Abstract
Atherosclerosis is an important medical and social problem, and the keys to solving this problem are still largely unknown. A common situation in real clinical practice is the comorbid course of atherosclerosis with chronic obstructive pulmonary disease (COPD). Diseases share some common risk factors and may be closely linked pathogenetically. Methods: Bioinformatics analysis of datasets from Gene Expression Omnibus (GEO) was performed to examine the gene ontology (GO) of common differentially expressed genes (DEGs) in COPD and peripheral arterial atherosclerosis. DEGs were identified using the limma R package with the settings p < 0.05, corrected using the Benjamini & Hochberg algorithm and ǀlog 2FCǀ > 1.0. The GO, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, and the protein–protein interaction (PPI) network analysis were performed with the detected DEGs. Results: The biological processes and signaling pathways involving common DEGs from airway epithelial datasets in COPD and tissue in peripheral atherosclerosis were identified. A total of 15 DEGs were identified, comprising 12 upregulated and 3 downregulated DEGs. The GO enrichment analysis demonstrated that the upregulated hub genes were mainly involved in the inflammatory response, reactive oxygen species metabolic process, cell adhesion, lipid metabolic process, regulation of angiogenesis, icosanoid biosynthetic process, and cellular response to a chemical stimulus. The KEGG pathway enrichment analysis demonstrated that the common pathways were Toll-like receptor signaling pathway, NF-kappa B signaling pathway, lipid and atherosclerosis, and cytokine–cytokine receptor interaction. Conclusions: Biological processes and signaling pathways associated with the immune response may link the development and progression of COPD and atherosclerosis.
Introduction
Atherosclerosis and chronic obstructive pulmonary disease (COPD) are among the most important medical and social problems of the modern society [1,2,3]. They are characterized by high prevalence and are associated with a high incidence of temporary and persistent disability and mortality. Both diseases often occur simultaneously in the same patient and are often not diagnosed on time, and it is not possible to determine which one developed first [4,5]. This may be due to several risk factors in common, with smoking playing a major role [5,6]. The pathogenesis of COPD is thought to be based on chronic diffuse inflammation in the bronchi, which involves many cells.
Interestingly, atherosclerosis is unevenly distributed among patients with different clinical variants of COPD. The full details of these relationships are not yet known, but it is thought that COPD may predispose patients to diffuse atherosclerotic lesions with worse clinical outcomes [7]. In addition, the bronchial variant of COPD is thought to be more often associated with atherosclerosis. The significance of the problem can be emphasized by the fact that cardiovascular diseases associated with atherosclerosis are the leading cause of death in COPD patients. The paradoxical relationship between the prognosis of COPD patients and obesity is also the subject of a separate discussion [8]. Studies suggest that overweight and obesity are associated with better clinical outcomes in COPD, which seems interesting given the known role of obesity in the course of cardiovascular disease. These findings suggest that despite intensive research into the pathogenesis of COPD and atherosclerosis, many links in the complex chain of events that link both diseases still require research. In addition, the clinical heterogeneity of COPD is both a cause and a consequence of the lack of research into the links with atherosclerosis, as most studies do not consider the clinical heterogeneity of COPD when conducting research.
In the complex history of the study of atherosclerosis, several theories of its pathogenesis have emerged, in which lipid metabolism disorders play a leading role, underpinning the current therapeutic strategies. The involvement of impaired immune processes also seems clear, given the involvement of immune cells such as macrophages and the production of many cytokines and chemokines by the cells. Low-density lipoproteins (LDL) accumulate in the arterial wall and are taken up by macrophages, leading to activation of innate immunity [9]. Moreover, macrophages play a crucial role in all stages of atherosclerotic lesion progression, releasing cytokines, chemokines, and matrix metalloproteinases (MMPs) that degrade extracellular matrix components, leading to plaque destabilization [10,11].
Research in recent years has significantly increased the understanding of the role of immune and metabolic links in the pathogenesis of both COPD and atherosclerosis. Indeed, the lung is an organ with unique lipid biology, and impaired lipid homeostasis in smoking is an important part of the pathogenesis of COPD.
Thus, closely intertwined immunometabolic pathways are characteristic of not only COPD but also atherosclerosis. In this regard, it is of interest that common biological processes and signaling pathways may link the development and progression of COPD and atherosclerosis.
The aim of this research was to investigate the gene ontology (GO) of common differentially expressed genes (DEGs) in COPD and peripheral arterial atherosclerosis using bioinformatics analysis. The data obtained in the present study may be of research and clinical interest, as the search for common links in pathogenesis will allow to better understand the mechanisms of comorbid disease course.
Materials and methods
Data collection
The analysis was performed on the previously studied data sets (gene sets) obtained from The Gene Expression Omnibus (GEO), The National Center for Biotechnology Information (NCBI) [12]. The gene expression levels in the GSE5058 [13,14], GSE11906 [15], GSE11784 [16,17], and GSE100927 [18] datasets were used for the analysis.
From the GSE5058 dataset, gene expression data in bronchoscopy bronchial epithelial samples from 12 healthy nonsmokers and 6 smokers with COPD were included in the analysis. Data were obtained using the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array. Data normalization was performed using a Mas5 normalization method.
From the GSE11906 dataset, gene expression data in bronchial epithelium samples of order 2–3 and bronchial epithelium samples of order 10–12 obtained by bronchoscopy in 42 healthy nonsmokers and 20 smokers with COPD were included in the analysis. Data were obtained using the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array platform. Mas5 normalization without log transformation was performed.
From the GSE11784 dataset, gene expression data in 10–12-order bronchial epithelium from 63 healthy nonsmokers and 22 patients with COPD were included in the analysis. Data were obtained using the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array platform. Mas5 normalization without log transformation was performed.
Gene expression data in 29 samples of atherosclerotic lesions of carotid arteries, 26 samples of atherosclerotic lesions of femoral, and 14 samples of atherosclerotic lesions of infra-popliteal arteries and samples from control arteries without atherosclerotic lesions from 12 carotid, 12 femoral, and 11 infra-popliteal from patients undergoing carotid, femoral, or infrapopliteal endarterectomy were included in the analysis from GSE100927 dataset. Data were obtained using a GPL17077 Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version). Data normalization was performed using a locally weighted scattered plot smoother analysis (LOWESS).
Data extraction
The following information was extracted for the COPD datasets: the number of COPD patients and healthy nonsmokers and preprocessed gene expression data. Comparison groups were formed to analyze the gene expression data: healthy nonsmokers and COPD patients.
For the data set with atherosclerosis, the following information was extracted: the number of samples obtained from patients with atherosclerosis and healthy individuals and preprocessed gene expression data. For gene expression data analysis, comparison groups were formed: samples obtained from patients with atherosclerosis of various localizations and a control group of samples from healthy individuals.
Differential expression analysis
To assess the DEGs, bioinformatic analysis was performed in the comparison groups using the limma package in R (v. 4.0.2) and the GEO2R and Phantasus application (v. 1.11.0) [19]. Normalization (log 2 transformation followed by quantile normalization) was performed in the analysis. The Benjamini & Hochberg (false discovery rate (FDR)) algorithm was used to adjust the statistical significance level for multiple comparisons. The conditions for screening for DEGs were an absolute value of ǀlogFCǀ > 1 and p values satisfying the FDR condition ≤5%. The Volcano plots were constructed using the SangerBox tool. Common DEGs were determined using the Functional Enrichment analysis tool (FunRich v. 3.1.3) with the construction of Venn diagrams [20]. DEGs that changed in the same direction in all data sets (were upregulated in all data sets or downregulated in all data sets) were considered to be common.
GO analysis
To further investigate the expression patterns of individual genes in the context of specific biological processes or molecular pathways, GO analysis and the identification of signaling pathways by the Kyoto Encyclopedia of Genes and Genomes (KEGG) were performed using GEO2Enrichr [21], ShinyGO (v. 0.741) [22], and GOnet [23], and their functional enrichment and visualization was done. The p < 0.05 value corrected using the Benjamini & Hochberg algorithm was set as the threshold for identifying biological processes and pathways.
Protein–protein interactions (PPIs) of protein products of common DEGs were evaluated using the online Search Tool for the Retrieval of Interacting Genes database (STRING v. 11.5). An interaction score of at least 0.4 (medium confidence score) was considered significant. PPIs were visualized using Cytoscape (v. 3.8.2). Interactions between DEGs were analyzed using the Network Analyzer plug-in module of Cytoscape software. In addition, molecular complex detection (MCODE) was used to search for gene clusters in the PPI network [24]. The cut-off criteria were “degree cutoff = 2,” “node score cutoff = 0.2,” “k-core = 2,” and “max depth = 100.”
In addition, the most important genes in the network were identified using the cytoHubba plugin tool in the Cytoscape software [25]. The Cytoscape cytoHubba plugin was used to rank the nodes in the network according to their network characteristics. The Maximal Clique Centrality (MCC) topological analysis algorithm was used to analyze, predict, and visualize key proteins in molecular PPI interaction networks.
Results
Identification of DEGs
The bioinformatics analysis performed showed the presence of DEGs in the comparison groups. Volcano plots were used to visualize the identified DEGs (Figure 1).

Volcano plots characterizing DEGs in selected sets. Colored dots in the graph indicate genes showing differential expression with |log 2FC| > 1 and p < 0.05, while black dots do not meet these criteria. The red dots indicate upregulated DEGs and are displayed on the right side of the graph, and the blue dots indicate downregulated DEGs – they are displayed on the opposite side.
In the GSE100927 set, 399 upregulated DEGs and 160 downregulated DEGs were identified using selected cut-off criteria (Figure 1, Table S1). Further analysis showed the presence of common DEGs in the GSE5058, GSE11784, and GSE11906 datasets studied (Figure 2). In the three COPD sets, 137 common upregulated DEGs and 92 downregulated DEGs were identified (Table S2).

Venn diagrams with common DEGs of the GSE5058, GSE11784, and GSE11906 datasets. (a) Upregulated common DEGs; (b) downregulated common DEGs.
Enrichment analysis of DEGs
Enrichment analysis of DEGs in COPD
The results of the GO biological process (BP) analysis revealed that the upregulated DEGs of COPD datasets were mainly enriched in the cellular response to chemical stimulus, cell adhesion, cell motility, unsaturated fatty acid metabolic process, arachidonic acid metabolic process, icosanoid metabolic process, reactive oxygen species (ROS) metabolic process, angiogenesis, and blood vessel morphogenesis (Figure 3, Table S3). Upregulated DEGs in COPD have been enriched by KEGG pathways such as arachidonic acid metabolism, metabolism of xenobiotics by cytochrome P450, ECM–receptor interaction, glycerolipid metabolism, phagosome, Toll-like receptor signaling pathway, lipid and atherosclerosis, chemical carcinogenesis, and metabolic pathways (Figure 3, Table S4). The findings confirm the importance of immune processes, including those associated with the Toll-like receptor signaling pathway, in the pathogenesis of COPD. Of interest is the involvement of metabolic pathways, including those related to the metabolism of arachidonic acid, which is at the intersection of pro- and anti-inflammatory pathways, being the metabolite for the synthesis of inflammatory mediators and specialized pro-resolving mediators [26,27].

GO enrichment analysis by biological processes (a) and KEGG pathways (b) for common upregulated DEGs from GSE5058, GSE11784, and GSE11906 datasets. Biological processes and KEGG pathways are ranked by fold enrichment values. The most significant processes are highlighted in red, and the less significant processes are highlighted in blue according to log10(FDR) values. Larger dots in the graph indicate a greater number of genes involved.
The results of the GO biological process (BP) analysis revealed that the downregulated DEGs of COPD datasets were mainly enriched in the lung-associated mesenchyme development, mesenchymal cell proliferation involved in lung development, positive regulation of transmembrane transport, positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis, response to mechanical stimulus, and cell differentiation (Figure 4, Table S5).

GO enrichment analysis by biological processes for common downregulated DEGs from GSE5058, GSE11784, and GSE11906 datasets. Biological processes are ranked by fold enrichment values. The most significant processes are highlighted in red, and the less significant processes are highlighted in blue according to log10(FDR) values. Larger dots in the graph indicate a greater number of genes involved.
Enrichment analysis of DEGs in atherosclerosis
The results of the GO biological process (BP) analysis revealed that the upregulated DEGs of atherosclerosis datasets were mainly enriched in the immune response, immune system process, cell activation, cell activation involved in immune response, cytokine production, leukocyte degranulation, leukocyte migration, myeloid leukocyte activation, myeloid leukocyte-mediated immunity, and myeloid cell activation involved in immune response (Figure 5, Table S6). Upregulated DEGs in atherosclerosis have been enriched by KEGG pathways such as chemokine signaling pathway, leukocyte transendothelial migration, phagosome, lipid and atherosclerosis, cell adhesion molecules, and cytokine–cytokine receptor interaction (Figure 5, Table S7). The findings confirm the known evidence that atherosclerosis is a disease with significant immune system involvement.

GO enrichment analysis by biological processes (a) and KEGG pathways (b) for common upregulated DEGs from data set GSE100927. Biological processes and KEGG pathways are ranked according to the fold enrichment values. The most significant processes and KEGG pathways are highlighted in red, and the less significant processes are highlighted in blue according to log10(FDR) values. Larger dots in the graph indicate a greater number of genes involved.
The results of the GO biological process (BP) analysis revealed that the downregulated DEGs of atherosclerosis datasets were mainly enriched in the regulation of the muscle system process, blood circulation, muscle structure development, smooth muscle cell proliferation, actomyosin structure organization, actin filament organization, regulation of cellular component movement, and anatomical structure morphogenesis (Figure 6, Table S8). The data obtained are in accordance with the known information about the role of impaired structure and the function of vascular smooth muscle cells in the pathogenesis of atherosclerosis. In addition, the role of actin filaments in providing structural and functional characteristics of endothelial cells and their association with the impaired nature of the blood flow in the development and progression of atherosclerosis is known [28].

GO enrichment analysis by biological processes for common downregulated DEGs from data set GSE100927. Biological processes are ranked according to the fold enrichment values. The most significant processes are highlighted in red, and the less significant ones in blue according to log10(FDR) values. Larger dots in the graph indicate a greater number of genes involved.
The data suggest an important role for impaired metabolic and immune processes in the pathogenesis of COPD and atherosclerosis.
Identification of the most important genes and pathways using DEG PPI network analysis
Identification of the most important genes in COPD
A PPI network was constructed for the DEGs GSE5058, GSE11784, and GSE11906 using the STRING online database. From the resulting PPI network, two clusters were identified using the MCODE plugin for Cytoscape (Figure 7). For the GO analysis, functionally related meaningful clusters from a common subnetwork were used with MCODE ≥5 and the number of nodes ≥6. The following hub genes were identified using the cytoHubba plugin (Figure 7).

The most important clusters selected from the PPI network (a) cluster 1; (b) cluster 2. A node in the PPI network denotes a protein and an edge denotes an interaction. The most important genes evaluated in the PPI network using the MCC algorithm in CytoHubba, Cytoscape (c) cluster 1; (d) cluster 2. The most important genes are ranked as follows: the most important genes are highlighted in red, the less important genes in orange, and even less important genes in yellow.
The functional enrichment analysis of these hub genes in the modules was mainly related to the immune system, including Toll-like receptor signaling pathway, NOD-like receptor signaling pathway, TNF signaling pathway, cytokine–cytokine receptor interaction, IL-17 signaling pathway, lipid metabolism disorders – lipid and atherosclerosis (Figure 8).

GO enrichment analysis. A hierarchical clustering tree summarizing the correlation among significant biological processes for DEGs from two clusters (a) cluster 1; (b) cluster 2 of data sets GSE5058, GSE11784, and GSE11906. Biological processes with many shared genes in common are grouped. Bigger dots indicate larger values of statistical differences (p).
Identification of the most important genes in atherosclerosis
A PPI network was constructed for DEGs from the GSE130927 dataset using the STRING online database. From the resulting PPI network, seven clusters were identified using the MCODE plugin for Cytoscape, of which four were selected for further analysis based on MCODE ≥5 and the number of nodes ≥6. (Figure 9). Hub genes for each cluster were identified using the cytoHubba plugin (Figure 9).

The most important clusters selected from the PPI network (a). A node in the PPI network denotes a protein and an edge denotes an interaction. The most important genes evaluated in the PPI network using the MCC algorithm in CytoHubba, Cytoscape (b). The most important genes are ranked as follows: the most important genes are highlighted in red, the less important in orange, and even less important in yellow.
The functional enrichment analysis of these hub genes in clusters was mainly related to the myeloid leukocyte-mediated immunity, macrophage activation, phagocytosis, neutrophil activation involved in immune response, immune response, cell surface receptor signaling pathway, interferon-gamma-mediated signaling pathway, antigen processing and presentation, activation of immune response, innate immune response, and response to cytokine (Figure 10, Table S9).

GO enrichment analysis by biological processes for common upregulated DEGs from the four most significant clusters of the GSE100927 dataset (a) cluster 1, (b) cluster 2, (c) cluster 3, (d) cluster 4. Biological processes are ranked according to the fold enrichment values. The most significant processes are highlighted in red, and the less significant processes are highlighted in blue according to log10(FDR) values. Larger dots in the graph indicate a greater number of genes involved.
Identification of common hub genes in COPD and atherosclerosis
Further analysis showed the presence of common DEGs in the GSE5058, GSE11784, GSE11906, and GSE100927 datasets. Twelve common upregulated DEGs and three downregulated DEGs were identified (Figure 11). As only three common downregulated DEGs (MT1M, ITLN1, and C3) were identified, they were not functionally annotated.

Venn diagrams with common DEGs from the GSE5058, GSE11784, GSE11906, and GSE100927 datasets: (a) upregulated DEGs and (b) downregulated DEGs.
The results of the GO biological process (BP) analysis revealed that the upregulated common DEGs were mainly enriched in the inflammatory response, cytokine production, cytokine-mediated signaling pathway, ROS metabolic process, regulation of response to stress, cellular response to chemical stimulus, lipid metabolic process, positive regulation of lipid localization, regulation of response to stress, regulation of cell adhesion (Figure 12, Table S10). Common upregulated DEGs GSE5058, GSE11784, GSE11906, and GSE100927 datasets have been enriched by KEGG pathways such as lipid and atherosclerosis, cytokine–cytokine receptor interaction, NF-kappa B signaling pathway, Toll-like receptor signaling pathway, NOD-like receptor signaling pathway, phagosome, necroptosis, and ECM–receptor interaction (Figure 12, Table S11). The findings confirm the significant role of immune processes and lipid metabolism in the pathogenesis of both COPD and atherosclerosis.

GO enrichment analysis by biological processes (a) and KEGG pathways (b) for common upregulated DEGs from GSE5058, GSE11784, GSE11906, and GSE100927 datasets. Biological processes and KEGG pathways are ranked according to fold enrichment values. The most significant processes are highlighted in red, and the less significant processes are highlighted in blue according to log10(FDR) values. Larger dots in the graph indicate a greater number of genes involved.
A network of PPIs was constructed for common upregulated DEGs using the STRING online database. The following hub genes were identified using the cytoHubba plugin in Cytoscape (Figure 13). The functional roles and general information about the nine upregulated hub genes are presented in Table 1.

The most important common upregulated DEGs (hub genes) of the GSE5058, GSE11784, GSE11906, and GSE100927 datasets. Hub genes are ranked as follows: the most important genes are highlighted in red, the less important in orange, and even less important in yellow.
Functional roles and general information of the upregulated hub genes
| Gene symbol | Other names | Full name | Protein | Function |
|---|---|---|---|---|
| ALOX5 | 5-LO, 5LPG, LOG5, 5-LOX | Arachidonate 5-lipoxygenase | Polyunsaturated fatty acid 5-lipoxygenase | Arachidonic acid metabolic process, leukotriene A4 biosynthetic process, regulation of inflammatory response |
| BCL2A1 | GRS, ACC1, ACC2, BFL1, ACC-1, ACC-2, HBPA1, BCL2L5 | BCL2-related protein A1 | Bcl-2-related protein A1 | Negative regulation of apoptotic process |
| CYBB | CGD, CGDX, NOX2, IMD34, AMCBX2, GP91-1, GP91PHOX, p91-PHOX, GP91-PHOX | Cytochrome b-245 beta chain | Cytochrome b-245 heavy chain | Innate immune response, inflammatory response, hypoxia-inducible factor-1alpha signaling pathway, superoxide metabolic process |
| IGSF6 | DORA | Immunoglobulin superfamily member 6 | Immunoglobulin superfamily member 6 | Immune response, cell surface receptor signaling pathway |
| IL1B | IL-1, IL1F2, IL1beta, IL1-BETA | Interleukin 1 beta | Interleukin-1 beta | Apoptotic process, cell–cell signaling, cellular response to lipopolysaccharide, immune response, inflammatory response |
| IL1RN | DIRA, IRAP, IL1F3, IL1RA, MVCD4, IL-1RN, IL-1ra, IL-1ra3, ICIL-1RA | Interleukin 1 receptor antagonist | Interleukin-1 receptor antagonist protein | Immune response, inflammatory response, inflammatory response to antigenic stimulus, lipid metabolic process |
| MMP12 | Matrix metallopeptidase 12 | ME, HME, MME, MMP-12 | Macrophage metalloelastase | Bronchiole development, extracellular matrix organization, lung alveolus development, negative regulation of endothelial cell–matrix adhesion via fibronectin |
| PLA2G7 | Phospholipase A2 group VII | PAFAD, PAFAH, LP-PLA2, LDL-PLA2 | Platelet-activating factor acetylhydrolase | Lipid oxidation, low-density lipoprotein particle remodeling, phosphatidylcholine catabolic process, positive regulation of inflammatory response, positive regulation of monocyte chemotaxis |
| SPP1 | Secreted phosphoprotein 1 | OPN, BNSP, BSPI, ETA-1 | Osteopontin | Cytokine activity, integrin binding |
The functional enrichment analysis by biological processes of these hub genes in the modules was mainly related to the regulation of response to external stimuli, regulation of angiogenesis, inflammatory response, ROS metabolic process, cell adhesion, lipid metabolic process, icosanoid biosynthetic process, cellular response to chemical stimulus, and anatomical structure morphogenesis (Figures 14 and 15, Table S12). For the GO molecular function analysis, the upregulated hub genes were significantly enriched in interleukin-1 binding, cytokine activity, cytokine receptor binding, arachidonate 5-lipoxygenase activity, and phospholipase A2 activity (Figure 14, Table S13). The analysis of the KEGG pathways involved in the most significant shared genes of GSE5058, GSE11784, GSE11906, and GSE100927 sets showed enrichment along NF-kappa B signaling pathway, Toll-like receptor signaling pathway, necroptosis, lipid and atherosclerosis, cytokine–cytokine receptor interaction (Figure 14, Table S14).
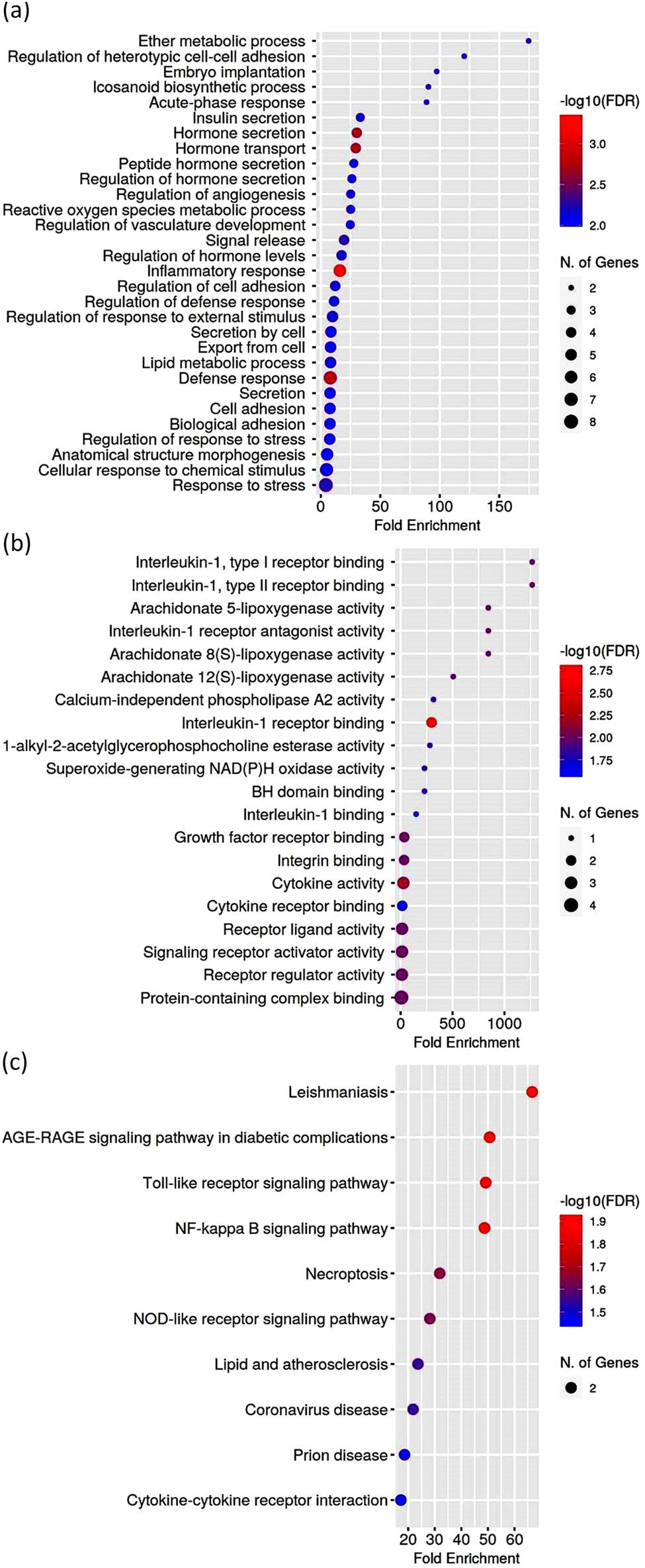
GO enrichment analysis by biological processes (a), molecular functions (b), KEGG pathways (c) for upregulated hub genes from the GSE5058, GSE11784, GSE11906, and GSE100927 datasets. Biological processes, molecular functions, and KEGG pathways are ranked according to fold enrichment values. The most significant processes are highlighted in red, and the less significant processes in blue according to log10(FDR) values. Larger dots in the graph indicate a greater number of genes involved.

Schematic showing GO term annotation on the biological processes of the common upregulated differentially expressed hub genes from the GSE5058, GSE11784, GSE11906, and GSE100927 datasets.
The findings showed that NF-kappa B signaling pathway, Toll-like receptor signaling pathway, NOD-like receptor signaling pathway, and lipid metabolism disorders are involved in the pathogenesis of both COPD and atherosclerosis.
Discussion
In the present study, an integrated bioinformatics analysis of human gene chip data in sets obtained from the GEO database was performed to examine key pathways associated with the comorbid development of COPD and atherosclerosis. Investigation of potential common mechanisms of COPD and atherosclerosis seems to be an urgent clinical task, given the wide prevalence of both diseases and the fact that diseases are often combined in one patient and mutually aggravate the clinical picture and prognosis.
DEGs were searched for and compared in the airway epithelium of COPD compared to normal controls and in atherosclerotic plaques of different localizations compared to the normal peripheral arterial wall. Statistically significant differences in gene expression levels were taken into account and corrected using the Benjamini & Hochberg algorithm. Common DEGs for COPD and peripheral atherosclerosis were subjected to the bioinformatics analysis. Their gene ontology was assessed for biological processes (GO-BP), molecular function (GO-MF), and involvement in the KEGG pathways. The analysis was then performed using STRING and Cytoscape tools to search for the most significant common genes and their functional analysis.
The most significant common upregulated DEGs were IL1b, CYBB, IL1RN, MMP12, BCL2A1, SPP1, ALOX5, IGSF6, and PLA2G7.
The findings support the evidence that the immune system is involved in the pathogenesis of both COPD and atherosclerosis [29,30,31,32,33]. A number of recent studies show that in a proportion of COPD patients, inflammation may generalize through the circulatory system and cause systemic inflammatory organ damage. In this context, COPD is often considered as a disease with not only pulmonary but also extrapulmonary clinical manifestations, which may have a major impact on severity and prognosis. According to this concept, the increased severity of systemic inflammation may contribute to concomitant chronic comorbidities often seen in patients with COPD, such as cardiovascular diseases associated with atherosclerosis and which are associated with poor clinical outcomes.
According to the current thinking, atherosclerosis is a chronic inflammatory disease of large and medium-sized arteries caused by an innate immune response involving myeloid cells such as monocytes and macrophages [34]. Atherosclerosis is characterized by a subendothelial accumulation of cholesterol, immune cells, and extracellular matrix, leading to the formation of atherosclerotic plaque [35,36,37,38]. Macrophages play a crucial role in all stages of atherosclerotic lesion progression [39,40]. Macrophages differentiate into foamy cells after recognition and internalization of oxidized LDL (oxLDL). In addition, oxLDL stimulates TLR in macrophages, exacerbating inflammation in the plaque. Activated macrophages release cytokines, chemokines, and MMPs, the latter destroying components of the extracellular matrix and thus leading to plaque destabilization.
Although the full details of comorbid disease development and course are still unknown, studies have shown that systemic inflammation and oxidative stress are considered to be key pathophysiological factors linking COPD and atherosclerosis.
The data from the study support the evidence that the immune system is involved in the pathogenesis of both COPD and atherosclerosis [30,31,32,33]. The most significant DEGs for both diseases was IL-1B. IL-1β is a typical cytokine of innate immunity that is involved in the initiation and maintenance of inflammation [41,42,43]. It is a key mediator of neutrophilic airway inflammation in COPD, and its levels are significantly elevated in small airway epithelial cells in COPD [43,44]. Smokers have elevated levels of IL-1β in bronchoalveolar lavage fluid compared with nonsmokers [45,46]. In addition, serum IL-1β levels are significantly increased during exacerbation of COPD and correlate positively with serum C-reactive protein levels, neutrophil counts, and smoking history, but correlate negatively with predicted FEV1%. These findings suggest that elevated serum IL-1β levels can be considered as a biomarker of persistent neutrophilic airway inflammation and COPD exacerbation [47].
IL-1β is known to be involved in the development of atherosclerosis by promoting increased expression of adhesion factors and chemokines [48,49,50]. It is involved in the inflammatory response in endothelial cells and contributes to the invasion and accumulation of inflammatory cells in the intima of blood vessels [50,51].
The significance of IL-1β in atherogenesis is demonstrated by the results of clinical trials of canakinumab, a therapeutic monoclonal antibody targeting interleukin-1β. The use of canakinumab resulted in a significantly lower incidence of recurrent cardiovascular events than placebo, regardless of the reduction in lipid levels [52].
The importance of IL-1β in the pathogenesis of these diseases may also be related to the fact that this cytokine may be associated with the induction of MMP production [50,53,54,55]. Increased production of MMP-12, MMP9, and various neutrophil chemoattractants was found in transgenic mice overexpressing IL-1β, which resulted in airway remodeling and emphysema [45]. Emphysema is one of the leading morphological and clinical features of COPD, and MMPs are involved in its development. MMP12 levels in sputum have been shown to be elevated in COPD patients compared to nonsmokers and healthy smokers [56]. Similar data were obtained in alveolar macrophages in the bronchoalveolar lavage fluid [57,58,59]. However, data on the role of MMP12 in the development of emphysema in humans require clarification.
ALOX5 encodes 5-lipoxygenase, a key enzyme in the biosynthesis of leukotrienes and lipoxins, thus contributing to the regulation of innate immunity. 5-Lipoxygenase metabolizes the arachidonic acid to 5-hydroperoxyacosatetraenoic acid (5-HpETE), providing the first steps in the synthesis of leukotriene A4 (LTA4), a powerful mediator of inflammation [60,61,62,63,64,65,66,67,68]. Hence, ALOX5 contributes to an increased acute inflammatory response and also promotes the development and progression of chronic inflammation. In contrast, ALOX5 is involved in the formation of some pro-resolving mediators, such as lipoxin and the resolvin subfamily, which provide the resolution phase of inflammation [69,70,71]. The finding that ALOX5 is among the most important DEGs is of interest, as it allows increased attention to the cross-linkages between lipid metabolism and the immune response. Arachidonic acid is an important participant in inflammation in COPD, as it is involved in the synthesis of both pro- and anti-inflammatory mediators. In addition, arachidonic acid metabolism is associated with infectious exacerbations of COPD. Arachidonic acid is stored in cell membranes as part of phospholipids, but when stimulated by phospholipase A2 (PLA2), it is mobilized for further metabolism into bioactive mediators involved in inflammation.
PLA2G7 (phospholipase A2 group VII) is a gene that encodes the protein platelet-activating factor acetylhydrolase (PAF-AH) or lipoprotein-associated phospholipase A2 (Lp-PLA 2). Platelet-activating factor (PAF) is a biologically active phospholipid mediator of inflammation, and PAF-AH is an enzyme that is involved in its inactivation [72]. In addition, PAF-AH can exhibit phospholipase A2 activity by hydrolyzing the ester complex of the fatty acyl group attached at the sn-2 position of phospholipids [73,74]. It can hydrolyze phospholipids with long fatty acyl chains only if they carry oxidized functional groups [73,75]. Most circulating PAF-AH is bound to LDL, and the remainder is bound to high-density lipoproteins (HDLs) [76]. Lp-PLA 2 levels are associated with atherosclerosis [76]. Its high activity is associated with an increased risk of coronary heart disease [77].
PLA2G7 levels correlate with BMI and lung function and were increased in blood and alveolar macrophages of patients with COPD compared with never smokers. In addition, Lp-PLA2 is considered a promising biomarker for predicting poor exercise tolerance in patients with COPD [78].
The BCL2A1 gene encodes Bcl-2-related protein A1, which is a highly regulated nuclear factor κB (NF-κB) target gene. BCL2A1 is an anti-apoptotic member of the Bcl-2 family that is predominantly expressed in hematopoietic and endothelial cells and promotes the cell survival. In endothelial cells, Bcl-2-related protein A1 is induced by inflammatory cytokines, tumor necrosis factor-alpha, and interleukin-1 beta. Given that Bcl-2-related protein A1 is induced by inflammatory cytokines, it is suggested that it may play a protective role during inflammation [79,80]. Monocytes can induce BCL2A1 upregulation in endothelial cells to protect them from death [81]. It seems possible that because inflammasome formation can induce BCL2A1 expression, this may contribute to the survival of pro-inflammatory cells during the immune response [82]. In chronic inflammatory disorders, regulation of BCL2A1 gene expression leads to myeloid cell recruitment and stabilization [83]. This mechanism may have a pro-inflammatory character by improving the survival of leukocytes, such as differentiating and mature neutrophils [82,84]. These data are of interest given the role of neutrophilic chronic inflammation in COPD.
Secreted phosphoprotein 1 (SPP1), also called osteopontin (OPN), is a secreted phosphorylated acidic glycoprotein. It can act as an extracellular matrix molecule and as a cytokine and plays an important role in physiological and pathophysiological processes [85,86,87]. SPP1 may be involved in the pathogenesis of COPD as it is associated with neutrophilic inflammation and emphysema [87,88]. It has been shown that cigarette smoking can increase SPP1 expression in induced sputum, and its levels are increased in induced sputum and lung tissue of patients with COPD [87,89,90]. Recent studies have shown that upregulation of SPP1 may be associated with an increased risk of lung cancer in patients with COPD [87].
Arteries under physiological conditions show low levels of SPP1 expression, which is essential for normal arterial biomechanics because SPP1 acts as a physiological inhibitor of vascular calcification [91]. This damage causes activation of SPP1, which promotes cell adhesion, proliferation, migration, and survival. These processes promote the healing process, and SPP1 expression usually decreases over time [92,93]. Activation of SPP1 is facilitated by ischemia, which is seen in stroke [94,95], myocardial infarction [96,97], and peripheral arterial disease [98,99]. Thus, OPN may be involved in the immune response under ischemic conditions, as evidenced by its involvement in macrophage infiltration in vivo [100,101]. The cells in the vascular wall that activate and secrete SPP1 are endothelial cells, vascular smooth muscle cells, and macrophages [85,92,102].
CYBB encodes the NOX2 protein (cytochrome b-245 β-chain), which constitutes the large transmembrane subunit of the gp91phox NADPH-oxidase complex. NOX2 is involved in the generation of large amounts of ROS in phagocytic cells such as neutrophils, monocytes, and macrophages [103]. NADPH oxidase, in turn, is an enzyme complex that plays a crucial role in innate immunity by generating ROS, which are key components of the bactericidal activity of phagocytes [104,105].
Thus, the findings indicate that COPD and peripheral atherosclerosis are closely linked to the immune system in their development. In addition, the increasing focus in recent years on the links between lipid metabolism and immune response is also reflected in the results of this analysis.
The novelty of the findings is related to the fact that the data obtained may expand the understanding of some issues related to the pathogenesis of the comorbid course of COPD and atherosclerosis. Both diseases have been shown to be characterized by the involvement of common DEGs related to the innate immune system and lipid metabolism. The reciprocal links between the innate immune system and disorders of lipid metabolism is a promising area for future research. Further studies may clarify the role of individual links in the common processes that are associated with the identified DEGs. These data will be useful for understanding the mechanisms underlying the comorbidity and clinical heterogeneity of COPD and atherosclerosis.
It should be noted that the present study has some limitations, which are generally characteristic of bioinformatics analysis. The limitations of this study are the insufficient sample due to the availability of data. In addition, the data were obtained at different times and using different platforms. Data on the clinical characteristics of the diseases, the presence of comorbidities, and the drug therapy received by the patients are not taken into account. In addition, the data are limited to gene expression only, without considering proteins, which does not allow to build a complete picture of the processes that occur in COPD and atherosclerosis. However, the present study has analyzed a large number of genes and may be of interest in planning future studies.
Conclusion
Thus, the results obtained in this study confirm the importance of the innate immune system in the pathogenesis of COPD and atherosclerosis, including those related to the Toll-like receptor signaling pathway. The findings on the involvement of several immune and metabolic processes may contribute to the increasing attention to the role of impaired lipid metabolism in the comorbid course of these diseases.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] May SM, Li JT. Burden of chronic obstructive pulmonary disease: healthcare costs and beyond. Allergy Asthma Proc. 2015;36(1):4–10.10.2500/aap.2015.36.3812Suche in Google Scholar PubMed PubMed Central
[2] Quaderi SA, Hurst JR. The unmet global burden of COPD. Glob Health Epidemiol Genom. 2018;3:e4-e.10.1017/gheg.2018.1Suche in Google Scholar PubMed PubMed Central
[3] Roquer J, Ois A. Atherosclerotic burden and mortality. In: Preedy VR, Watson RR, editors. Handbook of disease burdens and quality of life measures. New York, NY: Springer New York; 2010. p. 899–91810.1007/978-0-387-78665-0_51Suche in Google Scholar
[4] Sin DD, MacNee W. Chronic obstructive pulmonary disease and cardiovascular diseases: a “vulnerable” relationship. Am J Respiratory Crit Care Med. 2013;187(1):2–4.10.1164/rccm.201210-1953EDSuche in Google Scholar PubMed
[5] Vogelmeier CF, Criner GJ, Martinez FJ, Anzueto A, Barnes PJ, Bourbeau J, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report. GOLD executive summary. Am J Respir Crit Care Med. 2017;195(5):557–82.10.1164/rccm.201701-0218PPSuche in Google Scholar PubMed
[6] Tuleta I, Farrag T, Busse L, Pizarro C, Schaefer C, Pingel S, et al. High prevalence of COPD in atherosclerosis patients. Int J Chron Obstruct Pulmon Dis. 2017;12:3047–53.10.2147/COPD.S141988Suche in Google Scholar PubMed PubMed Central
[7] Enriquez JR, Parikh SV, Selzer F, Jacobs AK, Marroquin O, Mulukutla S, et al. Increased adverse events after percutaneous coronary intervention in patients with COPD: insights from the national heart, lung, and blood institute dynamic registry. Chest. 2011;140(3):604–10.10.1378/chest.10-2644Suche in Google Scholar PubMed PubMed Central
[8] Iyer AS, Dransfield MT. The “Obesity Paradox” in chronic obstructive pulmonary disease: can it be resolved? Ann Am Thorac Soc. 2018;15(2):158–9.10.1513/AnnalsATS.201711-901EDSuche in Google Scholar PubMed PubMed Central
[9] Hansson GK. The heart of immunology: immune mechanisms in cardiovascular medicine. Cardiovascular Res. 2021;117(13):e166–e8.Suche in Google Scholar
[10] Bobryshev YV, Ivanova EA, Chistiakov DA, Nikiforov NG, Orekhov AN. Macrophages and their role in atherosclerosis: pathophysiology and transcriptome analysis. Biomed Res Int. 2016;2016:9582430.10.1155/2016/9582430Suche in Google Scholar PubMed PubMed Central
[11] Barrett TJ. Macrophages in atherosclerosis regression. Arteriosclerosis, Thrombosis, Vasc Biol. 2020;40(1):20–33.10.1161/ATVBAHA.119.312802Suche in Google Scholar PubMed PubMed Central
[12] Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10.10.1093/nar/30.1.207Suche in Google Scholar PubMed PubMed Central
[13] Carolan BJ, Heguy A, Harvey BG, Leopold PL, Ferris B, Crystal RG. Up-regulation of expression of the ubiquitin carboxyl-terminal hydrolase L1 gene in human airway epithelium of cigarette smokers. Cancer Res. 2006;66(22):10729–40.10.1158/0008-5472.CAN-06-2224Suche in Google Scholar PubMed
[14] Tilley AE, Harvey BG, Heguy A, Hackett NR, Wang R, O'connor TP, et al. Down-regulation of the notch pathway in human airway epithelium in association with smoking and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2009;179(6):457–66.10.1164/rccm.200705-795OCSuche in Google Scholar PubMed PubMed Central
[15] Raman T, O'connor TP, Hackett NR, Wang W, Harvey B-G, Attiyeh MA, et al. Quality control in microarray assessment of gene expression in human airway epithelium. BMC genomics. 2009;10(1):1–14.10.1186/1471-2164-10-493Suche in Google Scholar PubMed PubMed Central
[16] Tilley AE, O'connor TP, Hackett NR, Strulovici-Barel Y, Salit J, Amoroso N, et al. Biologic phenotyping of the human small airway epithelial response to cigarette smoking. PLoS one. 2011;6(7):e22798.10.1371/journal.pone.0022798Suche in Google Scholar PubMed PubMed Central
[17] Gindele JA, Kiechle T, Benediktus K, Birk G, Brendel M, Heinemann F, et al. Intermittent exposure to whole cigarette smoke alters the differentiation of primary small airway epithelial cells in the air-liquid interface culture. Sci Rep. 2020;10(1):1–17.10.1038/s41598-020-63345-5Suche in Google Scholar PubMed PubMed Central
[18] Steenman M, Espitia O, Maurel B, Guyomarch B, Heymann MF, Pistorius MA, et al. Identification of genomic differences among peripheral arterial beds in atherosclerotic and healthy arteries. Sci Rep. 2018;8(1):3940.10.1038/s41598-018-22292-ySuche in Google Scholar PubMed PubMed Central
[19] Zenkova DKVSR, Artyomov M, Sergushichev A. Available from: https://genome.ifmo.ru/phantasus.Suche in Google Scholar
[20] Pathan M, Keerthikumar S, Chisanga D, Alessandro R, Ang CS, Askenase P, et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J Extracell Vesicles. 2017;6(1):1321455.10.1080/20013078.2017.1321455Suche in Google Scholar PubMed PubMed Central
[21] Gundersen GW, Jones MR, Rouillard AD, Kou Y, Monteiro CD, Feldmann AS, et al. GEO2Enrichr: browser extension and server app to extract gene sets from GEO and analyze them for biological functions. Bioinformatics. 2015;31(18):3060–2.10.1093/bioinformatics/btv297Suche in Google Scholar PubMed PubMed Central
[22] Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2019;36(8):2628–9.10.1093/bioinformatics/btz931Suche in Google Scholar PubMed PubMed Central
[23] Pomaznoy M, Ha B, Peters B. GOnet: a tool for interactive gene ontology analysis. BMC Bioinforma. 2018;19(1):470.10.1186/s12859-018-2533-3Suche in Google Scholar PubMed PubMed Central
[24] Bader GD, Hogue CWV. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinforma. 2003;4(1):2.10.1186/1471-2105-4-2Suche in Google Scholar PubMed PubMed Central
[25] Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11.10.1186/1752-0509-8-S4-S11Suche in Google Scholar PubMed PubMed Central
[26] Kotlyarov S, Kotlyarova A. Molecular mechanisms of lipid metabolism disorders in infectious exacerbations of chronic obstructive pulmonary disease. Int J Mol Sci. 2021;22(14):7634.10.3390/ijms22147634Suche in Google Scholar PubMed PubMed Central
[27] Kotlyarov S, Kotlyarova A. Anti-inflammatory function of fatty acids and involvement of their metabolites in the resolution of inflammation in chronic obstructive pulmonary disease. Int J Mol Sci. 2021;22(23):12803.10.3390/ijms222312803Suche in Google Scholar PubMed PubMed Central
[28] Kotlyarov S. Diversity of lipid function in atherogenesis: a focus on endothelial mechanobiology. Int J Mol Sci. 2021;22(21):11545.10.3390/ijms222111545Suche in Google Scholar PubMed PubMed Central
[29] Shen Y, Yang T, Guo S, Li X, Chen L, Wang T, et al. Increased serum ox-LDL levels correlated with lung function, inflammation, and oxidative stress in COPD. Mediators Inflamm. 2013;2013:972347.10.1155/2013/972347Suche in Google Scholar PubMed PubMed Central
[30] Herrero-Fernandez B, Gomez-Bris R, Somovilla-Crespo B, Gonzalez-Granado JM. Immunobiology of atherosclerosis: a complex net of interactions. Int J Mol Sci. 2019;20(21):5293.10.3390/ijms20215293Suche in Google Scholar PubMed PubMed Central
[31] Shimada K. Immune system and atherosclerotic disease: heterogeneity of leukocyte subsets participating in the pathogenesis of atherosclerosis. Circ J. 2009;73(6):994–1001.10.1253/circj.CJ-09-0277Suche in Google Scholar
[32] Hansson GK, Hermansson A. The immune system in atherosclerosis. Nat Immunology. 2011;12(3):204–12.10.1038/ni.2001Suche in Google Scholar PubMed
[33] Ammirati E, Moroni F, Magnoni M, Camici PG. The role of T and B cells in human atherosclerosis and atherothrombosis. Clin Exp Immunol. 2015;179(2):173–87.10.1111/cei.12477Suche in Google Scholar
[34] Wolf D, Ley K. Immunity and Inflammation in Atherosclerosis. Circ Res. 2019;124(2):315–27.10.1161/CIRCRESAHA.118.313591Suche in Google Scholar
[35] Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352(16):1685–95.10.1056/NEJMra043430Suche in Google Scholar
[36] Bäck M, Hansson GK. Anti-inflammatory therapies for atherosclerosis. Nat Rev Cardiol. 2015;12(4):199–211.10.1038/nrcardio.2015.5Suche in Google Scholar
[37] Galkina E, Ley K. Immune and inflammatory mechanisms of atherosclerosis (*). Annu Rev Immunol. 2009;27:165–97.10.1146/annurev.immunol.021908.132620Suche in Google Scholar
[38] Tanaka S, Matsumoto T, Matsubara Y, Harada Y, Kyuragi R, Koga JI, et al. BubR1 insufficiency results in decreased macrophage proliferation and attenuated atherogenesis in apolipoprotein E-deficient mice. J Am Heart Assoc. 2016;5(9):e004081.10.1161/JAHA.116.004081Suche in Google Scholar
[39] Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. 2013;13(10):709–21.10.1038/nri3520Suche in Google Scholar
[40] Moore KJ, Tabas I. Macrophages in the pathogenesis of atherosclerosis. Cell. 2011;145(3):341–55.10.1016/j.cell.2011.04.005Suche in Google Scholar
[41] Slack J, McMahan CJ, Waugh S, Schooley K, Spriggs MK, Sims JE, et al. Independent binding of interleukin-1 alpha and interleukin-1 beta to type I and type II interleukin-1 receptors. J Biol Chem. 1993;268(4):2513–24.10.1016/S0021-9258(18)53806-0Suche in Google Scholar
[42] Baines KJ, Simpson JL, Wood LG, Scott RJ, Gibson PG. Transcriptional phenotypes of asthma defined by gene expression profiling of induced sputum samples. J Allergy Clin Immunol. 2011;127(1):153–60, 60.e1-9.10.1016/j.jaci.2010.10.024Suche in Google Scholar PubMed
[43] Baines KJ, Fu JJ, McDonald VM, Gibson PG. Airway gene expression of IL-1 pathway mediators predicts exacerbation risk in obstructive airway disease. Int J Chron Obstruct Pulmon Dis. 2017;12:541–50.10.2147/COPD.S119443Suche in Google Scholar PubMed PubMed Central
[44] Yi G, Liang M, Li M, Fang X, Liu J, Lai Y, et al. A large lung gene expression study identifying IL1B as a novel player in airway inflammation in COPD airway epithelial cells. Inflamm Res. 2018;67(6):539–51.10.1007/s00011-018-1145-8Suche in Google Scholar PubMed
[45] Lappalainen U, Whitsett JA, Wert SE, Tichelaar JW, Bry K. Interleukin-1β Causes Pulmonary Inflammation, Emphysema, and Airway Remodeling in the Adult Murine Lung. Am J Respiratory Cell Mol Biol. 2005;32(4):311–8.10.1165/rcmb.2004-0309OCSuche in Google Scholar PubMed
[46] Ekberg-Jansson A, Andersson B, Bake B, Boijsen M, Enanden I, Rosengren A, et al. Neutrophil-associated activation markers in healthy smokers relates to a fall in DL(CO) and to emphysematous changes on high resolution CT. Respir Med. 2001;95(5):363–73.10.1053/rmed.2001.1050Suche in Google Scholar PubMed
[47] Zou Y, Chen X, Liu J, Zhou DB, Kuang X, Xiao J, et al. Serum IL-1β and IL-17 levels in patients with COPD: associations with clinical parameters. Int J Chron Obstruct Pulmon Dis. 2017;12:1247–54.10.2147/COPD.S131877Suche in Google Scholar PubMed PubMed Central
[48] Libby P. Interleukin-1 beta as a target for atherosclerosis therapy: biological basis of CANTOS and beyond. J Am Coll Cardiol. 2017;70(18):2278–89.10.1016/j.jacc.2017.09.028Suche in Google Scholar PubMed PubMed Central
[49] Abbate A, Van Tassell BW, Biondi-Zoccai GG. Blocking interleukin-1 as a novel therapeutic strategy for secondary prevention of cardiovascular events. BioDrugs. 2012;26(4):217–33.10.1007/BF03261881Suche in Google Scholar PubMed
[50] Mai W, Liao Y. Targeting IL-1β in the Treatment of Atherosclerosis. Front Immunol. 2020;11:589654.10.3389/fimmu.2020.589654Suche in Google Scholar PubMed PubMed Central
[51] Bevilacqua MP, Pober JS, Wheeler ME, Cotran RS, Gimbrone Jr MA. Interleukin-1 activation of vascular endothelium. Effects on procoagulant activity and leukocyte adhesion. Am J Pathol. 1985;121(3):394–403.Suche in Google Scholar
[52] Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377(12):1119–31.10.1056/NEJMoa1707914Suche in Google Scholar PubMed
[53] Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–50.10.1146/annurev.immunol.021908.132612Suche in Google Scholar PubMed
[54] Loppnow H, Libby P. Proliferating or interleukin 1-activated human vascular smooth muscle cells secrete copious interleukin 6. J Clin Invest. 1990;85(3):731–8.10.1172/JCI114498Suche in Google Scholar PubMed PubMed Central
[55] Beltrami-Moreira M, Vromman A, Sukhova GK, Folco EJ, Libby P. Redundancy of IL-1 Isoform Signaling and Its Implications for Arterial Remodeling. PLoS One. 2016;11(3):e0152474.10.1371/journal.pone.0152474Suche in Google Scholar PubMed PubMed Central
[56] Demedts IK, Morel-Montero A, Lebecque S, Pacheco Y, Cataldo D, Joos GF, et al. Elevated MMP-12 protein levels in induced sputum from patients with COPD. Thorax. 2006;61(3):196–201.10.1136/thx.2005.042432Suche in Google Scholar PubMed PubMed Central
[57] Babusyte A, Stravinskaite K, Jeroch J, Lötvall J, Sakalauskas R, Sitkauskiene B. Patterns of airway inflammation and MMP-12 expression in smokers and ex-smokers with COPD. Respir Res. 2007;8(1):81.10.1186/1465-9921-8-81Suche in Google Scholar PubMed PubMed Central
[58] Wallace AM, Sandford AJ, English JC, Burkett KM, Li H, Finley RJ, et al. Matrix metalloproteinase expression by human alveolar macrophages in relation to emphysema. COPD: J Chronic Obstr Pulmonary Dis. 2008;5(1):13–23.10.1080/15412550701817789Suche in Google Scholar PubMed
[59] Churg A, Zhou S, Wright JL. Matrix metalloproteinases in COPD. Eur Respiratory J. 2012;39(1):197–209.10.1183/09031936.00121611Suche in Google Scholar PubMed
[60] De Carolis E, Denis D, Riendeau D. Oxidative inactivation of human 5-lipoxygenase in phosphatidylcholine vesicles. Eur J Biochem. 1996;235(1–2):416–23.10.1111/j.1432-1033.1996.00416.xSuche in Google Scholar PubMed
[61] Gilbert NC, Bartlett SG, Waight MT, Neau DB, Boeglin WE, Brash AR, et al. The structure of human 5-lipoxygenase. Science. 2011;331(6014):217–9.10.1126/science.1197203Suche in Google Scholar PubMed PubMed Central
[62] Gilbert NC, Rui Z, Neau DB, Waight MT, Bartlett SG, Boeglin WE, et al. Conversion of human 5-lipoxygenase to a 15-lipoxygenase by a point mutation to mimic phosphorylation at Serine-663. Faseb J. 2012;26(8):3222–9.10.1096/fj.12-205286Suche in Google Scholar PubMed PubMed Central
[63] Horn T, Reddy Kakularam K, Anton M, Richter C, Reddanna P, Kuhn H. Functional characterization of genetic enzyme variations in human lipoxygenases. Redox Biol. 2013;1(1):566–77.10.1016/j.redox.2013.11.001Suche in Google Scholar PubMed PubMed Central
[64] Dincbas-Renqvist V, Pépin G, Rakonjac M, Plante I, Ouellet DL, Hermansson A, et al. Human Dicer C-terminus functions as a 5-lipoxygenase binding domain. Biochim Biophys Acta. 2009;1789(2):99–108.10.1016/j.bbagrm.2008.10.002Suche in Google Scholar
[65] Hofheinz K, Kakularam KR, Adel S, Anton M, Polymarasetty A, Reddanna P, et al. Conversion of pro-inflammatory murine Alox5 into an anti-inflammatory 15S-lipoxygenating enzyme by multiple mutations of sequence determinants. Arch Biochem Biophys. 2013;530(1):40–7.10.1016/j.abb.2012.11.015Suche in Google Scholar
[66] Petrich K, Ludwig P, Kühn H, Schewe T. The suppression of 5-lipoxygenation of arachidonic acid in human polymorphonuclear leucocytes by the 15-lipoxygenase product (15S)-hydroxy-(5Z,8Z,11Z,13E)-eicosatetraenoic acid: structure-activity relationship and mechanism of action. Biochem J. 1996;314(Pt 3):911–6.10.1042/bj3140911Suche in Google Scholar
[67] Smyrniotis CJ, Barbour SR, Xia Z, Hixon MS, Holman TR. ATP allosterically activates the human 5-lipoxygenase molecular mechanism of arachidonic acid and 5(S)-hydroperoxy-6(E),8(Z),11(Z),14(Z)-eicosatetraenoic acid. Biochemistry. 2014;53(27):4407–19.10.1021/bi401621dSuche in Google Scholar
[68] Ivanov I, Golovanov AB, Ferretti C, Canyelles-Niño M, Heydeck D, Stehling S, et al. Mutations of triad determinants changes the substrate alignment at the catalytic center of human ALOX5. ACS Chem Biol. 2019;14(12):2768–82.10.1021/acschembio.9b00674Suche in Google Scholar
[69] Haeggström JZ, Funk CD. Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem Rev. 2011;111(10):5866–98.10.1021/cr200246dSuche in Google Scholar
[70] Rossi AG, O’Flaherty JT. Bioactions of 5-hydroxyicosatetraenoate and its interaction with platelet-activating factor. Lipids. 1991;26(12):1184–8.10.1201/9781439832042.ch44Suche in Google Scholar
[71] Basil MC, Levy BD. Specialized pro-resolving mediators: endogenous regulators of infection and inflammation. Nat Rev Immunology. 2016;16(1):51–67.10.1038/nri.2015.4Suche in Google Scholar
[72] Khan MI, Hariprasad G. Human secretary phospholipase A2 mutations and their clinical implications. J Inflamm Res. 2020;13:551–61.10.2147/JIR.S269557Suche in Google Scholar
[73] Stremler KE, Stafforini DM, Prescott SM, McIntyre TM. Human plasma platelet-activating factor acetylhydrolase. Oxidatively fragmented phospholipids as substrates. J Biol Chem. 1991;266(17):11095–103.10.1016/S0021-9258(18)99132-5Suche in Google Scholar
[74] Min JH, Jain MK, Wilder C, Paul L, Apitz-Castro R, Aspleaf DC, et al. Membrane-bound plasma platelet activating factor acetylhydrolase acts on substrate in the aqueous phase. Biochemistry. 1999;38(39):12935–42.10.1021/bi991149uSuche in Google Scholar PubMed
[75] Tew DG, Southan C, Rice SQ, Lawrence MP, Li H, Boyd HF, et al. Purification, properties, sequencing, and cloning of a lipoprotein-associated, serine-dependent phospholipase involved in the oxidative modification of low-density lipoproteins. Arterioscler Thromb Vasc Biol. 1996;16(4):591–9.10.1161/01.ATV.16.4.591Suche in Google Scholar PubMed
[76] Stafforini DM, Tjoelker LW, McCormick SP, Vaitkus D, McIntyre TM, Gray PW, et al. Molecular basis of the interaction between plasma platelet-activating factor acetylhydrolase and low density lipoprotein. J Biol Chem. 1999;274(11):7018–24.10.1074/jbc.274.11.7018Suche in Google Scholar PubMed
[77] Packard CJ, O'reilly DS, Caslake MJ, McMahon AD, Ford I, Cooney J, et al. Lipoprotein-associated phospholipase A2 as an independent predictor of coronary heart disease. N Engl J Med. 2000;343(16):1148–55.10.1056/NEJM200010193431603Suche in Google Scholar PubMed
[78] Deng M, Yin Y, Zhang Q, Zhou X, Hou G. Identification of inflammation-related biomarker Lp-PLA2 for patients with COPD by comprehensive analysis. Front Immunol. 2021;12(1718):670971.10.3389/fimmu.2021.670971Suche in Google Scholar PubMed PubMed Central
[79] Smyth LA, Meader L, Xiao F, Woodward M, Brady HJ, Lechler R, et al. Constitutive expression of the anti-apoptotic Bcl-2 family member A1 in murine endothelial cells leads to transplant tolerance. Clin Exp Immunol. 2017;188(2):219–25.10.1111/cei.12931Suche in Google Scholar PubMed PubMed Central
[80] Karsan A, Yee E, Kaushansky K, Harlan JM. Cloning of human Bcl-2 homologue: inflammatory cytokines induce human A1 in cultured endothelial cells. Blood. 1996;87(8):3089–96.10.1182/blood.V87.8.3089.bloodjournal8783089Suche in Google Scholar
[81] Noble KE, Wickremasinghe RG, DeCornet C, Panayiotidis P, Yong KL. Monocytes stimulate expression of the Bcl-2 family member, A1, in endothelial cells and confer protection against apoptosis. J Immunol. 1999;162(3):1376–83.10.4049/jimmunol.162.3.1376Suche in Google Scholar
[82] Vogler M. BCL2A1: the underdog in the BCL2 family. Cell Death Differ. 2012;19(1):67–74.10.1038/cdd.2011.158Suche in Google Scholar PubMed PubMed Central
[83] Olsson Åkefeldt S, Ismail MB, Valentin H, Aricò M, Henter J-I, Delprat C. Targeting BCL2 family in human myeloid dendritic cells: a challenge to cure diseases with chronic inflammations associated with bone loss. Clin Developmental Immunology. 2013;2013:701305.10.1155/2013/701305Suche in Google Scholar PubMed PubMed Central
[84] Vier J, Groth M, Sochalska M, Kirschnek S. The anti-apoptotic Bcl-2 family protein A1/Bfl-1 regulates neutrophil survival and homeostasis and is controlled via PI3K and JAK/STAT signaling. Cell Death Dis. 2016;7(2):e2103-e.10.1038/cddis.2016.23Suche in Google Scholar PubMed PubMed Central
[85] Lok ZSY, Lyle AN. Osteopontin in vascular disease. Arteriosclerosis, Thrombosis, Vasc Biol. 2019;39(4):613–22.10.1161/ATVBAHA.118.311577Suche in Google Scholar PubMed PubMed Central
[86] Kohan M, Bader R, Puxeddu I, Levi-Schaffer F, Breuer R, Berkman N. Enhanced osteopontin expression in a murine model of allergen-induced airway remodelling. Clin Exp Allergy. 2007;37(10):1444–54.10.1111/j.1365-2222.2007.02801.xSuche in Google Scholar PubMed
[87] Miao T-W, Xiao W, Du L-Y, Mao B, Huang W, Chen X-M, et al. High expression of SPP1 in patients with chronic obstructive pulmonary disease (COPD) is correlated with increased risk of lung cancer. FEBS Open Bio. 2021;11(4):1237–49.10.1002/2211-5463.13127Suche in Google Scholar PubMed PubMed Central
[88] Shan M, Yuan X, Song LZ, Roberts L, Zarinkamar N, Seryshev A, et al. Cigarette smoke induction of osteopontin (SPP1) mediates T(H)17 inflammation in human and experimental emphysema. Sci Transl Med. 2012;4(117):117ra9.10.1126/scitranslmed.3003041Suche in Google Scholar PubMed PubMed Central
[89] Papaporfyriou A, Loukides S, Kostikas K, Simoes DCM, Papatheodorou G, Konstantellou E, et al. Increased levels of osteopontin in sputum supernatant in patients with COPD. Chest. 2014;146(4):951–8.10.1378/chest.13-2440Suche in Google Scholar PubMed
[90] Ali MN, Mori M, Mertens TCJ, Siddhuraj P, Erjefält JS, Önnerfjord P, et al. Osteopontin expression in small airway epithelium in copd is dependent on differentiation and confined to subsets of cells. Sci Rep. 2019;9(1):15566.10.1038/s41598-019-52208-3Suche in Google Scholar PubMed PubMed Central
[91] Myers DL, Harmon KJ, Lindner V, Liaw L. Alterations of arterial physiology in osteopontin-null mice. Arterioscler Thromb Vasc Biol. 2003;23(6):1021–8.10.1161/01.ATV.0000073312.34450.16Suche in Google Scholar PubMed
[92] Lyle AN, Joseph G, Fan AE, Weiss D, Landázuri N, Taylor WR. Reactive oxygen species regulate osteopontin expression in a murine model of postischemic neovascularization. Arterioscler Thromb Vasc Biol. 2012;32(6):1383–91.10.1161/ATVBAHA.112.248922Suche in Google Scholar PubMed PubMed Central
[93] Wang X, Louden C, Yue TL, Ellison JA, Barone FC, Solleveld HA, et al. Delayed expression of osteopontin after focal stroke in the rat. J Neurosci. 1998;18(6):2075–83.10.1523/JNEUROSCI.18-06-02075.1998Suche in Google Scholar
[94] Zhu Q, Luo X, Zhang J, Liu Y, Luo H, Huang Q, et al. Osteopontin as a potential therapeutic target for ischemic stroke. Curr Drug Deliv. 2017;14(6):766–2.10.2174/1567201814666161116162148Suche in Google Scholar PubMed
[95] Li Y, Dammer EB, Zhang-Brotzge X, Chen S, Duong DM, Seyfried NT, et al. Osteopontin is a blood biomarker for microglial activation and brain injury in experimental hypoxic-ischemic encephalopathy. eNeuro. 2017;4(1):0253-16.2016.10.1523/ENEURO.0253-16.2016Suche in Google Scholar
[96] Bjerre M, Pedersen SH, Møgelvang R, Lindberg S, Jensen JS, Galatius S, et al. High osteopontin levels predict long-term outcome after STEMI and primary percutaneous coronary intervention. Eur J Prev Cardiol. 2013;20(6):922–9.10.1177/2047487313487083Suche in Google Scholar
[97] Muller O, Delrue L, Hamilos M, Vercauteren S, Ntalianis A, Trana C, et al. Transcriptional fingerprint of human whole blood at the site of coronary occlusion in acute myocardial infarction. EuroIntervention. 2011;7(4):458–66.10.4244/EIJV7I4A75Suche in Google Scholar
[98] Koshikawa M, Aizawa K, Kasai H, Izawa A, Tomita T, Kumazaki S, et al. Elevated osteopontin levels in patients with peripheral arterial disease. Angiology. 2009;60(1):42–5.10.1177/0003319708314250Suche in Google Scholar
[99] Kapetanios D, Karkos C, Giagtzidis I, Papazoglou K, Kiroplastis K, Spyridis C. Vascular calcification biomarkers and peripheral arterial disease. Int Angiol. 2016;35(5):455–9.Suche in Google Scholar
[100] Lee GS, Salazar HF, Joseph G, Lok ZSY, Caroti CM, Weiss D, et al. Osteopontin isoforms differentially promote arteriogenesis in response to ischemia via macrophage accumulation and survival. Lab Invest. 2019;99(3):331–45.10.1038/s41374-018-0094-8Suche in Google Scholar
[101] Duvall CL, Weiss D, Robinson ST, Alameddine FM, Guldberg RE, Taylor WR. The role of osteopontin in recovery from hind limb ischemia. Arterioscler Thromb Vasc Biol. 2008;28(2):290–5.10.1161/ATVBAHA.107.158485Suche in Google Scholar
[102] O'brien ER, Garvin MR, Stewart DK, Hinohara T, Simpson JB, Schwartz SM, et al. Osteopontin is synthesized by macrophage, smooth muscle, and endothelial cells in primary and restenotic human coronary atherosclerotic plaques. Arterioscler Thromb. 1994;14(10):1648–56.10.1161/01.ATV.14.10.1648Suche in Google Scholar
[103] Sonkar VK, Kumar R, Jensen M, Wagner BA, Sharathkumar AA, Miller Jr FJ, et al. Nox2 NADPH oxidase is dispensable for platelet activation or arterial thrombosis in mice. Blood Adv. 2019;3(8):1272–84.10.1182/bloodadvances.2018025569Suche in Google Scholar
[104] Tarazona-Santos E, Machado M, Magalhães WC, Chen R, Lyon F, Burdett L, et al. Evolutionary dynamics of the human NADPH oxidase genes CYBB, CYBA, NCF2, and NCF4: functional implications. Mol Biol Evolution. 2013;30(9):2157–67.10.1093/molbev/mst119Suche in Google Scholar
[105] Heyworth PG, Cross AR, Curnutte JT. Chronic granulomatous disease. Curr Opin Immunol. 2003;15(5):578–84.10.1016/S0952-7915(03)00109-2Suche in Google Scholar
© 2022 Stanislav Kotlyarov, published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Research Articles
- Diagnostic accuracy of genetic markers for identification of the Lr46/Yr29 “slow rusting” locus in wheat (Triticum aestivum L.)
- NADPH-derived ROS generation drives fibrosis and endothelial-to-mesenchymal transition in systemic sclerosis: Potential cross talk with circulating miRNAs
- Effect of omega-3 fatty acids on the telomere length: A mini meta-analysis of clinical trials
- Analysis of differentially expressed genes and signaling pathways involved in atherosclerosis and chronic obstructive pulmonary disease
- The epigenetic dimension of protein structure
- Gamma-induced mutants of Bacillus and Streptomyces display enhanced antagonistic activities and suppression of the root rot and wilt diseases in pulses
- Corticosterone potentiates ochratoxin A-induced microglial activation
- Supercomplex supercomplexes: Raison d’etre and functional significance of supramolecular organization in oxidative phosphorylation
- Insights into functional connectivity in mammalian signal transduction pathways by pairwise comparison of protein interaction partners of critical signaling hubs
- The effects of supplementation of Nannochloropsis oculata microalgae on biochemical, inflammatory and antioxidant responses in diabetic rats
- Molecular epidemiology of human papillomavirus in pregnant women in Burkina Faso
- Review Articles
- Interaction of cervical microbiome with epigenome of epithelial cells: Significance of inflammation to primary healthcare
- Seaweeds’ pigments and phenolic compounds with antimicrobial potential
- The capture of host cell’s resources: The role of heat shock proteins and polyamines in SARS-COV-2 (COVID-19) pathway to viral infection
- Erratum
- Erratum to “Plant growth-promoting properties of bacterial endophytes isolated from roots of Thymus vulgaris L. and investigate their role as biofertilizers to enhance the essential oil contents”
- Special Issue on XXV Congress of the Italian Society for Pure and Applied Biophysics
- Low-temperature librations and dynamical transition in proteins at differing hydration levels
- The phosphoinositide PI(3,5)P2 inhibits the activity of plant NHX proton/potassium antiporters: Advantages of a novel electrophysiological approach
- Targeted photoimmunotherapy for cancer
- Calorimetry of extracellular vesicles fusion to single phospholipid membrane
- Calcium signaling in prostate cancer cells of increasing malignancy
- Oxygen diffusion pathways in mutated forms of a LOV photoreceptor from Methylobacterium radiotolerans: A molecular dynamics study
- A photosensitizing fusion protein with targeting capabilities
- Ion channels and neuronal excitability in polyglutamine neurodegenerative diseases
- Is styrene competitive for dopamine receptor binding?
- Diffusion of molecules through nanopores under confinement: Time-scale bridging and crowding effects via Markov state model
- Quantitative active super-resolution thermal imaging: The melanoma case study
- Innovative light sources for phototherapy
- Electrophysiological study of the effects of side products of RuBi-GABA uncaging on GABAA receptors in cerebellar granule cells
- Subcellular elements responsive to the biomechanical activity of triple-negative breast cancer-derived small extracellular vesicles
Artikel in diesem Heft
- Research Articles
- Diagnostic accuracy of genetic markers for identification of the Lr46/Yr29 “slow rusting” locus in wheat (Triticum aestivum L.)
- NADPH-derived ROS generation drives fibrosis and endothelial-to-mesenchymal transition in systemic sclerosis: Potential cross talk with circulating miRNAs
- Effect of omega-3 fatty acids on the telomere length: A mini meta-analysis of clinical trials
- Analysis of differentially expressed genes and signaling pathways involved in atherosclerosis and chronic obstructive pulmonary disease
- The epigenetic dimension of protein structure
- Gamma-induced mutants of Bacillus and Streptomyces display enhanced antagonistic activities and suppression of the root rot and wilt diseases in pulses
- Corticosterone potentiates ochratoxin A-induced microglial activation
- Supercomplex supercomplexes: Raison d’etre and functional significance of supramolecular organization in oxidative phosphorylation
- Insights into functional connectivity in mammalian signal transduction pathways by pairwise comparison of protein interaction partners of critical signaling hubs
- The effects of supplementation of Nannochloropsis oculata microalgae on biochemical, inflammatory and antioxidant responses in diabetic rats
- Molecular epidemiology of human papillomavirus in pregnant women in Burkina Faso
- Review Articles
- Interaction of cervical microbiome with epigenome of epithelial cells: Significance of inflammation to primary healthcare
- Seaweeds’ pigments and phenolic compounds with antimicrobial potential
- The capture of host cell’s resources: The role of heat shock proteins and polyamines in SARS-COV-2 (COVID-19) pathway to viral infection
- Erratum
- Erratum to “Plant growth-promoting properties of bacterial endophytes isolated from roots of Thymus vulgaris L. and investigate their role as biofertilizers to enhance the essential oil contents”
- Special Issue on XXV Congress of the Italian Society for Pure and Applied Biophysics
- Low-temperature librations and dynamical transition in proteins at differing hydration levels
- The phosphoinositide PI(3,5)P2 inhibits the activity of plant NHX proton/potassium antiporters: Advantages of a novel electrophysiological approach
- Targeted photoimmunotherapy for cancer
- Calorimetry of extracellular vesicles fusion to single phospholipid membrane
- Calcium signaling in prostate cancer cells of increasing malignancy
- Oxygen diffusion pathways in mutated forms of a LOV photoreceptor from Methylobacterium radiotolerans: A molecular dynamics study
- A photosensitizing fusion protein with targeting capabilities
- Ion channels and neuronal excitability in polyglutamine neurodegenerative diseases
- Is styrene competitive for dopamine receptor binding?
- Diffusion of molecules through nanopores under confinement: Time-scale bridging and crowding effects via Markov state model
- Quantitative active super-resolution thermal imaging: The melanoma case study
- Innovative light sources for phototherapy
- Electrophysiological study of the effects of side products of RuBi-GABA uncaging on GABAA receptors in cerebellar granule cells
- Subcellular elements responsive to the biomechanical activity of triple-negative breast cancer-derived small extracellular vesicles

