Abstract
Objective
The aim of this study was to identify biomarkers associated with immunity and prognosis in patients with cervical cancer.
Materials and methods
Data from patients with cervical squamous cell carcinoma (CESC) were retrieved from the UCSC Xena database and subjected to analysis. Gene sets representing 22 types of immunocytes were acquired, and immunocytes relevant to prognosis were identified. Weighted gene co-expression network analysis (WGCNA) was utilized to identify gene modules associated with prognosis-related immunocytes and to construct immune-related gene markers. Differentially expressed genes were then screened, and the association between immune score and biological function of immune-related gene markers was analyzed. Furthermore, tissue samples from cervical cancer patients in Northeast China were collected to validate the expression of two genes using real-time PCR and immunohistochemistry.
Results
This study identified 10 immunocytes significantly correlated with overall survival time in patients. Six gene modules were identified as significantly associated with prognosis-related immunocytes, with gene module 6 showing relevance to all prognosis-related immunocytes. Gene module 6 was related to all prognosis-related immunocytes. Moreover, two genes (including PLA2G2D and CHIT1) were found to be significantly associated with overall survival in cancer patients. Patients with CESC were classified into high and low immune score groups based on the median score of gene markers. Correlation analysis of the immune score and biological function was performed. Immunohistochemistry and real-time PCR results revealed high expression of CHIT1 and PLA2G2D in CESC tumor tissues.
Conclusion
PLA2G2D and CHIT1 show promise as biomarkers for evaluating immune infiltration and prognosis in patients with cervical cancer.
1 Introduction
Cervical cancer ranks fourth among female malignant tumors worldwide, following breast cancer, colorectal cancer, and lung cancer. Every year, more than 500 thousand women are diagnosed with cervical cancer around the world, with over 300 thousand deaths[1]. The incidence of cervical cancer is highest among women aged 40 to 50, with another peak occurring between 60 and 70 years old. It is rare among women under 20 years old. Cervical cancer primarily comprises cervical squamous cell carcinoma, adenocarcinoma, adenosquamous carcinoma, and a few other types, with squamous cell carcinoma being the predominant histological type[2].
Immunocytes, also known as immune cells, are cells involved in or related to immune response, constituting a crucial component of the tumor microenvironment. They possess dual functions within this environment, capable of either inhibiting tumor formation or promoting tumor occurrence. In terms of tumor immunity, cytotoxic T cells serve as vital effector cells in anti-tumor immunity[3]. CD4+ T cells exhibit the capacity to differentiate into numerous subtypes and collaborate in a wide array of immune responses, including possessing a cytotoxicity program that enables direct killing of tumor cells[4]. CD8+ T cells play an important role in tumor immunomodulation, with the diminished response of tumor-specific CD8+ T cells representing the primary mechanism of tumor evasion[5]. B cells contribute by presenting antigens to both CD4+ and CD8+ T cells, thereby forming a tumor microenvironment characterized by antigen-specific immune responses[6]. NK cells, meanwhile, demonstrate the ability to recognize tumor cells at an early stage and subsequently lyse them upon activation[7]. Tumor immunotherapy aims to stimulate the body’s anti-tumor immune response.
The functions of the immune system are intricately tied to T cells, B cells, and natural killer cells. Decreased levels of CD4+ and CD8+ T cells can impair immune function, potentially leading to the failure to timely clear infected cells, which may contribute to the development of cervical cancer[8]. Conversely, an increase in CD4+ and CD8+ T cell signaling has been closely associated with the efficacy of neoadjuvant chemotherapy[9]. B cells play a crucial role in immune protection against cervical cancer by effectively eliminating invading human papillomavirus[10]. Utilizing TCGA gene expression data, we identified and constructed immune-related prognostic markers for cervical cancer that significantly predict longer overall survival in patients. In addition, we conducted an analysis to discern differences in immune infiltration scores and biological functions between groups with high and low immune prognosis scores.
2 Materials and Methods
2.1 Data preparation and preprocessing
Gene expression data and clinical phenotype data of 308 patients with cervical squamous cell carcinoma (CESC) and endocervical adenocarcinoma were obtained from the UCSC Xena database (https://xena.ucsc.edu/). The ethical statement of human use and welfare was approved by the Institutional Review Board of Harbin Medical University Cancer Hospital (IRB: No. KY2022-68).
2.2 Gene Set of immunocytes
The gene set of 22 types of immunocytes was obtained from CIBERSORT (http://cibersort.stanford.edu).
TIDE score was used to evaluate the potential clinical efficacy of immunotherapy across various risk groups, serving as an indicator of tumor immune escape potential. A higher TIDE score indicates diminished efficacy of immune checkpoint inhibitors (ICI). To obtain immune-related scores for CESC patients, we utilized an online tool for calculating the TIDE score (http://tide.dfci.harvard.edu/login/).
2.3 Construction of immune-related prognostic markers
Firstly, we employed single-sample gene set enrichment analysis (ssGSEA) to calculate and score 22 kinds of immunocytes within each CESC sample based on the gene expression data. Subsequently, genes were subjected to univariate Cox regression analysis to determine hazard ratio (HR) and prognostic significance. Genes with a significance level of P < 0.05 were identified as prognosis-related immunocytes. Secondly, a gene set comprising prognosis-related immunocytes was obtained using weighted correlation network analysis (WGCNA) (Reference: WGCNA: an R package for weighted correlation network analysis) as the candidate gene set. Thirdly, based on the candidate gene set, immune-related prognostic genes were selected using the lasso-cox method (P < 0.05), and immune-related prognostic markers were constructed. The scoring formula for the prognostic marker is as follows: Score = -0.011118867*exp(PLA2G2D) - 0.004579813*exp(CHIT1). Finally, the samples were categorized into high and low groups based on the median score, and the correlation between these two types and CESC was further analyzed. In addition, multivariate Cox analysis was performed to explore the independent prognostic value of the score.
2.4 Screening of differentially expressed genes and functional enrichment analyses
Differential expression analysis was carried out on two groups of patients using fold change values and the Wilcox test. Differentially expressed genes were screened using a specific filter condition, requiring an absolute value of log2 (FC) greater than 1 and P-value less than 0.05. KEGG and GO enrichment analyses were performed on differentially expressed genes using the DAVID tool, and the results were visually presented with a false discovery rate (FDR < 0.05).
2.5 Patients and tissue specimens
After obtaining approval from the Institutional Review Committee and obtaining informed consent from patients, we collected samples from 35 CESC patients residing in cold regions. These patients were recruited from the Cancer Hospital affiliated with Harbin Medical University in China from January 2015 to December 2018. Among the cohort, 20 patients were diagnosed with cervical cancer, while 15 had normal cervical epithelium. Patients with normal cervical epithelia received hysterectomy to address benign conditions, while those diagnosed with cervical cancer patients underwent radical hysterectomy and pelvic lymphadenectomy. Patients identified with high-risk factors received postoperative radiotherapy.
2.6 Immunohistochemical staining
Paraffin sections were first dewaxed, hydrated, and then inactivated with 3% hydrogen peroxide. Subsequently, different paraffin sections were incubated with PLA2G2D and CHIT1 specific antibodies (1 : 1000 dilution, Abcam) at 4°C for 12 hours. Following this, the paraffin sections were mixed with the second antibody mixture and incubated for 30 minutes. The sections were then stained using 3, 3 '-diaminoamphetamine (DAB) solution. After staining, hematoxylin counterstaining and 1% hydrochloric acid ethanol differentiation were performed. The stained sections were observed under a microscope (Olympus, Tokyo, Japan).
2.7 Real-time PCR
Total RNA from the cervical cancer tumor and control groups was extracted by TRIpure lysis solution (BioTeke, Beijing, China). These RNAs were then reverse-transcribed into cDNA using BeyoRT II M-MLV reverse transcriptase (Beyotime China). Then, real-time PCR reaction was performed using the cDNA as a template using the fluorescence quantitative realtime PCR kit (Takara). Fluorescence quantitative analysis was performed using the ExicyclerTM 96 fluorescence quantitative analyzer (BIONEER, Korea). The relative mRNA expression was calculated using 2–ΔΔCt methods. The primer pair sequences used are as follows: For CHIT1: 5'-CAGCGAACTCATCTTTGCCAG' (forward) and 5 '-GGACACCTGGAATTCGTTGC-3' (reverse); for PLA2G2D: 5'-ACTTTTCCCAGGGGAACATCC-3' (forward) and 5'-GCAGTCGCTTCTGGTAGGTG-3' (reverse); for β-actin: 5'-GGCACCCAGCACAATGAA-3' (forward) and 5'-TAGAAGCATTTGCGGTGGG-3' (reverse).
2.8 Statistical analysis
R software (version 4.2.1) was used for all statistical analyses. The Wilcox test was used to identify statistically significant differences in gene expression and infiltrating immunocytes. Kaplan-Meier curves were generated, and differences in overall survival (OS) between groups were assessed using the log-rank test. Multivariate Cox proportional hazard regression analysis was conducted to explore the relationship between scores and OS.
Receiver operating characteristic (ROC) analysis was performed to evaluate the sensitivity and specificity of the score in predicting prognosis, with the area under the ROC curve (AUC) as an indicator of prognostic accuracy. A significance threshold of P < 0.05 was applied to all analyses.
3 Results
3.1 Recognition of prognosis-related immunocytes
Based on gene expression data, scores representing the abundance of 22 types of immunocytes in each CESC sample were calculated using the single-sample gene set enrichment analysis (ssGSEA) method. Univariate Cox regression analysis was performed to identify immunocytes associated with prognosis. Among the immunocytes analyzed, a total of 10 types were found to be significantly correlated with the OS in the patients. These included B cells memory, B cells naïve, NK cells resting, plasma cells, T cells CD4 memory resting, T cells CD4 naïve, T cells CD8, T cells follicular helper, T cells gamma delta, and T cells regulatory Tregs (Fig. 1).
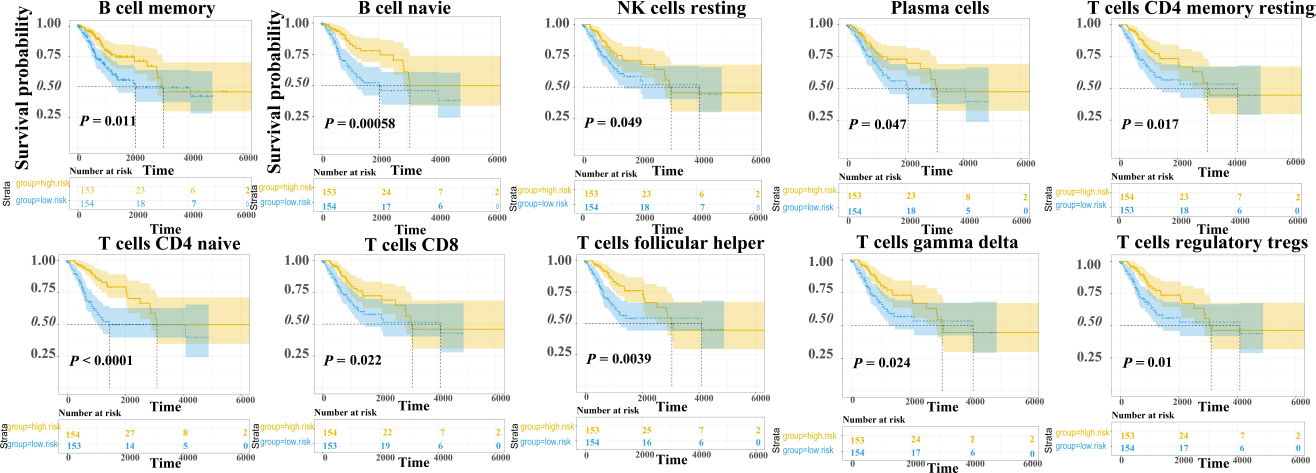
Survival curve of immune cell score
3.2 Recognition of gene modules of prognosis-related immunocytes
Based on gene expression data, the WGCNA method was employed to identify gene modules significantly associated with the prognosis of immunocytes. Among the the identified modules, gene module 6 exhibited a significant correlation with all prognostic immunocytes (P < 0.05). This module comprises 47 genes, including CCL18 and IFNG, which are closely related to the immune processes (Fig. 2).
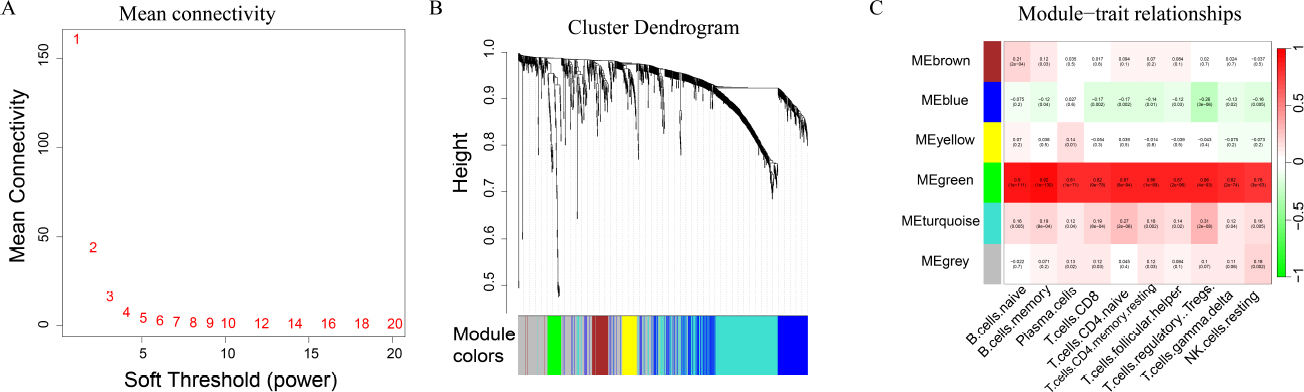
WGCNA analysis
(A) WGCNA determines optimal threshold; (B) WGCNA hierarchical clustering; (C) Correlation analysis map between gene module and immune cell score.
3.3 Construction of immune-related gene markers
Using the 47 genes identified in module 6 as the core, the lasso-cox method was used to analyze gene expression data, leading to the identification of two genes PLA2G2D and CHIT1, that are significantly associated with OS time of cancer patients (Fig. 3A-B). CESC patients were then stratified into groups based on the median score of these gene markers. The study revealed a longer OS time in CESC patients with a high immune score (log-rank test, P = 0.0027, Fig. 3C).
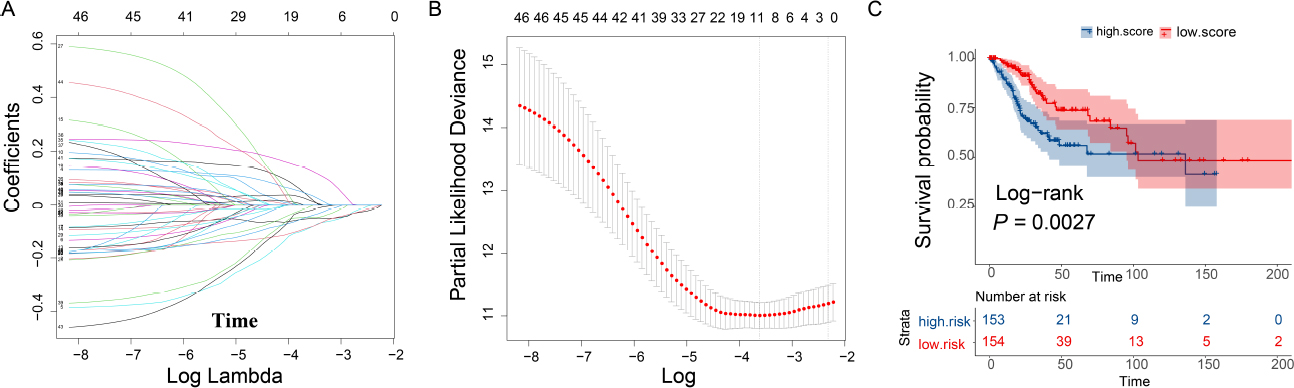
LASSO regression analysis
(A) The larger the lambda is, the closer the regression coefficient tends to zero; (B) The smaller the likelihood deviation is, the better the model fittingis; (C) Survival curve of immune-related gene marker scores.
Furthermore, to account for the effects of age and AJCC stage, a multifactor cox proportional hazard model was employed. It was observed that the scores of the immune-related genetic markers, along with AJCC stage IV, independently and significantly predicted OS in CESC patients. Moreover, utilizing the clinical phenotype data of CESC patients, a nomogram was constructed to quantitatively predict the survival rate of patients. Furthermore, by plotting the ROC curve, it was demonstrated that the immune-related gene markers’ score could predict patients’ 5-year survival time, with an area under the curve (AUC) of 0.818 (Fig. 4).
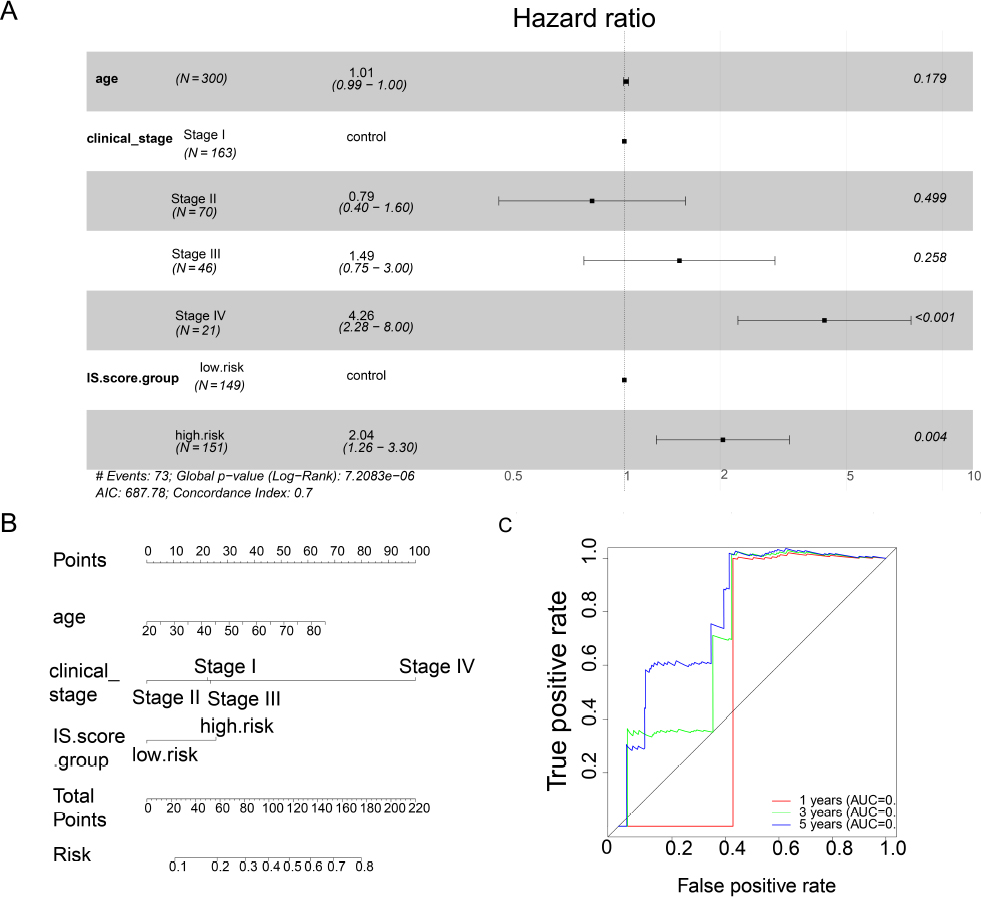
Survival association analysis of immune-related gene markers
(A) Multivariate cox proportional risk model analysis; (B) Nomogram of survival prediction of cervical squamous cell carcinoma (CESC) patients; (C) ROC curve analysis.
In addition, we analyzed the prognostic value of this gene marker in CESC patients stratified by the four AJCC stages. Our analysis revealed that the gene marker could predict shorter OS time in patients with stage II and stage III CESC patients (logrank test, P = 0.05 and 0.035. Fig. 5).
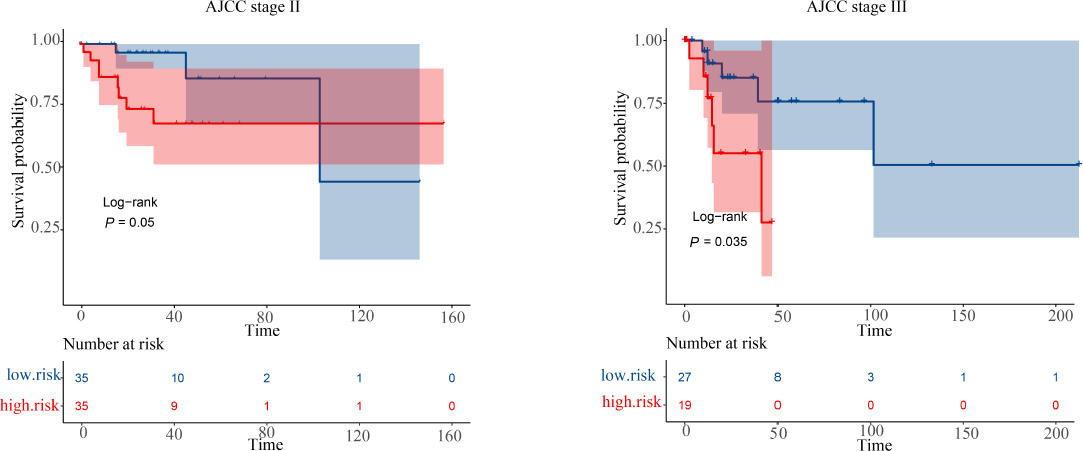
The overall survival of the gene markers in stage II and stage III cervical squamous cell carcinoma (CESC) patients
3.4 Association analysis between immune-related gene markers and immune scores
Initially, the stromal score, immune score, and ESTIMATE score of CESC patients were obtained from the ESTIMATE data resource. Through the Wilcox rank sum test, it was discovered that patients with low scores of immune-related gene markers exhibited significantly higher stromal score, immune score, and ESTIMATE score compared to those with high scores (P < 0.05, Fig. 6A). Additionally, other immune-related scores were retrieved from the TIDE data resources. With the exception of the TIDE score, all other immune-related scores demonstrated a significant correlation with immune-related gene markers. Notably, in patients with high scores of immune-related gene markers, the scores of cancer-associated fibroblasts (CAF), exclusion, myeloid-derived suppressor cells (MDSC), and M2 macrophages (TAM.M2) were notably higher compared to those in patients with low scores (P < 0.05). Conversely, the scores of CD8, CD274, dysfunction, IFNG, Merck18 and MSI in patients with low scores of immune-related gene markers were significantly higher than those in patients with high scores (P < 0.05, Fig. 6B).
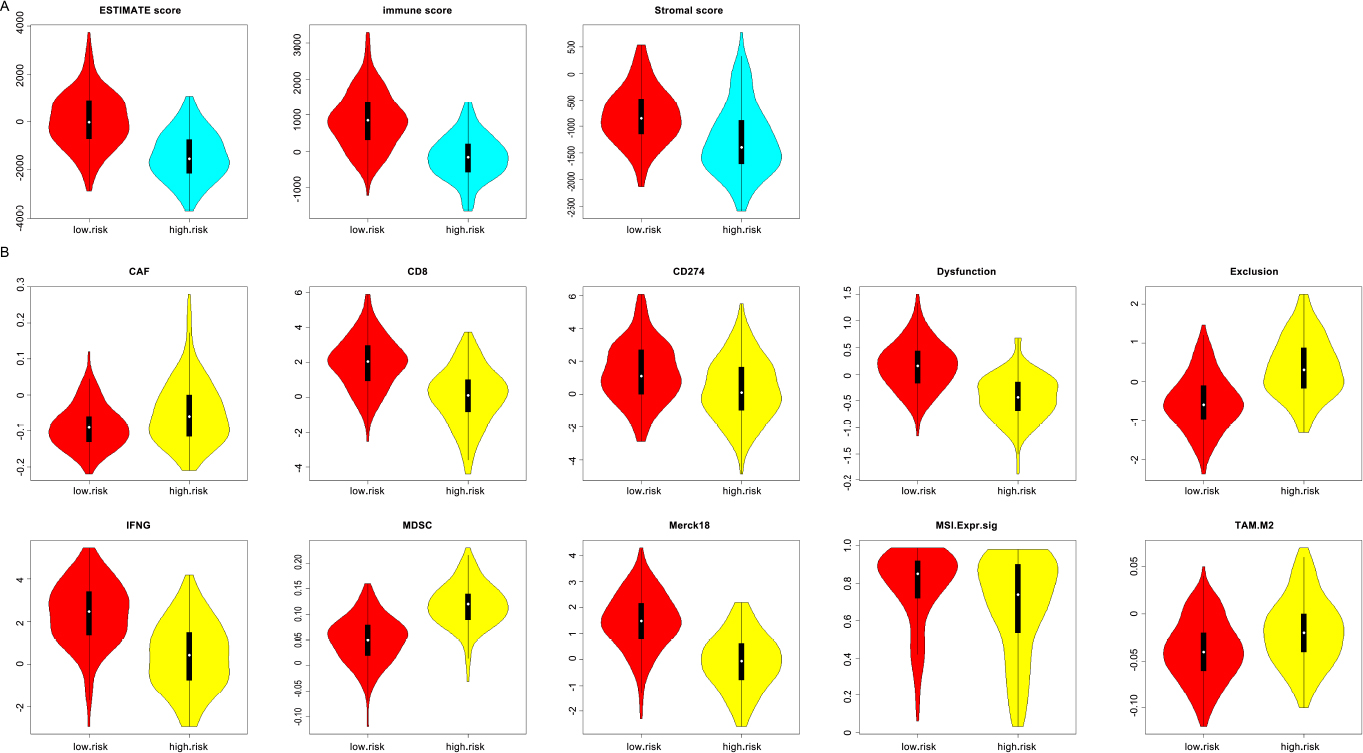
Immune score differences between the high immune-related gene marker score group and the low immune-related gene marker score group
(A) Difference of immune score from ESTIMATE data resource; (B) Difference of immune score from TIDE data resource.
3.5 Biological functions of immune-related genetic markers
To delve deeper into the potential biological behavior of immunerelated gene markers, fold change values and the Wilcox rank sum test were employed to analyze differential expression between patients with high and low scores. Under the condition of |log2FC| > 1 and FDR < 0.05, a total of 857 upregulated genes and 979 downregulated genes were identified. GO and KEGG enrichment analyses were carried out by the DAVID software on differentially expressed genes (Fig. 7). The results revealed that immune-related gene markers were significantly associated with immune functions such as immune response, cellular response to interleukin (IL)-1, and positive regulation of IL-12 production (Fig. 7). Meanwhile, immune-related gene markers exhibited significant correlations with key cancer pathways such as the chemokine signaling pathway and NF-κB signaling pathway (FDR < 0.05, Fig. 7).
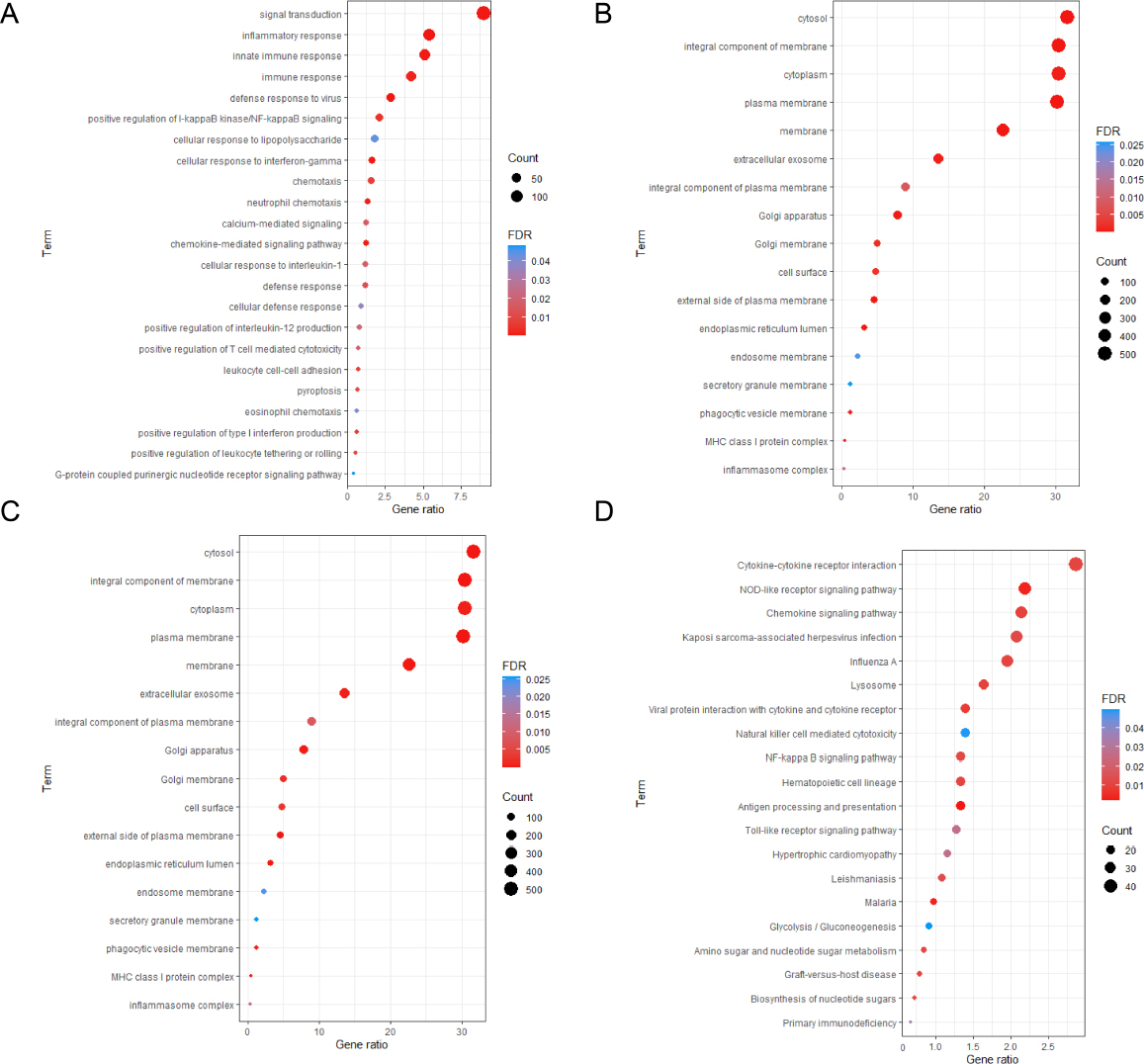
Analysis of biological function differences between the high immune-related gene marker score group and the low immune-related gene marker score group (A) Biologic process function of differentially expressed genes; (B) Cellular components function of differentially expressed genes; (C) Molecular functions of differentially expressed genes; (D) KEGG pathway enrichment analysis of differentially expressed genes.
According to the GEPIA2 database, we also found that PLA2G2D and CHIT1 were abundantly expressed in CESC patients compared to the normal samples (Fig. 8).
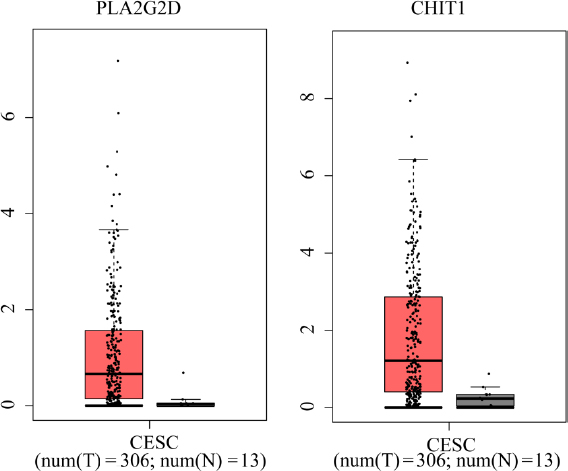
The expression of PLA2G2D and CHIT1 between cervical squamous cell carcinoma (CESC) patients and normal samples
3.6 Up-regulated expression of PLA2G2D protein and CHIT1 protein in cervical cancer
Immunohistochemical staining was used to detect the expression of PLA2G2D protein and CHIT1 protein, respectively, in 20 cases of cervical cancer left over from surgery and 15 cases of normal control group. The results showed that compared with the control group, the expression levels of PLA2G2D protein and CHIT1 protein in cervical cancer tissues were significantly up-regulated (Fig. 9A-J) and were mainly located in the cytoplasm, with statistically significant differences (Fig. 9K-L). Real-time PCR was used to detect the expression levels of PLA2G2D and CHIT1 genes in cervical cancer tissues and control group. The results showed that: Compared with the control group, mRNA expressions of PLA2G2D and CHIT1 genes in cervical cancer tissues were significantly increased, with statistically significant differences (P < 0.001) (Fig. 10).
Immunohistochemical staining was employed to detect the expression of the PLA2G2D and CHIT1 proteins in 20 cases of cervical cancer samples remaining from surgery and 15 cases from the normal control group. The findings revealed a significant up-regulation of both PLA2G2D and CHIT1 protein expression in cervical cancer tissues compared to the control group (Fig. 9A-J). These proteins were predominantly located in the cytoplasm, exhibiting statistically significant variances (Fig. 9K-L). Furthermore, real-time PCR was conducted to assess the expression levels of the PLA2G2D and CHIT1 genes in cervical cancer tissues and the control group. The results demonstrated a marked increase in mRNA expressions of both PLA2G2D and CHIT1 genes in cervical cancer tissues compared to the control group, with statistically significant differences (P < 0.001) (Fig. 10).
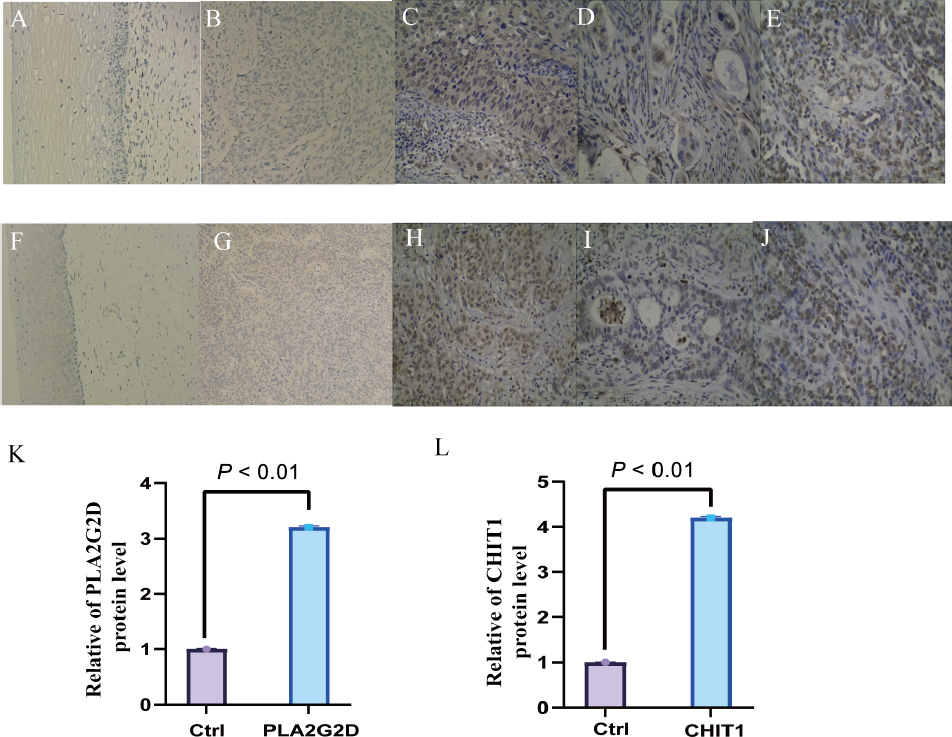
Expression changes of PLA2G2D and CHIT1 in tumor tissues of cervical squamous cell carcinoma (CESC) patients and normal controls
(A) Negative expression of PLA2G2D in normal cervical epithelium; (B-C) Low and high expression of PLA2G2D in cervical squamous cell carcinoma; (D-E) Low and high expression of PLA2G2D in cervical adenocarcinoma; (F) Negative expression of CHIT1 in normal cervical epithelium; (G-H) Low and high expression of CHIT1 in cervical squamous cell carcinoma; (I-J) Low and high expression of CHIT1 in cervical adenocarcinoma (100x); (K-L) Immunohistochemical results of expression of PLA2G2D and CHIT1 in tumor tissues of CESC patients and normal controls.
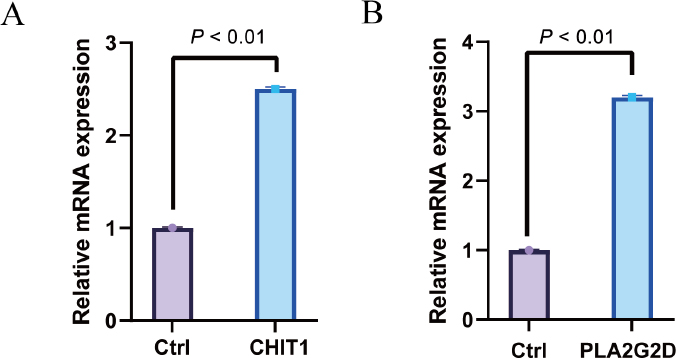
The expression levels of CHIT1 and PLA2G2D genes in the cervical squamous cell carcinoma (CESC) and normal control group were detected by Real-time PCR
4 Discussion
Based on the gene expression data, the cell score of CESC samples was initially calculated using the ssGSEA method. Univariate Cox regression analysis revealed that 10 immunocytes tended to be closely associated with the overall survival time of patients. Subsequently, WGCNA identified 6 gene modules significantly correlated with prognostic immunocytes. Module 6, comprising 47 genes, was identified as pivotal, among which PLA2G2D and CHIT1 emerged as core genes significantly linked to the overall survival time of cancer patients. Based on the median score of gene markers, CESC patients were stratified into two groups. Patients with low immune scores, Patients with high immune scores demonstrated significantly prolonged OS compared to those with low immune scores, indicating the potential utility of immune scores in survival analysis.
This study highlights the potential relevance of two key immune-related genes, PLA2G2D and CHIT1, to the prognosis and survival time of patients with cervical cancer. PLA2G2D, a member of the secretory phospholipase A2 (Spla2) family, exhibits wide expression across various tissues, including squamous epithelium, spleen, and lymph nodes[11]. Upon stimulation by pro-inflammatory mediators, immunocytes, including dendritic cells and macrophages, secrete PLA2G2A, promoting the production of pro-inflammatory lipids and cytokines[12]. Depending on the context of inflammation and cancer, PLA2G2D may exert either beneficial or detrimental effects. Studies have shown that PLA2G2D deficiency enhances anti-tumor immunity, reducing tumor-promoting M2-like macrophages while augmenting tumor-suppressing M1-like macrophages and cytotoxic T cells[13-14]. Similarly, CHIT1, a highly conserved and regulated chitinase belonging to the similarly, CHIT1, a highly conserved and regulated chitinase belonging to the 18-glycosylase family (GH18)[15-16], is secreted by activated macrophages and plays a crucial role in the body’s immune response, closely associated with inflammation, infection, tissue damage and tumor[17]. Both immunocytes, such as activated macrophages and neutrophils, and structural cells, such as epithelial cells, can secrete CHIT1[18]. Elevated expression of CHIT1 has been observed in macrophages of patients with idiopathic pulmonary fibrosis[17], and therapeutic inhibition of CHIT1 has shown promise in reducing pulmonary fibrosis[19]. Moreover, Yu et al.[20] identified four core genes, including CHIT1, GTSF1L, PLA2G2D, and GNG8, by analyzing TCGA transcriptome data in conjunction with the infiltration levels of 28 immunocytes. They stratified cervical cancer patients into high-score and low-score subgroups, revealing distinct infiltration landscapes, survival outcomes, and responses to ICI between the two subgroups. This finding underscores the potential clinical implications of immune-related genes in cervical cancer management.
To delve deeper into the predictive significance of two key immune-related genes (PLA2G2D and CHIT1) in cervical cancer prognosis, a correlation analysis was conducted between immune-related gene markers and immune scores. It was observed that there tended to be a negative correlation between the scores of immune-related gene markers and stromal score, immune score, and ESTIMATE score. In other words, patients with low scores of immune-related gene markers exhibited significantly higher stromal score, immune score, and ESTIMATE score. This finding aligns with the research by Wang et al.[21] who reported an inverse relationship between ESTIMATE score and tumor purity in cervical cancer. Moreover, the group with lower immune marker scores displayed lower tumor purity and a more favorable prognosis. Conversely, the scores of MDSC, exclusion, CAF, and TAM. M2 in the group with high scores of immune-related gene markers were notably elevated compared to those in the low-score group. This observation resonates with the findings of Yu et al.[20] who identified a positive association between MDSC score and CAF score with tumor-promoting immunocytes and tumor prognosis.
However, contrasting results were reported by Wen et al.[22] who demonstrated that lung adenocarcinoma patients with high MDSC scores experienced more pronounced immune escape, leading to a poorer prognosis. Further analysis revealed that patients with low scores of immune-related gene markers exhibited significantly higher scores of CD8, CD274 (PD-L1), dysfunction, IFNG, Merck18, and MSI compared to those with high scores. This trend is consistent with the findings of Zhou et al.[23] who noted increased scores of Merck18, CD274, and CAF in the low-risk group of breast cancer, suggesting lower sensitivity to chemotherapy drugs. Consequently, patients in the low-risk group may derive greater benefits from immunotherapy and chemotherapy interventions, as supported by our results.
This study delves further into the biological behavior of immune-related gene markers, revealing their significant associations with immune functions such as immune response, cellular response to IL-1, and positive regulation of IL-12 production. IL-1 has been implicated in promoting tumor growth and metastasis in both mouse and human tumors, while also dampening effective adaptive anti-tumor immune responses[24]. Conversely, IL-12 functions as an anti-tumor cytokine primarily acting on lymphocytes, promoting the differentiation of Th1 cells with antitumor activity while inhibiting cytokines such as IL-4 produced by Th2 cells, which have tumor-promoting activity[25,26,27]. Through KEGG enrichment analysis, IL-12 was found to be significantly associated with crucial cancer pathways including the chemokine signaling pathway and the NF-κB signaling pathway. The chemokine signaling system plays a role in tumor growth and progression through various mechanisms[28]. Chemokine receptors expressed by cancer cells can interact with ligands secreted by tumor cells, tumor-associated fibroblasts, and immunocytes infiltrating the tumor microenvironment. This interaction activates signaling pathways such as PI3K/AKT and ERK 1/2 leading to tumor cell proliferation[29]. Additionally, the NF-κB signal transduction pathway is known to promote tumorigenesis by responding to inflammatory factors, carcinogens, cancer promoters, and the tumor microenvironment. Continuously activated NF-κB is associated with various aspects of tumor biology including occurrence, proliferation, anti-apoptosis, invasion, angiogenesis, and metastasis. NF-κB is continuously activated in many tumors[30]. Finally, this article validates the upregulation of proteins and nucleic acids, specifically CHIT1 and PLA2G2D, in cervical cancer tumor tissues of patients from cold regions of northern China. This finding underscores the potential significance of CHIT1 and PLA2G2D as biomarkers in cervical cancer prognosis and suggests their possible involvement in tumor progression in this geographical context.
5 Conclusion
In conclusion, the bioinformatics analysis presented in this paper highlights the close association between immune-related markers PLA2G2D and CHIT1 and the prognosis of CESC. While the study conducted bioinformatics analysis alongside immunohistochemical and real-time PCR tests, it remains essential to supplement these findings with further in vivo and in vitro experiments in future research endeavors. Nonetheless, PLA2G2D and CHIT1 emerge as promising biomarkers for evaluating immune infiltration and prognosis outcomes in patients with cervical cancer. Their potential significance warrants continued investigation to elucidate their role in cervical cancer progression and their utility in clinical settings.
Funding statement: This work was supported by the Haiyan Foundation Youth Project JJQN2022-12.
-
Author contributions Wang J and Jiang L L: designed the experimental protocol, Jiang L L and Qiao Q S: carried out the experiments and wrote the main manuscript text, prepared figures, and Wang J: revised the manuscript. All authors reviewed the manuscript.
-
Ethical approval The ethical statement of human use and welfare was approved by the Institutional Review Board of Harbin Medical University Cancer Hospital (IRB: No. KY2022-68).
-
Conflicts of interest All authors disclosed no relevant relationships.
-
Data availability statement The data are all included in this paper. The original data are available from the corresponding author upon reasonable request.
References
[1] Cohen P, Jhingran A, Oaknin A, et al. Cervical cancer. Lancet, 2019; 393(10167): 169-182.10.1016/S0140-6736(18)32470-XSuche in Google Scholar PubMed
[2] Mileshkin L, Moore K, Barnes E, et al. Adjuvant chemotherapy following chemoradiotherapy as primary treatment for locally advanced cervical cancer versus chemoradiotherapy alone: an international, openlabel, randomised, phase 3 trial. Lancet Oncol, 2023; 24(5): 468-482.10.1016/S1470-2045(23)00147-XSuche in Google Scholar PubMed PubMed Central
[3] Xiao M, Xie L, Cao G, et al. CD4 T-cell epitope-based heterologous prime-boost vaccination potentiates anti-tumor immunity and PD-1/PD-L1 immunotherapy. J Immunother Cancer, 2022; 10(5): e004022.10.1136/jitc-2021-004022Suche in Google Scholar PubMed PubMed Central
[4] Oh D, Fong L. Cytotoxic CD4 T cells in cancer: Expanding the immune effector toolbox. Immunity, 2021; 54(12): 2701-2711.10.1016/j.immuni.2021.11.015Suche in Google Scholar PubMed PubMed Central
[5] Philip M, Schietinger A. CD8 T cell differentiation and dysfunction in cancer. Nat Rev Immunol, 2022; 22(4): 209-223.10.1038/s41577-021-00574-3Suche in Google Scholar PubMed PubMed Central
[6] Sharonov G, Serebrovskaya E, Yuzhakova D, et al. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat Rev Immunol, 2020; 20(5): 294-307.10.1038/s41577-019-0257-xSuche in Google Scholar PubMed
[7] Huntington N, Cursons J, Rautela J. The cancer-natural killer cell immunity cycle. Nat Rev Cancer, 2020; 20(8): 437-454.10.1038/s41568-020-0272-zSuche in Google Scholar PubMed
[8] Kurmyshkina O, Kovchur P, Schegoleva L, et al.T- and NK-cell populations with regulatory phenotype and markers of apoptosis in circulating lymphocytes of patients with CIN3 or microcarcinoma of the cervix: evidence for potential mechanisms of immune suppression. Infect Agent Cancer, 2017; 12: 56.10.1186/s13027-017-0166-1Suche in Google Scholar PubMed PubMed Central
[9] Zhang Y, Yu M, Jing Y, et al. Baseline immunity and impact of chemotherapy on immune microenvironment in cervical cancer. Br J Cancer, 2021; 124(2): 414-424.10.1038/s41416-020-01123-wSuche in Google Scholar PubMed PubMed Central
[10] Zhang L, Zhang H, Huang Y, et al. Expression of immune cell markers and tumor markers in patients with cervical cancer. Int J Gynecol Cancer, 2020; 30(7): 969-974.10.1136/ijgc-2020-001254Suche in Google Scholar PubMed
[11] Sato H, Taketomi Y, Miki Y, et al. Secreted phospholipase PLA2G2D contributes to metabolic health by mobilizing ω3 polyunsaturated fatty acids in WAT. Cell Rep, 2020; 31(5): 107579.10.1016/j.celrep.2020.107579Suche in Google Scholar PubMed
[12] Murakami M, Yamamoto K, Miki Y, et al. The roles of the secreted phospholipase A gene family in immunology. Adv Immunol, 2016; 132: 91-134.10.1016/bs.ai.2016.05.001Suche in Google Scholar PubMed PubMed Central
[13] Miki Y, Yamamoto K, Taketomi Y, et al. Lymphoid tissue phospholipase A2 group IID resolves contact hypersensitivity by driving antiinflammatory lipid mediators. J Exp Med, 2013; 210(6): 1217-1234.10.1084/jem.20121887Suche in Google Scholar PubMed PubMed Central
[14] Miki Y, Kidoguchi Y, Sato M, et al. Dual roles of group IID phospholipase A2 in inflammation and cancer. J Biol Chem, 2016; 291(30): 15588-155601.10.1074/jbc.M116.734624Suche in Google Scholar PubMed PubMed Central
[15] Liu H, Xu R, Gao C, et al. Metabolic molecule PLA2G2D is a potential prognostic biomarker correlating with immune cell infiltration and the expression of immune checkpoint genes in cervical squamous cell carcinoma. Front Oncol, 2021; 11: 755668.10.3389/fonc.2021.755668Suche in Google Scholar PubMed PubMed Central
[16] Di Francesco A, Verrecchia E, Manna S, et al. The chitinases as biomarkers in immune-mediate diseases. Clin Chem Lab Med, 2022; 61(8): 1363-1381.10.1515/cclm-2022-0767Suche in Google Scholar PubMed
[17] Chang D, Sharma L, Dela Cruz C. Chitotriosidase: a marker and modulator of lung disease. Eur Respir Rev, 2020; 29(156): 190143.10.1183/16000617.0143-2019Suche in Google Scholar PubMed PubMed Central
[18] Lee C, Herzog E, Ahangari F, et al. Chitinase 1 is a biomarker for and therapeutic target in scleroderma-associated interstitial lung disease that augments TGF-β1 signaling. J Immunol. 2012; 189(5): 2635-2644.10.4049/jimmunol.1201115Suche in Google Scholar PubMed PubMed Central
[19] Koralewski R, Dymek B, Mazur M, et al. Discovery of, a First-in-Class chitinase inhibitor as potential new therapeutics for idiopathic pulmonary fibrosis. J Med Chem, 2020; 63(24): 15527-15540.10.1021/acs.jmedchem.0c01179Suche in Google Scholar PubMed
[20] Yu J, Zhang W, Ding D, et al. Bioinformatics analysis combined with experiments predicts PUDP as a potential prognostic biomarker for hepatocellular carcinoma through its interaction with tumor microenvironment. Front Oncol, 2022; 12: 830174.10.3389/fonc.2022.830174Suche in Google Scholar PubMed PubMed Central
[21] Wang Q, Xu Y. Comprehensive analysis of cuproptosis-related lncRNAs model in tumor immune microenvironment and prognostic value of cervical cancer. Front Pharmacol, 2022; 13: 1065701.10.3389/fphar.2022.1065701Suche in Google Scholar PubMed PubMed Central
[22] Wen H, Chen H, Xie L, et al. Macrophage-related molecular subtypes in lung adenocarcinoma identify novel tumor microenvironment with prognostic and therapeutic implications. Front Genet, 2022; 13: 1012164.10.3389/fgene.2022.1012164Suche in Google Scholar PubMed PubMed Central
[23] Zhou N, Kong D, Lin Q, et al. Unfolded protein response signature unveils novel insights into breast cancer prognosis and tumor microenvironment. Cancer Genet, 2023; 276-277: 17-29.10.1016/j.cancergen.2023.06.001Suche in Google Scholar PubMed
[24] Garlanda C, Mantovani A. Interleukin-1 in tumor progression, therapy, and prevention. Cancer Cell, 2021; 39(8): 1023-1027.10.1016/j.ccell.2021.04.011Suche in Google Scholar PubMed
[25] Verstockt B, Salas A, Sands B, et al. IL-12 and IL-23 pathway inhibition in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol, 2023; 20(7): 433-446.10.1038/s41575-023-00768-1Suche in Google Scholar PubMed PubMed Central
[26] Lewis N, Sia C, Kirwin K, et al. Exosome surface display of IL12 results in tumor-retained pharmacology with superior potency and limited systemic exposure compared with recombinant IL12. Mol Cancer Ther, 2021; 20(3): 523-534.10.1158/1535-7163.MCT-20-0484Suche in Google Scholar PubMed
[27] Jia Z, Ragoonanan D, Mahadeo K, et al. IL12 immune therapy clinical trial review: novel strategies for avoiding CRS-associated cytokines. Front Immunol, 2022; 13: 952231.10.3389/fimmu.2022.952231Suche in Google Scholar PubMed PubMed Central
[28] Goenka A, Khan F, Verma B, et al. Tumor microenvironment signaling and therapeutics in cancer progression. Cancer Commun (Lond), 2023; 43(5): 525-561.10.1002/cac2.12416Suche in Google Scholar PubMed PubMed Central
[29] Bule P, Aguiar S, Aires-Da-Silva F, et al. Chemokine-Directed tumor microenvironment modulation in cancer immunotherapy. Int J Mol Sci, 2021; 22(18): 9804.10.3390/ijms22189804Suche in Google Scholar PubMed PubMed Central
[30] Ren X, Chen C, Luo Y, et al. lncRNA-PLACT1 sustains activation of NF-κB pathway through a positive feedback loop with IκBα/E2F1 axis in pancreatic cancer. Mol Cancer, 2020; 19(1): 35.10.1186/s12943-020-01153-1Suche in Google Scholar PubMed PubMed Central
© 2024 Liangliang Jiang, Qiushuang Qiao, Jing Wang, published by De Gruyter on behalf of Heilongjiang Health Development Research Center
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Frontmatter
- Review
- Impacts of cold exposure on energy metabolism
- Cryoablation techniques in bladder cancer: A review
- Cold environments and human metabolism: A traditional chinese medicine perspective
- Original Article
- Toll-like receptors 2 polymorphism is associated with psoriasis: A case-control study in the northern Chinese population
- Vitamin D deficiency and increased inflammatory factor intercellular cell adhesion molecule-1 indicate severe leukoaraiosis in northern China
- PLA2G2D and CHIT1: Potential biomarkers for immune infiltration and prognosis in cervical squamous cell carcinoma
- Characteristics of gut microbiota in anastomotic leakage patients in cold zones post-colorectal cancer surgery: A high-throughput sequencing and propensity-score matching study
Artikel in diesem Heft
- Frontmatter
- Review
- Impacts of cold exposure on energy metabolism
- Cryoablation techniques in bladder cancer: A review
- Cold environments and human metabolism: A traditional chinese medicine perspective
- Original Article
- Toll-like receptors 2 polymorphism is associated with psoriasis: A case-control study in the northern Chinese population
- Vitamin D deficiency and increased inflammatory factor intercellular cell adhesion molecule-1 indicate severe leukoaraiosis in northern China
- PLA2G2D and CHIT1: Potential biomarkers for immune infiltration and prognosis in cervical squamous cell carcinoma
- Characteristics of gut microbiota in anastomotic leakage patients in cold zones post-colorectal cancer surgery: A high-throughput sequencing and propensity-score matching study

