Abstract
CCTG repeat expansion in intron 1 of the cellular nucleic acid-binding protein (CNBP) gene has been identified to be the genetic cause of myotonic dystrophy type 2 (DM2). Yet the underlying reasons for the genetic instability in CCTG repeats remain elusive. In recent years, CCTG repeats have been found to form various types of unusual secondary structures including mini-dumbbell (MDB), hairpin and dumbbell, revealing that there is a high structural diversity in CCTG repeats intrinsically. Upon strand slippage, the formation of unusual structures in the nascent strand during DNA replication has been proposed to be the culprit of CCTG repeat expansions. On the one hand, the thermodynamic stability, size, and conformational dynamics of these unusual structures affect the propensity of strand slippage. On the other hand, these structural properties determine whether the unusual structure can successfully escape from DNA repair. In this short overview, we first summarize the recent advances in elucidating the solution structures of CCTG repeats. We then discuss the potential pathways by which these unusual structures bring about variable sizes of repeat expansion, high strand slippage propensity and efficient repair escape.
Introduction
Tetranucleotide CCTG repeats belong to a class of short tandem repeats or microsatellites containing one to nine nucleotides that are repeated in a head-to-tail manner. In the human genome, expansions of CCTG repeats in intron 1 of the cellular nucleic acid-binding protein (CNBP) gene on chromosome 3q21, which is previously known as the zinc finger protein 9 (ZNF9) gene, bring about myotonic dystrophy type 2 (DM2) (1). DM2 is a complex multisystem disorder characterized by myotonia and muscle dysfunction including weakness, pain and stiffness (2), (3). Some DM2 patients also show cardiac conduction defects, iridescent posterior subcapsular cataracts, insulin insensitive type 2 diabetes mellitus and testicular failure. Apart from DM2, there is another subtype of myotonic dystrophy, namely myotonic dystrophy type 1 (DM1), which is caused by trinucleotide CTG repeat expansions in the 3′ untranslated region of the dystrophia myotonica protein kinase (DMPK) gene (4). According to the data provided in the European Neuromuscular Centre International Workshop, there has been an increasing trend of DM2 and the incidence has already been very close to that of DM1 (5), (6).
Although DM2 is considered to be milder than DM1 in terms of their clinical phenotypes (3), (7), CCTG repeat expansions are extremely unstable and variable (8). In intron 1 of the CNBP gene, there are different numbers (n) of CCTG repeats located in a part of the complex repetitive motif (TG)14−25(TCTG)4−10(CCTG)n (1), (9), (10). In DM2 patients, the repeat tract is uninterrupted and the repeat size can vary between 55 and ~11 000 repeats (11). In normal individuals, the CCTG repeat tract generally contains less than 30 repeats and is interrupted by one or more A/G/TCTG motifs (Figure 1). The unprecedented large-size of expansion yields variable clinical phenotypes, making the diagnosis of DM2 difficult (12).
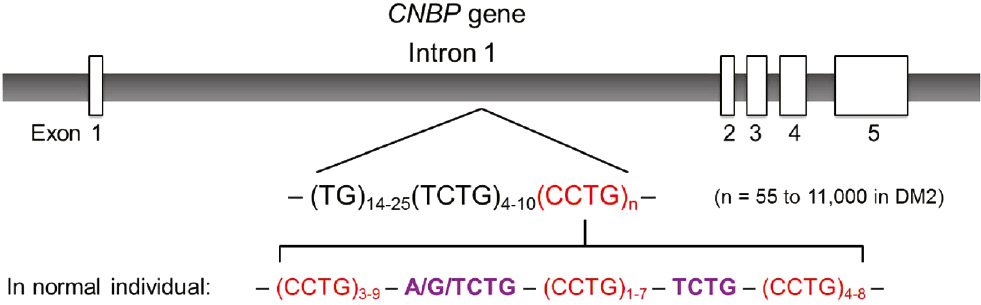
CCTG repeats in intron 1 of the CNBP gene.
For DM2 patients, the CCTG repeat tract is uninterrupted and the repeat length can vary between 55 and ~11 000 repeats. For normal individuals, the CCTG repeat tract is interrupted by A/G/TCTG motifs, making the tract contains short segments of CCTG repeats.
Currently, the underlying causes for CCTG repeat expansions remain unclear although the pathogenic mechanism of DM2 has been suggested to associate with the long CCUG-containing RNA transcripts that recruit the muscleblind-like (MBNL) proteins. Upon binding with the RNA transcripts, the MBNL proteins lose their normal functions to splice pre-mRNAs (13). As the RNA-protein binding complexes cannot be transported from the cell nucleus to cytoplasm, there will be an accumulation of these complexes as discrete nuclear foci, thus increasing the level of RNA toxicity (3). Despite the overwhelming evidence showing the critical roles of MBNL proteins in the DM2 pathology, the downstream pathways by which these proteins cause muscle wasting and weakness are not well understood. Recently, the molecular pathways by which mutations may cause muscle atrophy have been reviewed (14). Although there have been some progress in developing therapeutic interventions for muscle wasting and weakness in DM1 (15), (16), no significant achievement has been made for DM2. As a result, a better understanding of the genetic instability of CCTG repeats will benefit the development of DM2 therapy.
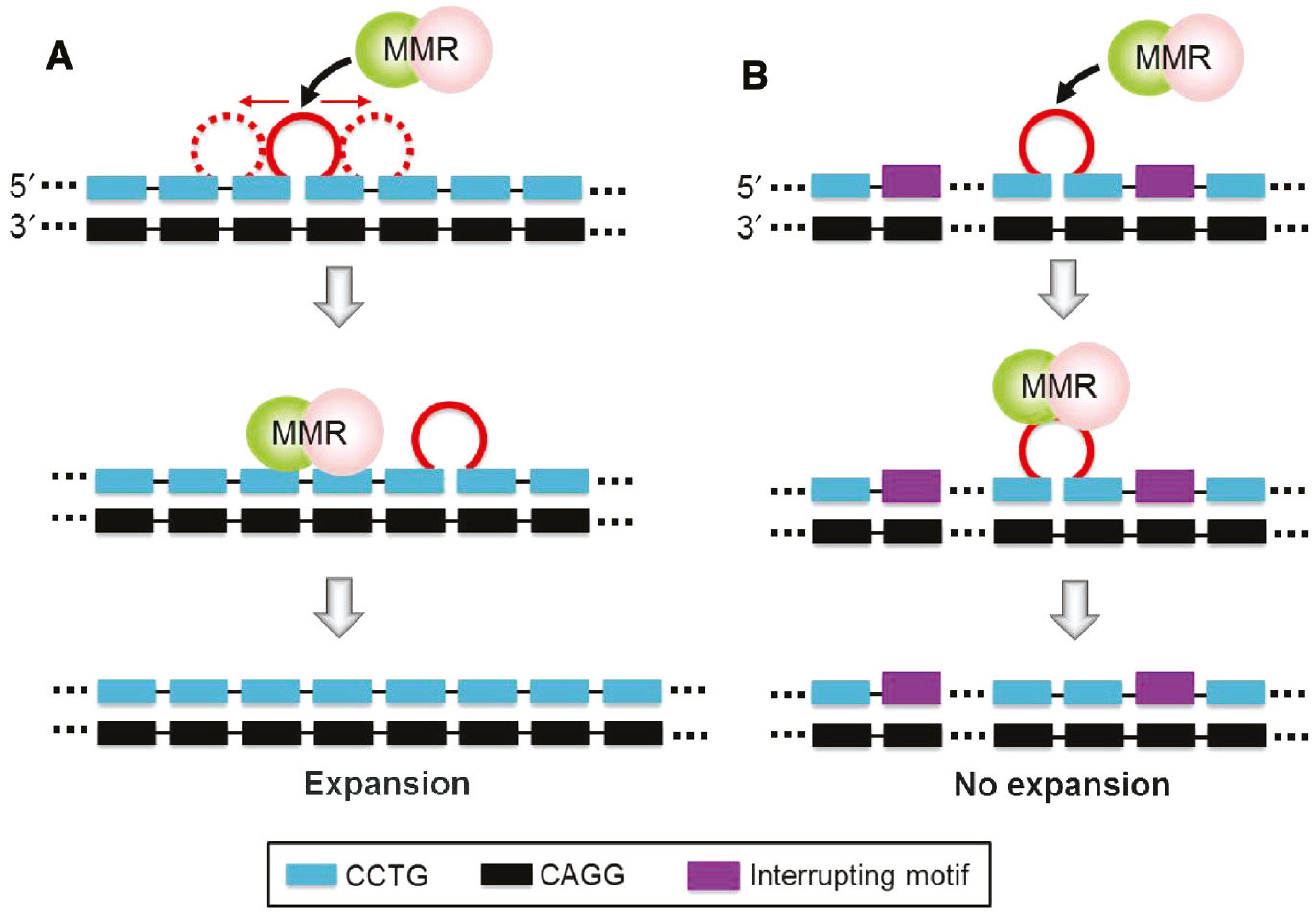
Over the past three decades, extensive studies have been carried out to investigate trinucleotide (17), (18), (19), (20), (21), (22), (23) and tetranucleotide (12), (24), (25), (26), (27), (28), (29) repeat expansions. One commonly accepted expansion pathway involves the formation of an unusual structure in the nascent strand during DNA replication (30), (31), (32) although this can also occur in DNA repair or recombination (20), (28), (33), (34), (35), (36). The formation of unusual structure promotes strand slippage via stabilizing the slipped strand (Figure 2A). In general, the unusual structure formed in the nascent strand can be recognized and removed by mismatch repair (MMR), which is a post-replication repair system to maintain the fidelity of DNA replication. If the unusual structure successfully escapes from MMR, the nascent strand will be lengthened (Figure 2B). As a result, repeat expansions will occur if there are both (i) strand slippage and (ii) repair escape.
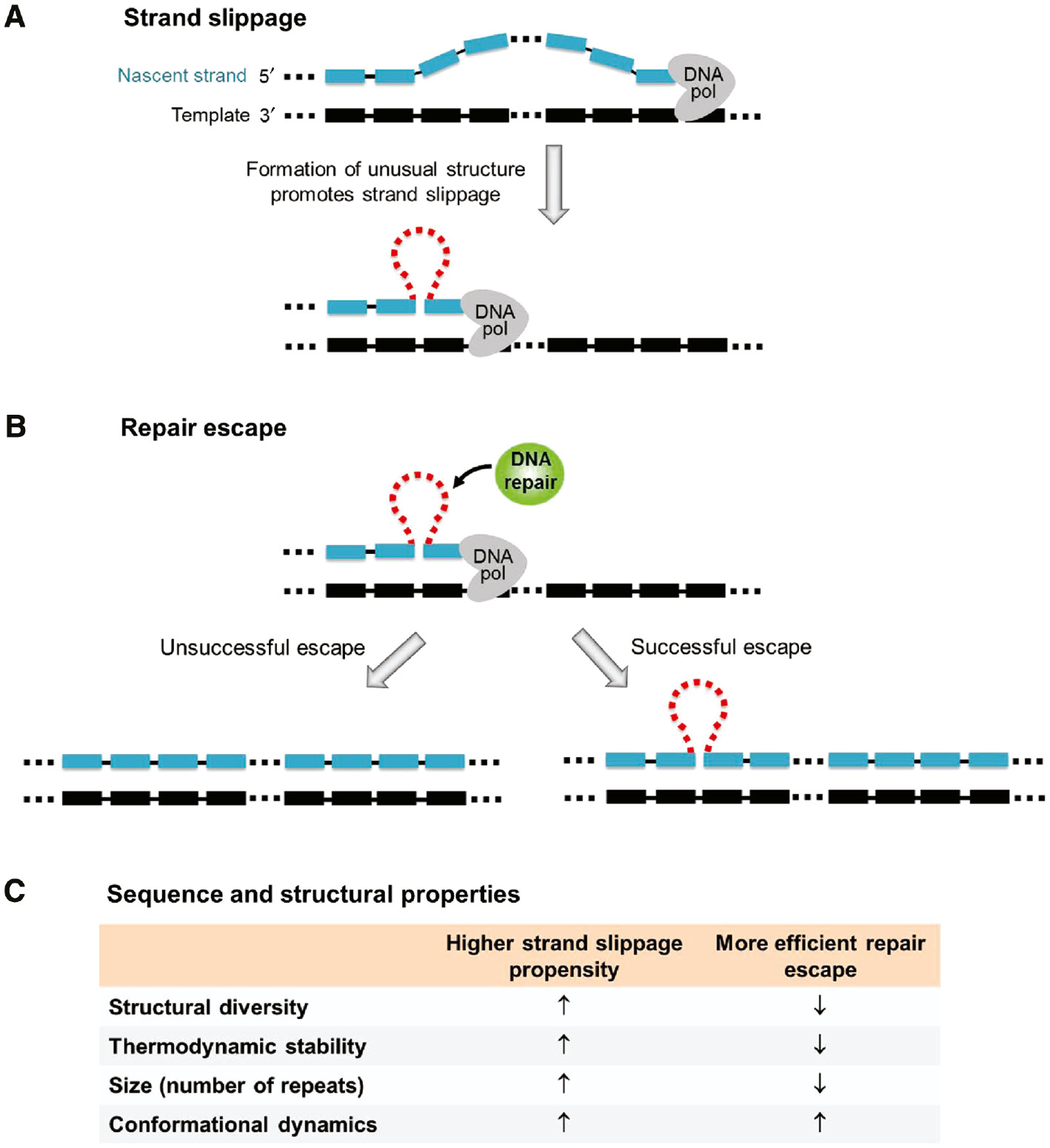
A proposed pathway of repeat expansion during DNA replication.
(A) Slippage occurs at the 3′-terminal of the nascent strand. The formation of an unusual structure promotes strand slippage via stabilizing the slipped strand. (B) Repeat expansion occurs if the unusual structure successfully escapes from DNA repair. (C) This table summarizes the sequence and structural properties that lead to a higher strand slippage propensity and more efficient repair escape.
Despite the torsional stress resulting from supercoiling induced by the replication activity of downstream genes (37), the formation of unusual structure is largely dependent on its intrinsic sequence and structural properties (38), (39). These include (i) the structural diversity of the repeating sequence, i.e. the ability of the sequence to form different types of structures, (ii) the thermodynamic stability of the unusual structure, (iii) the size of the unusual structure, i.e. the number of repeats involved, and (iv) the conformational dynamics, i.e. the feasibility of exchange between different structural conformers (Figure 2C). A higher propensity of strand slippage will be expected if the sequence has a higher level of structural diversity, the unusual structure formed is more stable, contains more repeats, or undergoes conformational exchange more feasibly. However, less efficient repair escape will be expected for the above properties except the one related to conformational exchange. Therefore, these sequence and structural properties play critical roles in determining whether there will be repeat expansion mutations.
At present, the hairpins formed by CTG repeats (19), tetraplexes formed by GCC and CGG repeats (40), (41), and triplexes formed by GAA repeats (23) have been proposed to be the culprits of different types of trinucleotide repeat expansions. In the following sections, we first summarize the recent findings on the unusual structures of DNA sequences containing CCTG repeats. Then we discuss how these structures participate in the potential pathways that lead to variable sizes of repeat expansion, and different levels of strand slippage and repair escape.
Detailed structural information of CCTG repeats
About a decade ago, it was shown by enzymatic and chemical probing experiments that a DNA sequence containing 26 CCTG repeats did not adopt any stable secondary structures (29). As suggested by the presence of multiple cleavage sites, CCTG repeats were proposed to adopt hairpin structures containing different sizes of loops and various lengths of overhangs. Conformational exchanges among these different hairpins were thought to be feasible because there were unpaired residues in the loops and also T·T and C·T mismatches in the stems. Since then, no significant advance has been made in understanding the genetic instability of CCTG repeats. For studying nucleic acid structures, there are two high resolution techniques, namely, X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy, that can provide atomic level structural information. However, owing to the intrinsic conformational dynamics, growing DNA crystals containing CCTG repeats is expected to be difficult, thus limiting the use of X-ray diffraction technique. For NMR investigations, there are challenges due to (i) the repetitive nature of CCTG repeating sequence which leads to serious spectral overlap and ambiguous resonance assignment, and (ii) the presence of conformational exchange which brings about peak broadening (42).
Despite the abovementioned limitations and challenges, it has been demonstrated that single-site substitution experiments could successfully differentiate the NMR signals from different DNA conformers of CTG repeats (19). Therefore, single-site substitution experiments have recently been attempted on CCTG repeats and high resolution structural information have been obtained (25), (27). Figure 3 shows a summary of the detailed structural information successfully obtained in CCTG repeats with different repeat lengths.
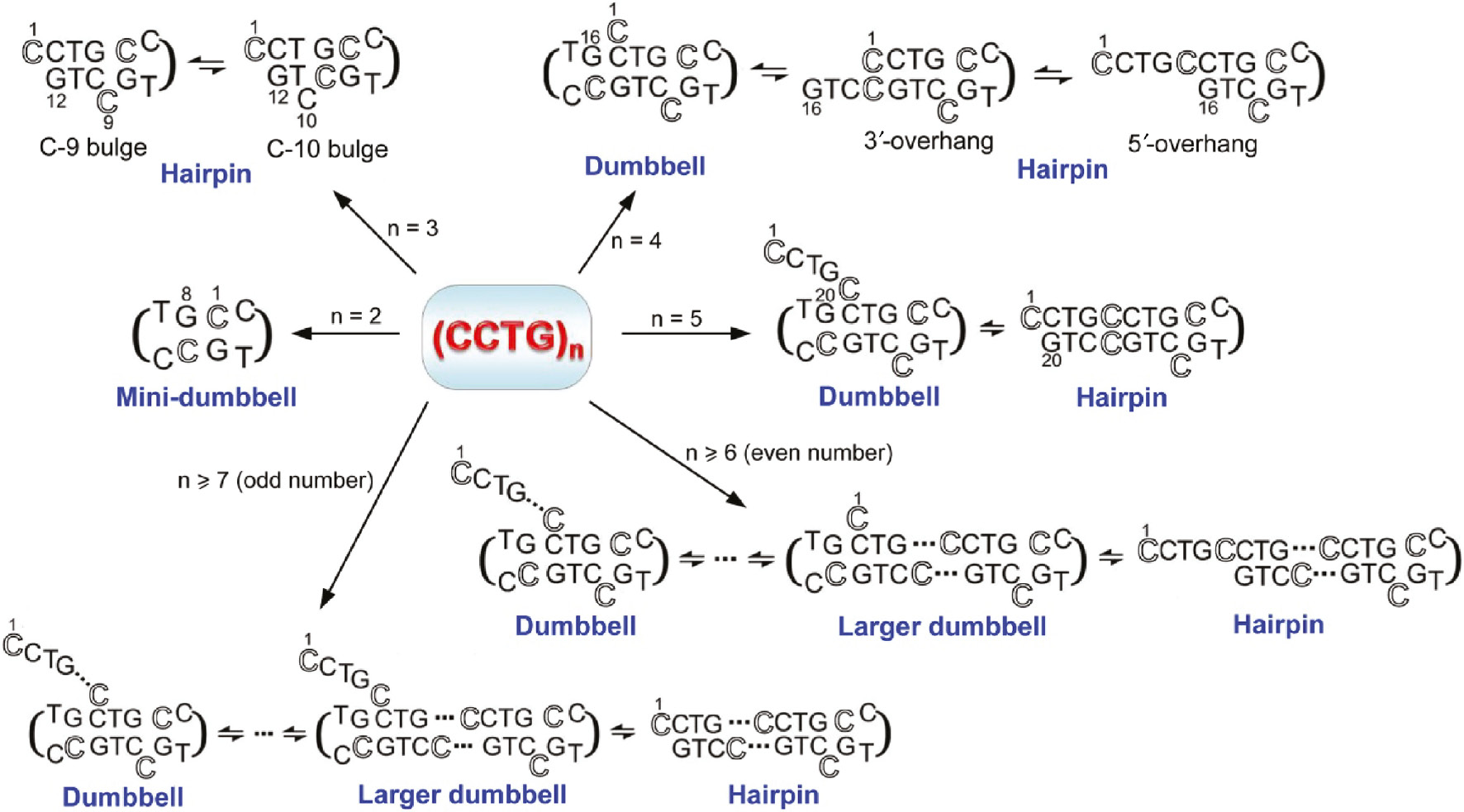
A summary of the diverse unusual structures formed by CCTG repeats.
Depending on the repeat length, CCTG repeats can adopt MDB, hairpin and/or dumbbell structures. For better illustrating the participation of each repeat in the unusual structures, an outlined font is used for the first cytosine residue of each repeat.
In 2011, the first piece of high resolution structural information was reported (27). Based on the solution NMR results, a DNA sequence containing three CCTG repeats was found to adopt an hairpin which comprises (i) a CCTG type II loop, and (ii) a flexible stem containing a shifting C-bulge between C9 and C10, and a T·T mismatch. For the CCTG type II loop, the structure was found to be similar to that formed in the DNA hairpin d(CG CCTG CG) (43) in which the third loop residue, thymine, stacks over the base plane of C-G loop-closing base pair, while the second loop residue, cytosine, is located in the minor groove. The C9-bulge conformer was found to be more populated than the C10-bulge conformer because a cytosine bulge flanked by two Watson-Crick C-G base pairs is less destabilizing than the one flanked by a T·T mismatch and a C-G base pair.
For a DNA sequence containing four CCTG repeats, it was found to adopt a predominant dumbbell structure exchanging with an hairpin structure (27). The dumbbell comprises two CCTG type II loops formed by the second and fourth repeats and a stem containing a C-bulge and a T·T mismatch. For the hairpin, it contains a CCTG type II loop formed by the third repeat. Conformational exchange between the dumbbell and hairpin structures reveals the second, third and fourth repeats to have the ability to fold into type II loops.
As the conformational space of CCTG repeats increases with the repeat length, it is possible that multiple forms of hairpins and dumbbells can be adopted by longer CCTG repeats. For DNA sequences containing 5–10 repeats, they also showed characteristic 1H and 31P NMR signals of a typical CCTG type II loop. The results from 31P–31P exchange spectroscopy provide important pieces of evidence supporting the presence of conformational exchange in these longer repeats (27). With the aid of single-site substitution samples, dynamic exchange was found between the dumbbell and hairpin conformers (27). Depending on the repeat length, the dumbbell conformer can be formed by the last four repeats with a 5′-overhang containing different number of repeat(s). Alternatively, a realignment of the repetitive strand can occur, leading to the formation of larger dumbbell conformers with shorter 5′-overhangs.
In addition to the hairpin and dumbbell conformers, a mini-dumbbell (MDB) structure containing only two CCTG type II loops has recently been reported (25), revealing the high structural diversity of CCTG repeats. In this MDB structure, the first (C1 and C5) and fourth (G4 and G8) loop residues of the two CCTG repeats form two loop-closing base pairs. The second loop residues (C2 and C6) fold into the minor groove and form hydrogen bond(s) with each other. The third loop residues T3 and T7 stack on C1-G4 and C5-G8, respectively. From the detailed MDB structural features (44), various types of stabilizing interactions were found (Figure 4A). These include (i) the 3′–5′ terminal stacking between the two C-G Watson-Crick loop-closing base pairs, (ii) the stacking between the third loop residues and the loop-closing base pairs, (iii) the hydrophobic interactions between the third loop residues and their two preceding residues, (iv) the hydrogen bond(s) between the two minor groove residues C2 and C6, and (v) the hydrogen bonds between the minor groove residues and the loop-closing base pairs/phosphodiester backbone. Among these, there are extensive loop-loop interactions in this MDB structure (Figure 4B), which are absent in the larger dumbbell structures formed by four or more repeats (27).
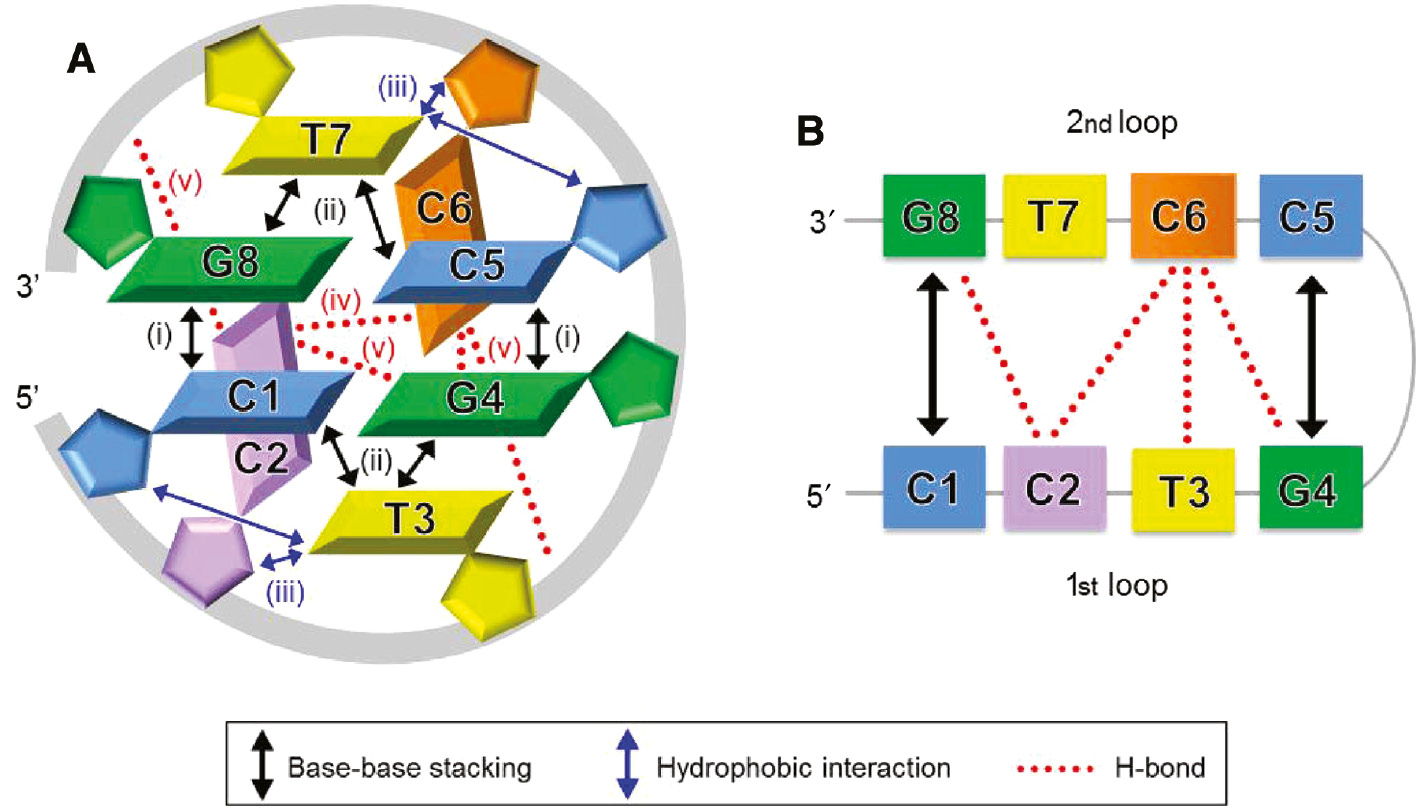
A schematic representation of the MDB structure formed by two CCTG repeats.
(A) Five types of stabilizing interactions are present in this MDB, including (i) the stacking between C1-G4 and C5-G8 base pairs, (ii) the stacking between T3/T7 and its loop-closing base pair, (iii) the hydrophobic interactions between T3/T7 and its two preceding residues, (iv) the hydrogen bond(s) between C2 and C6, and (v) the hydrogen bonds between C2/C6 and the loop-closing base pairs/phosphodiester backbone. (B) Extensive loop-loop interactions, including the stacking between the two loop-closing base pairs, the hydrogen bonds between C2 and C6/G8, and between C6 and T3/G4, are present in this MDB.
Based on the above structural features, any two adjacent repeats in a CCTG repeat tract are capable of forming an MDB. Yet no MDB was observed in the NMR studies of three or longer CCTG repeats (27). As revealed by the single-site substitution experiments (25), the formation of MDB was hindered by the formation of a more competitive hairpin in the sequence containing three CCTG repeats. This hairpin can be formed due to a feasible realignment of the 5′ and 3′-terminal repeats which maximizes the stabilizing interactions in the stem region. Upon weakening these stabilizing interactions through substituting C10 with T10 in the observed hairpin, the formation of an MDB with a 5′-overhang was shown to become possible (Figure 5A). In intron 1 of the CNBP gene, CCTG repeats do not appear in a 5′ or 3′-terminal position. In addition, the 5′-terminal of CCTG repeats in the nascent strand is hybridized with the template strand, making it less flexible to interact with the 3′-terminal repeat. Therefore, it is expected that there will be the formation of MDB in the slipped nascent strand. In principle, the formation of an MDB with a 3′-overhang is also possible. Since the formation of these two types of MDBs involves the folding of the middle repeat, fast exchange between these MDBs will result in the formation of a mini-loop (Figure 5B). Such exchange between two competing MDBs has recently been observed in a DNA sequence containing three TTTA repeats (24).

MDB can possibly be formed in a DNA sequence containing three CCTG repeats.
(A) Substituting C10 with T10 in the hairpin promotes the formation of an MDB with a 5′-overhang. (B) Two types of MDB conformers can be formed in a DNA segment containing three CCTG repeats. Fast exchange between them results in a mini-loop structure (red box) because the middle repeat remains folded as a type II loop in both conformers.
Participation of unusual structures in variable sizes of repeat expansion
Although the genetic cause of DM2 has been identified (1), the underlying reasons for the extremely unstable CCTG repeat expansions remain elusive. The sizes of CCTG repeat expansion can vary from one repeat in an in vitro primer extension assay (45) to ~11 000 repeats in DM2 patients (11). Owing to the high structural diversity of CCTG repeats, the MDB, hairpin and dumbbell structures can serve as structural intermediates leading to the occurrence of two, three and four-repeat expansions, respectively, in a single strand slippage event (25), (27). In addition, fast exchange between two adjacent competing MDBs results in a mini-loop that can lead to one-repeat expansion. Re-occurrence of the mini-loop in additional strand slippage events is also possible, providing a pathway for any larger sizes of repeat expansion. Alternatively, the presence of one or more of these unusual structures in the nascent strand can also bring about larger sizes of repeat expansion (Figure 6). It has been shown that the amount and number of different types of slipped structures were proportional to the repeat tract length and the homogeneity of sequence (46), (47), (48). Therefore, for the long and uninterrupted CCTG repeats in DM2 patients, it is likely that there will be multiple unusual structures in the nascent CCTG strand during DNA replication.
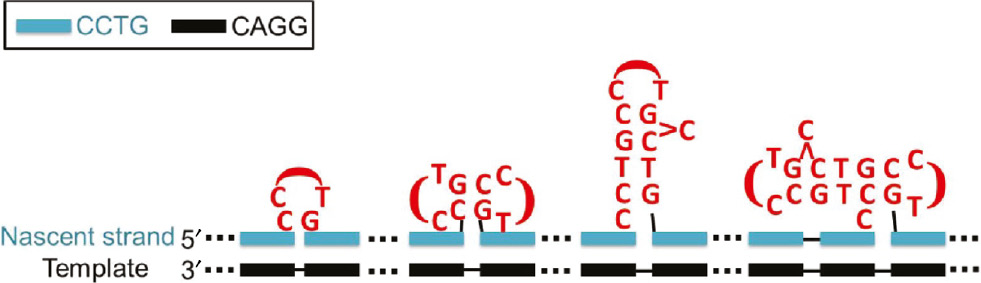
Multiple unusual CCTG structures can co-exist in the nascent strand.
The presence of one or more of these unusual CCTG structures can lead to any sizes of repeat expansion.
Interconversion between different structures promotes both strand slippage and repair escape
As revealed in Figure 3, CCTG repeats can adopt the MDB, hairpin and dumbbell structures. It has also been demonstrated that conformational exchange between two adjacent competing MDBs can lead to the formation of a mini-loop (25). Indeed, conformational exchanges between the (i) mini-loop and MDB, (ii) MDB and hairpin, and (iii) hairpin and dumbbell are also possible via the participation of an additional CCTG repeat (Figure 7A), thus increasing the conformational dynamics of CCTG repeats. On the one hand, interconversion between different structures promotes strand slippage as this involves a realignment process that requires the nascent strand to partially dissociate from the template. On the other hand, conformational exchange also enhances the efficiency of repair escape as the recognition of a dynamically exchanging structure is more difficult.
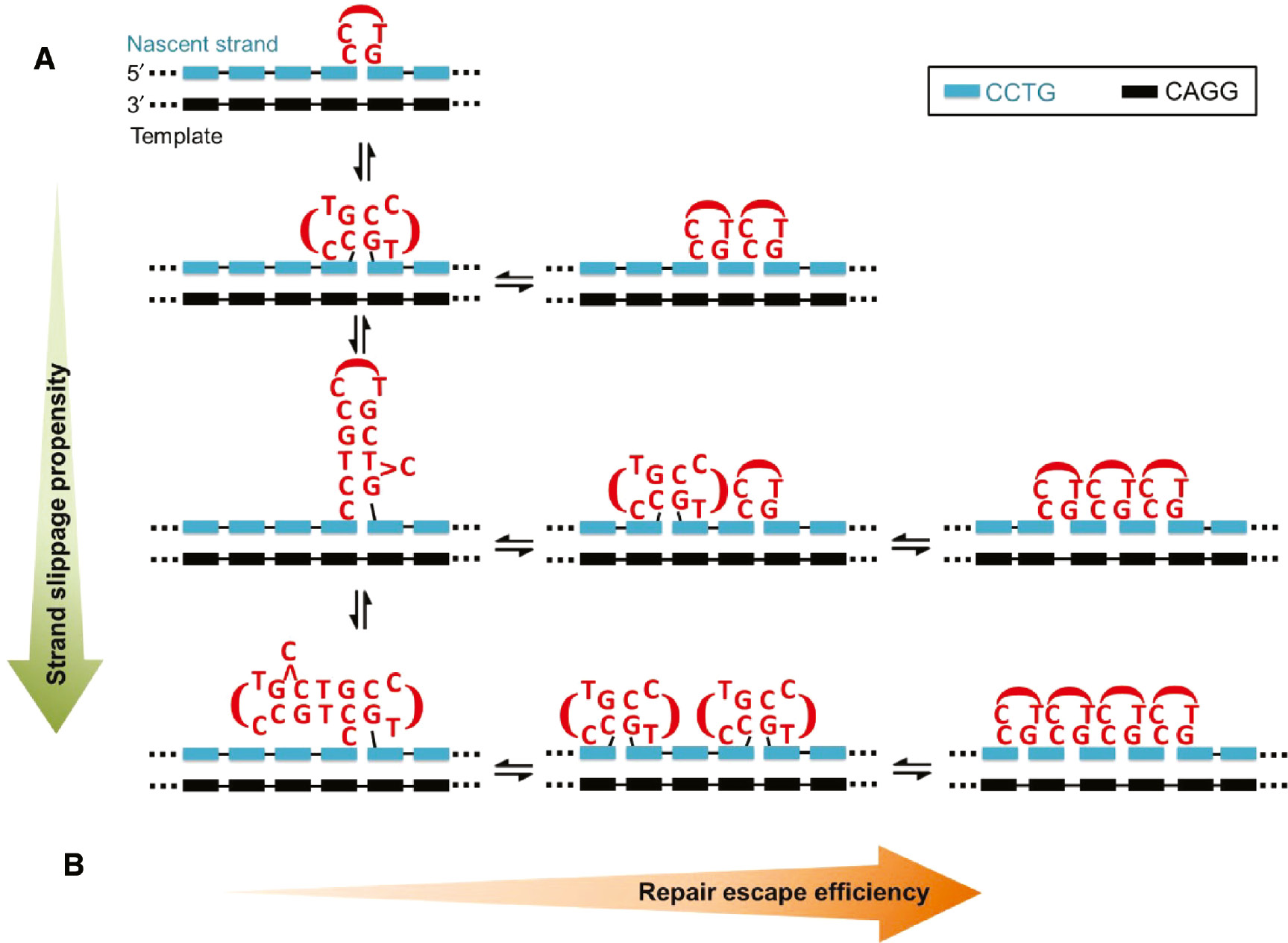
Interconversion between different types of unusual CCTG structures.
(A) Conversion to a larger and more stable unusual structure will lead to a higher propensity of strand slippage. (B) Conversion from a larger unusual structure to multiple smaller structures will bring about more efficient repair escape.
In the uninterrupted CCTG repeat tract of DM2 patients, the high propensity of strand slippage may originate from the formation of any unusual structure that will subsequently be converted to larger unusual structures. To increase the chance of successful escape from MMR, these larger unusual structures can be converted back to multiple smaller unusual structures (Figure 7B). Previously, it has been shown that multiple short loops containing one to three CTG repeats along a single repeat tract can escape from MMR (17). Therefore, we believe that the extremely unstable and variable CCTG repeat expansions in DM2 patients originate from (i) the high strand slippage propensity of uninterrupted CCTG repeats and (ii) the efficient repair escape due to the feasible conversion of a large unusual structure to multiple smaller structures.
Shifting of mini-loop provides a more efficient repair escape pathway
As revealed by the structural features of CCTG repeats, a mini-loop structural intermediate can be formed due to fast exchange between two adjacent competing MDBs. In the uninterrupted CCTG repeat tract of DM2 patients, MDBs can be formed by any two adjacent repeats in the nascent CCTG strand during DNA replication. Therefore, we expect the resulting mini-loop can shift in both the 5′ and 3′-directions along the tract, enhancing the capability of the mini-loop to escape from MMR (Figure 8A). In normal individuals, intron 1 of the CNBP gene contains only a short CCTG repeat tract interrupted by A/G/TCTG motifs. As a consequence, shifting of the mini-loop will be hindered and thereby the mini-loop can be recognized and removed more efficiently by MMR (Figure 8B).
Shifting of the mini-loop provides more efficient escape from MMR.
(A) In the uninterrupted CCTG repeat tract of DM2 patients, the mini-loop can shift in both the 5′ and 3′ directions, making it more easily to escape from MMR. (B) In normal individuals, the presence of interrupting motif(s) in the CCTG repeat tract will hinder the shifting of mini-loop. Thereby, the mini-loop can be recognized and removed more efficiently by the MMR proteins.
Outlook
The high structural diversity of CCTG repeats brings about the unusual conformational dynamics which plays critical roles in various potential pathways of CCTG repeat expansions. In addition to the research efforts that have been devoted to understand the intrinsic structural behaviors of CCTG repeats, we foresee that there will be extensive works to understand (i) the environmental effect on the unusual structures of CCTG repeats, and (ii) how exactly these unusual CCTG structures bring about repeat expansions.
First, it is well known that solution structures of DNA are affected by crowding agents (49), salt (50) and pH (51). In the nucleus of a human cell, there are abundant macromolecules which exhibit molecular crowding effect on DNA. In addition, variations of nuclear salt concentration and pH are not uncommon during different phases of the cell cycle (52), (53). Therefore, the effects of crowding agents, salt and pH on the unusual structure formation propensities of CCTG repeats need to be studied in order to obtain more reliable structural information in a condition mimicking the cell nucleus. In addition to in vitro experiments, a further step is to reveal the presence of unusual CCTG structures in vivo.
Second, as interconversion between different unusual CCTG structures has been proposed to bring about different levels of strand slippage and repair escape, it will be interesting to know how exactly different types of unusual CCTG structures participate in repeat expansions. Through using different lengths of CCTG repeats and/or single site substitution sample, a specific type of unusual structures can be built on the nascent strand of a DNA primer-template model. Therefore, the impact of each type of unusual structures can be investigated via in vitro replication extension assays.
Third, in order to understand the underlying reasons for the efficient repair escape of unusual CCTG structures, the specificity and repair efficiency of different types of MMR heterodimer proteins including hMSH2-hMSH3 and hMSH2-hMSH6 on different types of unusual CCTG structures need to be investigated. As we have proposed that more efficient repair escape can be achieved through conversions to smaller or less stable unusual structures or a shifting of mini-loop, it will be worthwhile to investigate whether the presence of interrupting motifs in the CCTG repeat tract in normal individuals can really reduce the formation of larger or more stable unusual structures and/or hinder the shifting of mini-loop.
Acknowledgments
We would like to thank Professor H.N.C. Wong for his continuing support on the research activities of our group. The work described in this paper was supported by General Research Fund (Project No. CUHK14302915) from the Research Grants Council of the Hong Kong Special Administrative Region and a direct grant (Project ID: 3132672) from the Faculty of Science of The Chinese University of Hong Kong.
- List of abbreviations
- CNBP
nucleic acid-binding protein
- DM1
myotonic dystrophy type 1
- DM2
myotonic dystrophy type 2
- DMPK
dystrophia myotonica protein kinase
- MBNL
muscleblind-like
- MDB
mini-dumbbell
- MMR
mismatch repair
- NMR
nuclear magnetic resonance
- ZNF9
zinc finger protein 9.
References
1. Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science 2001; 293: 864–7.10.1126/science.1062125Search in Google Scholar
2. Udd B, Krahe R. The myotonic dystrophies: molecular, clinical, and therapeutic challenges. Lancet Neurol 2012; 11: 891–905.10.1016/S1474-4422(12)70204-1Search in Google Scholar
3. Cho DH, Tapscott SJ. Myotonic dystrophy: emerging mechanisms for DM1 and DM2. Biochim Biophys Acta 2007; 1772: 195–204.10.1016/j.bbadis.2006.05.013Search in Google Scholar PubMed
4. Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, Neville C, Narang M, Barcelo J, O’Hoy K, Leblond S, Earle-Macdonald J, de Jong PJ, Wieringa B, Korneluk RG. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science 1992; 255: 1253–5.10.1126/science.1546325Search in Google Scholar PubMed
5. Udd B, Meola G, Krahe R, Thornton C, Ranum LP, Bassez G, Kress W, Schoser B, Moxley R. 140th ENMC International Workshop: Myotonic Dystrophy DM2/PROMM and other myotonic dystrophies with guidelines on management. Neuromuscul. Disord. 2006; 16: 403–13.10.1016/j.nmd.2006.03.010Search in Google Scholar PubMed
6. Udd B, Meola G, Krahe R, Wansink DG, Bassez G, Kress W, Schoser B, Moxley R. Myotonic dystrophy type 2 (DM2) and related disorders report of the 180th ENMC workshop including guidelines on diagnostics and management 3–5 December 2010, Naarden, The Netherlands. Neuromuscul Disord 2011; 21: 443–50.10.1016/j.nmd.2011.03.013Search in Google Scholar PubMed
7. Vihola A, Bassez G, Meola G, Zhang S, Haapasalo H, Paetau A, Mancinelli E, Rouche A, Hogrel JY, Laforet P, Maisonobe T, Pellissier JF, Krahe R, Eymard B, Udd B. Histopathological differences of myotonic dystrophy type 1 (DM1) and PROMM/DM2. Neurology 2003; 60: 1854–7.10.1212/01.WNL.0000065898.61358.09Search in Google Scholar PubMed
8. Salisbury E, Schoser B, Schneider-Gold C, Wang GL, Huichalaf C, Jin B, Sirito M, Sarkar P, Krahe R, Timchenko NA, Timchenko LT. Expression of RNA CCUG repeats dysregulates translation and degradation of proteins in myotonic dystrophy 2 patients. Am J Pathol 2009; 175: 748–62.10.2353/ajpath.2009.090047Search in Google Scholar PubMed PubMed Central
9. Bachinski LL, Czernuszewicz T, Ramagli LS, Suominen T, Shriver MD, Udd B, Siciliano MJ, Krahe R. Premutation allele pool in myotonic dystrophy type 2. Neurology 2009; 72: 490–7.10.1212/01.wnl.0000333665.01888.33Search in Google Scholar PubMed PubMed Central
10. Bachinski LL, Udd B, Meola G, Sansone V, Bassez G, Eymard B, Thornton CA, Moxley RT, Harper PS, Rogers MT, Jurkat-Rott K, Lehmann-Horn F, Wieser T, Gamez J, Navarro C, Bottani A, Kohler A, Shriver MD, Sallinen R, Wessman M, Zhang S, Wright FA, Krahe R. Confirmation of the type 2 myotonic dystrophy (CCTG)n expansion mutation in patients with proximal myotonic myopathy/proximal myotonic dystrophy of different European origins: a single shared haplotype indicates an ancestral founder effect. Am J Hum Genet 2003; 73: 835–48.10.1086/378566Search in Google Scholar PubMed PubMed Central
11. Meola G, Cardani R. Myotonic dystrophies: An update on clinical aspects, genetic, pathology, and molecular pathomechanisms. Biochim Biophys Acta 2015; 1852: 594–606.10.1016/j.bbadis.2014.05.019Search in Google Scholar PubMed
12. Kurosaki T, Ueda S, Ishida T, Abe K, Ohno K, Matsuura T. The unstable CCTG repeat responsible for myotonic dystrophy type 2 originates from an AluSx element insertion into an early primate genome. PLoS One 2012; 7: e38379.10.1371/journal.pone.0038379Search in Google Scholar PubMed PubMed Central
13. Holt I, Jacquemin V, Fardaei M, Sewry CA, Butler-Browne GS, Furling D, Brook JD, Morris GE. Muscleblind-like proteins: similarities and differences in normal and myotonic dystrophy muscle. Am J Pathol 2009; 174: 216–27.10.1016/j.nmd.2008.06.258Search in Google Scholar
14. Timchenko L. Molecular mechanisms of muscle atrophy in myotonic dystrophies. Int J Biochem Cell Biol 2013; 45: 2280–2287.10.1016/j.biocel.2013.06.010Search in Google Scholar PubMed PubMed Central
15. Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science 2009; 325: 336–9.10.1126/science.1173110Search in Google Scholar PubMed PubMed Central
16. Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature 2012; 488: 111–5.10.1038/nature11362Search in Google Scholar PubMed PubMed Central
17. Panigrahi GB, Slean MM, Simard JP, Gileadi O, Pearson CE. Isolated short CTG/CAG DNA slip-outs are repaired efficiently by hMutSbeta, but clustered slip-outs are poorly repaired. Proc Natl Acad Sci USA 2010; 107: 12593–8.10.1073/pnas.0909087107Search in Google Scholar PubMed PubMed Central
18. Panigrahi GB, Lau R, Montgomery SE, Leonard MR, Pearson CE. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat Struct Mol Biol 2005; 12: 654–62.10.1038/nsmb959Search in Google Scholar PubMed
19. Chi LM, Lam SL. Structural roles of CTG repeats in slippage expansion during DNA replication. Nuc Acids Res 2005; 33: 1604–17.10.1093/nar/gki307Search in Google Scholar PubMed PubMed Central
20. van den Broek WJ, Nelen MR, Wansink DG, Coerwinkel MM, te Riele H, Groenen PJ, Wieringa B. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet 2002; 11: 191–8.10.1093/hmg/11.2.191Search in Google Scholar PubMed
21. Panigrahi GB, Cleary JD, Pearson CE. In vitro (CTG)*(CAG) expansions and deletions by human cell extracts. J Biol Chem 2002; 277: 13926–34.10.1074/jbc.M109761200Search in Google Scholar
22. Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet 2001; 27: 407–11.10.1038/86906Search in Google Scholar
23. Gacy AM, Goellner GM, Spiro C, Chen X, Gupta G, Bradbury EM, Dyer RB, Mikesell MJ, Yao JZ, Johnson AJ, Richter A, Melançon SB, McMurray CT. GAA instability in Friedreich’s Ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol Cell 1998; 1: 583–93.10.1016/S1097-2765(00)80058-1Search in Google Scholar
24. Guo P, Lam SL. Unusual structures of TTTA repeats in icaC gene of Staphylococcus aureus. FEBS Lett. 2015; 589: 1296–300.10.1016/j.febslet.2015.04.031Search in Google Scholar PubMed
25. Guo P, Lam SL. New insights into the genetic instability in CCTG repeats. FEBS Lett 2015; 589: 3058–63.10.1016/j.febslet.2015.09.007Search in Google Scholar PubMed
26. Radvanszky J, Surovy M, Polak E, Kadasi L. Uninterrupted CCTG tracts in the myotonic dystrophy type 2 associated locus. Neuromuscul. Disord 2013; 23: 591–8.10.1016/j.nmd.2013.02.013Search in Google Scholar
27. Lam SL, Wu F, Yang H, Chi LM The origin of genetic instability in CCTG repeats. Nuc. Acids Res 2011; 39: 6260–8.10.1093/nar/gkr185Search in Google Scholar PubMed PubMed Central
28. Dere R, Wells RD. DM2 CCTG*CAGG repeats are crossover hotspots that are more prone to expansions than the DM1 CTG*CAG repeats in Escherichia coli. J Mol Biol 2006; 360: 21–36.10.1016/j.jmb.2006.05.012Search in Google Scholar PubMed
29. Dere R, Napierala M, Ranum LP, Wells RD. Hairpin structure-forming propensity of the (CCTG·CAGG) tetranucleotide repeats contributes to the genetic instability associated with myotonic dystrophy type 2. J Biol Chem 2004; 279: 41715–26.10.1074/jbc.M406415200Search in Google Scholar PubMed
30. Strand M, Prolla TA, Liskay RM, Petes TD. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 1993; 365: 274–6.10.1038/365274a0Search in Google Scholar PubMed
31. Richard GF, Paques F. Mini- and microsatellite expansions: the recombination connection. EMBO Rep 2000; 1: 122–6.10.1093/embo-reports/kvd031Search in Google Scholar PubMed PubMed Central
32. Mirkin SM. Expandable DNA repeats and human disease. Nature 2007; 447: 932–40.10.1038/nature05977Search in Google Scholar PubMed
33. Savouret C, Brisson E, Essers J, Kanaar R, Pastink A, te Riele H, Junien C, Gourdon G. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J 2003; 22: 2264–2273.10.1093/emboj/cdg202Search in Google Scholar PubMed PubMed Central
34. Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet 1999; 23: 471–3.10.1038/70598Search in Google Scholar PubMed
35. Entezam A, Lokanga AR, Le W, Hoffman G, Usdin K. Potassium bromate, a potent DNA oxidizing agent, exacerbates germline repeat expansion in a fragile X premutation mouse model. Hum Mutat 2010; 31: 611–6.10.1002/humu.21237Search in Google Scholar PubMed PubMed Central
36. Goula AV, Berquist BR, Wilson DM, 3rd, Wheeler VC, Trottier Y, Merienne K. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington’s disease transgenic mice. PLoS Genet 2009; 5: e1000749.10.1371/journal.pgen.1000749Search in Google Scholar PubMed PubMed Central
37. Levens D, Benham CJ. DNA stress and strain, in silico, in vitro and in vivo. Phys Biol 2011; 8: 035011.10.1088/1478-3975/8/3/035011Search in Google Scholar PubMed
38. Zhabinskaya D, Benham CJ. Competitive superhelical transitions involving cruciform extrusion. Nuc Acids Res 2013; 41: 9610–9621.10.1093/nar/gkt733Search in Google Scholar PubMed PubMed Central
39. Du X, Gertz, EM, Wojtowicz D, Zhabinskaya D, Levens D, Benham CJ, Schaffer AA, Przytycka TM. Potential non-B DNA regions in the human genome are associated with higher rates of nucleotide mutation and expression variation. Nuc Acids Res 2014; 42: 12367–79.10.1093/nar/gku921Search in Google Scholar PubMed PubMed Central
40. Fry M, Loeb LA. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc Natl Acad Sci USA 1994; 91: 4950–4.10.1073/pnas.91.11.4950Search in Google Scholar PubMed PubMed Central
41. Fojtik P, Vorlickova M. The fragile X chromosome (GCC) repeat folds into a DNA tetraplex at neutral pH. Nuc Acids Res 2001; 29: 4684–90.10.1093/nar/29.22.4684Search in Google Scholar PubMed PubMed Central
42. Lam SL. Overcoming the challenges in solution structure studies of CTG and CCTG repeats. Chinese J Magn Reson 2012; 29; 1–20.Search in Google Scholar
43. Ippel HH, van den Elst H, van der Marel GA, van Boom JH, Altona C. Structural similarities and differences between H1- and H2-family DNA minihairpin loops: NMR studies of octameric minihairpins. Biopolymers 1998; 46: 375–93.10.1002/(SICI)1097-0282(199811)46:6<375::AID-BIP3>3.0.CO;2-6Search in Google Scholar
44. Guo P, Lam SL. Minidumbbell: a new form of native DNA structure. J Am Chem Soc 2016; 138: 12534–40.10.1021/jacs.6b06897Search in Google Scholar
45. Heidenfelder BL, Topal MD. Effects of sequence on repeat expansion during DNA replication. Nuc Acids Res 2003; 31: 7159–64.10.1093/nar/gkg920Search in Google Scholar
46. Sinden RR, Potaman VN, Oussatcheva EA, Pearson CE, Lyubchenko YL, Shlyakhtenko LS. Triplet repeat DNA structures and human genetic disease: dynamic mutations from dynamic DNA. J Biosci 2002; 27: 53–65.10.1007/BF02703683Search in Google Scholar
47. Pearson CE, Sinden RR. Trinucleotide repeat DNA structures: dynamic mutations from dynamic DNA. Curr Opin Struct Biol 1998; 8: 321–30.10.1016/S0959-440X(98)80065-1Search in Google Scholar
48. Fry M, Usdin K. Human nucleotide expansion disorders. Berlin, Heidelberg, Germany: Springer-Verlag, 2006.10.1007/3-540-33336-3Search in Google Scholar
49. Nakano S, Miyoshi D, Sugimoto N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem Rev 2014; 114: 2733–58.10.1021/cr400113mSearch in Google Scholar
50. Kim BG, Evans HM, Dubins DN, Chalikian TV. Effects of salt on the stability of a G-quadruplex from the human c-MYC promoter. Biochemistry 2015; 54: 3420–30.10.1021/acs.biochem.5b00097Search in Google Scholar
51. Jin KS, Shin SR, Ahn B, Rho Y, Kim SJ, Ree M. pH-dependent structures of an i-motif DNA in solution. J Phys Chem B 2009; 113: 1852–56.10.1021/jp808186zSearch in Google Scholar
52. Ismail A, Takeda S, Nick P. Life and death under salt stress: same players, different timing? J Exp Bot 2014; 65: 2963–79.10.1093/jxb/eru159Search in Google Scholar
53. Seksek O, Bolard J. Nuclear pH gradient in mammalian cells revealed by laser microspectrofluorimetry. J Cell Sci 1996; 109: 257–62.10.1242/jcs.109.1.257Search in Google Scholar PubMed
©2016 Walter de Gruyter GmbH, Berlin/Boston
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Articles in the same Issue
- Frontmatter
- Reviews
- Coupling de novo protein folding with subunit exchange into pre-formed oligomeric protein complexes: the ‘heritable template’ hypothesis
- Role of non-motile microtubule-associated proteins in virus trafficking
- Advanced glycation end products mediated cellular and molecular events in the pathology of diabetic nephropathy
- Short Conceptual Overviews
- The microRNA-200 family: still much to discover
- Matrix metalloproteinases: an emerging role in regulation of actin microfilament system
- Unusual structures of CCTG repeats and their participation in repeat expansion
Articles in the same Issue
- Frontmatter
- Reviews
- Coupling de novo protein folding with subunit exchange into pre-formed oligomeric protein complexes: the ‘heritable template’ hypothesis
- Role of non-motile microtubule-associated proteins in virus trafficking
- Advanced glycation end products mediated cellular and molecular events in the pathology of diabetic nephropathy
- Short Conceptual Overviews
- The microRNA-200 family: still much to discover
- Matrix metalloproteinases: an emerging role in regulation of actin microfilament system
- Unusual structures of CCTG repeats and their participation in repeat expansion

