Immunochemical Recognition and its Diagnostic and Therapeutic Applications
-
Douglas Templeton
The October 2014 issue of Pure and Applied Chemistry includes several papers arising from an IUPAC project on immunochemistry that describe interactions between molecules of the immune system and their ligands [1,2] and consider how a knowledge of this chemistry is furthering the development of both the analytical applications of immune-based sensors [3], and of better therapies for a variety of human medical conditions [4]. Each paper is structured to provide some historical context, followed by a critical presentation of our current perspective on the role chemistry is playing in the basic understanding of the immune system and its potential for exploitation.
The intricacies of the human immune system represent one of the most complex areas of current biology. In a forum in Scientific American (April 2012), Stuart Firestein noted that, as a neurobiologist, he can “understand the questions that drive immunology”, but though he “can’t grasp much of immunology ... the wonderful thing is that most immunologists can’t either”. The point was, of course, the increasing specialization of all fields of science, but the example was well chosen. It is a challenge to discuss developments in immunology in a narrative that is comprehensible to chemists. Nevertheless, by including information boxes in the first two papers that are intended as a minimal introduction to the immune system, and by setting developments in a historical context, the PAC papers attempt to do so.
The immune system manages the body’s reactions to invading microbes, allergens, and other foreign substances. It has two subsystems. The first, called “acquired” (or “adaptive”) immunity, recognizes non-self-structures (antigens; Ag) of the invader and counteracts them with molecular responses that reflect a memory of previous exposure. This is what happens when, for example, we are vaccinated, become immune to recurrent disease, or develop allergies. This subsystem relies upon immunoglobulin protein antibodies (Ab’s) and receptor proteins produced by two classes of specialized blood cells called lymphocytes. B lymphocytes, or “B cells”, secrete Ab’s that must recognize and combine with the foreign chemical structures into body fluids. T lymphocytes, or “T cells”, express the recognition proteins as surface receptors on their cell membranes. Each of these cell types proliferates when the target molecule is detected, expanding to produce a large number of cells of a single clone. The B cell clones express monoclonal Ab’s, and the T cell clones express identical copies of the T cell receptor (TCR), in each case recognizing the triggering Ag.
The second subsystem of immunity, described as “innate”, recognizes and then counteracts microorganisms and/or viruses based on certain specific chemical structures that do not occur in vertebrates. This involves molecules with names like NOD-like, Toll-like, and RIG-like helicase receptors that are said to serve their function through pattern recognition; they are termed pattern recognition receptors (PRRs). Both subsystems of immunity involve intricate processes of molecular recognition; Ag-Ab and peptide-TCR in the first case, and ligand-receptor in the second. A number of crystal structures have been presented [1,2] to demonstrate how the recognition process occurs at a structural level. Examples have been chosen that reflect current research in the field, recognizing that the focus of much of this research is on infectious and autoimmune diseases and cancer. They also illustrate the beauty in the common themes and subtle variations of these interactions.
Antigen binding
Human immunoglobulins consist of disulfide-bonded dimeric structures with two so-called heavy chains (H chains) and two light chains (L chains) arranged as shown in Fig. 1. Antigen-binding sites are found in the regions between the L chain and the short arm of the H chain. The minimal recognition and binding structure consists of this fragment only, called a Fab fragment. For simplicity of crystallization and analysis, most structures have been determined with Ag-Fab complexes. Complex gene shuffling for coding regions of variable sequence in the binding pocket of Fab gives rise to the vast variety of sequences necessary to recognize seemingly any antigenic sequence the environment can present. These variable amino acid sequences are found in six complementarity-determining regions (CDR), three on each of the H and L chains, that dominate molecular interactions with Ag. In the case of a peptide/protein Ag, interaction occurs with a specific region of the Ag’s peptide surface, called an epitope, that may involve several adjacent amino acids or, more commonly, amino acids widely separated in the primary peptide sequence that are in close proximity in the folded Ag protein.

Fig. 1: Depictions of immunoglobulin structure. (a) Conventional schematic of a bivalent immunoglobulin showing disulfide-bonded H chains (blue) and L chains (red), presenting two Ag-binding sites (antigens shown as green circles). Upon digestion with papain, cleavage of the H chains releases two Fab and one Fc fragments. (b) A more realistic depiction based on crystallographic data, showing the two L chains in blue and the H chains in green. [1]
Initial structural studies with abundant model proteins—hen egg white lysozyme was historically one of the most important—revealed that the Ag-Ab contact surface was relatively flat and reasonably constant in area. Energetic studies suggested that multiple contacts were involved and that the exclusion of water molecules from the contact region could provide a significant entropic contribution to binding. When a refined structure of a lysozyme-Ab complex at 0.20 nm resolution became available in 2000, several generalities were established. First, all six CDRs were in contact with the Ag epitope, and provided multiple hydrogen bonds, salt bridges, and van der Waals contacts. Second, small conformational movements in the CDR loops and flexibility in side chains of amino acids in the interface region enhanced complementarity of binding—an interplay of lock-and-key fit and conformational flexibility provide the required enthalpic and entropic contributions to binding energy. Third, far from excluding water, the interface region included ordered water molecules that were not observed at lower resolution. These water molecules fill gaps in the interface region, improving fit and accommodating unpaired hydrogen-bonding residues.
Since the detailed analysis of the structure of the lysozyme-Ab complex was presented, several thousand Ag-Ab structures have been solved, and a structure is now pretty much expected as part of Ab characterization. The field of greatest activity is in studying viral epitopes, notably of influenza and the human immunodeficiency virus, although many other proteins are of interest. An attractive structure occurs, for example, in an Ab binding to a lethal venom from the scorpion Androctonus australis hector (Fig. 2). Less planar than most interactions, the toxin embeds into the Ab in a structure the authors describe as an “egg in a cup”. The “cup” consists of a binding pocket 1.3 nm deep and 1.2 nm wide formed by the six CDRs. An extended L-chain CDR1 and an anionic H-chain CDR2, together with the long H-chain CDR3, form a boundary that “clamps” the cationic toxin, with additional anchoring points on the remaining CDRs. Strong complementarity buries about 25 % (10 nm2) of the venom surface at the interface. A detailed understanding of such Ag-Ab interaction suggests strategies for a molecular approach to both Ab blocking and Ab design.
Homologous to the immunoglobulin H and L chains, the TCR has α and β chains that also use six CDRs for recognition and binding. Here, interaction is not with the complete antigen protein, but with a protein fragment presented to the T cell by another component of the immune system, a cell-bound molecule of the major histocompatibility complex (MHC). Although fewer structures have been presented, in part because of the complexity of crystallization and structural analysis of tertiary complexes of MHC-peptide-TCR, not surprisingly the recognition and binding principles of the TCR resemble those of the immunoglobulin Ab. In addition to studies of viral recognition, the involvement of T cells in autoimmune diseases has stimulated intensive effort to understand binding of TCR to self-molecules.
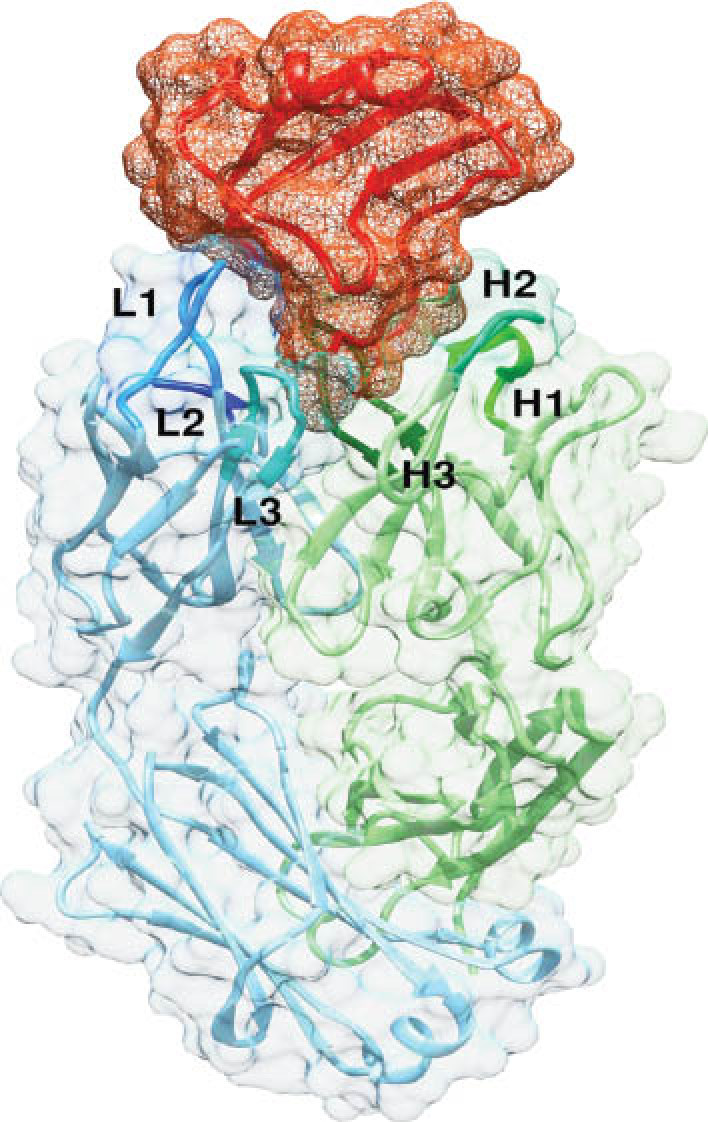
Fig. 2: The fitting of a scorpion toxin (red) into Fab4C1 makes contacts with all six CDR loops of the H chain (green) and the L chain (blue). The authors have described this as “an egg inserted small-end first in the egg cup” (see text for details). The basket around the toxin represents the molecular surface of the protein in mesh style, and the surface around Fab antibody is rendered as a transparent solid. [1]
Interactions with pathogen-associated molecular patterns
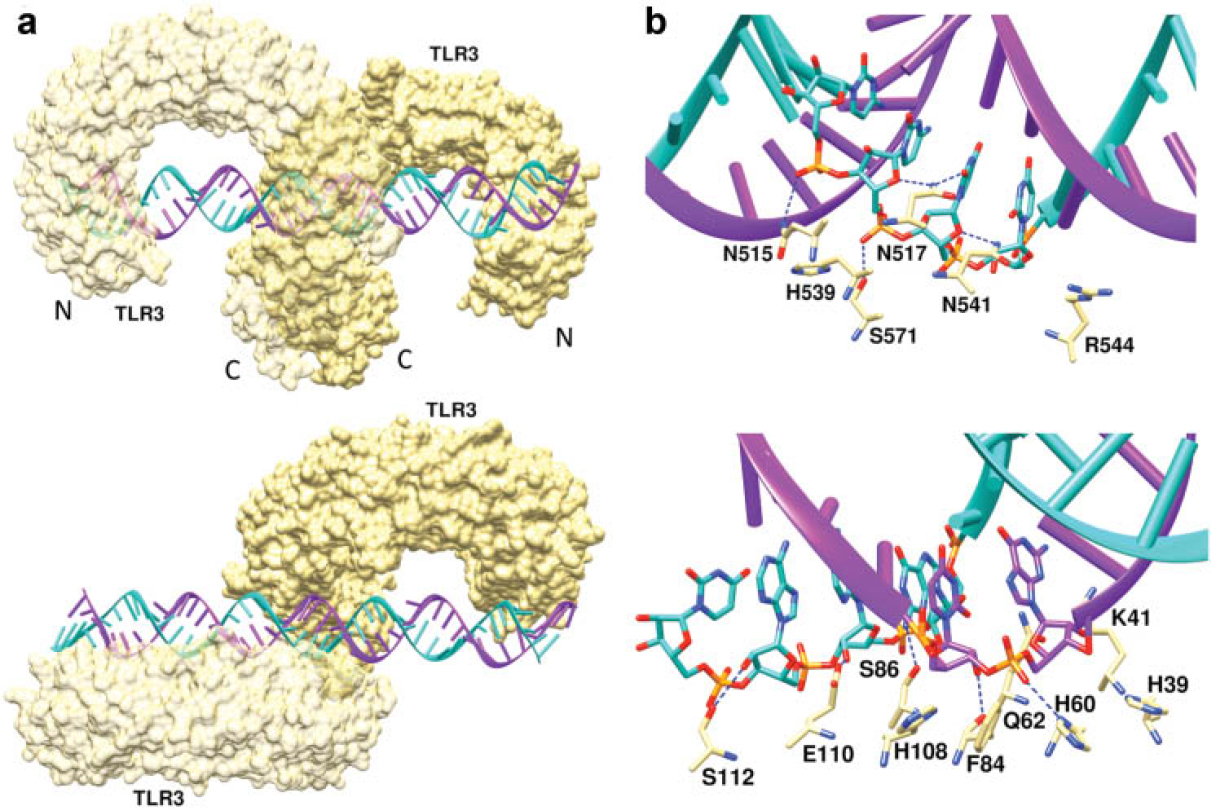
Fig. 3: The double-stranded (ds) RNA:TLR3 signaling complex. (a) Mouse TLR3 ectodomains form a dimer on the dsRNA. (b) The dsRNA binding sites of TLR3. TLR residues within contact distance of the dsRNA include, at the C-terminal site, Asn515, Asn517, His539, Asn541 and Arg544, which are all well conserved in vertebrates. The N-terminal interaction site is formed by His39, His60, Arg64, Phe84, Ser86, His108 and Glu110. [2]
The PRRs on cells of the innate immune system, such as dendritic cells, detect a variety of molecular signatures referred to as pathogen-associated molecular patterns (PAMPs) from a broad range of different invading pathogens. A number of PAMPs are known that range in size from relatively small molecules, to molecules of intermediate size such as bacterial lipopolysaccharides, lipopeptides, and oligosaccharides, to macromolecules such as viral DNA, RNA, and pathogen-associated proteins such as flagellin. A major class of PRRs is the Toll-like receptors (TLR) that consist of a large extracellular binding domain, a transmembrane helix, and an intracellular signaling domain. The nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs) are cytosolic proteins and detect PAMPs that have gained access to the cell. Both NLRs and TLR extracellular domains contain multiple leucine-rich repeats that form crescent structures responsible for PAMP recognition. Upon binding to the PAMP, both types of PRRs initiate signals that ultimately lead to selective expansion and activation of appropriate B- and T-cell populations with specificity for the infectious agent presenting the PAMP. The structure of a mouse TLR3 homodimer binding to viral double-stranded (ds)RNA is shown in Fig. 3. Two other major classes of the PRRs are the C-type lectin receptors that contain lectin-like carbohydrate binding domains, and RIG-like helicase receptors that recognize viral dsRNA. Upon binding to viral DNA, RIG-1 initiates a signaling cascade that induces innate immune defenses. However, aberrant RIG-1 signaling can also lead to apoptosis, inflammation, autoimmune disease, and cancer.
Diagnostic applications
Biochemists have long exploited the specificity of Ag-Ab recognition to design selective separation and detection techniques, and Gubala et al. [3] have surveyed these in the context of recent developments in immunosensor design and diagnostic Ab production and use. Realization that fragments (e.g., Fab noted above) carry the recognition sites means that production of such fragments by a technique called phage display now often replaces the lengthier traditional approaches of animal injections and hybridoma technology. Interest has also focused on camelid Abs, as these naturally lack the L chain; they are quite stable and are themselves of low immunogenicity in humans. Small molecules become antigenic when conjugated to a larger protein, but then conventional means of Ab production yield mixtures of Abs that also react to epitopes on the macromolecular carrier, and purification can be tedious. One direction where small molecule Ab technology is finding increased use is in generating Ab’s that can simultaneously measure multiple congeners of a drug or metabolite. Other advances are being made in introducing chemical reactivity into engineered Ab’s that facilitate immobilization, and amplification with secondary Ab’s against a structure chemically attached to the first. Such recognition or reactive sites may be introduced either by genetic engineering or chemical synthesis.
Traditional approaches of immunoseparation and immunodetection have included immunoprecipitation and agglutination, immunodiffusion, immunochromatography and immunoelectrophoresis. These approaches have seen advances from improved chemistries for coupling to substrates, the development of many commercial kits for miniaturized sandwich-based assay systems, and microfluidic chromatography. Improvements in Ab manipulation are also responsible for increased applications of flow cytometry to diagnostic examination of cell populations. Knowledge of how primary structure and conformational flexibility affect Ab affinities and specificities has allowed these advances to come together in immunosensor design and implementation of chip technology. New coupling chemistries are providing more robust surface coverage, as well as optimal Ab orientation and flexibility to maximize affinity to Ag (Fig. 4). The most favourable orientation of the immobilized biomolecules is generally that in which the Ag-binding site is oriented away from the surface itself. Approaches include engineering glycan moieties into the Ab, or metal-binding sites to exploit with metal ion affinity methods. New areas where these advances are being applied include drug monitoring and detection of markers for organ-based diseases, autoimmune diseases, infectious diseases, cancer, food hygiene, and environmental chemical exposures.
Therapeutic applications
Further promise for revolutionary advances in health care by exploiting Ab chemistry comes in the field of vaccinology. Molecular details of how an immune response is triggered in the organism and how an Ab recognizes its target are fertile ground for manipulation. Chemical principles are being applied to enhance immune system activation and Ab effectiveness through a number of strategies [4].
Traditional procedures for vaccination against a virus involve techniques for cultivation of the virus, identification of the infectious principle, selection and purification of appropriate Ag, cloning, animal testing, and vaccine development—a process typically taking 5-15 years. Today, vaccination with a DNA plasmid encoding the Ag is common, and predicting epitopes in silico (“reverse vaccinology”) can further reduce the time of vaccine development to 1-2 years. DNA vaccines expressed from plasmids may enter cells for subsequent expression and release of the Ag, which may then be taken up by antigen-presenting cells and processed for presentation by MHC molecules to the TCR of T cells (see above) (Fig. 5). Instead of inserting the plasmid into a viral vector, against which some recipients may already have immunity, delivery now is often via particulate carriers, where charge and size can be altered to increase efficiency of delivery (e.g., plasmid DNA binding to carboxylated polystyrene particles via a cationic poly-L-lysine-linker). Rapid progress is also being made using biocompatible, biodegradable nanoparticles such as polycaprolactone (PCL), polyvinylpyrrolidone (PVP), and polyesters—particularly polylactic acid (PLA) and poly(lactic-co-glycolic acid) (PLGA)—and those based on a core of iron oxide such as maghemite (γ-Fe2O3) or magnetide (Fe3O4). Furthermore, encapsulation in nanoparticles not only protects the plasmid DNA from degradation, but controlled DNA delivery systems can be designed to exhibit varying degradation times and release kinetics of DNA for prolonged gene expression. Co-administration of adjuvants enhances the immunogenicity of the Ag. The traditional ajuvant was alum, although its mechanism of action was not really understood. Today, adjuvants are being designed to selectively enhance certain aspects of the immune response by increasing cytokine production, cell recruitment, endocytic pathways, etc., and these may be encoded in the same plasmid that carries the Ag sequence.

Fig. 4: Possible orientations of immunoglobulin G on a surface. The schematic shows that some of the orientations will lead to partial or total blocking of the antigen-binding sites of the antibody. Therefore, it is important to be aware of this problem when designing an assay using immobilized antibody. [3]

Fig. 5: Mechanisms of antigen expression and presentation upon vaccination with plasmid DNA (pDNA). pDNA is taken up by cells via endocytosis. After endosomal escape, cytosolic trafficking, and nuclear entry, the pDNA can be transcribed into mRNA, followed by intracellular translation of the antigen peptide. For T-lymphocyte activation, antigens must be presented in the context of MHC molecules in the presence of co-stimulatory molecules. Although non-antigen-presenting cells (non-APC) cannot induce T-cell activation, extracellular release or cell death can lead to an uptake of the antigen by APC. Antigens produced by direct transfection of APCs are presented by MHC class I. Antigen present in the cytoplasm of APC probably because of endosomal escape can also enter the MHC class I pathway (“cross presentation”). [4]
Structural and computational chemistry is facilitating the development of epitope mimetics. With knowledge of the receptor for a particular virus on a cell surface, Ab’s can be designed to fit structural epitopes without building the epitope itself. These structural studies have also revealed that many of the protective epitopes of human pathogens contain loop, β-hairpin, or α-helical motifs. Conformationally constrained (e.g., cyclized) synthetic epitope mimetics based on these structures are proving useful in vaccine design. Epitope screening of random peptides can be followed by computational modeling to derive a consensus sequence for the target epitope.
Chemistry is playing a central role in engineering effective Ab’s. Structural studies such as those described above [1,2] are proving useful in tailoring designed Ab’s for maximum affinity and specificity by site-directed mutagenesis, CDR shuffling, computational approaches to affinity maturation, etc. Engineering is also directed towards improving pharmacokinetics and pharmaceutical properties, increasing thermal and chemical stability with amino acid analogs, improving solubility and viscosity of the delivery system, reducing non-specific and Ag-mediated clearance, modifying protein aggregation, and improving heterogeneity (e.g., with the presence of glycosylation sites in the Ag-binding regions).
Molecular genetics comes into play in reducing antigenicity of mouse-derived monoclonal Ab’s by replacing all or part of the Ab with human sequence. In this way, ‘humanizing’ all but some murine residues in the CDR recognition sequences has produced several therapeutic Ab’s now in clinical use, including alemtuzumab (an Ab against a lymphocyte surface Ag, used in treating chronic lymphocytic leukemia), trastuzumab (targeting the HER2 receptor in some breast cancers), and daclizumab (an Ab against a cytokine receptor on T cells used in managing rejection following organ transplant). Fully humanized Ab’s are now being produced in mice, as has been achieved with the XenoMouse™ (Fig. 6). This mouse was created by engineering yeast artificial chromosomes (YACs) and then producing chimeric mice by fusion of yeast spheroblasts with mouse embryonic stem cells. These chimeras were then crossed with mice carrying a series of targeted disruptions of mouse heavy and light chain genes. However, even fully humanized Ab’s may carry the risk of eliciting an immune response in the recipient, and although several are in clinical trial, they will not represent the final answer to the ‘perfect’ therapeutic Ab.

Fig. 6: Steps in the production of fully human monoclonal antibodies using the XenoMouse technology. [4]
Advances in biology and structural chemistry are identifying new targets for vaccine research—not only in infectious diseases but also in cancer, allergies, autoimmune diseases and other chronic inflammatory disorders. A critical question for the development of successful vaccines in the future will be which of many available technologies will elicit the best protective or therapeutic response toward a specific pathogen or antigen.
References
1. Templeton, D.M. and K. Moehle, Structural aspects of molecular recognition in the immune system. Part I: Acquired immunity. Pure Appl. Chem. 86(10):1435-1481, 2014. http://dx.doi.org/10.1515/pac-2013-1020Search in Google Scholar
2. Robinson, J.A. and K. Moehle, Structural aspects of molecular recognition in the immune system. Part II: Pattern recognition receptors. Pure Appl. Chem. 86(10):1483-1538, 2014. http://dx.doi.org/10.1515/pac-2013-1026Search in Google Scholar
3. Gubala, V., R. Klein, D.M. Templeton, and M. Schwenk, Immunodiagnostics and immunosensor design. Pure Appl. Chem. 86(10):1539-1571, 2014. http://dx.doi.org/10.1515/pac-2013-1027Search in Google Scholar
4. Klein, R., D.M. Templeton, and M. Schwenk, Applications of immunochemistry in human health: Advances in vaccinology and antibody design. Pure Appl. Chem. 86(10):1573-1617, 2014. http://dx.doi.org/10.1515/pac-2013-1028Search in Google Scholar
©2015 by Walter de Gruyter Berlin/Boston
Articles in the same Issue
- Masthead - Full issue pdf
- Oficer’s Column
- The First IUPAC Congress in Korea
- Features
- Books of Secrets: Writing & Reading Alchemy
- What's in a Name? Quite a Lot, as it Happens!
- Systematic Flexibility and the History of the IUPAC Nomenclature of Organic Chemistry
- IUPAC Wire
- Election of IUPAC Officers and Bureau Members – Call for Nominations
- Measurements and Light
- ICSU announces Dr. Heide Hackmann to be Executive Director and Dr. Lucilla Spini to be Head of Science Programmes
- The 2014 AAAS Award for Science Diplomacy goes to Zafra M. Lerman
- OPCW-The Hague Award Presented at 19th Conference of States Parties
- IUPAC Physical Chemistry Cartoon Competition 2015
- IUPAC Office move
- IUPAC Provisional Recommendations
- Glossary of Terms Used in Neurotoxicology
- Stamps International
- Otto Wichterle: An Eye for Hydrogels
- Making an ImPACt
- Immunochemical Recognition and its Diagnostic and Therapeutic Applications
- NOTeS
- On the Use of Quantity Calculus
- Conference Call
- Pesticide Chemistry
- Photobiology
- Solubility Phenomena and Related Equilibrium Processes
- Data Sharing for Sustainability
- Where 2B & Y
- Mark Your Calendar
Articles in the same Issue
- Masthead - Full issue pdf
- Oficer’s Column
- The First IUPAC Congress in Korea
- Features
- Books of Secrets: Writing & Reading Alchemy
- What's in a Name? Quite a Lot, as it Happens!
- Systematic Flexibility and the History of the IUPAC Nomenclature of Organic Chemistry
- IUPAC Wire
- Election of IUPAC Officers and Bureau Members – Call for Nominations
- Measurements and Light
- ICSU announces Dr. Heide Hackmann to be Executive Director and Dr. Lucilla Spini to be Head of Science Programmes
- The 2014 AAAS Award for Science Diplomacy goes to Zafra M. Lerman
- OPCW-The Hague Award Presented at 19th Conference of States Parties
- IUPAC Physical Chemistry Cartoon Competition 2015
- IUPAC Office move
- IUPAC Provisional Recommendations
- Glossary of Terms Used in Neurotoxicology
- Stamps International
- Otto Wichterle: An Eye for Hydrogels
- Making an ImPACt
- Immunochemical Recognition and its Diagnostic and Therapeutic Applications
- NOTeS
- On the Use of Quantity Calculus
- Conference Call
- Pesticide Chemistry
- Photobiology
- Solubility Phenomena and Related Equilibrium Processes
- Data Sharing for Sustainability
- Where 2B & Y
- Mark Your Calendar

