 2072–2087, https://doi.org/10.1016/j.jmb.2015.03.004.Search in Google Scholar PubMed PubMed Central
Valm, A.M., Cohen, S., Legant, W.R., Melunis, J., Hershberg, U., Wait, E., Cohen, A.R., Davidson, M.W., Betzig, E., and Lippincott-Schwartz, J. (2017). Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546: 162–167, https://doi.org/10.1038/nature22369.Search in Google Scholar PubMed PubMed Central
Vance, J.E. (1990). Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265: 7248–7256.10.1016/S0021-9258(19)39106-9Search in Google Scholar
Voloboueva, L.A., Duan, M., Ouyang, Y., Emery, J.F., Stoy, C., and Giffard, R.G. (2008). Overexpression of mitochondrial Hsp70/Hsp75 protects astrocytes against ischemic injury in vitro. J Cereb. Blood Flow Metab. 28: 1009–1016, https://doi.org/10.1038/sj.jcbfm.9600600.Search in Google Scholar PubMed PubMed Central
Wadhwa, R., Kaul, S.C., Mitsui, Y., and Sugimoto, Y. (1993). Differential subcellular distribution of mortalin in mortal and immortal mouse and human fibroblasts. Exp. Cell Res. 207: 442–448, https://doi.org/10.1006/excr.1993.1213.Search in Google Scholar PubMed
Walter, P. and Ron, D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334: 1081–1086, https://doi.org/10.1126/science.1209038.Search in Google Scholar PubMed
Westermann, B. (2008). Molecular machinery of mitochondrial fusion and fission. J. Biol. Chem. 283: 13501–13505, https://doi.org/10.1074/jbc.r800011200.Search in Google Scholar PubMed
Wong, Y.C., Ysselstein, D., and Krainc, D. (2018). Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554: 382–386, https://doi.org/10.1038/nature25486.Search in Google Scholar PubMed PubMed Central
Wu, W., Lin, C., Wu, K., Jiang, L., Wang, X., Li, W., Zhuang, H., Zhang, X., Chen, H., Li, S., et al. (2016). FUNDC1 regulates mitochondrial dynamics at the ER-mitochondrial contact site under hypoxic conditions. EMBO J. 35: 1368–1384, https://doi.org/10.15252/embj.201593102.Search in Google Scholar PubMed PubMed Central
Wu, H., Carvalho, P., and Voeltz, G.K. (2018). Here, there, and everywhere: the importance of ER membrane contact sites. Science 361: eaan5835, https://doi.org/10.1126/science.aan5835.Search in Google Scholar PubMed PubMed Central
Xue, Y., Schmollinger, S., Attar, N., Campos, O.A., Vogelauer, M., Carey, M.F., Merchant, S.S. and Kurdistani, S.K. (2017). Endoplasmic reticulum-mitochondria junction is required for iron homeostasis. J. Biol. Chem. 292: 13197–13204, https://doi.org/10.1074/jbc.m117.784249.Search in Google Scholar
Zhang, X., Gibhardt, C.S., Will, T., Stanisz, H., Korbel, C., Mitkovski, M., Stejerean, I., Cappello, S., Pacheu-Grau, D., Dudek, J., et al. (2019). Redox signals at the ER-mitochondria interface control melanoma progression. EMBO J. 38: e100871, https://doi.org/10.15252/embj.2018100871.Search in Google Scholar PubMed PubMed Central
© 2020 Walter de Gruyter GmbH, Berlin/Boston
Articles in the same Issue
- Frontmatter
- Highlight: In Honor of Walter Neupert: Mitochondria
- Editorial
- Mitochondria and friends – a special issue in honor of Walter Neupert (1939–2019)
- Early steps in mitochondrial protein translocation
- From cytosol to mitochondria: the beginning of a protein journey
- Evolution of mitochondrial protein import – lessons from trypanosomes
- Protein import: crossing the outer membrane
- Biogenesis pathways of α-helical mitochondrial outer membrane proteins
- The structure of the TOM core complex in the mitochondrial outer membrane
- Porins as helpers in mitochondrial protein translocation
- Protein translocation beyond the outer membrane
- From TOM to the TIM23 complex – handing over of a precursor
- How to get to the other side of the mitochondrial inner membrane – the protein import motor
- The biogenesis of mitochondrial intermembrane space proteins
- Protein import by the mitochondrial disulfide relay in higher eukaryotes
- Mitochondrial ultrastructure and dynamics
- The MICOS complex, a structural element of mitochondria with versatile functions
- Asymmetric inheritance of mitochondria in yeast
- Lipid transport and mitochondrial contact sites
- New horizons in mitochondrial contact site research
- The endoplasmic reticulum-mitochondria encounter structure: coordinating lipid metabolism across membranes
- Lipid homeostasis in mitochondria
- The biogenesis of enzymes
- Modular assembly of yeast mitochondrial ATP synthase and cytochrome oxidase
- From the discovery to molecular understanding of cellular iron-sulfur protein biogenesis
- Mitochondrial quality control
- Regulation of mitochondrial plasticity by the i-AAA protease YME1L
- PINK1 and Parkin: team players in stress-induced mitophagy
Articles in the same Issue
- Frontmatter
- Highlight: In Honor of Walter Neupert: Mitochondria
- Editorial
- Mitochondria and friends – a special issue in honor of Walter Neupert (1939–2019)
- Early steps in mitochondrial protein translocation
- From cytosol to mitochondria: the beginning of a protein journey
- Evolution of mitochondrial protein import – lessons from trypanosomes
- Protein import: crossing the outer membrane
- Biogenesis pathways of α-helical mitochondrial outer membrane proteins
- The structure of the TOM core complex in the mitochondrial outer membrane
- Porins as helpers in mitochondrial protein translocation
- Protein translocation beyond the outer membrane
- From TOM to the TIM23 complex – handing over of a precursor
- How to get to the other side of the mitochondrial inner membrane – the protein import motor
- The biogenesis of mitochondrial intermembrane space proteins
- Protein import by the mitochondrial disulfide relay in higher eukaryotes
- Mitochondrial ultrastructure and dynamics
- The MICOS complex, a structural element of mitochondria with versatile functions
- Asymmetric inheritance of mitochondria in yeast
- Lipid transport and mitochondrial contact sites
- New horizons in mitochondrial contact site research
- The endoplasmic reticulum-mitochondria encounter structure: coordinating lipid metabolism across membranes
- Lipid homeostasis in mitochondria
- The biogenesis of enzymes
- Modular assembly of yeast mitochondrial ATP synthase and cytochrome oxidase
- From the discovery to molecular understanding of cellular iron-sulfur protein biogenesis
- Mitochondrial quality control
- Regulation of mitochondrial plasticity by the i-AAA protease YME1L
- PINK1 and Parkin: team players in stress-induced mitophagy

