Abstract
The synthesis of a biotinylated bivalent zanamivir analog as a probe for influenza viruses is reported. The compound was used in a ‘glycan’ based sandwich assay; where glycans were immobilized on glass slides to capture strains of influenza A H1N1, A/Brisbane/59/2007 virus; the biotinylated bivalent zanamivir analog-labeled streptavidin complex was used as reporter. This research strongly suggests that glycans can be used for capturing and reporting influenza viruses and the biotinylated compounds can be used as probes for capturing and isolating influenza viruses from complex mixtures.
Introduction
Influenza virus, a highly transmissible respiratory pathogen, infects several millions of people annually. Influenza-related deaths ranked 8th among the leading causes of death in the United States in 2014 according to the Centers for Disease Control and Prevention, Atlanta, GA, USA [1], [2]. While seasonal influenza can and is damaging, emerging strains can cause widespread and significant economic harm with entire school systems forced to shut down during a pandemic [3]. Vaccines are highly recommended for the most vulnerable, however, the effectiveness of vaccines in any given season varies widely; in most years it is ≤50% effective [4]. Moreover, it is logistically not possible to vaccinate all vulnerable populations. Antivirals that include tamiflu and relenza represent an alternate countermeasure to counter the harmful effects of the virus, but they must be provided to patients 24–48 h before onset of infection for optimal efficacy [5]. A third and important countermeasure is precise and rapid diagnostics to detect the virus because early detection can be used to quarantine patients to arrest the spread of a rapidly evolving and transmissible strain.
Diagnostics for influenza virus include traditional plaque assays [6], rRT-PCR [7] and antibody based assays [8], [9]. There are advantages and disadvantages to all these assays; cell culture methods require trained personnel and a laboratory equipment, rRT-PCR methods such as the recently approved alere™ influenza tests are expensive and antibody based tests are not preferred as they are not very sensitive [9], [10], [11]. We recently reported an alternative diagnostic method where the surface glycoproteins are targeted [12], [13], [14], [15]. Briefly, influenza virus has two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA) that play an important role in the viral lifecycle. HA binds to a nine-carbon glycan, N-acetylneuraminic acid or sialic acid, present on the termini of glycolipids and glycoproteins of the epithelial cells of the respiratory tract [16], [17]. This is followed by cell entry and infection of the host cell. After utilizing the host cell machinery for virus propagation, the progeny escapes the host cell to infect neighboring cells and tissues. Interestingly, sialic acid present on the infected host cell prevents the viral progeny from escaping. Mucins containing sialic acid also inhibit the virus from propagating [18]. The enzyme NA cleaves sialic acid on the infected host cell and mucins freeing viral progeny to infect other cells. We choose to target these glycoproteins for three reasons: First, they are vital proteins of the virus requiring sialic acids. Second, there are approximately 300–400 copies of HA and 50–100 copies of NA on a single viral particle. HA exists as a trimer and NA as a tetramer, and a substantial number of copies on a single virion provides an adequate number of protein targets for synthetic or natural sialic acids to capture [19], [20]. Third, this receptor based approach is unique in comparison to antibody based diagnostics. All strains of influenza virus are expected to bind to sialic acid analogs and each strain would yield a ‘fingerprint’ pattern of recognition that can be used to detect the virus [14], [15]. In previous studies, we developed microarrays comprised on synthetic sialic acid analogs that could bind to HA and/or NA. After exposure of the focused microarray to different virus strains, a fluorescently labeled antibody was used as the readout [14], [15] (Figure 1A). These studies were important to establish proof of principle that sialic acid analogs could be used to detect influenza viruses; however, a limitation of the studies was that antibody based reporters are required. Following a report by Wong and coworkers [21], we now report the synthesis, characterization and initial binding studies of ‘universal’ glycan based reporter molecules that could bind to all virus strains. Since the capture and reporter molecules are made of glycans, antibodies are not required (Figure 1B).
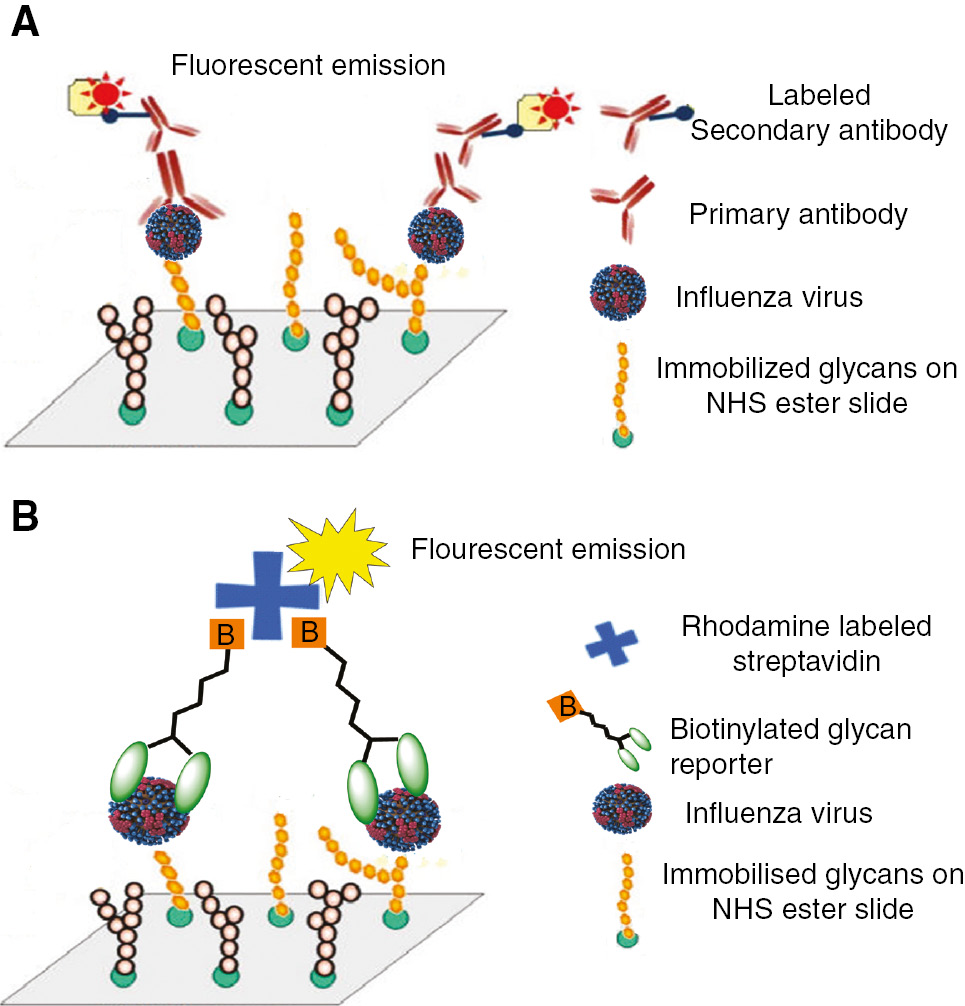
(A) Cartoon representation of the sandwich assay using glycans as capture and labeled antibody as reporters. (B) Cartoon representation of the sandwich assay using glycans as capture and biotinylated glycan-labeled streptavidin as reporters.
Results and discussion
The design and synthesis of the universal reporter is shown in Scheme 1 and it follows our previous publications [15]. We choose to use zanamivir analogs as the glycan headgroups as the binding affinity of zanamivir to NA is in the nanomolar range. The active site of influenza NA is also highly conserved and, furthermore, resistance to zanamivir is not as high as to oseltamivir. In fact, very few cases of zanamivir resistance has been reported [22]. We also choose to develop a dimeric scaffold as dimers improve the avidity effect [23]. We designed the molecule in which two zanamivir analogs fit into two active sites of a NA tetramer or two adjacent NA tetramers [24]. We attached biotin at the opposite end of the scaffold because biotin can bind to fluorescently labeled avidin yielding the desired reporter. The final construct is expected to possess eight zanamivir analogs as one avidin binds to four biotin molecules. The azido compound 1 and the scaffold 2 were synthesized as described previously. 1,3 Dipolar cycloaddition [25] reaction under standard conditions was performed to yield the biotinylated bivalent compound 3 in 60% yield. Deprotection of the tert-butoxycarbonyl and methyl ester groups resulted in the desired reporter molecule AD-R. This compound was purified using size exclusion Biogel P2 columns to yield a foamy solid. All intermediates and the final compound were characterized using NMR and MS.
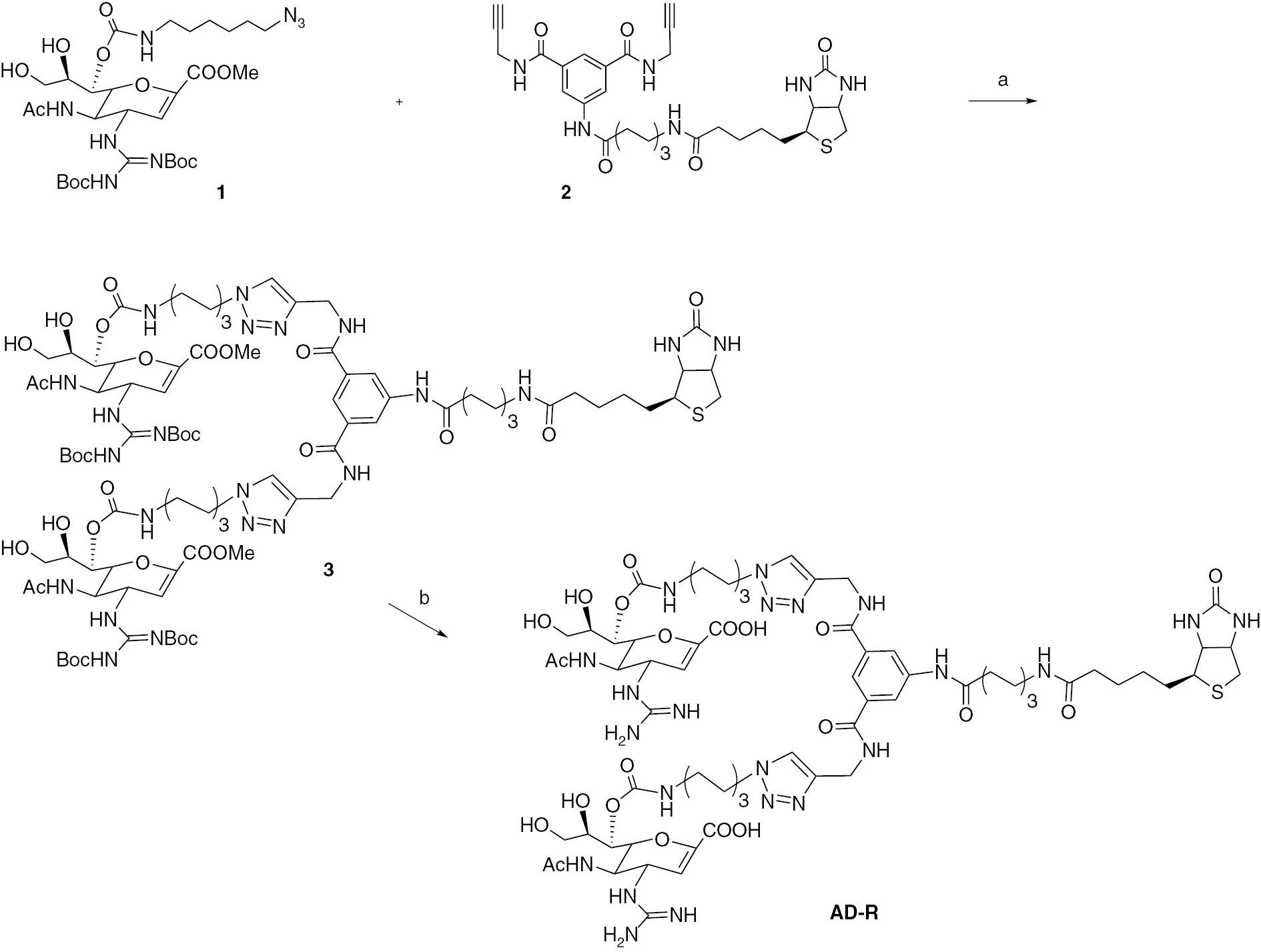
Reagents and conditions: (a) tert-BuOH:H20 (7:3), CuSO4, sodium L-ascorbate, rt, 36 h, yield 60%; (b) 50mm NaOH, rt, 2 h, then TFA:DC (1:1), rt 1 h, yield 80% (over two steps).
Next, we used the biotinylated compound as a reporter in microarray studies. We printed four bivalent compounds on glass slides for these exploratory studies [14], [15] (Figure 2). These compounds have been shown in our previous studies to capture different strains of influenza viruses [15]. After incubating with Influenza A H1N1, A/Brisbane/59/2007 virus at rt for 1 h, the slides were washed extensively, and exposed to AD-R. This step was followed by extensive washing and addition of labeled streptavidin. While we choose a two-step procedure of adding the biotinylated compound followed by the labeled streptavidin, an alternate method of premixing one equivalent of the avidin with four equivalents of the biotinylated compound could also be used with the caveat that the latter method requires purification of the avidin-biotin complex. The slides were scanned using a Genepix400B scanner at 557 nm. It was gratifying to observe that all four printed compounds capture the virus when the labeled streptavidin-AD-R conjugate is used as the reporter; non-specific binding to PEG is negligible (Figure 3). The increase in relative fluorescence units (RFU) from background is not as high in comparison with antibody based reporters [15], presumably because capture and reporter ligands that primarily bind to NA are used. The number of NAs on a single virion is not as high as the number of HAs [19], [20]. Secondly, NAs have been shown to be present in clusters or scattered on the virus surface [19], [20] which may limit the number of available sites for the reporter to bind when the virus is captured by the glycans on the glass slides. This may lead to a lower RFU value. We plan to develop the use of ligands that capture HA, which should give a higher RFU value. It was also determined that the limit of detection of this virus is 104 PFU (plaque forming units).
Structures of the four glycans immobilized on glass slides for capturing influenza virus.
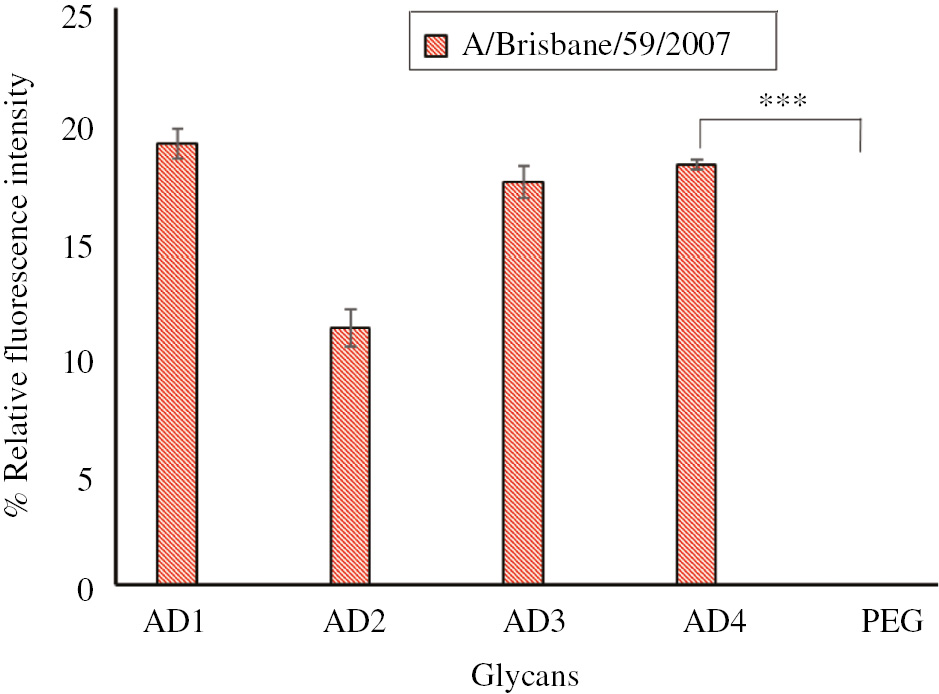
Influenza virus binding studies.
Fluorescence detection of H1N1 (A/Brisbane/59/2007) influenza A virus using synthetic glycans as capture and biotinylated glycan-label streptavidin as reporter. Glycans and PEG (negative control) were printed at 200 μm. Virus concentration was 105 PFU. Fluorescence intensity was measured by the Genepix400B scanner using tetramethylrhodamine (TRITC) labeled streptavidin and scanned at 557 nm. All experiments were performed in triplicate (*p<0.05, **p<0.01, ***p<0.001, ns>0.05).
Conclusions
A biotinylated bivalent zanamivir analog for the specific capture of influenza viruses was synthesized. This molecule was used as a reporter in the microarray studies and it was demonstrated that this compound could also be used as a reporter in a sandwich assay with an influenza A H1N1 virus strain. The biotinylated molecule could be used as a probe to capture influenza viruses as the biotin can readily be attached to streptavidin coated magnetic beads [26], nanoparticles [27] or surfaces [23].
Experimental
All reactions were performed under argon atmosphere with dried solvents. All chemical reagents were of analytical grade and used as supplied. Tetramethylrhodamine (TRITC) labeled streptavidin was purchased from Thermo Scientific as a lyophilized powder. Compounds AD1-4 were synthesized as described previously [15]. The acidic ion exchange resin used was Amberlite® IR 120 (H+) resin. Analytical thin layer chromatography (TLC) was performed on silica gel 230–400 mesh (Sicicycle). Plates were visualized under UV light, and/or by staining with CeH8Mo3N2O12 followed by heating. Column chromatography was performed on silica gel (230–400 mesh). 1H NMR (400 MHz) and 13C NMR (100 MHz) spectra were recorded on a Bruker 400 spectrometer. Electrospray ionization mass spectra were recorded on a Micromass Q\T 2 (Waters) and data were analyzed with MassLynx® 4.0 (Waters) software. Yields refer to spectroscopically and chromatographically pure compounds that were dried under reduced pressure (10−2 mbar).
Synthesis of compound 3
A mixture of compound 1 (0.03 g, 0.05 mmol), 2 (0.01 g, 0.02 mmol), CuSO4 (4.6 mg, 0.03 mmol) and sodium L-ascorbate (7.3 mg, 0.02 mmol) in t-BuOH/H2O (1.0 mL, 7:3) was stirred at rt for 12 h with progress of the reaction monitored by TLC. Upon completion, solvent was removed under reduced pressure and the residue was purified by column chromatography eluting with dichloromethane/methanol (10:1) to yield 60 mg (60%) of 3: 1H NMR (CDCl3): δ 11.40 (s, 2H), 9.74 (s, 1H), 8.62 (s,1H), 8.48 (d, J=4.0 Hz, 2H), 8.20 (s, 1H), 8.05 (s, 2H), 7.80 (s, 1H), 7.39 (s, 2H), 6.90 (s, 1H), 6.52 (m, 3H), 5.90 (s, 2H), 5.42–5.33 (m, 3H), 5.14 (s, 1H), 5.10–5.08 (m, 1H), 5.02–4.99 (m, 1H), 4.75–4.51 (m, 9H), 4.38–4.22 (m, 10H), 4.03–4.01 (m, 3H), 3.77 (s, 6H), 3.70–3.66 (m, 3H), 3.58–3.50 (m, 3H), 3.37 (s, 1H), 3.20–3.05 (m, 6H), 2.99–2.84 (m, 5H), 2.33–2.15 (m, 16 H), 1.92–1.78 (m, 10H), 1.66–1.59 (m, 8H), 1.47 (s, 18H), 1.27 (s, 18H); 13C NMR (CDCl3): δ 170.9, 166.9, 164.2, 164.1, 163.0, 162.5, 156.9, 152.6, 144.9, 139.4, 122.9, 110.6, 100.1, 83.6, 79.6, 73.9, 73.0, 71.7, 69.9, 68.6, 62.6, 60.4, 55.9, 52.5, 50.1, 46.9, 31.9, 29.7, 29.7, 29.8, 29.6, 29.4, 29.1, 28.9, 28.3, 28.0, 25.6, 25.6, 25.5, 22.9, 22.7, 14.1, 1.0. ESI-HRMS. Calcd for C91H140N22O29S: m/z 2038.29. Found: m/z 1020.00 (m=M++2H; z=2).
Synthesis of compound AD-R
A solution of compound 3 (0.01 g, 0.004 mmol) in dichloromethane/trifluoroacetic acid (1 mL, 1:1) was stirred at rt for 1 h, then neutralized with H+ DOWEX 50WX8-200 ion-exchange resin to pH 7. The resin was filtered, washed with methanol and the solution was concentrated under reduced pressure. The solid residue was re-dissolved in methanol, the solution was treated with NaOH (50 mm, 1 mL) and the mixture was stirred at rt for 2 h. After neutralization with H+ DOWEX 50WX8-200 ion-exchange resin, the resin was filtered and the product was purified with Bio-Gel P-2 Gel eluting with deionized water to give 5.2 mg (80%) of AD-R: 1H NMR (D2O): δ 7.91–7.85 (m, 5H), 5.55–5.51 (m, 2H), 4.79–4.77 (m, 4H), 4.57 (s, 3H), 4.41–4.40 (m, 1H), 4.39 (d, J=8 Hz, 1H), 4.36–4.35 (m, 1H), 4.31–4.28 (m, 5H), 4.03–3.90 (m, 4H), 3.61 (s, 3H), 3.54–3.44 (m, 3H), 3.36–3.34 (m, 1H), 3.08–2.99 (m,3H), 2.88–2.86 (m, 3H), 2.63 (s, 1H), 2.30–2.26 (m, 2H), 2.12–2.06 (m, 2H), 2.01–1.99 (m, 1H) 1.90 (s, 1H), 1.84 (s, 6H), 1.77–1.73 (m, 2H), 1.67–1.64 (m, 2H), 1.55–1.53 (m, 2H), 1.48–1.45 (m, 2H), 1.39–1.37 (m, 2H), 1.31–1.29 (m, 4H), 1.23–1.20 (m, 6H), 1.15–1.12 (m, 6H), 0.80–0.78 (m,2H); 13C NMR (D2O): δ 175.8, 171.2, 168.4, 163.5, 163.2, 162.9, 156.2, 135.8, 134.9, 129.3, 123.9, 122.5, 115.3, 110.0, 104.8, 100.0, 78.7, 75.2, 71.9, 69.6, 68.5, 62.5, 51.8, 50.0,47.3, 35.0, 29.1, 28.4, 28.0, 27.8, 27.5, 26.0, 25.6, 25.1, 21.8, 20.0. ESI-HRMS. Calcd for C69H104N22O21S: m/z 1609.7639. Found: m/z 1610.7600 (M++H).
Biological assays
Immobilization of glycans
Synthetic glycans were covalently immobilized onto Nexterion NHS slides using a DIGILAB OmniGrid Micro printer in 300 mm phosphate buffer with 0.005% Tween-20 at pH 8.5 [15]. Each glycan was printed 20 times in quintuplicate at 200 μm concentration. Following printing, the glycans were allowed to react for 30 min at 60% humidity. After overnight desiccation, the slides were blocked for 60 min with 50 mm ethanolamine in 50 mm boric acid buffer (pH 9.5), washed 3 times with deionized water, dried and stored at −20°C.
Binding studies
A serial 10-fold dilution of A/Brisbane/59/2007, starting with a concentration of 2.4×106 plaque forming units (PFU) was prepared in a buffer consisting of PBS, 2% BSA and 0.05% Tween-20. A sample of 200 μL of this solution was applied to the microarray for 60 min. Following virus incubation and wash (3 times with PBS and 0.05% Tween-20 and 2 times with PBS), 200 μL of 20 mm of AD-reporter was applied to the microarray for 60 min in the same buffer. TRITC labelled streptavidin (1 mg) was re-suspended into 1 mL of the buffer. A 20 000-fold dilution of the fluorescently labelled streptavidin was applied to the microarray after the slides were washed extensively and incubated for 60 min. The slides were washed followed by a rinse with deionized water (3 times). The slides were dried and scanned using a GenePix4000B scanner at a wavelength of 557 nm. All experiments were performed in triplicate.
Acknowledgments
We are grateful NIH-NIAID (R01-AI089450) for funding and thank BEI Resources, NIAID, NIH for the viral strains. We also thank Dr. Binghe Wang, GSU for use of the microarray printing facilities.
References
[1] Heron, M. Deaths: leading causes for 2013. Natl. Vital Stat. Rep.2016, 65, 1–95.Search in Google Scholar
[2] Short, K. R.; Habets, M. N.; Hermans, P. W. M.; Diavatopoulos, D. A. Interactions between Streptococcus pneumoniae and influenza virus: a mutually beneficial relationship. Future Microbiol.2012, 7, 609–624.10.2217/fmb.12.29Search in Google Scholar
[3] Cauchemez, S.; Van Kerkhove, M. D.; Archer, B. N.; Cetron, M.; Cowling, B. J.; Grove, P.; Hunt, D.; Kojouharova, M.; Kon, P.; Ungchusak, K.; et al. School closures during the 2009 influenza pandemic: national and local experiences. BMC Infect. Dis.2014, 14, 207.10.1186/1471-2334-14-207Search in Google Scholar
[4] Osterholm, M. T.; Kelley, N. S.; Sommer, A.; Belongia, E. A. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect. Dis.2012, 12, 36–44.10.1016/S1473-3099(11)70295-XSearch in Google Scholar
[5] Fiore, A. E.; Fry, A.; Shay, D.; Gubareva, L.; Bresee, J. S.; Uyeki, T. M. Centers for Disease Control and Prevention (CDC). Antiviral agents for the treatment and chemoprophylaxis of influenza – recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm. Rep.2011, 60, 1–24.Search in Google Scholar
[6] Baer, A.; Kehn-Hall, K. Viral concentration determination through plaque assays: using traditional and novel overlay systems. J. Vis. Exp.2014, e52065.10.3791/52065Search in Google Scholar PubMed PubMed Central
[7] Novak-Weekley, S.; Marlowe, E.; Poulter, M.; Dwyer, D.; Speers, D.; Rawlinson, W.; Baleriola, C.; Robinson, C. Evaluation of the Cepheid® Xpert® Flu Assay for rapid identification and differentiation of influenza A, influenza A 2009 H1N1, and influenza B. J. Clin. Microbiol.2012, 50, 1704–1710.10.1128/JCM.06520-11Search in Google Scholar PubMed PubMed Central
[8] Hurt, A. C.; Alexander, R.; Hibbert, J.; Deed, N.; Barr, I. G. Performance of six influenza rapid tests in detecting human influenza in clinical specimens. J. Clin. Virol.2007, 39, 132–135.10.1016/j.jcv.2007.03.002Search in Google Scholar PubMed
[9] Smit, M.; Beynon, K. A.; Murdoch, D. R.; Jennings, L. C. Comparison of the NOW Influenza A & B, NOW Flu A, NOW Flu B, and Directigen Flu A+B assays, and immunofluorescence with viral culture for the detection of influenza A and B viruses. Diagn. Microbiol. Infect. Dis.2007, 57, 67–70.10.1016/j.diagmicrobio.2006.11.003Search in Google Scholar PubMed
[10] Nie, S.; Roth, R. B.; Stiles, J.; Mikhlina, A.; Lu, X.; Tang, Y.-W.; Babady, N. E. Evaluation of Alere i Influenza A&B for rapid detection of influenza viruses A and B. J. Clin. Microbiol.2014, 52, 3339–3344.10.1128/JCM.01132-14Search in Google Scholar PubMed PubMed Central
[11] Bell, J.; Bonner, A.; Cohen, D. M.; Birkhahn, R.; Yogev, R.; Triner, W.; Cohen, J.; Palavecino, E.; Selvarangan, R. Multicenter clinical evaluation of the novel Alere™ i Influenza A&B isothermal nucleic acid amplification test. J. Clin. Virol.2014, 61, 81–86.10.1016/j.jcv.2014.06.001Search in Google Scholar PubMed
[12] Cui, X.; Das, A.; Dhawane, A. N.; Sweeney, J.; Zhang, X.; Chivukula, V.; Iyer, S. S. Highly specific and rapid glycan based amperometric detection of influenza viruses. Chem. Sci.2017, 8, 3628.10.1039/C6SC03720HSearch in Google Scholar
[13] Zhang, X.; Dhawane, A. N.; Sweeney, J.; He, Y.; Vasireddi, M.; Iyer, S. S. Electrochemical assay to detect influenza viruses and measure drug susceptibility. Angew. Chem. Int. Edit. Engl.2015, 54, 5929–5932.10.1002/anie.201412164Search in Google Scholar PubMed PubMed Central
[14] He, Y.; Yang, Y.; Iyer, S. S. Neuraminidase resistant sialosides for the detection of influenza viruses. Bioconjug. Chem.2016, 27, 1509–1517.10.1021/acs.bioconjchem.6b00150Search in Google Scholar PubMed
[15] Dinh, H.; Zhang, X.; Sweeney, J.; Yang, Y.; He, Y.; Dhawane, A.; Iyer, S. S. Glycan based detection and drug susceptibility of influenza virus. Anal. Chem.2014, 86, 8238–8244.10.1021/ac501624vSearch in Google Scholar PubMed PubMed Central
[16] von Itzstein, M. The war against influenza: discovery and development of sialidase inhibitors. Nat. Rev. Drug Discov.2007, 6, 967–974.10.1038/nrd2400Search in Google Scholar PubMed
[17] Kulkarni, A. A.; Weiss, A. A.; Iyer, S. S. Glycan-based high-affinity ligands for toxins and pathogen receptors. Med. Res. Rev.2010, 30, 327–393.10.1002/med.20196Search in Google Scholar PubMed
[18] Yang, X.; Steukers, L.; Forier, K.; Xiong, R.; Braeckmans, K.; Van Reeth, K.; Nauwynck, H. A beneficiary role for neuraminidase in influenza virus penetration through the respiratory mucus. PLoS One2014, 9, e110026.10.1371/journal.pone.0110026Search in Google Scholar PubMed PubMed Central
[19] Harris, A.; Cardone, G.; Winkler, D. C.; Heymann, J. B.; Brecher, M.; White, J. M.; Steven, A. C. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sc.i USA2006, 103, 19123–19127.10.1073/pnas.0607614103Search in Google Scholar PubMed PubMed Central
[20] Wasilewski, S.; Calder, L. J.; Grant, T.; Rosenthal, P. B. Distribution of surface glycoproteins on influenza A virus determined by electron cryotomography. Vaccine2012, 30, 7368–7373.10.1016/j.vaccine.2012.09.082Search in Google Scholar PubMed PubMed Central
[21] Cheng, T. J.; Wang, S. Y.; Wen, W. H.; Su, C. Y.; Lin, M.; Huang, W. I.; Liu, M. T.; Wu, H. S.; Wang, N. S.; Cheng, C. K.; et al. Chemical probes for drug-resistance assessment by binding competition (RABC): oseltamivir susceptibility evaluation. Angew. Chem. In.t Ed. Engl.2013, 52, 366–370.10.1002/anie.201204062Search in Google Scholar PubMed
[22] Thorlund, K.; Awad, T.; Boivin, G.; Thabane, L. Systematic review of influenza resistance to the neuraminidase inhibitors. BMC Infect. Dis.2011, 11, 134.10.1186/1471-2334-11-134Search in Google Scholar PubMed PubMed Central
[23] Dhawane, A. N.; Diez-Valcarce, M.; Gurale, B. P.; Dinh, H.; Vinje, J.; Iyer, S. S. Synthesis and evaluation of biotinylated bivalent histoblood group antigens for capturing human noroviruses. Bioconjug Chem.2016, 27, 1822–1829.10.1021/acs.bioconjchem.6b00226Search in Google Scholar
[24] Yang, Y.; He, Y.; Li, X.; Dinh, H.; Iyer, S. S. Bifunctional thiosialosides inhibit influenza virus. Bioorg. Med. Chem. Lett.2014, 24, 636–643.10.1016/j.bmcl.2013.11.077Search in Google Scholar
[25] Rostovtsev, V. V.; Green, L. G.; Fokin, V. V.; Sharpless, K. B. A stepwise huisgen cycloaddition process: copper (I)-catalyzed regioselective ‘ligation’ of azides and terminal alkynes. Angew. Chem.2002, 114, 2708–2711.10.1002/1521-3757(20020715)114:14<2708::AID-ANGE2708>3.0.CO;2-0Search in Google Scholar
[26] Hatch, D. M.; Weiss, A. A.; Kale, R. R.; Iyer, S. S. Biotinylated bi- and tetra-antennary glycoconjugates for Escherichia coli detection. ChemBioChem2008, 9, 2433–2442.10.1002/cbic.200800188Search in Google Scholar
[27] Garcia, I.; Marradi, M.; Penades, S. Glyconanoparticles: multifunctional nanomaterials for biomedical applications. Nanomedicine (London)2010, 5, 777–792.10.2217/nnm.10.48Search in Google Scholar
©2017 Walter de Gruyter GmbH, Berlin/Boston
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Articles in the same Issue
- Frontmatter
- Editorial
- Carbohydrate chemistry/glycoscience
- Reviews
- Boron-based small molecules in disease detection and treatment (2013–2016)
- Impact of modified ribose sugars on nucleic acid conformation and function
- Preliminary Communication
- Crystallization-induced amide bond formation creates a boron-centered spirocyclic system
- Research Articles
- Anion and sugar recognition by 2,6-pyridinedicarboxamide bis-boronic acid derivatives
- Synthesis of biotinylated bivalent zanamivir analogs as probes for influenza viruses
- Synthesis of silodosin glucuronide and its deuterated counterpart: solving a problematic O-glycosylation of a nitrogen-containing molecule
- Synthesis and antimicrobial activity of 4-trifluoromethylpyridine nucleosides
- Synthesis of anti-inflammatory 2,3-unsaturated O-glycosides using conventional and microwave heating techniques
- Synthesis of various β-D-glucopyranosyl and β-D-xylopyranosyl hydroxybenzoates and evaluation of their antioxidant activities
- Synthesis of carbohydrate-substituted isoxazoles and evaluation of their antitubercular activity
- Synthesis and bioactivity of novel C2-glycosyl triazole derivatives as acetylcholinesterase inhibitors
- Modification of bovine serum albumin with aminophenylboronic acid as glycan sensor based on surface plasmon resonance and isothermal titration calorimetry
Articles in the same Issue
- Frontmatter
- Editorial
- Carbohydrate chemistry/glycoscience
- Reviews
- Boron-based small molecules in disease detection and treatment (2013–2016)
- Impact of modified ribose sugars on nucleic acid conformation and function
- Preliminary Communication
- Crystallization-induced amide bond formation creates a boron-centered spirocyclic system
- Research Articles
- Anion and sugar recognition by 2,6-pyridinedicarboxamide bis-boronic acid derivatives
- Synthesis of biotinylated bivalent zanamivir analogs as probes for influenza viruses
- Synthesis of silodosin glucuronide and its deuterated counterpart: solving a problematic O-glycosylation of a nitrogen-containing molecule
- Synthesis and antimicrobial activity of 4-trifluoromethylpyridine nucleosides
- Synthesis of anti-inflammatory 2,3-unsaturated O-glycosides using conventional and microwave heating techniques
- Synthesis of various β-D-glucopyranosyl and β-D-xylopyranosyl hydroxybenzoates and evaluation of their antioxidant activities
- Synthesis of carbohydrate-substituted isoxazoles and evaluation of their antitubercular activity
- Synthesis and bioactivity of novel C2-glycosyl triazole derivatives as acetylcholinesterase inhibitors
- Modification of bovine serum albumin with aminophenylboronic acid as glycan sensor based on surface plasmon resonance and isothermal titration calorimetry

