The impact of exercise on mitochondrial biogenesis in skeletal muscle: A systematic review and meta-analysis of randomized trials
-
Diana Marisol Abrego-Guandique
and Roberto Cannataro
Abstract
The interaction between exercise and mitochondrial biogenesis in skeletal muscle is fundamental to human physiology, with important implications for health and athletic performance. While exercise is known to stimulate mitochondrial biogenesis, the effectiveness of varying-intensity exercise remains unclear. This systematic review and meta-analysis aimed to evaluate the impact of physical activity on mitochondrial biogenesis pathways in skeletal muscle and identify key biomolecular markers in healthy individuals. Among these, PGC-1α emerged as the most consistently reported marker. The meta-analysis showed a significant increase in PGC-1α expression following endurance exercise, with a pooled effect size of Hedge’s g = 1.17 (95% confidence interval: 0.14–2.19, I 2 = 84.5%), indicating a large effect with substantial heterogeneity. Subgroup analyses revealed that both interval and continuous endurance training produced large effects (Hedge’s g = 1.29 and 1.01, respectively), with no significant difference between modalities (p > 0.05). These findings confirm that exercise induces significant molecular and structural mitochondrial adaptations, with responses influenced by exercise type, intensity, and duration. This underscores exercise as a potent stimulus for mitochondrial biogenesis, supporting its role in promoting metabolic health and physical performance.
Abbreviations
- AMPK
-
AMP-activated protein kinase
- ATP
-
Adenosine triphosphate
- CaMK
-
Calcium/calmodulin-dependent protein kinase
- CREB
-
cAMP response element-binding protein
- MDPs
-
Mitochondrial-derived peptides
- MOTS-c
-
Mitochondrial open reading frame of the 12S rRNA-c
- mtDNA
-
Mitochondrial DNA
- NRF-1
-
Nuclear respiratory factor 1
- NRF-2
-
Nuclear respiratory factor 2
- p38MAPK
-
p38 mitogen-activated protein kinase
- p53
-
Tumor protein p53
- PGC-1α
-
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- PKA
-
Protein kinase A
- SIRT1
-
Silent mating type information regulation 2 homolog 1
- TFAM
-
Mitochondrial transcription factor A
- VO₂peak
-
Peak oxygen uptake
Introduction
Skeletal muscle is one of the human body’s most dynamic and plastic tissues [1]. Motor neurons’ movement is under voluntary control, integrating central nervous system stimulus and focus, to produce force and enable locomotion [1,2]. Most cellular functions that regulate movement depend significantly on mitochondrial activity [3]. These organelles are critical for regulating overall metabolic status [4] and play a crucial role in maintaining cellular homeostasis [5]. As the primary site for cellular respiration, these organelles employ oxidative phosphorylation to convert the reducing equivalent coming from nutrient metabolism into adenosine triphosphate (ATP), the coin of cellular energy [6]. During exercise, the demand for ATP surges in skeletal muscles, intensifying during contractions [7], and an inflammation grade is reported [8]. This prompts the mitochondria to an adaptive response, leading to an upregulation of mitochondrial mediator linked to their biogenesis. Mitochondrial biogenesis is the process of generating new mitochondria within cells [9], which involves joining and separating the mitochondrial network through fusion and fission, respectively. It plays an important role in cellular adaptation to exercise [10]. The main regulators of mitochondrial biogenesis, are transcription factors nuclear respiratory factors 1 and 2 (NRF-1 and NRF-2), the mitochondrial transcription factor A (TFAM), the p53, and the transcriptional coactivator peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) [11,12,13]. This latter is present in tissues with a high capacity of mitochondrial systems [14]. The expression of the PGC-1α gene is rapidly induced by exposure to cold [15], acute exercise, and fasting [16]. In all these physiological situations, the demand for energy in the form of heat or ATP is increased [17]. It has been shown that skeletal muscle contains certain levels of PGC-1α in the cytoplasm. Through the stimulus of exercise, these levels are activated and mobilized into the nucleus, where they act as coactivators of the transcription factors NRF-1/2, facilitating the synthesis of mitochondrial proteins. This is the first phase of mitochondrial biogenesis [18]. Some studies have reported that an increase in aerobic capacity due to training is in part due to improvements in mitochondrial biogenesis and function [19,20,21,22]; furthermore, in a recent meta-analysis, the majority of the results suggested that exercise improves mitochondrial morphology and biogenesis in cardiovascular diseases patients [23]. This systematic review and meta-analysis aimed to investigate the key biomolecular markers of mitochondrial biogenesis in healthy individuals.
Materials and methods
The systematic review was conducted according to the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines [24] and registered at the PROSPERO International Prospective Registry (CRD42024522994). The methodological approach is based on three steps: (i) paper location and selection, (ii) paper analysis, and (iii) results presentation. We adopted a semi-automated approach using the mySLR platform [25], a robust and affordable platform successfully used previously by our group [26,27,28].
Paper location and selection
Two investigators (N.M.A.R. and D.M.A.-G.) independently searched the PubMed and Web of Science databases to identify publications in peer-reviewed journals published before March 30, 2025. The search was conducted using the Boolean operators “AND” and “OR” to combine the following terms: (“NRF” OR “Nuclear Respiratory Factor” OR “TFAM” OR “Mitochfondrial transcription factor A” OR “PGC-1α” OR “PGC-1 ALPHA” OR “p-53” OR “Transcription Factor p53” OR “SIRT1” OR “silent mating type information regulation homolog 1”) AND (“mitochondria” OR “mitochondrial biogenesis” OR “mitochondrion biogenesis” OR “mitochondria autophagy” OR “mitochondrial fission/fusion” OR “mitochondrial content” OR “Mitochondrial transcription” OR “mitochondria signaling”) AND (“skeletal muscle” OR “muscular tissue” OR “skeleton”) AND (“physical activity” OR “physical exercise” OR “training” OR “aerobic training” OR “anaerobic training” OR “endurance” OR “exercise”).
Study selection and data extraction
Studies included assessed mitochondrial biogenesis in skeletal muscle during different types of exercise, all of which were published before March 2025. The inclusion criteria for this systematic review were as follows:
Population: studies in humans conducted with participants ≥18 years of age.
Exposure: studies that reported skeletal muscle mitochondrial biogenesis during exercise.
Outcomes: studies that provided sufficient information about mitochondrial biogenesis signaling in muscle biopsies.
This review includes randomized trials (RT) or randomized crossover trials published in English over the last 10 years (data: January 2014–March 2025). Articles were excluded from the review for the following reasons: studies not published in English or prior to March 2014; reviews, meta-analyses, letters, conference papers, comments, or book chapters; and studies on animal models or in vitro experiments. Data from all included articles were extracted by one author (N.M.A.R.) and checked by two authors (D.M.A.-G. and E.C.). The following information was recorded: authors’ names, publication year, study country, study design, participant characteristics (sample size, gender), physical activity, outcomes of interest, and results.
Quality assessment
The possibility of bias in the design and analysis of each study was assessed by two different evaluators (N.M.A.R. and D.M.A.-G.) using the NIH Study Quality Assessment Tool (https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools, accessed March 12, 2025), and three additional reviewers were consulted when necessary (R.C., E.C., and A.C.). Specifically, the Quality Assessment of Controlled Intervention Studies was used; both forms have 14 questions designed to help focus on the key concepts for evaluating the internal validity of a study, such as the risk of potential selection, method, information, measurement, and confounding bias.
Statistical analysis
Data for PGC-1α mRNA expression were extracted. Values were digitized from figures using PlotDigitizer PRO v3.3.9 (2025) when not reported. Effect sizes (Cohen’s d) were calculated to compare pre- and post-exercise in interval and continuous endurance conditions. Cohen’s d values were converted to Hedge’s g to correct for small sample bias. Standard errors and 95% confidence intervals were computed for each effect size. Individual Hedge’s g values were then pooled to estimate the overall effect of exercise on PGC-1α expression. All statistical tests in this study were carried out using fixed and random effect designs implemented in the R studio, version 3.2.4.
Results
In Figure 1, a total of 1,104 records were identified through the database search. After removing 441 duplicates, 663 records remained for screening. A total of 92 full-text articles were assessed for eligibility, of which 79 were excluded for the following reasons: animal studies (n = 15), not aligned with the topic (n = 29), published before 2014 (n = 15), or not RT (n = 20). Thus, a total of 13 studies were included in the final systematic review. Although the primary aim of this review was to assess the impact of exercise on mitochondrial biogenesis in healthy individuals, studies involving participants with obesity were included when no additional metabolic disorders were present [29], as obesity alone does not inherently constitute a diseased state.
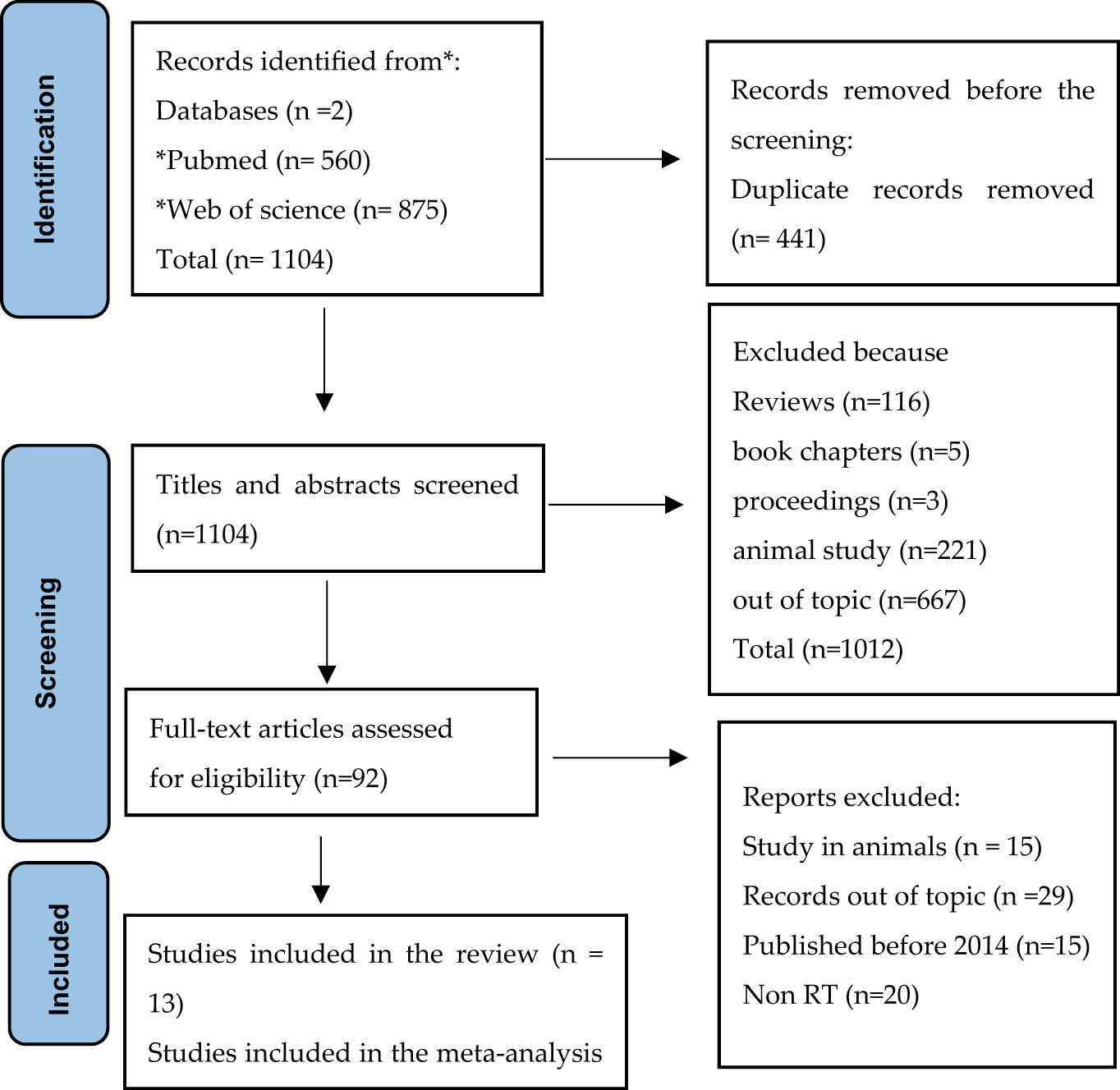
PRISMA flow diagram showing the algorithm for selecting eligible studies. RT, randomized trial.
Characteristics of the studies included
Table 1 presents the characteristics of the included studies. Twelve randomized controlled trials were eligible for this systematic review. Of these, eight were parallel-group randomized controlled trials [29,30,31,32,33,34,35,36,37] and four were randomized crossover trials [38,39,40,41]. These studies were published between 2014 and 2021 and conducted in eight countries: three in Sweden [30,35,40], three in Australia [31,33,36], two in Denmark [32,38], and one in Canada, the United States, the United Kingdom, Brazil, and South Africa [29,34,37,39,41].
Summary of studies on exercise and mitochondrial biogenesis
| Study | Location | Study design | n | Condition | Exercise type | Mitochondrial pathway | Results |
|---|---|---|---|---|---|---|---|
| Von Walden et al. [30] | Sweden | Randomized controlled trial | 30 | Recreationally active adults (both sexes) | Acute endurance vs resistance exercise (cycling, leg press/knee extension) | PGC-1α Humanin, MOTS-c | Acute EE and RE promoted changes in skeletal muscle gene expression (PGC-1α) typically seen in response to each exercise modality |
| Acute endurance exercise increased circulating MDPs (humanin and MOTS-c), indicating systemic mitochondrial signaling; resistance training did not alter levels | |||||||
| Mendham et al. [29] | South Africa | Randomized controlled trial | 35 | Obese sedentary women | Combined aerobic and resistance exercise (12 weeks) | Mitochondrial respiration, PGC-1α | Exercise increased mitochondrial respiration; however, the mitochondrial changes were content-driven and not associated with insulin sensitivity. No changes in PGC-1α |
| Almquist et al. [40] | Sweden | Randomized crossover trial | 10 | Healthy men | Repeated sprint exercise | Mitochondrial proteins, gene expression PGC-1α | Repeated sprint exercise led to upregulating mitochondrial biogenesis markers, including gene expression related to energy metabolism |
| Andrade-Souza et al. [39] | Brazil | Randomized crossover trial | 8 | Healthy active men | Twice-a-day vs once-daily cycling | PGC-1α, TFAM, nuclear and cytosolic | Twice-a-day exercise enhanced transcription of PGC-1α, TFAM, more than once-daily or control, suggesting potentiated mitochondrial biogenic signaling |
| Fiorenza et al. [38] | Denmark | Randomized crossover trial | 12 | Endurance-trained men | SE, RS, and CM | PGC-1α, TFAM, NRF-2 | The exercise-induced PGC-1α mRNA response was higher in SE and CM than in RS, with no difference between SE and CM. Muscle NRF-2, TFAM, MFN2, DRP1, and SOD2 mRNA content was elevated to the same extent by SE and CM, while RS did not affect these mRNAs |
| Taylor et al. [41] | UK | Randomized crossover trial | 8 | Trained male cyclists | All-out sprint cycling interval (INT) vs continuous cycling (2 min total) (CON) | PGC-1α, AMPK | Interval and continuous sprint cycling matched for the duration, similarly increased AMPK phosphorylation and mRNA expression of PGC-1α |
| Tachtsis et al. [31] | Australia | Randomized controlled trial | 16 | Healthy untrained men | Single bout of endurance exercise vs control | p53, PGC-1α | Acute endurance exercise increased nuclear p53 but not PGC-1α expression, suggesting p53 may initiate autophagy prior to mitochondrial biogenesis |
| Skovgaard et al. [32] | Denmark | Randomized controlled trial | 17 | Young healthy men; trained | Cycling speed endurance exercise (S), endurance exercise (E), and speed endurance followed by endurance exercise (S + E) | PGC-1α | Combined speed endurance and endurance exercise resulted in greater increases in muscle PGC-1α mRNA than either exercise alone. Speed endurance alone increased PGC-1α, whereas endurance exercise alone did not |
| Mendham et al. [33] | Australia | Randomized controlled trial | 33 | Middle-aged inactive men | Small-sided games (SSG) vs cycling (CYC) or control (CON) (12 w) | PGC-1α, SIRT1, p53, TFAM, NRF-1, NRF-2, Complexes I–V | No changes were observed in the skeletal muscle protein content of mitochondrial biogenesis markers despite improvements in metabolic health indicators with both SSG and CYC training |
| Irving et al. [34] | USA | Randomized controlled trial | 65 | Healthy young and older sedentary adults | Endurance (ET), resistance (RT), and combination (CT) training | PGC-1α, TFAM, SIRT3, NRF-1, NRF-2, Complexes III-IV mRNA and proteins | PGC-1α ET and CT significantly increased total PGC-1α, SIRT3 expression increased with all training types. TFAM CT showed a trend toward increased expression, though changes were not at the mRNA level. COX3 & COX4 ET and CT significantly upregulated both genes. Protein Expression. ET significantly increased PGC-1α, and CT showed a trend. SIRT3 protein levels increased with ET, RT, and CT. TFAM protein tended to increase with CT. COX3 and COX4 proteins, as part of total OXPHOS content, also increased with ET and CT |
| Gidlund et al. [35] | Sweden | Randomized controlled trial | 20 | Healthy untrained (both sexes) | Single session of endurance cycling or control | PGC-1α-b protein | PGC-1α-b protein levels increased rapidly post-exercise, indicating its role as an early regulator of mitochondrial biogenesis |
| Granata et al. [36] | Australia | Randomized controlled trial | 29 | Healthy men, moderately training | Different intensities of cycling SIT, HIIT, and STCT (12 cycling sessions, 4 weeks) | PGC-1α, p53, TFAM | The maximal mitochondrial respiration in permeabilized muscle fibers increased significantly only after SIT. Similarly, the protein content of PGC-1α, p53, increased only after SIT. Conversely, citrate synthase activity, the protein content of TFAM, and subunits of the electron transport system complexes remained unchanged |
| MacNeil et al. [37] | Canada | Randomized controlled trial | 18 | Young adults untrained (both sexes) | Sessions at gradually increasing intensities, endurance exercise preceding (END.RES) or following (RES.END) resistance exercise (3 times per week/6 weeks) | PGC-1α, NRF-1, TFAM | A single high-intensity interval session increased PGC-1α and TFAM mRNA levels; repeated sessions augmented mitochondrial protein content |
PGC-1α, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; SIRT, sirtuin; NRF-1, nuclear respiratory factor 1; NRF-2, nuclear factor erythroid 2 like 2; p53, tumor protein p53; TFAM, mitochondrial transcription factor A; END, endurance; RES, resistance; CON, control; CYC, cycling; SIT, interval training; HIIT, high-intensity interval training; STCT, continuous training; SE, speed endurance.
Three studies included both sexes [30,37,42], while the remaining seven focused solely on male participants [31,32,33,36,38,39,40,41]. Only one study was conducted exclusively in women [29].
The systematic review included studies reporting outcomes related to different types of exercise or training in terms of biochemical signals of mitochondrial biogenesis. Based on existing literature, Figure 2 illustrates the potential pathways that may be modulated by physical activity. All 13 studies assessed markers of mitochondrial biogenesis. The most frequently measured signal was PGC-1α. Other key markers included TFAM [33,34,36–39], NRF-1/NRF-2 [33,37,38], SIRT1 [33], and p53 [31,33,36]. Von Walden et al. reported that mitochondrial-derived peptides (MDPs), including human and MOTS-c, serve as systemic markers of mitochondrial signaling [30]. Some studies also include other signals, such as SOD2, complex I–IV, and citrate synthase.
![Figure 2
Molecular pathways are potentially modulated by physical activity and associated with mitochondrial biogenesis. This conceptual model illustrates the central signaling cascades and transcriptional regulators activated by various forms of physical activity, such as endurance and resistance training. Key pathways include AMPK, p38MAPK, CaMK, and SIRT1, which converge on the activation of PGC-1α, leading to the expression of nuclear-encoded mitochondrial genes (e.g., TFAM, NRF-1/2) and promoting mitochondrial biogenesis [43]. Created with BioRender. PGC-1α, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; SIRT, sirtuin; NRF-1, nuclear respiratory factor 1; NRF-2, nuclear factor erythroid 2 like 2; p53, tumor protein p53; TFAM, mitochondrial transcription factor A; mtDNA, mitochondrial DNA; P, phosphate; NAD/NADH, nicotinamide adenine dinucleotide/nicotinamide adenine dinucleotide hydrogenated; CREB, cAMP response element-binding protein; PKA, protein kinase A; p38MAPK, p38 mitogen-activated protein kinase; CaMK, calcium/calmodulin-dependent protein kinase; AMPK, AMP-activated protein kinase; AMP/ATP, adenosine monophosphate/adenosine triphosphate.](/document/doi/10.1515/bmc-2025-0055/asset/graphic/j_bmc-2025-0055_fig_002.jpg)
Molecular pathways are potentially modulated by physical activity and associated with mitochondrial biogenesis. This conceptual model illustrates the central signaling cascades and transcriptional regulators activated by various forms of physical activity, such as endurance and resistance training. Key pathways include AMPK, p38MAPK, CaMK, and SIRT1, which converge on the activation of PGC-1α, leading to the expression of nuclear-encoded mitochondrial genes (e.g., TFAM, NRF-1/2) and promoting mitochondrial biogenesis [43]. Created with BioRender. PGC-1α, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; SIRT, sirtuin; NRF-1, nuclear respiratory factor 1; NRF-2, nuclear factor erythroid 2 like 2; p53, tumor protein p53; TFAM, mitochondrial transcription factor A; mtDNA, mitochondrial DNA; P, phosphate; NAD/NADH, nicotinamide adenine dinucleotide/nicotinamide adenine dinucleotide hydrogenated; CREB, cAMP response element-binding protein; PKA, protein kinase A; p38MAPK, p38 mitogen-activated protein kinase; CaMK, calcium/calmodulin-dependent protein kinase; AMPK, AMP-activated protein kinase; AMP/ATP, adenosine monophosphate/adenosine triphosphate.
Considering the type of exercise used and the specificity of training, resistance, and endurance, or a combination of both training types, were evaluated in our review. Exercise modalities varied considerably across the included studies. A total of 12 studies incorporated endurance-based protocols, either as stand-alone interventions or in combination with other training types [30–39]. Only one study exclusively implemented sprint exercise without an endurance component [40]. Resistance training was explicitly included in two studies [34,37], while three studies applied combined endurance and resistance training protocols [32,33,37]. While the majority of interventions were acute, focusing on responses to single bouts of exercise, four studies employed longer-term (chronic) training protocols ranging from 4 to 12 weeks [29,33,36,37], providing insight into sustained adaptations in mitochondrial biogenesis.
All the studies selected in our review use muscle biopsy as a parameter to evaluate biogenesis after exercise, and protocols include biopsies collected pre- and post-training. Biopsies provide an important tool for studying and diagnosing different mitochondrial biogenesis signaling pathways in skeletal muscle, as they are not easily measured or detected in other tissues. Considering that evaluating several parameters, such as mitochondrial DNA deletion or mitochondrial content/volume, is more readily detectable in muscle tissue than in other tissues or serum, including blood.
Meta-analysis of the effects of endurance exercise on PGC-1α mRNA expression
Seven studies were included in the meta-analysis to evaluate the effect of endurance exercise on PGC-1α mRNA expression. When analyzed by exercise modality, interval training showed a pooled effect size of Hedge’s g = 1.29 (95% confidence interval [CI]: −0.18 to 2.76), indicating a large but statistically non-significant increase in PGC-1α expression, with moderate heterogeneity (I 2 = 72%, p = 0.0065). Continuous endurance exercise yielded a similar pooled effect size of Hedge’s g = 1.01 (95% CI: −1.17 to 3.19), though with very high heterogeneity (I 2 = 90.8%, p < 0.0001), reflecting substantial variability across studies. The overall pooled effect across all studies was Hedge’s g = 1.17 (95% CI: 0.14–2.19), suggesting a statistically significant and large increase in PGC-1α mRNA expression following endurance exercise. However, the wide prediction interval (−1.62 to 3.95) indicates considerable variation in potential future study outcomes. No significant differences were found between exercise modalities (p > 0.05), suggesting that both interval and continuous endurance exercise similarly enhance PGC-1α expression (Figure 3).

Forest plot of the meta-analysis examining the effects of endurance exercise on PGC-1α mRNA expression. Effect sizes (Hedge’s g) and 95% CI are presented for each study, grouped by exercise modality (interval vs continuous). Diamonds represent pooled effect sizes using a random-effects model (HK method). The vertical line indicates the null effect. Square sizes reflect the weight of each study. Heterogeneity is reported as I 2 and τ 2. No significant differences were found between exercise modalities (p > 0.5). The red line represents the prediction interval, indicating the expected range of effects in future studies.
Discussion
The present systematic review and meta-analysis aimed to assess the potential impact of exercise on mitochondrial biogenesis in subjects with apparent healthy status focusing on PGC1α as a biomarker. This enables us to organize the outcomes systematically and comprehensively. Exercise is widely recognized as a potent stimulus for activating key signaling pathways that promote mitochondrial adaptations in skeletal muscle [43]. These adaptations are crucial for enhancing muscular endurance and metabolic efficiency. Exercise protocols must be carefully designed to effectively induce such physiological changes, particularly in intensity and duration. A central player in the adaptive process is PGC-1α, a transcriptional coactivator that regulates mitochondrial biogenesis and muscle remodeling [18,44]. Its activation through exercise highlights the importance of structured training programs designed to optimize muscular and metabolic health. Several studies have demonstrated that PGC-1α is highly sensitive to physical activity’s intensity, frequency, and metabolic demands. Gidlund and co-workers observed a rapid increase in PGC-1α protein levels in 2 h post-exercise, indicating a swift molecular response to endurance-type activity [35]. Similarly, Granata and co-workers reported increased levels of both PGC-1α and the tumor suppressor protein p53 following high-intensity interval training (HIIT), underlining the importance of exercise intensity in activating mitochondrial signaling pathways [36]. Supporting this, Fiorenza and co-workers demonstrated that low-volume, high-intensity exercise induces significant transcription of PGC-1α, highlighting the transcriptional sensitivity of this gene to both intensity and metabolic stress [38]. In addition, Bishop et al. describe that HIIT has also been shown to induce more remarkable mitochondrial protein synthesis, and this latter may best reflect exercise-induced mitochondrial biogenesis [45]. Two groups by Tachtsis and Andrade-Souza emphasized that PGC-1α expression is also influenced by factors such as training frequency and nutrient availability, particularly in “train-low” strategies protocols [31,39]. Similar results were obtained by Conceição and co-workers supporting that low-intensity exercise has lower effectiveness in stimulating mitochondrial biogenesis [46]. However, the response to exercise is not uniform across all populations or training contexts. Notably, Mendham et al. reported no significant change in PGC-1α protein content after 8 weeks of either cycling or small-sided games training in previously inactive men, suggesting that baseline metabolic health or training status may modulate the exercise response [33]. These findings align with the current meta-analysis, which revealed a large and statistically significant overall increase in PGC-1α expression following endurance exercise (Hedge’s g = 1.17, 95% CI: 0.14–2.19), although with substantial heterogeneity (I 2 = 84.5%). While both interval and continuous endurance training elicited large effect sizes (1.29 and 1.01, respectively), the differences between modalities were not statistically significant (p > 0.05), reinforcing the view that various forms of endurance exercise can effectively promote mitochondrial gene expression, depending on contextual and individual factors.
TFAM plays a crucial role in the transcription and replication of mitochondrial DNA, and its expression appears to be influenced by both the type and context of exercise. However, research has shown that the response of TFAM to physical activity can vary depending on several factors. For example, Andrade-Souza et al. and Fiorenza et al. observed increased TFAM mRNA expression following high-intensity exercise, indicating that acute bouts of intense physical stress can stimulate the transcriptional activity of mitochondrial genes [38,39]. This aligns with the broader understanding of intensity as a key driver of mitochondrial adaptation. Extending these findings, Irving et al. reported significant elevations in TFAM and other mitochondrial regulators, such as NRF-1 and SIRT3, after an 8-week combined endurance and resistance training program [34]. Importantly, these adaptations were evident in both young and older adults, suggesting that age does not necessarily blunt the mitochondrial response when an appropriate training stimulus is applied. Conversely, Mendham et al. found no change in TFAM protein content following a similar training duration, consistent with their earlier observations regarding PGC-1α [33]. These findings imply that population-specific factors, such as initial fitness level, metabolic health, or training modality, may influence the extent of mitochondrial gene activation in response to exercise.
The type of exercise performed plays a crucial role in determining the extent and nature of mitochondrial adaptations. The training modality can significantly impact mitochondrial biogenesis and function, whether endurance, resistance, or a combination of both. Irving and co-workers demonstrated that combined endurance and resistance training elicited greater mitochondrial adaptations compared to either modality alone [34]. Supporting this, Mendham and co-workers found that combined training significantly improved mitochondrial respiration in obese women, underscoring its potential effectiveness even in metabolically compromised populations [29]. Combining exercise upregulates PGC-1α and enhances mitochondrial enzyme activity, muscle mass, strength, and VO₂peak. However, exercise order only affected complex II protein content [37]; other training strategies may elicit more pronounced molecular responses. In particular, speed endurance training, especially when combined with traditional endurance exercise, induces a more robust transcriptional response in pathways related to mitochondrial function, metabolism, and vascular adaptation than endurance training alone [32]. Concurrent resistance and endurance training in different sequences may alter mitochondrial biogenesis signals, likely due to prior activation of PGC-1α pathways, with a mechanism involving mTOR signaling [47]. Beyond skeletal muscle, systemic effects of endurance exercise have also been observed. Von Walden et al. reported increased circulating levels of MDPs such as MOTS-c and humanin following acute endurance activity, suggesting that mitochondrial signaling extends beyond local tissue responses [30]. At the same time, a growing body of research is linking molecular adaptations with functional performance outcomes. High-intensity training elevated mitochondrial gene expression and resulted in measurable improvements in endurance capacity [32,38]. Similarly, Irving and co-workers associated increased mitochondrial protein expression with gains in VO₂peak and positive shifts in body composition, demonstrating these molecular changes’ clinical and physiological relevance [34]. This systematic review with meta-analysis highlights the significant role of exercise in stimulating mitochondrial biogenesis, particularly through the activation of PGC1α. Evidence consistently supports that exercise intensity, frequency, and modality are critical factors influencing the extent of mitochondrial adaptations. While endurance and resistance exercises each offer benefits, their combination tends to elicit more robust and comprehensive adaptations, enhancing both muscular and systemic responses. However, individual variability such as baseline fitness, age, and training history can modulate the molecular response to exercise, suggesting the need for personalized training approaches. Importantly, improvements in mitochondrial density may benefit for healthy individuals and offer therapeutic potential in pathological conditions. In particular, disorders like sarcopenia, characterized by a decline in muscle mass, and conditions like lipedema [48]. Since biopsy is an invasive procedure, a promising biomarker candidate can be represented by microRNAs [47], which are also used in diagnostic procedures [49].
Conclusions
Overall, exercise emerges as a powerful, non-pharmacological tool for enhancing mitochondrial function via PCG1α expression, with significant implications for both metabolic health and disease management. Future research should continue to refine optimal training strategies and explore pathological factors influence mitochondrial responsiveness to exercise.
-
Funding information: This research was funded by “The Next Generation EU – Italian NRRP, Mission 4, Component 2, Investment 1.5, call for the creation and strengthening of ‘Innovation Ecosystems’, building ‘Territorial R&D Leaders’ (Directorial Decree n. 2021/3277) – project Tech4You – Technologies for climate change adaptation and quality of life improvement, n. ECS0000009” to Erika Cione.
-
Author contributions: Conceptualization, E.C. and R.C.; methodology, N.M.A.R. and D.M.A.-G.; writing – original draft preparation, N.M.A.R. and D.M.A.-G.; writing – review and editing A.C., E.C., and R.C.; visualization A.C. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References
[1] Mukund K, Subramaniam S. Skeletal muscle: A review of molecular structure and function, in health and disease. WIREs Syst Biol Med. 2020;12(1):e1462. 10.1002/wsbm.1462.Search in Google Scholar PubMed PubMed Central
[2] Elechi JOG, Guandique DMA, Cannataro R. Creatine in cognitive performance: a commentary. Curr Mol Pharmacol. 2024;17:e18761429272915.10.2174/0118761429272915231122112748Search in Google Scholar PubMed
[3] Dong H, Tsai S-Y. Mitochondrial properties in skeletal muscle fiber. Cells. 2023;12(17):2183. 10.3390/cells12172183.Search in Google Scholar PubMed PubMed Central
[4] Hood DA, Memme JM, Oliveira AN, Triolo M. Maintenance of skeletal muscle mitochondria in health, exercise, and aging. Annu Rev Physiol. 2019;81(1):19–41. 10.1146/annurev-physiol-020518-114310.Search in Google Scholar PubMed
[5] Chen X, Ji Y, Liu R, Zhu X, Wang K, Yang X, et al. Mitochondrial dysfunction: roles in skeletal muscle atrophy. J Transl Med. 2023;21(1):503. 10.1186/s12967-023-04369-z.Search in Google Scholar PubMed PubMed Central
[6] Casanova A, Wevers A, Navarro-Ledesma S, Pruimboom L. Mitochondria: It is all about energy. Front Physiol. 2023;14:1114231, https://www.frontiersin.org/articles/10.3389/fphys.2023.1114231.10.3389/fphys.2023.1114231Search in Google Scholar PubMed PubMed Central
[7] Baker JS, McCormick MC, Robergs RA. Interaction among skeletal muscle metabolic energy systems during intense exercise. J Nutr Metab. 2010;2010:905612. 10.1155/2010/905612.Search in Google Scholar PubMed PubMed Central
[8] Cannataro R, Abrego-Guandique DM, Straface N, Cione E. Omega-3 and sports: focus on inflammation. Life. 2024;14(10):1315. 10.3390/life14101315.Search in Google Scholar PubMed PubMed Central
[9] Chen W, Zhao H, Li Y. Mitochondrial dynamics in health and disease: mechanisms and potential targets. Signal Transduct Target Ther. 2023;8(1):333. 10.1038/s41392-023-01547-9.Search in Google Scholar PubMed PubMed Central
[10] Russell AP, Foletta VC, Snow RJ, Wadley GD. Skeletal muscle mitochondria: A major player in exercise, health and disease. Biochim Biophys Acta - Gen Subj. 2014;1840(4):1276–84. 10.1016/J.BBAGEN.2013.11.016.Search in Google Scholar
[11] Gordon JW, Rungi AA, Inagaki H, Hood DA. Selected contribution: effects of contractile activity on mitochondrial transcription factor A expression in skeletal muscle. J Appl Physiol. 2001;90(1):389–96. 10.1152/jappl.2001.90.1.389.Search in Google Scholar PubMed
[12] Hood DA. Mechanisms of exercise-induced mitochondrial biogenesis in skeletal muscle. Appl Physiol Nutr Metab. 2009;34(3):465–72. 10.1139/H09-045.Search in Google Scholar PubMed
[13] Mesquita P, Vann CG, Phillips SM, McKendry J, Young KC, Kavazis AN, et al. Skeletal muscle ribosome and mitochondrial biogenesis in response to different exercise training modalities. Front Physiol. 2021;12:725866, https://www.frontiersin.org/journals/physiology/articles/10.3389/fphys.2021.725866.10.3389/fphys.2021.725866Search in Google Scholar PubMed PubMed Central
[14] Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, et al. Exercise stimulates Pgc-1α transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem. 2005;280(20):19587–93. 10.1074/jbc.M408862200.Search in Google Scholar PubMed
[15] Larson C, Opichka M, McGlynn ML, Collins CW, Slivka D. Exercise- and cold-induced human PGC-1α mRNA isoform specific responses. Int J Environ Res Public Health. 2020;17(16):5740. 10.3390/ijerph17165740.Search in Google Scholar PubMed PubMed Central
[16] Kristensen CM, Olsen MA, Jessen H, Brandt N, Meldgaard JN, Pilegaard H. PGC-1α in exercise and fasting-induced regulation of hepatic UPR in mice. Pflügers Arch - Eur J Physiol. 2018;470(10):1431–47. 10.1007/s00424-018-2159-3.Search in Google Scholar PubMed PubMed Central
[17] Finck BN, Kelly DP. PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116(3):615–22. 10.1172/JCI27794.Search in Google Scholar PubMed PubMed Central
[18] Jung S, Kim K. Exercise-induced PGC-1α transcriptional factors in skeletal muscle. Integr Med Res. 2014;3(4):155–60. 10.1016/j.imr.2014.09.004.Search in Google Scholar PubMed PubMed Central
[19] Di Donato DM, West DWD, Churchward-Venne TA, Breen L, Baker SK, Phillips SM. Influence of aerobic exercise intensity on myofibrillar and mitochondrial protein synthesis in young men during early and late postexercise recovery. Am J Physiol Metab. 2014;306(9):E1025–32.10.1152/ajpendo.00487.2013Search in Google Scholar PubMed PubMed Central
[20] Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, et al. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16(14):1879–86. 10.1096/fj.02-0367com.Search in Google Scholar PubMed
[21] Bori Z, Zhao Z, Koltai E, Fatouros IG, Jamurtas AZ, Douroudos II, et al. The effects of aging, physical training, and a single bout of exercise on mitochondrial protein expression in human skeletal muscle. Exp Gerontol. 2012;47(6):417–24. 10.1016/J.EXGER.2012.03.004.Search in Google Scholar PubMed PubMed Central
[22] Holloszy JO. Biochemical adaptations in muscle: effects of exercise on mitochondrial oxygen uptake and respiratory enzyme activity in skeletal muscle. J Biol Chem. 1967;242(9):2278–82.10.1016/S0021-9258(18)96046-1Search in Google Scholar
[23] Lim AY, Chen Y-C, Hsu C-C, Fu T-C, Wang J-S. The effects of exercise training on mitochondrial function in cardiovascular diseases: a systematic review and meta-analysis. Int J Mol Sci. 2022;23(20):CD013544. 10.3390/ijms232012559.Search in Google Scholar PubMed PubMed Central
[24] Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol. 2009;62(10):1006–12. 10.1016/j.jclinepi.2009.06.005.Search in Google Scholar PubMed
[25] Ammirato S, Felicetti AM, Rogano D, Linzalone R, Corvello V. Digitalising the Systematic Literature Review process: the MySLR platform. Knowl Manag Res Pract. 2022;21:777–94. 10.1080/14778238.2022.2041375.Search in Google Scholar
[26] Saraceno GF, Abrego-Guandique DM, Cannataro R, Caroleo MC, Cione E. Machine learning approach to identify case-control studies on ApoE gene mutations linked to Alzheimer’s disease in Italy. BioMedInformatics. 2024;4(1):600–22. 10.3390/biomedinformatics4010033.Search in Google Scholar
[27] Abrego-Guandique DM, Saraceno GF, Cannataro R, de Burnside MM, Caroleo MC, Cione E. Apolipoprotein E and Alzheimer’s disease in Italian population: systematic review and meta-analysis. Brain Sci. 2024;14(9):908.10.3390/brainsci14090908Search in Google Scholar PubMed PubMed Central
[28] Radbakhsh S, Abrego-Guandique DM, Bacchetti T, Aghaee-Bakhtiari SH, Mahmoudi A, Manteghi AA, et al. Direct hybridization and bioinformatics analysis of circulating microRNAs in patients with Alzheimer’s disease under intravenous trehalose treatment. Brain Res. 2025;1857:149607. 10.1016/j.brainres.2025.149607.Search in Google Scholar PubMed
[29] Mendham AE, Goedecke JH, Zeng Y, Larsen S, George C, Hauksson J, et al. Exercise training improves mitochondrial respiration and is associated with an altered intramuscular phospholipid signature in women with obesity. Diabetologia. 2021;64:1642–59. 10.1007/s00125-021-05430-6/Published.Search in Google Scholar
[30] Von Walden F, Fernandez-Gonzalo R, Norrbom J, Emanuelsson EB, Figueiredo VC, Gidlund EK, et al. Acute endurance exercise stimulates circulating levels of mitochondrial-derived peptides in humans. J Appl Physiol. 2021;131(3):1035–42. 10.1152/japplphysiol.00706.2019.Search in Google Scholar PubMed
[31] Tachtsis B, Smiles WJ, Lane SC, Hawley JA, Camera DM. Acute endurance exercise induces nuclear p53 abundance in human skeletal muscle. Front Physiol. 2016;7:144. 10.3389/fphys.2016.00144.Search in Google Scholar PubMed PubMed Central
[32] Skovgaard C, Brandt N, Pilegaard H, Bangsbo J. Combined speed endurance and endurance exercise amplify the exercise-induced PGC-1α and PDK4 mRNA response in trained human muscle. Physiol Rep. 2016;4(14):e12864. 10.14814/phy2.12864.Search in Google Scholar PubMed PubMed Central
[33] Mendham AE, Duffield R, Coutts AJ, Marino F, Boyko A, Bishop DJ. Rugby-specific small-sided games training is an effective alternative to stationary cycling at reducing clinical risk factors associated with the development of type 2 diabetes: A randomized, controlled trial. PLoS One. 2015;10(6):0127548. 10.1371/journal.pone.0127548.Search in Google Scholar PubMed PubMed Central
[34] Irving BA, Lanza IR, Henderson GC, Rao RR, Spiegelman BM, Sreekumaran Nair K. Combined training enhances skeletal muscle mitochondrial oxidative capacity independent of age. J Clin Endocrinol Metab. 2015;100(4):1654–63. 10.1210/jc.2014-3081.Search in Google Scholar PubMed PubMed Central
[35] Gidlund E-K, Ydfors M, Appel S, Rundqvist H, Sundberg CJ, Norrbom J. Rapidly elevated levels of PGC-1-b protein in human skeletal muscle after exercise: exploring regulatory factors in a randomized controlled trial. J Appl Physiol. 2015;119:374–84. 10.1152/japplphysiol.01000.2014.-In.Search in Google Scholar
[36] Granata C, Oliveira RSF, Little JP, Renner K, Bishop DJ. Training intensity modulates changes in PGC-1α and p53 protein content and mitochondrial respiration, but not markers of mitochondrial content in human skeletal muscle. FASEB J. 2016;30(2):959–70. 10.1096/fj.15-276907.Search in Google Scholar PubMed
[37] MacNeil LG, Glover E, Bergstra TG, Safdar A, Tarnopolsky MA. The order of exercise during concurrent training for rehabilitation does not alter acute genetic expression, mitochondrial enzyme activity or improvements in muscle function. PLoS One. 2014;9:10. 10.1371/journal.pone.0109189.Search in Google Scholar PubMed PubMed Central
[38] Fiorenza M, Gunnarsson TP, Hostrup M, Iaia FM, Schena F, Pilegaard H, et al. Metabolic stress-dependent regulation of the mitochondrial biogenic molecular response to high-intensity exercise in human skeletal muscle. J Physiol. 2018;596(14):2823–40. 10.1113/JP275972.Search in Google Scholar PubMed PubMed Central
[39] Andrade-Souza VA, Ghiarone T, Sansonio A, Santos Silva KA, Tomazini F, Arcoverde L, et al. Exercise twice-a-day potentiates markers of mitochondrial biogenesis in men. FASEB J. 2020;34(1):1602–19. 10.1096/fj.201901207RR.Search in Google Scholar PubMed
[40] Almquist NW, Ellefsen S, Sandbakk Ø, Rønnestad BR. Effects of including sprints during prolonged cycling on hormonal and muscular responses and recovery in elite cyclists. Scand J Med Sci Sport. 2021;31(3):529–41. 10.1111/sms.13865.Search in Google Scholar PubMed PubMed Central
[41] Taylor CW, Ingham SA, Hunt JEA, Martin NRW, Pringle JSM, Ferguson RA. Exercise duration-matched interval and continuous sprint cycling induce similar increases in AMPK phosphorylation, PGC-1α and VEGF mRNA expression in trained individuals. Eur J Appl Physiol. 2016;116(8):1445–54. 10.1007/s00421-016-3402-2.Search in Google Scholar PubMed PubMed Central
[42] Gidlund E, Ydfors M, Appel S, Rundqvist H, Sundberg CJ, Norrbom J. Rapidly elevated levels of PGC-1α-b protein in human skeletal muscle after exercise: exploring regulatory factors in a randomized controlled trial. J Appl Physiol. 2015;119(4):374–84. 10.1152/japplphysiol.01000.2014.Search in Google Scholar PubMed
[43] Memme JM, Erlich AT, Phukan G, Hood DA. Exercise and mitochondrial health. J Physiol. 2021;599(3):803–17. 10.1113/JP278853.Search in Google Scholar PubMed
[44] Norheim F, Langleite TM, Hjorth M, Holen T, Kielland A, Stadheim HK, et al. The effects of acute and chronic exercise on PGC-1α, irisin and browning of subcutaneous adipose tissue in humans. FEBS J. 2014;281(3):739–49. 10.1111/febs.12619.Search in Google Scholar PubMed
[45] Bishop DJ, Botella J, Genders AJ, Lee MJ, Saner NJ, Kuang J, et al. High-intensity exercise and mitochondrial biogenesis: current controversies and future research directions. Physiology. 2019;34(1):56–70.10.1152/physiol.00038.2018Search in Google Scholar PubMed
[46] Conceição MS, Chacon-Mikahil MP, Telles GD, Libardi CA, Júnior EM, Vechin FC, et al. Attenuated PGC-1α isoforms following endurance exercise with blood flow restriction. Med Sci Sports Exerc. 2016;48(9):1699–707. 10.1249/MSS.0000000000000970.Search in Google Scholar PubMed
[47] Zhao Y-C, Gao B. Integrative effects of resistance training and endurance training on mitochondrial remodeling in skeletal muscle. Eur J Appl Physiol. 2024;124(10):2851–65. 10.1007/s00421-024-05549-5.Search in Google Scholar PubMed
[48] Patton L, Ricolfi L, Bortolon M, Gabriele G, Zolesio P, Cione E, et al. Observational study on a large Italian population with lipedema: Biochemical and hormonal profile, anatomical and clinical evaluation, self-reported history. Int J Mol Sci. 2024;25(3):1599. 10.3390/ijms25031599.Search in Google Scholar PubMed PubMed Central
[49] Peronace C, Cione E, Abrego-Guandique DM, Fazio M, Panduri G, Caroleo MC, et al. FAM19A4 and hsa-miR124-2 double methylation as screening for ASC-H- and CIN1 HPV-positive women. Pathogens. 2024;13(4):312. 10.3390/pathogens13040312.Search in Google Scholar PubMed PubMed Central
© 2025 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Research Articles
- Anti-arthritic potential of crude sulfated polysaccharide from marine macroalgae Sargassum ilicifolium (Turner) C. Agardh: Regulation of cytokine cascade
- Bio-based hydrogel patches made of κ-carrageenan enriched with degalactosylated xyloglucan for wound dressing applications
- Clinical spectrum of COVID-19 patients and decreased serum level of miR-146a as a sign of inflammation
- Review Articles
- Quest for space: Tenacity of DNA, Protein, and Lipid macromolecules in intracellular crowded environment
- The impact of exercise on mitochondrial biogenesis in skeletal muscle: A systematic review and meta-analysis of randomized trials
- Communication
- Entropy and mechanistic concepts after intraovarian platelet-rich plasma: Experimental considerations for local tissue responses mediated by NF-κB and TNF-α
Articles in the same Issue
- Research Articles
- Anti-arthritic potential of crude sulfated polysaccharide from marine macroalgae Sargassum ilicifolium (Turner) C. Agardh: Regulation of cytokine cascade
- Bio-based hydrogel patches made of κ-carrageenan enriched with degalactosylated xyloglucan for wound dressing applications
- Clinical spectrum of COVID-19 patients and decreased serum level of miR-146a as a sign of inflammation
- Review Articles
- Quest for space: Tenacity of DNA, Protein, and Lipid macromolecules in intracellular crowded environment
- The impact of exercise on mitochondrial biogenesis in skeletal muscle: A systematic review and meta-analysis of randomized trials
- Communication
- Entropy and mechanistic concepts after intraovarian platelet-rich plasma: Experimental considerations for local tissue responses mediated by NF-κB and TNF-α

