Clinical utility of liquid biopsy for the diagnosis and monitoring of EML4-ALK NSCLC patients
Abstract
Background
Genomic rearrangement in anaplastic lymphoma kinase (ALK) gene occurs in 3−7% of patients with non-small-cell lung cancer (NSCLC). The detection of this alteration is crucial as ALK positive NSCLC patients benefit from ALK inhibitors, which improve both the patient's quality of life and overall survival (OS) compared to traditional chemotherapy.
Content
In routine clinical practice, ALK rearrangements are detected using tissue biopsy. Nevertheless, the availability of tumor tissue is compromised in NSCLC patients due to surgical complications or difficult access to the cancer lesion. In addition, DNA quality and heterogeneity may impair tumor biopsies testing. These limitations can be overcome by liquid biopsy, which refers to non-invasive approaches for tumor molecular profiling. In this paper we review currently available technology for non-invasive ALK testing, in NSCLC patients, based on the analysis of circulating tumor DNA (ctDNA), circulating tumor RNA (ctRNA), circulating tumor cells (CTCs), tumor-educated platelets (TEPs) and extracellular vesicles (EVs) such as exosomes.
Summary and outlook
Non-invasive tumor molecular profiling is crucial to improve outcomes and quality of life of NSCLC patients whose tumors harbor a translocation involving ALK locus.
Introduction
Lung cancer is the most commonly diagnosed cancer along with breast cancer, contributing to more than 8.8 million deaths every year [1]. Non-small-cell lung cancer (NSCLC), the most common subtype of lung cancer, is diagnosed in most cases in advanced stages, making not possible a curative treatment [2].
The anaplastic lymphoma kinase (ALK) gene, that was discovered in 1994 in anaplastic large cell lymphoma (ALCL), encodes a transmembrane receptor tyrosine kinase [3], whose alteration leads to constitutive ALK activation and generates oncogenic activity [4], [5]. Nearly 30 different ALK-fusion protein partners have been described and the most prevalent fusion partner in NSCLC patients is echinoderm microtubule-associated protein-like 4 (EML4-ALK) [6], [7] (Figure 1), leading to multiple fusions variants of which variant 1 (E13;A20, 33%), variant 2 (E20;A20, 10%), and variants 3 a/b (E6;A20, 29%) are the most frequent fusions [8].
![Figure 1:
EML4-ALK translocation.
ALK (red) and EML4 (purple) genes are located in the short arm of chromosome 2 and they are oriented in opposite directions. Arrows indicate the orientation of the genes. The translocation occurs through a paracentric inversion [inv(2)(p21p23)] leading a fusion transcript that contains ALK catalytic domain and EML4 amino-terminal half. (Figure designed by https://app.biorender.com/).](/document/doi/10.1515/almed-2019-0019/asset/graphic/j_almed-2019-0019_fig_001.jpg)
EML4-ALK translocation.
ALK (red) and EML4 (purple) genes are located in the short arm of chromosome 2 and they are oriented in opposite directions. Arrows indicate the orientation of the genes. The translocation occurs through a paracentric inversion [inv(2)(p21p23)] leading a fusion transcript that contains ALK catalytic domain and EML4 amino-terminal half. (Figure designed by https://app.biorender.com/).
EML4-ALK rearrangement occurs in 3−7% of NSCLC patients, which defines a specific molecular subtype of NSCLC [9], [10]. Interestingly, ALK rearrangement is observed predominantly in younger patients and never or light smokers with adenocarcinoma.
ALK+ NSCLC treatment
The development of tyrosine kinase inhibitors (TKIs) against ALK-gene has dramatically improved the patient's outcomes in terms of quality of life and prognosis, with significant higher progression-free survival (PFS), overall survival (OS) and objective response rates (ORRs) compared with chemotherapy [11], [12]. Thus, the identification of patients whose tumors harbor an ALK translocation is essential. Crizotinib, an ATP analog inhibitor of ALK that received approval from the US FDA (Food and Drug Administration) in 2011, is recommended as the first-line standard therapy for EML4-ALK advanced or metastatic NSCLC by the National Comprehensive Cancer Network (NCCN) guideline with an ORR of 74% and a PFS of 10.9 months [11]. However, despite its initial efficacy, resistance develops in approximately 73% of patients within 1–2 years of treatment [13]. Nevertheless, several second-generation ALK inhibitors have become available, such as ceritinib, alectinib and brigatinib, which received US FDA approval in 2014, 2015 and 2017, respectively. These therapies presented higher ORRs and PFS compared to platinum-pemetrexed based chemotherapy or crizotinib (ORRs of 73%, 83%, and 71%, and PFS of 16.6, 34.8 and not reported* months respectively [*median follow-up 11 months with brigatinib] [14], [15], [16]. Moreover, third-generation ALK inhibitors such as lorlatinib (a selective brain-penetrant ALK inhibitor) received FDA approval in 2018 [17]. Although next-generation ALK inhibitors are now available being more potent and effective than crizotinib against central nervous system metastases, little is known about the potential mechanisms of resistance to these agents. Several mechanisms of ALK-TKI resistance have been described (Figure 2), including ALK dependent mechanisms such as resistance mutations in the tyrosine kinase domain of ALK (G1202R, G1269A, F1174L or L1196M are the most frequent) and amplification of the ALK fusion gene, and ALK-independent mechanisms, as for example, dysregulation of bypass signalling pathways (EGFR, c-KIT, RAS-MAPK, PI3K-Akt activation, etc.) and epithelial-mesenchymal transition (EMT) [18].
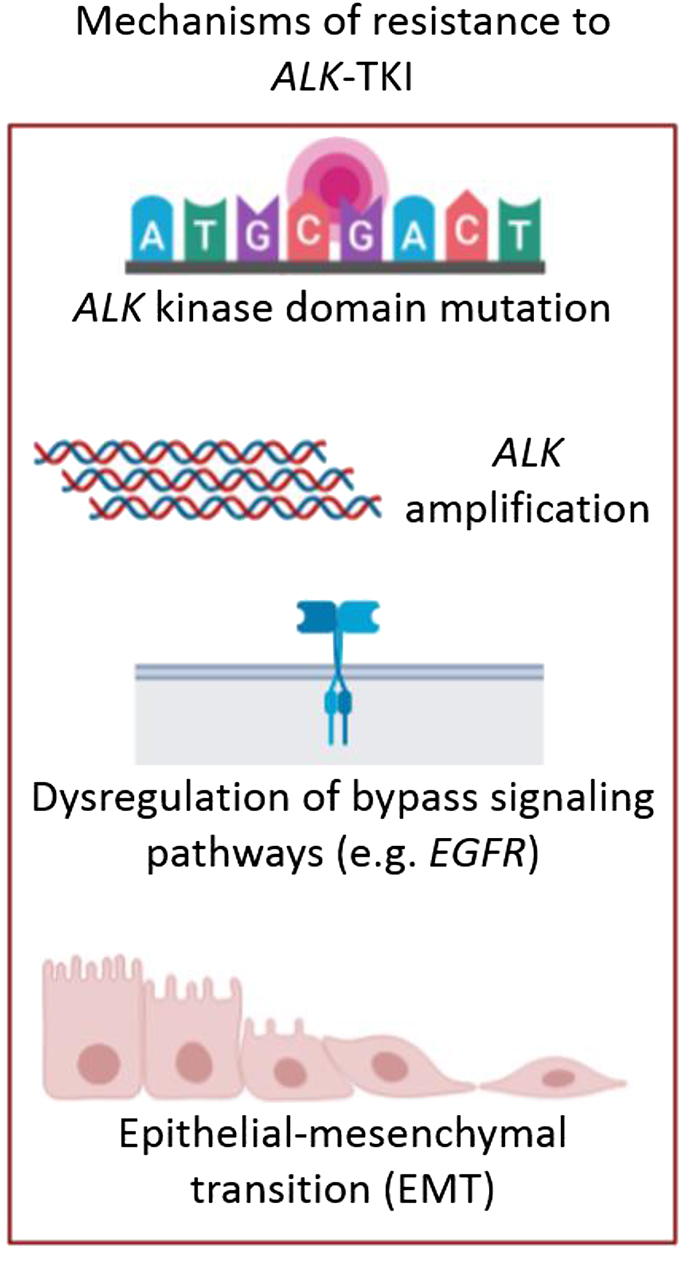
Treatment of ALK+ NSCLC patients.
Mechanisms of resistance to ALK-TKIs: ALK kinase domain mutations (such as G1202R, G1269A, F1174L or L1196M), ALK amplification, dysregulation of bypass signalling pathways (such as EGFR, c-KIT, RAS-MAPK or PI3K-Akt activation) and epithelial-mesenchymal transition (EMT). (Figure designed by https://app.biorender.com/).
However, the re-biopsy of the tumor is seldom performed in routine clinical practice in these patients, due to lung tumor difficult access. Thus, in most cases, ALK-TKIs resistance mechanisms are unknown, leading to an empirical prescription of subsequent lines of treatment, without knowing the tumor molecular profile upon disease progression. Noteworthy, it's known that ALK inhibitors have different binding affinities in the context of different resistance mutations; for example, early pre-clinical evidence suggests that the L1196M or S1206Y mutations confer resistance to crizotinib but not to ceritinib [19]. Thus, an optimal ALK-TKI sequence based on ALK mutations upon disease progression is necessary to allow clinicians to personalize ALK-targeted therapies, which will definitively improve the patient's outcome.
Clinical practice in EML4-ALK patients
Different methods can be used to detect ALK rearrangements in tumor tissues. In routine clinical practice, the determination of EML4-ALK translocation is made by fluorescent in situ hybridization (FISH), immunohistochemistry (IHC) or next-generation sequence (NGS) [20]. Furthermore, it is important to highlight that when EML4-ALK translocation is identified by IHC and/or FISH, it is not possible to identify which variant is present. Although, all variants are oncogenic and induce ALK dependency [21], some studies have demonstrated that “short onco-proteins” resulting from EML4-ALK fusion variants such as variant 3 and variant 5 confer poorer outcome [22], [23]. Conversely, “long onco-protein” products of EML4-ALK, such as variant 2, are associated with better outcome [24]. Moreover, different EML4-ALK variants could have distinct sensitivities against the wide variety of ALK-TKI that are nowadays available. In this way, as in the case of EGFR mutations, in which tumors with exon 19 deletions are known to be more sensitive to TKIs than tumors harboring other alterations such as exon insertions in exon 20 [25].
In addition, methods such as immunocytochemistry (ICC), reverse transcriptase-polymerase chain reaction (RT-PCR), NGS and commercial kits are frequently employed. Finally, nCounter is a new technology capable of highly multiplexed analysis of different molecules such as RNA, miRNA, proteins and DNA. This methodology can detect ALK fusions using different starting material such as FFPE, fresh frozen tissues [16] or lung cancer patient-derived xenograft (PDX)-derived tumors [26].
Nevertheless, diagnosis based on tumor biopsies have some limitations such as the availability of samples for tumor molecular profiling, especially in lung cancer patients upon disease progression. Therefore, the lack of tissue biopsy condemns many patients with rearrangement in ALK to receive chemotherapy instead of ALK-TKIs, with a median overall survival (OS) of ∼12 months instead of ∼50 months from time of diagnosis of metastatic disease as reported by several observational studies using sequential ALK inhibitors [27], [28], [29], [30]. Moreover, the diagnosis based on the molecular profiling of a single biopsy may not reflect the overall situation of the entire tumor due to its heterogeneity [31]. Therefore, the development of methodologies that allow the non-invasive identification of EML4-ALK translocation, its variants and ALK-TKIs resistance mechanisms remains an unmet clinical need.
Liquid biopsy
The term liquid biopsy refers to different methodological approaches including the study of circulating tumor DNA (ctDNA), circulating tumor RNA (ctRNA), circulating tumor cells (CTCs), tumor-educated platelets (TEPs) and extracellular vesicles (EVs; exosomes, microvesicles, microparticles, oncosomes) (Figure 3) [32].
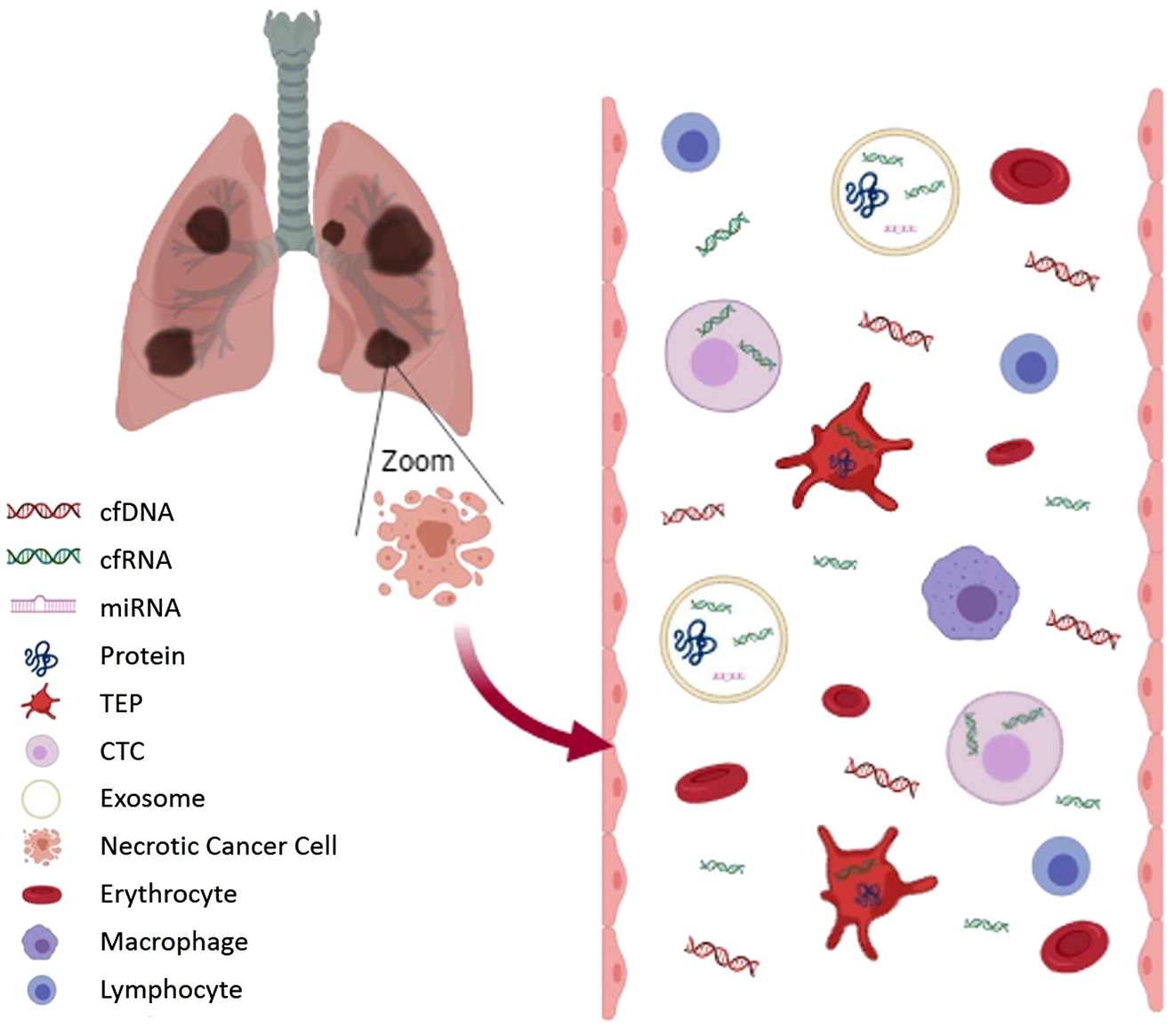
Components of liquid biopsy sample.
cfDNA and cfRNA in blood come from dying cells (necrosis or apoptosis). A small fraction of cfDNA is ctDNA. Wild type DNA from blood cells can dilute ctDNA and therefore pre-analytical conditions should avoid cell lysis. CTCs, platelets (TEPs) and exosomes can capture these molecules and others, such as miRNA or cancer cell proteins. This genetic information is better protected in these compartments than in the bloodstream. (Figure designed by https://app.biorender.com/).
Liquid biopsy, as a minimally invasive, safe and sensitive method, can overcome the aforementioned tissue biopsy limitations.
Tumor cells shed circulating tumor DNA (ctDNA) into the bloodstream, which can be subjected to molecular analysis. However, healthy cells also shed nucleic acids into bloodstream, as is the case of the erythrocytes, macrophages or lymphocytes. Therefore, it is important to extreme caution with pre-analytical conditions in order to avoid contamination of ctDNA with DNA that do not actually come from tumor cells (3). Importantly, the study of ctDNA requires methodologies with a very low limit of detection, 0.1% or lower, such as digital polymerase chain reaction (dPCR) [33], BEAMing [34], or NGS.
cfDNA profiling has proved to be useful to guide personalized treatments based on the molecular profile, for early detection of resistance mechanisms [35], [36], early detection of disease recurrences, as well as tumor response to therapy monitoring [37] by means of sequential liquid biopsies. Furthermore, several studies comparing the detection of EML4-ALK translocation by formalin-fixed paraffin-embedded (FFPE) tissues and liquid biopsy have reported that indeed cfDNA can be used as a surrogate of FFPE DNA [38].
Nevertheless, ALK fusion testing using liquid biopsies is rather challenging. First, genomic rearrangement identification using cfDNA is difficult as genomic brake-points are usually unknown and the rearrangement usually involves thousands of bp. Fusion transcripts although being more easy to detect requires to employ cfRNA which, unlike cfDNA, degrades very quickly due to the presence of RNases in the bloodstream. Therefore, it is plausible to hypothesize that a good strategy fusion testing would be the analysis of RNA that is contained in vesicles such as exosomes or platelets where it is protected from RNases.
EML4-ALK detection strategies by liquid biopsy
Circulating free DNA (cfDNA)
According to recent recommendations, liquid biopsy NGS panels using cfDNA is an adequate approach for the detection of EML4-ALK NSCLC patients resistance mutations when re-biopsy of the progression site is not feasible [19]. NGS seems to be the optimal method as it reaches levels of sensitivity and specificity around 80% of 100% respectively [39] and can provide information not only on resistance mutations of ALK gene but also on other molecular resistance mechanisms which may be target of a treatment either through a clinical trial or expanded access. However, the wide application of liquid NGS is currently limited owing to the need for specialized equipment and the high costs.
Conversely, alterations such as EML4-ALK translocation, can hardly be detected in the cfDNA since the brake-points at the DNA level is frequently unknown. Also, these alterations involve a large number of base pairs and cfDNA is fragmented (typically has a peak of approximately 150 bp). Thus, in contrast to EGFR mutations, ALK rearrangement detection from cfDNA is scarcely implemented in daily oncology.
Despite all of the above, EML4-ALK gene fusions have been detected by analyzing cfDNA, using a NGS liquid biopsy assay designed to detect the genomic breakpoint junctions. The assay was validated using custom cell lines with 10 EML4-ALK gene fusions and 26 synthetic fusions designed by computationally joining of the InVisionFirst Assay [40]. In addition, ALK fusion status has been analyzed in plasma cfDNA from NSCLC patients by using capture based NGS with a specificity of 100% [41], indicating that rearrangement detection through cfDNA sequencing is possible. However, due to the aforementioned limitations a negative result should be interpreted with caution.
Circulating free RNA (cfRNA)
Complex aberrations such as large genomic rearrangements, including translocations, can be easily detected at the RNA level as fusion transcripts. Nevertheless, unlike cfDNA, free circulating RNA (cfRNA) degrades very quickly which constitutes an important limitation. Nonetheless, it has been reported that the optimization of the pre-analytical conditions of liquid biopsy samples can improve the sensitivity of RT-PCR based on cfRNA for the detection of EML4-ALK fusion transcripts [42].
Part of this cfRNA that circulates in the bloodstream is captured by diverse compartments, where it is more protected and functionally active [43]. Currently, the most frequently studied compartments include circulating tumor cells (CTCs), tumor-educated platelets (TEPs) and extracellular vesicles (EVs; exosomes, microvesicles, microparticles, oncosomes).
Circulating tumor cells (CTCs)
Circulating tumor cells (CTCs) are viable circulating cells in the bloodstream of cancer patients that have been shed by a primary tumor. CTCs take part in metastasis as they are able to adhere to the wall of capillaries and penetrate a new tissue [44]. CTCs presence and counting has been reported to be associated with worse prognosis [45], [46]. Nevertheless, since CTCs are a minor fraction of cell population in the peripheral blood of cancer patients, both in terms of absolute (<10 cell/mL) and relative numbers compared to other blood cells (1 CTC per 106–107 leukocytes) [47], their use for the detection of EML4-ALK rearrangements must rely on highly efficient detection strategies. In addition, due to the heterogeneity of tumors, CTCs could present genomic variability, which implies a clonal heterogeneity of CTCs that must be taken into account [48]. Moreover, CTCs may present differences respect to primary and metastatic tumors [49].
Besides these limitations, EML4-ALK rearrangement has been detected in CTCs from peripheral blood of NSCLC cancer patients [45], [46], [47]. A variety of methods have been developed to isolate and identify CTCs; however, currently the only FDA-approved method for enumerating CTCs in blood samples is the CELLSEARCH® system [44]. Other methods used to detect ALK rearrangements rely on an initial enrichment process followed by CTCs detection, or vice versa, to increase the sensitivity and specificity [45].
Size of Epithelial Tumor Cells (ISET) technology is another method to isolate CTCs which has demonstrated greater effectiveness with respect to CELLSEARCH® system [50]. Using ISET followed by FISH and IHC, CTCs can be a reliable source for the detection of ALK-gene rearrangements in lung cancer patients, with ≥90% concordance with the tissue biopsy [51]. In addition, resistance mutations in ALK gene, such as L1196M, have been detected in CTCs [52] and can be expanded ex vivo for drug testing. Therefore, this source has clinical utility not only for diagnosis, but also a potential tool for drug sensitivity testing and for personalized precision medicine.
Tumor-educated platelets (TEPs)
Platelets are cell fragments without nucleus originating from megakaryocytes in the bone marrow. These blood elements can sequester tumor related RNA by a microvesicle dependent mechanism, resulting in a change of its RNA and protein content [53]. As a result, these tumor-educated platelets (TEPs) have an altered function and can promote tumor cell survival and metastasis [53]. Furthermore, new evidence suggest that platelets are implicated in the immune responses and inflammatory diseases of the lungs [54]. Besides platelet content, count and size of platelets and platelet protein markers, such as Pselectin, are used for cancer diagnostics and prognostics [55]. However, platelets study has limitations such as a reducing count or potential activation due to certain therapies, which may affect the interpretation of the results.
EML4-ALK rearrangement has been identified by the analysis of RNA platelets of NSCLC patients before starting treatment with an ALK-TKI and upon disease progression, reappearing even before positron emission tomography- computed tomography (PET-CT) will show disease progression [56]. On the other hand, when patients respond to the treatment, EML4-ALK translocation was not detected in platelets. The sensitivity and specificity for the detection of EML4-ALK rearrangements in RNA isolated from platelets in patients has been reported to be between 65% and 100% respectively [56]. Even with the optimization of the pre-analytical conditions of liquid biopsy samples, the sensitivity of the detection of EML4-ALK rearrangements using cfRNA was lower compared to platelet RNA (21% and 65% respectively) [36].
Platelets can also capture extracellular vesicles (EVs), released by cancer cells, harboring tumor-specific RNA [53] such as tumor-derived EML4-ALK rearranged RNA. EVs are another starting material for nucleic acid isolation which are used for the study of EML4-ALK translocation by liquid biopsy samples.
Exosomes
Exosomes are a type of EVs and refers to nanovesicles (30–200 nm) released after fusion of multivesicular bodies with the plasma membrane at the end of the endocytic recycling path. Exosomes with significant changes in their composition, are released by cancer cells and are able to act as a vehicle for the exchange of genetic and protein material between cells, which causes modifications such as angiogenesis, the acquisition of therapeutic resistance, the formation of metastases and an increase in proliferation [57].
Many potential non-invasive biomarkers that have been studied by liquid biopsy are commonly located in exosomes [58], [59]. Exosomal biomarkers could achieve a higher diagnostic and prognostic efficiency than using cfDNA alone [60].
Exosomes have not only been isolated from blood, but also from other biological fluids such as urine. However, the EVs concentration in urine is lower than in blood [48]. Many methods of exosomes isolation have been reported, being ultracentrifugation and some commercial kits the most commonly used. However, the efficiency and purity of the exosomes obtained by both strategies are different; ultracentrifugation has less efficiency but provides high purity, while commercial kits have lower purity due to improved efficiency [61], [62]. On the other hand, ultracentrifugation is a tedious method that takes a long time, so the improvement of new technologies such as commercially available kits or NGS-based methods by capture instead of by amplification could increase the sensitivity of ALK-fusion detection [63], [64]. Moreover, these new technologies may be easier and faster, so they could be implemented in routine clinical practice.
Few studies have analyzed fusion genes in exosomes. However, EML4-ALK translocation has been detected by the analysis of exosomal RNA, isolated from plasma samples from NSCLC patients, with a specificity of 100% and a sensitivity of 64–70% regarding tissue analysis [63], [65].
Future approaches
Several studies have shown an excellent specificity for EML4-ALK detection by the different starting materials for nucleic acid isolation available in liquid biopsy considering tissue biopsy as the gold standard. Sensitivity, however, is rather low and negative results should be regarded with caution [41], [45], [56], [63]. Therefore, the challenge now is to increase the sensitivity of liquid biopsy for the detection of ALK rearrangements, improving both the isolation and detection technologies in order to establish robust and reproducible protocols. PCR based methods, dPCR, BeAMing, nCounter and NGS methodologies reach higher levels of sensitivity in the detection of ALK rearrangements compared with other strategies, although there are still some limitations.
Finally, evaluating EML4-ALK detection by different methods using liquid biopsy approaches in prospective cohorts is needed in order to establish diagnostic accuracy of different methodologies.
Conclusions
Liquid biopsy is a non-invasive method, which can improve EML4-ALK translocation and ALK-TKIs resistance mechanisms detection, which will significantly improve ALK+ NCSCL diagnosis and patients management, leading to a better prognosis and quality of life of ALK+ NSCLC patients.
Funding source: Carlos III Institute of Health
Funding source: Spanish Ministry of Science and Innovation
Funding source: European Regional Development Fund
Award Identifier / Grant number: PIE17/01977
Funding statement: This study was supported by the Carlos III Institute of Health, the Spanish Ministry of Science and Innovation and the European Regional Development Fund (grant number: PIE17/01977). ES was financed by the Consejería de Educación, Juventud y Deporte of Comunidad de Madrid and by the Fondo Social Europeo (Programa Operativo de Empleo Juvenil, and Iniciativa de Empleo Juvenil, PEJ-2017-AI/SAL-6478).
-
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Competing interests: The funding organization(s) played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the report for publication.
-
Article Note: La versión traducida del artículo puede encontrarse aquí: https://doi.org/10.1515/almed-2020-0007
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics in USA. CA Cancer J Clin 2018;68:7-30.10.3322/caac.21442Search in Google Scholar PubMed
2. Hanna N, Neubauer M, Yiannoutsos C, McGarry R, Arseneau J, Ansari R, et al. Phase III study of cisplatin, etoposide, and concurrent chest radiation with or without consolidation docetaxel in patients with inoperable stage III non-small-cell lung cancer: the hoosier oncology group and U.S. Oncology. J Clin Oncol 2008;26:5755-60.10.1200/JCO.2008.17.7840Search in Google Scholar PubMed
3. Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247-53.10.1200/JCO.2009.22.6993Search in Google Scholar PubMed PubMed Central
4. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6.10.1038/nature05945Search in Google Scholar PubMed
5. Chia PL, Dobrovic A, Dobrovic A, John T. Prevalence and natural history of ALK positive non-small-cell lung cancer and the clinical impact of targeted therapy with ALK inhibitors. Clin Epidemiol Dove Medical Press Ltd; 2014;6:423-32.10.2147/CLEP.S69718Search in Google Scholar PubMed PubMed Central
6. Jemal A, Clegg LX, Ward E, Ries LAG, Wu X, Jamison PM, et al. Annual report to the nation on the status of cancer, 1975-2001, with a special feature regarding survival. Cancer 2004;101:3-27.10.1002/cncr.20288Search in Google Scholar PubMed
7. Takeuchi K, Choi YL, Soda M, Inamura K, Togashi Y, Hatano S, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Canc Res 2008;14:6618-24.10.1158/1078-0432.CCR-08-1018Search in Google Scholar PubMed
8. Woo CG, Seo S, Kim SW, Jang SJ, Park KS, Song JY, et al. Differential protein stability and clinical responses of EML4-ALK fusion variants to various ALK inhibitors in advanced ALK-rearranged non-small cell lung cancer. Ann Oncol 2017;28:791-7.10.1093/annonc/mdw693Search in Google Scholar PubMed
9. Sasaki T, Rodig SJ, Chirieac LR, Jänne PA. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Canc 2010;46:1773-80.10.1016/j.ejca.2010.04.002Search in Google Scholar PubMed PubMed Central
10. Yang L, Ling Y, Guo L, Ma D, Xue X, Wang B, et al. Detection of ALK translocation in non-small cell lung carcinoma (NSCLC) and its clinicopathological significance using the Ventana immunohistochemical staining method: a single-center large-scale investigation of 1,504 Chinese Han patient. Chin J Canc Res 2016;28:495-502.10.21147/j.issn.1000-9604.2016.05.04Search in Google Scholar PubMed PubMed Central
11. Solomon BJ, Mok T, Kim D-W, Wu Y-L, Nakagawa K, Mekhail T, et al. First-line crizotinib versus chemotherapy in ALK -positive lung cancer. N Engl J Med 2014;371:2167-77.10.1056/NEJMoa1408440Search in Google Scholar PubMed
12. Shaw AT, Kim TM, Crinò L, Gridelli C, Kiura K, Liu G, et al. Ceritinib versus chemotherapy in patients with ALK-rearranged non-small-cell lung cancer previously given chemotherapy and crizotinib (ASCEND-5): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol 2017;18:874-86.10.1016/S1470-2045(17)30339-XSearch in Google Scholar PubMed
13. Zhou J, Zheng J, Zhang X, Zhao J, Zhu Y, Shen Q, et al. Crizotinib in patients with anaplastic lymphoma kinase-positive advanced non-small cell lung cancer versus chemotherapy as a first-line treatment. BMC Canc 2018;18:10.10.1186/s12885-017-3720-8Search in Google Scholar PubMed PubMed Central
14. Soria J-C, Tan DSW, Chiari R, Wu Y-L, Paz-Ares L, Wolf J, et al. First-line Ceritinib versus platinum-based chemotherapy in advanced ALK -rearranged non-small-cell lung cancer (ASCEND-4): a randomised, open-label, phase 3 study. Lancet 2017;389:917-29.10.1016/S0140-6736(17)30123-XSearch in Google Scholar PubMed
15. Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim D-W, et al. Alectinib versus crizotinib in untreated ALK -positive non-small-cell lung cancer. N Engl J Med 2017;377:829-38.10.1056/NEJMoa1704795Search in Google Scholar PubMed
16. Camidge DR, Kim HR, Ahn M-J, Yang JC-H, Han J-Y, Lee J-S, et al. Brigatinib versus crizotinib in ALK -positive non-small-cell lung cancer. N Engl J Med 2018;379:2027-39.10.1056/NEJMoa1810171Search in Google Scholar PubMed
17. Rapoport B, Arani RB, Mathieson N, Krendyukov A. Meta-analysis comparing incidence of grade 3-4 neutropenia with ALK inhibitors and chemotherapy in patients with non-small-cell lung cancer. Future Oncol 2019;15:2163-74.10.2217/fon-2018-0863Search in Google Scholar PubMed
18. Friboulet L, Katayama R, Digumarthy S, Le LP, Yoda S, Gadgeel S, et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Canc Discov 2016;6:1118-33.10.1158/2159-8290.CD-16-0596Search in Google Scholar PubMed PubMed Central
19. Rolfo C, Mack PC, Scagliotti GV, Baas P, Barlesi F, Bivona TG, et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): a statement paper from the iaslc. J Thorac Oncol Elsevier Inc; 2018;13:1248-68.10.1016/j.jtho.2018.05.030Search in Google Scholar PubMed
20. Karachaliou N, Rosell R. Optimal detection of ALK rearranged lung adenocarcinomas. J Thorac Oncol 2013;8:255-6.10.1097/JTO.0b013e318282ddc3Search in Google Scholar PubMed
21. Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010;363:1734-9.10.1056/NEJMoa1007478Search in Google Scholar PubMed
22. O'Regan L, Barone G, Adib R, Woo CG, Jeong HJ, Richardson EL, et al. EML4-ALK V3 drives cell migration through NEK9 and NEK7 kinases in non-small-cell lung cancer. BioRxiv 2019;567305.10.1101/567305Search in Google Scholar
23. Heuckmann JM, Balke-Want H, Malchers F, Peifer M, Sos ML, Koker M, et al. Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Canc Res 2012;18:4682-90.10.1158/1078-0432.CCR-11-3260Search in Google Scholar PubMed
24. Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, WistubaII, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. J Am Med Assoc 2014;311:1998-2006.10.1001/jama.2014.3741Search in Google Scholar PubMed PubMed Central
25. Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol 2012;13:e23-31.10.1016/S1470-2045(11)70129-2Search in Google Scholar PubMed
26. Fang DD, Zhang B, Gu Q, Lira M, Xu Q, Sun H, et al. HIP1-ALK, A novel ALK fusion variant that responds to crizotinib. J Thorac Oncol 2014;9:285-94.10.1097/JTO.0000000000000087Search in Google Scholar PubMed
27. Kayaniyil S, Hurry M, Wilson J, Wheatley-Price P, Melosky B, Rothenstein J, et al. Treatment patterns and survival in patients with ALK-positive non-small-cell lung cancer: a Canadian retrospective study. Curr Oncol 2016;23:589.10.3747/co.23.3273Search in Google Scholar PubMed PubMed Central
28. Gainor JF, Tan DSW, De Pas T, Solomon BJ, Ahmad A, Lazzari C, et al. Progression-free and overall survival in ALK-positive NSCLC patients treated with sequential Crizotinib and Ceritinib. Clin Canc Res 2015;21:2745-52.10.1158/1078-0432.CCR-14-3009Search in Google Scholar PubMed PubMed Central
29. Watanabe S, Hayashi H, Okamoto K, Fujiwara K, Hasegawa Y, Kaneda H, et al. Progression-free and overall survival of patients with ALK rearrangement-positive non-small cell lung cancer treated sequentially with crizotinib and alectinib. Clin Lung Canc 2016;17:528-34.10.1016/j.cllc.2016.05.001Search in Google Scholar PubMed
30. Shaw AT, Yeap BY, Solomon BJ, Riely GJ, Iafrate AJ, Shapiro G, et al. Impact of crizotinib on survival in patients with advanced, ALK-positive NSCLC compared with historical controls. J Clin Oncol 2011;29:7507-7507.10.1200/jco.2011.29.15_suppl.7507Search in Google Scholar
31. García-Saenz JA, Ayllón P, Laig M, Acosta-Eyzaguirre D, García-Esquinas M, Montes M, et al. Tumor burden monitoring using cell-free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC Canc 2017;17:210.10.1186/s12885-017-3185-9Search in Google Scholar PubMed PubMed Central
32. Bardelli A, Pantel K. Liquid biopsies, what we do not know (yet). vol. 31, Cancer Cell. Cell Press; 2017. pp. 172-9.10.1016/j.ccell.2017.01.002Search in Google Scholar PubMed
33. Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA 1999;96:9236-41.10.1073/pnas.96.16.9236Search in Google Scholar PubMed PubMed Central
34. Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA 2003;100:8817-22.10.1073/pnas.1133470100Search in Google Scholar PubMed PubMed Central
35. Provencio M, Torrente M, Calvo V, Gutiérrez L, Pérez-Callejo D, Pérez-Barrios C, et al. Dynamic circulating tumor DNA quantificaton for the individualization of non-small-cell lung cancer patients treatment. Oncotarget 2017;8:60291-8.10.18632/oncotarget.20016Search in Google Scholar PubMed PubMed Central
36. Provencio M, Torrente M, Calvo V, Pérez-Callejo D, Gutiérrez L, Franco F, et al. Prognostic value of quantitative ctDNA levels in non small cell lung cancer patients. Oncotarget 2018;9:488-94.10.18632/oncotarget.22470Search in Google Scholar PubMed PubMed Central
37. Pérez-Barrios C, Nieto-Alcolado I, Torrente M, Jiménez-Sánchez C, Calvo V, Gutierrez-Sanz L, et al. Comparison of methods for circulating cell-free DNA isolation using blood from cancer patients: impact on biomarker testing. Transl Lung Cancer Res 2016;5:665-72.10.21037/tlcr.2016.12.03Search in Google Scholar PubMed PubMed Central
38. Park CK, Kim JE, Kim MS, Kho BG, Park HY, Kim TO, et al. Feasibility of liquid biopsy using plasma and platelets for detection of anaplastic lymphoma kinase rearrangements in non-small cell lung cancer. J Canc Res Clin Oncol 2019;145:2071-82.10.1007/s00432-019-02944-wSearch in Google Scholar PubMed PubMed Central
39. Wang Y, Tian P-W, Wang W-Y, Wang K, Zhang Z, Chen B-J, et al. Noninvasive genotyping and monitoring of anaplastic lymphoma kinase (ALK) rearranged non-small cell lung cancer by capture-based next-generation sequencing. Oncotarget 2016;7:65208-17.10.18632/oncotarget.11569Search in Google Scholar PubMed PubMed Central
40. Plagnol V, Woodhouse S, Howarth K, Lensing S, Smith M, Epstein M, et al. Analytical validation of a next generation sequencing liquid biopsy assay for high sensitivity broad molecular profiling. PLoS One 2018;13:e0193802.10.1371/journal.pone.0193802Search in Google Scholar PubMed PubMed Central
41. Cui S, Zhang W, Xiong L, Pan F, Niu Y, Chu T, et al. Use of capture-based next-generation sequencing to detect ALK fusion in plasma cell-free DNA of patients with non-small-cell lung cancer. Oncotarget 2017;8:2771-80.10.18632/oncotarget.13741Search in Google Scholar PubMed PubMed Central
42. Aguado C, Giménez-Capitán A, Karachaliou N, Pérez-Rosado A, Viteri S, Morales-Espinosa D, et al. Fusion gene and splice variant analyses in liquid biopsies of lung cancer patients. Transl Lung Cancer Res 2016;5:525-31.10.21037/tlcr.2016.09.02Search in Google Scholar PubMed PubMed Central
43. Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2011;2. https://doi.org/10.1038/ncomms1285.10.1038/ncomms1285Search in Google Scholar PubMed PubMed Central
44. Yap TA, Lorente D, Omlin A, Olmos D, De Bono JS. Circulating tumor cells: a multifunctional biomarker. Clin Canc Res 2014;20:2553-8.10.1158/1078-0432.CCR-13-2664Search in Google Scholar PubMed
45. Hofman V, Bonnetaud C, Ilie MI, Vielh P, Vignaud JM, Fléjou JF, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Canc Res 2011;17:827-35.10.1158/1078-0432.CCR-10-0445Search in Google Scholar PubMed
46. Provencio M, Pérez-Callejo D, Torrente M, Martin P, Calvo V, Gutiérrez L, et al. Concordance between circulating tumor cells and clinical status during follow-up in anaplastic lymphoma kinase (ALK) non-small-cell lung cancer patients. Oncotarget 2017;8:59408-16.10.18632/oncotarget.19722Search in Google Scholar PubMed PubMed Central
47. Ross AA, Cooper BW, Lazarus HM, Mackay W, Moss TJ, Ciobanu N, et al. Detection and viability of tumor cells in peripheral blood stem cell collections from breast cancer patients using immunocytochemical and clonogenic assay techniques. Blood 1993;82:2605-10.10.1182/blood.V82.9.2605.bloodjournal8292605Search in Google Scholar
48. Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92.10.1056/NEJMoa1113205Search in Google Scholar PubMed PubMed Central
49. Flores LM, Kindelberger DW, Ligon AH, Capelletti M, Fiorentino M, Loda M, et al. Improving the yield of circulating tumour cells facilitates molecular characterisation and recognition of discordant HER2 amplification in breast cancer. Br J Canc 2010;102:1495-502.10.1038/sj.bjc.6605676Search in Google Scholar PubMed PubMed Central
50. Krebs MG, Hou JM, Sloane R, Lancashire L, Priest L, Nonaka D, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol 2012;7:306-15.10.1097/JTO.0b013e31823c5c16Search in Google Scholar PubMed
51. Ilie M, Long E, Butori C, Hofman V, Coelle C, Mauro V, et al. ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol 2012;23:2907-13.10.1093/annonc/mds137Search in Google Scholar PubMed
52. Zhang Z, Shiratsuchi H, Palanisamy N, Nagrath S, Ramnath N. Expanded circulating tumor cells from a patient with ALK-positive lung cancer present with EML4-ALK rearrangement along with resistance mutation and enable drug sensitivity testing: a case study. J Thorac Oncol 2017;12:397-402.10.1016/j.jtho.2016.07.027Search in Google Scholar PubMed PubMed Central
53. Nilsson RJA, Balaj L, Hulleman E, Van Rijn S, Pegtel DM, Walraven M, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood 2011;118:3680-3.10.1182/blood-2011-03-344408Search in Google Scholar PubMed PubMed Central
54. Middleton EA, Weyrich AS, Zimmerman GA. Platelets in pulmonary immune responses and inflammatory lung diseases. Physiol Rev 2016;96:1211-59.10.1152/physrev.00038.2015Search in Google Scholar PubMed PubMed Central
55. Matowicka-Karna J, Kamocki Z, Poli?ska B, Osada J, Kemona H. Platelets and inflammatory markers in patients with gastric cancer. Clin Dev Immunol 2013;2013:401623.10.1155/2013/401623Search in Google Scholar PubMed PubMed Central
56. Nilsson RJA, Karachaliou N, Berenguer J, Gimenez-Capitan A, Schellen P, Teixido C, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016;7. https://doi.org/10.18632/oncotarget.6279.10.18632/oncotarget.6279Search in Google Scholar PubMed PubMed Central
57. Sundararajan V, Sarkar FH, Ramasamy TS. The versatile role of exosomes in cancer progression: diagnostic and therapeutic implications. Cell Oncol Springer Netherlands; 2018;41:223-52.10.1007/s13402-018-0378-4Search in Google Scholar PubMed
58. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:1-4.10.1038/cr.2015.82Search in Google Scholar PubMed PubMed Central
59. Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One 2012;7:e30679.10.1371/journal.pone.0030679Search in Google Scholar PubMed PubMed Central
60. Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao B, et al. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther 2017;10:3843-51.10.2147/OTT.S140062Search in Google Scholar PubMed PubMed Central
61. Helwa I, Cai J, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML, et al. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. Camussi G, editor. PLoS One. 2017;12:e0170628.10.1371/journal.pone.0170628Search in Google Scholar PubMed PubMed Central
62. Tang Y-T, Huang Y-Y, Zheng L, Qin S-H, Xu X-P, An T-X, et al. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int J Mol Med 2017;40:834-44.10.3892/ijmm.2017.3080Search in Google Scholar PubMed PubMed Central
63. Reclusa P, Laes J-F, Malapelle U, Valentino A, Rocco D, Gil-Bazo I, et al. EML4-ALK translocation identification in RNA exosomal cargo (ExoALK) in NSCLC patients: a novel role for liquid biopsy. Transl Cancer Res 2019;8:S76-8.10.21037/tcr.2018.11.35Search in Google Scholar PubMed PubMed Central
64. Patel GK, Khan MA, Zubair H, Srivastava SK, Khushman M, Singh S, et al. Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci Rep 2019;9:5335.10.1038/s41598-019-41800-2Search in Google Scholar PubMed PubMed Central
65. Brinkmann K, Enderle D, Koestler T, Bentink S, Emenegger J, Spiel A, et al. Abstract 545: plasma-based diagnostics for detection of EML4-ALK fusion transcripts in NSCLC patients. In: Clinical Research (Excluding Clinical Trials). American Association for Cancer Research; 2015. p. 545-545.10.1158/1538-7445.AM2015-545Search in Google Scholar
© 2020 Estela Sánchez-Herrero et al., published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Editorial
- A new journal for progress in Laboratory Medicine
- Nace una revista para avanzar en la Medicina de Laboratorio
- Review / Artículo de Revisión
- Towards personalized prostate cancer screening
- Hacia un cribado personalizado del cáncer de próstata
- Clinical utility of liquid biopsy for the diagnosis and monitoring of EML4-ALK NSCLC patients
- Utilidad clínica de la biopsia líquida para el diagnóstico y seguimiento de los pacientes con CPNM y EML4-ALK
- Original Article / Artículo Original
- Impact of implementing a category 1 external quality assurance scheme for monitoring harmonization of clinical laboratories in Spain
- Impacto de la introducción de un programa externo de categoría 1 en la vigilancia de la estandarización entre laboratorios clínicos en España
- Performance evaluation of Siemens Atellica enhanced estradiol assay
- Evaluación del ensayo enhaced estradiol (eE2) en el analizador Atellica IM 1600 de Siemens
- Utility of recombinant human TSH stimulation test in the follow-up of patients with differentiated thyroid cancer depending on basal thyroglobulin results
- Utilidad del test de TSH recombinante en el seguimiento de pacientes con cáncer diferenciado de tiroides según los resultados de tiroglobulina basal
- Case Report / Caso Clínico
- IgG4-related disease: a case report
- Enfermedad relacionada con IgG4: a propósito de un caso
Articles in the same Issue
- Editorial
- A new journal for progress in Laboratory Medicine
- Nace una revista para avanzar en la Medicina de Laboratorio
- Review / Artículo de Revisión
- Towards personalized prostate cancer screening
- Hacia un cribado personalizado del cáncer de próstata
- Clinical utility of liquid biopsy for the diagnosis and monitoring of EML4-ALK NSCLC patients
- Utilidad clínica de la biopsia líquida para el diagnóstico y seguimiento de los pacientes con CPNM y EML4-ALK
- Original Article / Artículo Original
- Impact of implementing a category 1 external quality assurance scheme for monitoring harmonization of clinical laboratories in Spain
- Impacto de la introducción de un programa externo de categoría 1 en la vigilancia de la estandarización entre laboratorios clínicos en España
- Performance evaluation of Siemens Atellica enhanced estradiol assay
- Evaluación del ensayo enhaced estradiol (eE2) en el analizador Atellica IM 1600 de Siemens
- Utility of recombinant human TSH stimulation test in the follow-up of patients with differentiated thyroid cancer depending on basal thyroglobulin results
- Utilidad del test de TSH recombinante en el seguimiento de pacientes con cáncer diferenciado de tiroides según los resultados de tiroglobulina basal
- Case Report / Caso Clínico
- IgG4-related disease: a case report
- Enfermedad relacionada con IgG4: a propósito de un caso

