Peptidomic and proteomic analysis of stool for diagnosing IBD and deciphering disease pathogenesis
-
Daniela Basso
, Andrea Padoan
, Renata D’Incà
Abstract
Background
The sensitivities and specificities of C-reactive protein (CRP) and faecal calprotectin (fCal), as recommended for inflammatory bowel diseases (IBD) diagnosis and monitoring, are low. Our aim was to discover new stool protein/peptide biomarkers for diagnosing IBD.
Methods
For peptides, MALDI-TOF/MS (m/z 1000–4000) was performed using stools from an exploratory (34 controls; 72 Crohn’s disease [CD], 56 ulcerative colitis [UC]) and a validation (28 controls, 27 CD, 15 UC) cohort. For proteins, LTQ-Orbitrap XL MS analysis (6 controls, 5 CD, 5 UC) was performed.
Results
MALDI-TOF/MS spectra of IBD patients had numerous features, unlike controls. Overall, 426 features (67 control-associated, 359 IBD-associated) were identified. Spectra were classified as control or IBD (absence or presence of IBD-associated features). In the exploratory cohort, the sensitivity and specificity of this classification algorithm were 81% and 97%, respectively. Blind analysis of the validation cohort confirmed 97% specificity, with a lower sensitivity (55%) paralleling active disease frequency. Following binary logistic regression analysis, IBD was independently correlated with MALDI-TOF/MS spectra (p < 0.0001), outperforming fCal measurements (p = 0.029). The IBD-correlated m/z 1810.8 feature was a fragment of APC2, homologous with APC, over-expressed by infiltrating cells lining the surface in UC or the muscularis-mucosae in CD (assessed by immunohistochemistry). IBD-associated over-expressed proteins included immunoglobulins and neutrophil proteins, while those under-expressed comprised proteins of the nucleic acid assembly or those (OLFM4, ENPP7) related to cancer risk.
Conclusions
Our study provides evidence for the clinical utility of a novel proteomic method for diagnosing IBD and insight on the pathogenic role of APC. Moreover, the newly described IBD-associated proteins might become tools for cancer risk assessment in IBD patients.
Introduction
The incidence of inflammatory bowel diseases (IBD) is increasing worldwide [1], [2], [3]. At onset, the symptoms of the two main clinical IBD subtypes, Crohn’s disease (CD) and ulcerative colitis (UC), are often common to functional disorders such as irritable bowel syndrome (IBS), thus causing delay in diagnosis. Intestinal inflammation with flares and remissions is a hallmark of IBD, with different pro-inflammatory cells that release inflammatory cytokines and chemokines abundantly present within the intestinal mucosa [4], [5], [6]. The sensitivities and specificities of blood inflammatory biomarkers, mainly C-reactive protein (CRP), erythrocyte sedimentation rate (ESR), white blood cells and platelet counts are very limited [7]. Faecal inflammatory biomarkers, mainly calprotectin (fCal), are more reliable indices of the intestinal inflammation typical of IBD [8], [9], [10], [11]. fCal is currently recommended for distinguishing between IBD from IBS and also for IBD monitoring [12], [13], [14], [15], [16]. However, neither the sensitivity nor specificity of fCal exceed 80% for IBD diagnosis [17], with further reductions in reliability in the presence of mild forms of the disease [18]. Moreover, none of the blood and faecal inflammatory biomarkers studied so far allow CD to be distinguished from UC. In this setting, ASCA and pANCA determinations in blood have sensitivities that do not exceed 65% and specificities of only around 80% [19].
The approaches reported in the literature in the search for new biomarkers for IBD diagnosis include microRNA expression profiling of circulating blood mononuclear cells, metabolomic profiling, microbiota characterisation as well as proteomic studies focusing on the search for differentially expressed proteins in tissue samples [20], [21], [22]. Among the novel biomarkers proposed, however, none has proven to outperform the established inflammatory indices in distinguishing between IBD and IBS nor in distinguishing between CD and UC [23].
A complex inflammatory condition, such as IBD, is expected to be associated with an increased mucosal accumulation and release of inflammatory biomarkers into the faecal stream. This includes, but is not limited to, fCal. In agreement with this, Lehmann et al. demonstrated that the most significant alterations of the human faecal proteome between IBD patients and healthy individuals included not only inflammatory proteins but also immunoglobulins, antimicrobial and cell integrity proteins [24]. Proteases are also involved in the inflammatory process, and, in line with this, increased matrix metalloproteases, such as MMP9, have been found in IBD diseased mucosa and in stool [25], [26]. Therefore, in IBD it might be expected that an increased mucosal accumulation of inflammatory proteins and proteases that enhance protein fragmentation occurs, with a consequent accumulation of peptides in stool. An integrated approach aimed to evaluate both peptides and proteins in stool might enhance the chance to discover not only new biomarkers for IBD diagnosis but also to identify new molecular pathways involved in disease pathogenesis and outcome. Therefore, the aim of the present prospective study was to analyse the faecal peptidome and proteome in IBD patients, comparing them with values in healthy subjects by using high throughput techniques, namely, MALDI-TOF/MS and LTQ-Orbitrap XL MS.
Materials and methods
Study population
This prospective study was approved by the local Ethics Committee (Prot. 3756/AO/16), and fully informed consent was obtained in writing from all cases and controls. A total of 175 IBD patients (74 F, 101 M; age range 17–83 years) and 62 healthy controls (23 F, 39 M; age range 30–63 years) were consecutively enrolled from September 2016 to February 2019. No statistically significant differences between patients and controls were found for sex and age (Table 1). Patients with histologically confirmed IBD were recruited during outpatient monitoring visits. None of the subjects in the control group, which comprised University-Hospital of Padova employees already included in the standard occupational program, had intestinal symptoms or a history of intestinal disease. The demographic, clinical and biochemical characteristics of patients and controls are reported in Table 1. Blood indices were classified as follows: (1) white blood cells count (WBC): normal if comprised within the reference range, reduced if below the lower (4.4×109/L) and increased if above the upper (11.0×109/L) limits of the reference range; (2) haemoglobin (Hb): normal if within and reduced if below the lower reference range limit (123–153 g/L for women and 140–175 g/L for men); (3) serum CRP: normal if within and increased if above the upper reference range limit (0–6 mg/L); (4) plasma alanine aminotransferase (ALT): normal if within and increased if above the upper reference range limit (7–35 U/L for women and 10–50 U/L for men); (5) faecal occult blood test (FOBT): negative or positive if below or above 100 ng/mL, respectively; (6) fCal: negative or positive if below/equal to or above 50 μg/g, respectively. The characterisation of IBD patients is detailed in Table 2. Disease location was classified according to the Montreal Classification [27]. Disease activity was evaluated on the basis of the disease activity indices (Harvey-Bradshow index [HBI] for CD and partial Mayo score [PMS] for UC), the inflammatory markers CRP and fCal and endoscopic and histological findings. Complete remission (CR) was defined by HBI <3 points in CD and PMS <2 points in UC. Deep remission (DR) was defined on the basis of the association of CR with endoscopic remission and/or fCal <70 μg/g and CRP levels of ≤6 mg/L [28].
Descriptive statistics of controls and IBD patients.
| Controls | Crohn’s disease | Ulcerative colitis | Unclassified | Statisticsa | |
|---|---|---|---|---|---|
| Cases number (exploratory/validation cohort) | 62 (34/28) | 99 (72/27) | 71 (56/15) | 5 (5/0) | |
| Sex, females/males | 23/39 | 40/59 | 31/40 | 3/2 | p=0.712 |
| Age years, mean+SD | 49±8 | 47±15 | 50±15 | 48±27 | F=0.70, p=0.5555 |
| Family history of Crohn’s disease, positive cases/total, % | 1/62 (2) | 11/99 (11) | 3/71 (4) | 0/5 (0) | p=0.153 |
| Family history of ulcerative colitis, positive cases/total, % | 1/62 (2) | 8/99 (8) | 8/71 (11) | 0/5 (0) | p=0.152 |
| Smoking habits, positive cases/total, % | 5/51 (10) | 19/95 (20) | 12/66 (18) | 0/5 (0) | p=0.356 |
| Alcohol consumption, positive cases/total, % | 13/51 (25) | 14/95 (15) | 9/66 (14) | 0/5 (0) | p=0.267 |
| WBC, positive cases/total, % | |||||
| ≤4.4×103/L | 6/61 (10) | 6/85 (7) | 2/60 (3) | 0/5 (0) | p=0.671 |
| 4.4–11.0×103/L | 54/61 (89) | 75/85 (88) | 57/60 (95) | 5/5 (100) | |
| >11.0×103/L | 1/61 (1) | 4/85 (5) | 1/60 (2) | 0/5 (0) | |
| Haemoglobin <123 g/L for females and 140 g/L for males, positive cases/total, % | 4/61 (7) | 25/92 (27) | 16/66 (24) | 3/5 (60) | p=0.001 |
| CRP >6 g/L, positive cases/total, % | 0/62 (0) | 9/86 (10) | 11/63 (17) | 0/2 (0) | p=0.002 |
| ALT >35 U/L for females and 50 U/L for males, positive cases/total, % | 4/58 (7) | 4/65 (6) | 2/47 (4) | 0/3 (0) | p=0.933 |
| FOBT, positive cases/total, % | 1/60 (2) | 20/96 (21) | 19/68 (28) | 1/5 (20) | p<0.0001 |
| Faecal calprotectin μg/g, mean+SEM | 77±19 | 410±62b | 527±81b | 672±389b | F=7.88, p=0.0001 |
| Faecal calprotectin >50 μg/g, positive cases/total, % | 27/60 (45%) | 75/99 (76%) | 58/71 (82%) | 4/5 (80%) | p<0.0001 |
aFisher’s exact test and one-way ANOVA; bBonferroni’s test for pairwise comparisons: p<0.001 with respect to controls.
Clinical characteristic of IBD patients with Crohn’s disease or ulcerative colitis.
| Crohn’s disease (n=99) | Ulcerative colitis (n=71) | Statistical analysis | |
|---|---|---|---|
| HBI and PMS | HBI 0=57 HBI 1=17 HBI 2=11 HBI 3=2 HBI 4=4 HBI >4=8 |
PMS 0=41 PMS 1=9 PMS 2=6 PMS 3=4 PMS 4=5 PMS >4=6 |
|
| Disease location | L1=25 (25%) L2=29 (29%) L3=45 (46%) |
E1=14 (20%) E2=10 (14%) E3=47 (66%) |
|
| Disease duration, years (mean±SE) | 13±0.9 | 14±1.3 | t=−0.5612, p=0.575 |
| Previous surgery | Yes=37 (37%) No=62 (63%) |
Yes=3 (4%) No=68 (96%) |
Fisher’s exact test: p<0.001 |
| Remission | DR=22 (22%) CR=51 (52%) Active=26 (26%) |
DR=14 (20%) CR=26 (37%) Active=31 (44%) |
Fisher’s exact test: p=0.053 |
| Therapy | AZA/6MP=14 (14%) | AZA/6MP=11 (15%) | Fisher’s exact test: p=0.829 |
| Mesalazine=73 (74%) | Mesalazine=57 (80%) | Fisher’s exact test: p=0.363 | |
| MTX=2 (2%) | MTX=1 (1%) | Fisher’s exact test: p=0.999 | |
| Steroids=4 (4%) | Steroids=6 (8%) | Fisher’s exact test: p=0.323 | |
| Biological agents=41/99 (41%) Infliximab=24/41 (59%) Golimumab=0/41 (0) Adalimumab=17/41 (41%) |
Biological agents=18/71 (25%) Infliximab=10/18 (56%) Golimumab=3/18 (17%) Adalimumab=5/18 (27%) |
Fisher’s exact test: p=0.001 |
L1, ileal; L2, colonic; L3, ileocolonic; E1, ulcerative proctitis; E2, left sided UC (distal UC); E3, extensive UC (pancolitis); CR, complete remission; DR, deep remission; AZA, azathioprine; 6MP, 6-mercaptopurine; MTX, methotrexate.
Biochemical markers analyses
Blood, plasma, serum and faecal (FOBT and fCal) biochemical data were analysed by the routine procedures of the Department of Laboratory Medicine, University-Hospital of Padova, which has obtained the ISO 9001 certification in 1997, the ISO 15189:2012 and the CPA-UK accreditations in 2016 and 1995, respectively. They are detailed in Supplementary Materials and Methods.
Faecal MALDI-TOF/MS peptidomic analyses
Faecal samples from the first bowel movement of the day, obtained from all cases and controls at enrolment, were refrigerated for up to 4 h before delivery to the laboratory. Following arrival in the laboratory, a total of 100 mg of each faecal sample was resuspended in 100 μL of ultrapure water (100% w/v), vigorously mixed for 5 min and then centrifuged for 30 min at 15,000 g. Supernatants containing peptide extracts were immediately frozen at −80°C and stored for no more than 3 months until MALDI-TOF/MS analyses, performed by means of the patented method (Italian Economic Development Ministry Number 102018000005689, 24 May 2018; WO 2019/224663 A1, 28 November 2019) as detailed in Supplementary Materials and Methods. Within laboratory reproducibility and repeatability of MALDI-TOF/MS were evaluated prior to stool sample analyses of IBD patients and controls. Two stool samples were analysed in batch four times for repeatability, one representative result being shown in Supplementary Figure 1. Within laboratory reproducibility was evaluated by replicating the assays of two samples in three consecutive days (representative results in Supplementary Figure 1).
MS-Tag bioinformatic analyses
MALDI-TOF/MS-MS spectra were analysed by the MS-Tag database search program of ProteinProspector (http://prospector.ucsf.edu/prospector/mshome.htm). Searches were performed against SwissProt.2016.9.6 human database using the following parameters: no enzyme, methionine oxidation, instrument MALDI-TOF-TOF and variable modifications.
SDS-PAGE electrophoresis
SDS-PAGE electrophoresis was performed by loading a total of 8 μg of protein for each sample onto a precast gel (NuPAGE, 4%–12% Bis-Tris, Life Technologies, Monza, Italy), as detailed in Supplementary Materials and Methods.
Faecal LTQ-Orbitrap proteomic analyses
Three pools of faecal samples were prepared. The first pool was made of six healthy control faecal samples (final protein concentration: 5 μg/μL). The second and third pools were made of five UC and five CD faecal samples (final protein concentration: 4.4 μg/μL and 4.6 μg/μL, respectively). LTQ-Orbitrap XL mass spectrometry was performed as detailed in Supplementary Materials and Methods.
Immunohistochemistry
We tested the anti-APC antibody in formalin fixed, paraffin embedded tissue samples obtained from colonic wall of patients having undergone surgical procedures for IBD (CD and UC), diverticulitis or colorectal cancer. From each paraffin embedded specimen, one histological section (2 μm thick) was obtained. All histological slides were automatically stained with anti-APC (rabbit antibody; working dilution 1:50, Sigma-Aldrich) using a LEICA Bond III immunostainer.
Statistical analyses
Descriptive statistics were used to explore data by using mean and standard deviation or median and interquartile range (IRQ), where appropriate. Fishers’ exact and chi square tests were used with categorical data. One way analysis of variance (ANOVA) was used with continuous data to evaluate differences among groups. Logistic regression analyses were performed for predicting the diagnosis condition by different independent variables. Stata v13.1 (Statacorp, LakeWay Drive, TX, USA) was used for the analyses.
Results
The clinical characteristics of patients and controls are detailed in Tables 1 and 2. IBD patients comprised 99 CD, 71 UC and five patients with unclassified IBD. The exploratory cohort comprised patients and controls enrolled from September 2016 to September 2017 (34 controls, 72 CD, 56 UC and 5 unclassified IBD). The validation cohort comprised patients enrolled from October 2017 to February 2019 (28 controls, 27 CD and 15 UC). Exploratory cohort MALDI-TOF/MS analysis of faecal peptidomic profiles in the m/z range 1000–4000 identified a total of 438 features. Some features were common to patients and controls, while others were almost exclusively measured in either patients or controls (Supplementary Table 1). The spectra of control subjects were devoid of features (flat spectra) in 13/34 (38%) cases, a representative result being shown in Figure 1A. Similar flat spectra were found in a lower percentage (17/133, 13%) of patient samples (Pearson X2=11.904, p=0.001). In 21/34 control samples, MALDI-TOF/MS spectra were characterised by the presence of one or more features (representative example in Figure 1B), that belonged to the 79 control features or shared features list reported in Supplementary Table 1. The spectra of patient samples appeared much more numerous in features (representative examples in Figure 1C–F). On the basis of the high enrichment in features found in stool samples from IBD patients, we verified whether parallel variations could be observed in stool proteins by performing SDS-PAGE electrophoresis in a subset of 109 IBD patients (65 males, 44 females; mean age±DS=49±16 years) and 81 controls (48 males, 33 females; mean age±SD=50±8 years). Supplementary Figure 2 shows the results of a representative SDS-PAGE experiment.
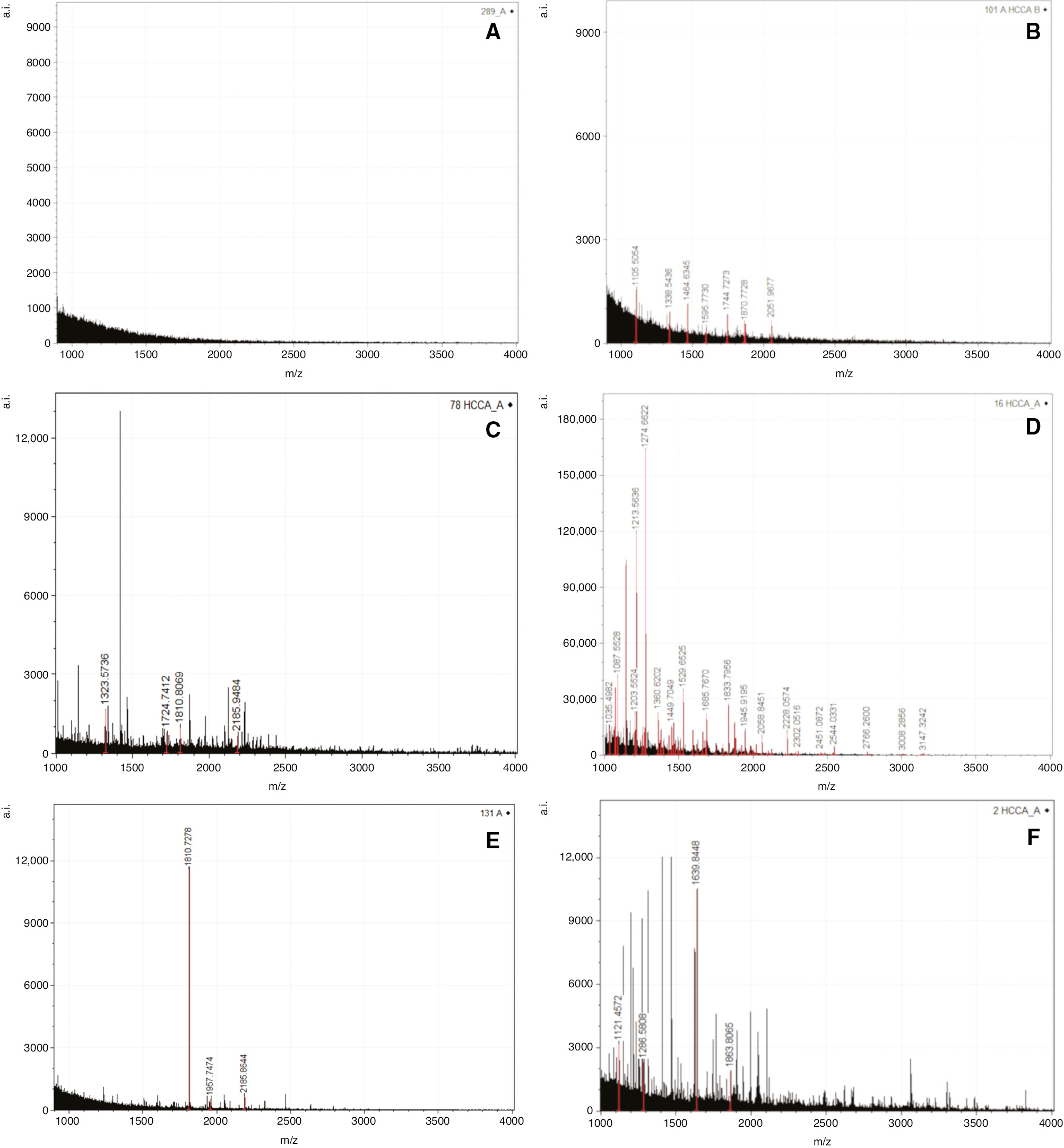
Faecal peptidomic profiling.
Representative MALDI-TOF/MS spectra in the m/z 1000–4000 range. Control spectra without features (A) or with control-associated and shared features (B). (C and D) Crohn’s disease spectra. (E and F) Ulcerative colitis spectra.
The MALDI-TOF/MS features collected from the exploratory cohort included 67/438 (15.3%) features shared between IBD patients and controls (shared features listed in Supplementary Table 1) and 359/438 (81.9%) disease-associated features (IBD features listed in Supplementary Table 1). Twelve out of 438 features (2.8%) were found only in control samples (control features listed in Supplementary Table 1). MALDI-TOF/MS spectra were evaluated and classified as control or IBD spectra on the basis of the following criteria: (1) control spectra if flat or showing only control or shared features; (2) IBD spectra if showing at least one IBD associated feature. The same criteria were then adopted to classify in blind MALDI-TOF/MS spectra obtained from samples of the validation cohort. Table 3 reports sensitivity, specificity and positive and negative predictive values of the above described MALDI-TOF/MS spectra classification in distinguishing controls from IBD patients in the exploratory and in the validation cohorts. For comparison fCal results at 50 μg/g cutoff are reported in the two cohorts. IBD MALDI-TOF/MS spectra were correlated with the absence of SDS-PAGE protein bands at a MW higher than 45 kDa, and with the presence of protein bands at a MW lower than 43 kDa (Supplementary Table 2).
Sensitivity, specificity and positive (PPV) and negative (NPV) predictive values of MALDI-TOF/MS spectra classification obtained in the exploratory and validation cohorts.
| C (n=34) | CD (n=72) | UC (n=56) | Un-IBD (n=5) | Sensitivity % (95% CI) | Specificity % (95% CI) | PPV % (95% CI) | NPV % (95% CI) | |
|---|---|---|---|---|---|---|---|---|
| Exploratory cohort | ||||||||
| Control/IBD spectra, cases nr. |
33/1 | 13/59 | 12/44 | 0/5 | 81.20 (73.52–87.45) |
97.06 (84.67–99.93) |
99.08 (93.99–99.87) |
56.90 (47.99–65.38) |
| Negative/Positive fCal, cases, nr. |
22/12 | 16/56 | 12/44 | 1/4 | 78.20 (70.21–84.88) |
64.71 (46.49–80.25) |
89.66 (84.50–93.24) |
43.14 (33.57–53.25) |
| C (n=28) | CD (n=27) | UC (n=15) | Un-IBD (n=0) | Sensitivity % (95% CI) | Specificity % (95% CI) | PPV % (95% CI) | NPV % (95% CI) | |
| Validation cohort | ||||||||
| Control/IBD spectra, cases nr. |
27/1 | 13/14 | 6/9 | 0/0 | 54.76 (38.67–70.15) |
96.43 (81.65–99.91) |
95.83 (76.70–99.38) |
58.70 (50.28–66.63) |
| Negative/Positive fCal, cases, nr. |
12/16 | 8/19 | 1/14 | 0/0 | 78.57 (63.19–89.70) |
42.86 (24.46–62.82) |
67.35 (59.06–74.68) |
57.14 (39.36–73.26) |
For comparative purposes, faecal calprotectin (fCal) results obtained in the same cohorts are reported. fCal findings were considered positive if above 50 μg/g. C, controls; CD, Crohn’s disease; UC, ulcerative colitis, Un-IBD, unclassified IBD; CI, confidence interval.
IBD spectra were compared with clinical and biochemical parameters considering all patients and controls of the exploratory and validation cohorts. IBD spectra were correlated with positive fCal (>50 μg/g) when considering patient and control samples overall (Pearson X2=16.527, p<0.0001) but not when controls (Fisher’s exact test, p=0.407), CD (Fisher’s exact test, p=0.791) or UC (Fisher’s exact test, p=0.079) patients were considered singly. Neither in CD nor UC were IBD spectra and fCal correlated with clinical score (HBI or PMS), disease location, previous surgery or therapy. IBD spectra correlated with disease activity both in CD (Fisher’s exact test, p<0.0001) and UC (Fisher’s exact test, p=0.010). Positive fCal results were also correlated with disease activity (Fisher’s exact test, p<0.0001 for CD and UC), but this finding was expected since fCal was included in the classification of DR.
Binary logistic regression analysis was performed to ascertain which of the parameters associated with the presence of IBD is more effective in making a diagnosis. The results, shown in Table 4, demonstrate that IBD spectra were independently and highly significantly correlated with IBD. Of the 359 IBD-associated features, 34 were almost exclusively observed in CD, while 25 were almost exclusively observed in UC stool samples. With the aim of ascertaining whether these features might aid the classification of disease type, IBD spectra were classified as IBD-CD or IBD-UC when only CD or only UC associated features were found. IBD-CD spectra were significantly correlated with CD in the exploratory (Fisher’s exact, p<0.0001) and the validation cohorts (Fisher’s exact, p=0.028), as IBD-UC spectra were correlated with UC in the exploratory (Fisher’s exact, p<0.0001) and validation cohorts (Fisher’s exact, p=0.008).
Binary logistic regression analysis.
| Coefficient | SE | Z | p-Value | 95% CI |
||
|---|---|---|---|---|---|---|
| IBD spectra | 4.132194 | 0.7528157 | 5.49 | 0.000 | 2.656702 | 5.607685 |
| fCal | 0.9724741 | 0.4449761 | 2.19 | 0.029 | 0.1003369 | 1.844611 |
| Hb | 1.253761 | 0.6734565 | 1.86 | 0.063 | −0.0661896 | 2.573711 |
| FOBT | 1.713719 | 1.162158 | 1.47 | 0.140 | −0.5640698 | 3.991507 |
| Age | 0.0033127 | 0.0191778 | 0.17 | 0.863 | −0.034275 | 0.0409005 |
| Sex | −0.2414579 | 0.4408126 | −0.55 | 0.584 | −1.105435 | 0.622519 |
| Constant | −1.144366 | 1.11641 | −1.03 | 0.305 | −3.332489 | 1.043757 |
The outcome variable was presence or absence of IBD. Predictor variables were IBD MALDI-TOF/MS spectra (IBD spectra), faecal calprotectin (fCal), blood haemoglobin (Hb), faecal occult blood (FOBT), age and sex.
MALDI-TOF/MS stool features were further characterised by MALDI-TOF/MS–MS, and fragmentation results were evaluated using the MS-Tag bioinformatics tool of ProteinProspector v 5.22.1 (available at http://prospector.ucsf.edu/prospector/mshome.htm) in the attempt to identify the candidate proteins (full molecule). A significant pattern was obtained for 14 features (Supplementary Table 3). We focused on the feature at m/z 1810.8, the most frequently present in IBD patients’ stool, that matched with the adenomatous polyposis coli gene 2, a homologue of the adenomatous polyposis coli (APC) tumour suppressor. In no cases was the entire APC protein detected in stool samples when western blot analyses were performed (data not shown). On the contrary, positive staining was obtained at immunohistochemistry of tissue samples. Figure 2 shows representative results obtained at immunohistochemistry, performed using intestinal tissue samples from CD and UC patients and from the healthy mucosa of one patient with colorectal cancer. In colorectal cancer, a few sparse interstitial cells were positively stained; in UC and CD an increasing number of infiltrating positive cells were observed in the colon mucosa, these cells being polarised towards the lumen in UC or towards the muscularis mucosae in CD. In CD patients, a diffuse infiltrate of APC positive cells was also detected in the ileum mucosa.
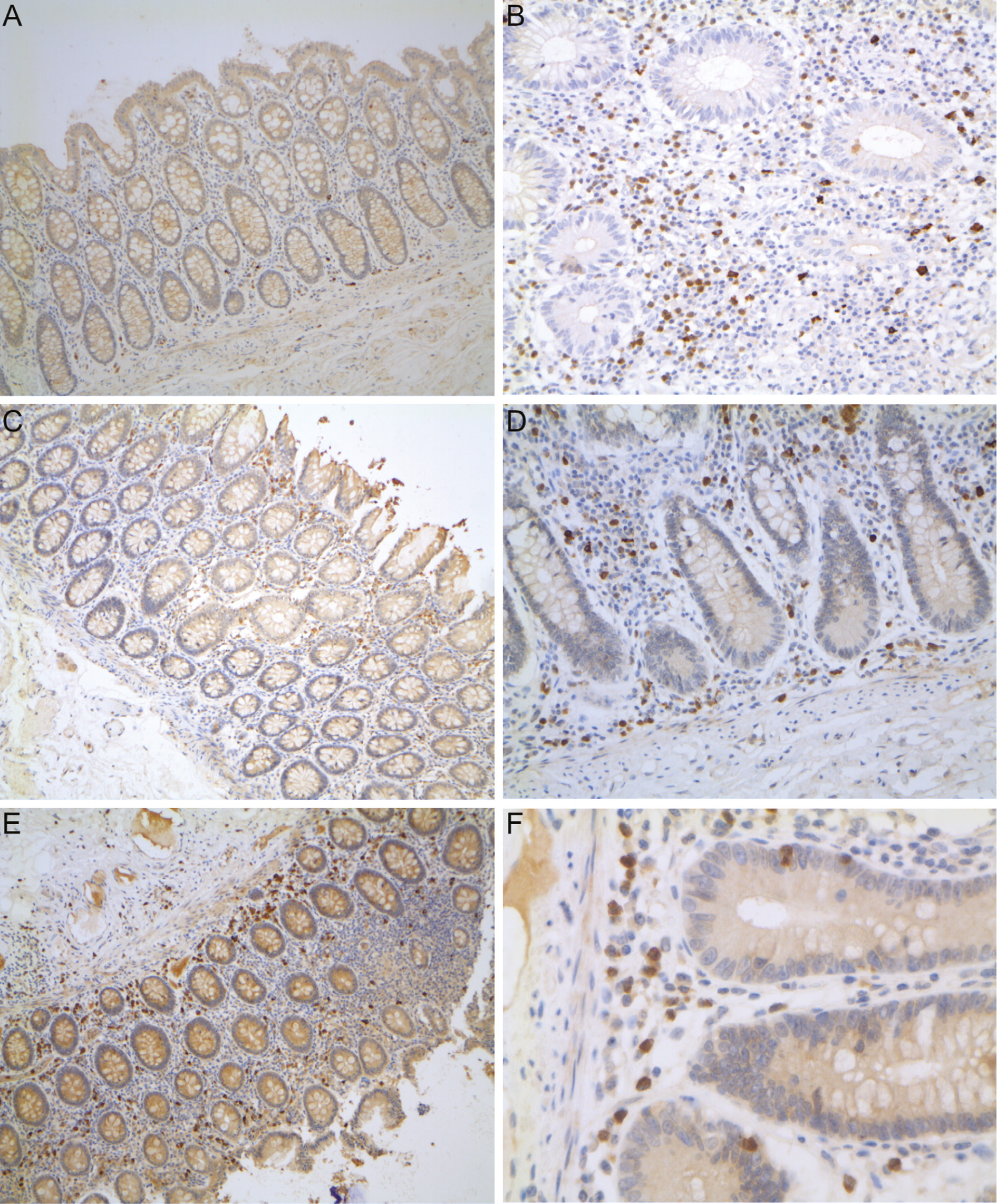
Adenomatous polyposis coli (APC) immunohistochemistry.
In the left side the colon mucosa of a patient with colorectal cancer (A), a patient with UC (C) and a patient with CD (E) are shown. In the right side of the figure the ileum mucosa of CD patients are shown. In CD, APC positively stained cells (in brown) are highly represented in the stroma of the inflamed ileum (B, D and F), but also in the non-inflamed colon (C), and they are preferentially localised towards the muscolaris mucosae. In UC the same APC positive cells are mainly localised towards the surface epithelium.
To detect new IBD diagnostic protein biomarkers, we analysed three stool samples pools, made up of six control, five CD and five UC samples. After SDS-PAGE electrophoresis, proteins were gel extracted and analysed by LC-MS using an Orbitrap mass analyser. A total of 193 proteins were identified. Proteins were considered differentially expressed between patients and controls when the ratio between absolute intensity of the quantification values were above 2.0 (over-expressed proteins in patients, ↑) or below 0.5 (under-expressed proteins in patients, ↓). This left 161 differentially expressed proteins that are reported in Supplementary Table 4. A comprehensive analysis was made of the differentially expressed proteins using David software. GO terms for biological processes were significant (Benjamini and Hochberg adjustment for multiple comparisons, p<0.05) in the following comparisons: (1) increased in IBD with respect to controls, with no difference between CD and UC; (2) increased in IBD with respect to controls, with higher levels in UC than in CD; (3) reduced in IBD with respect to controls. Supplementary Table 5 reports the complete list of the significant GO terms for proteins found to be increased in faeces of IBD patients with respect to controls. Figure 3 shows the significant GO biological processes associated with over-represented and under-represented stool proteins in IBD patients with respect to controls.
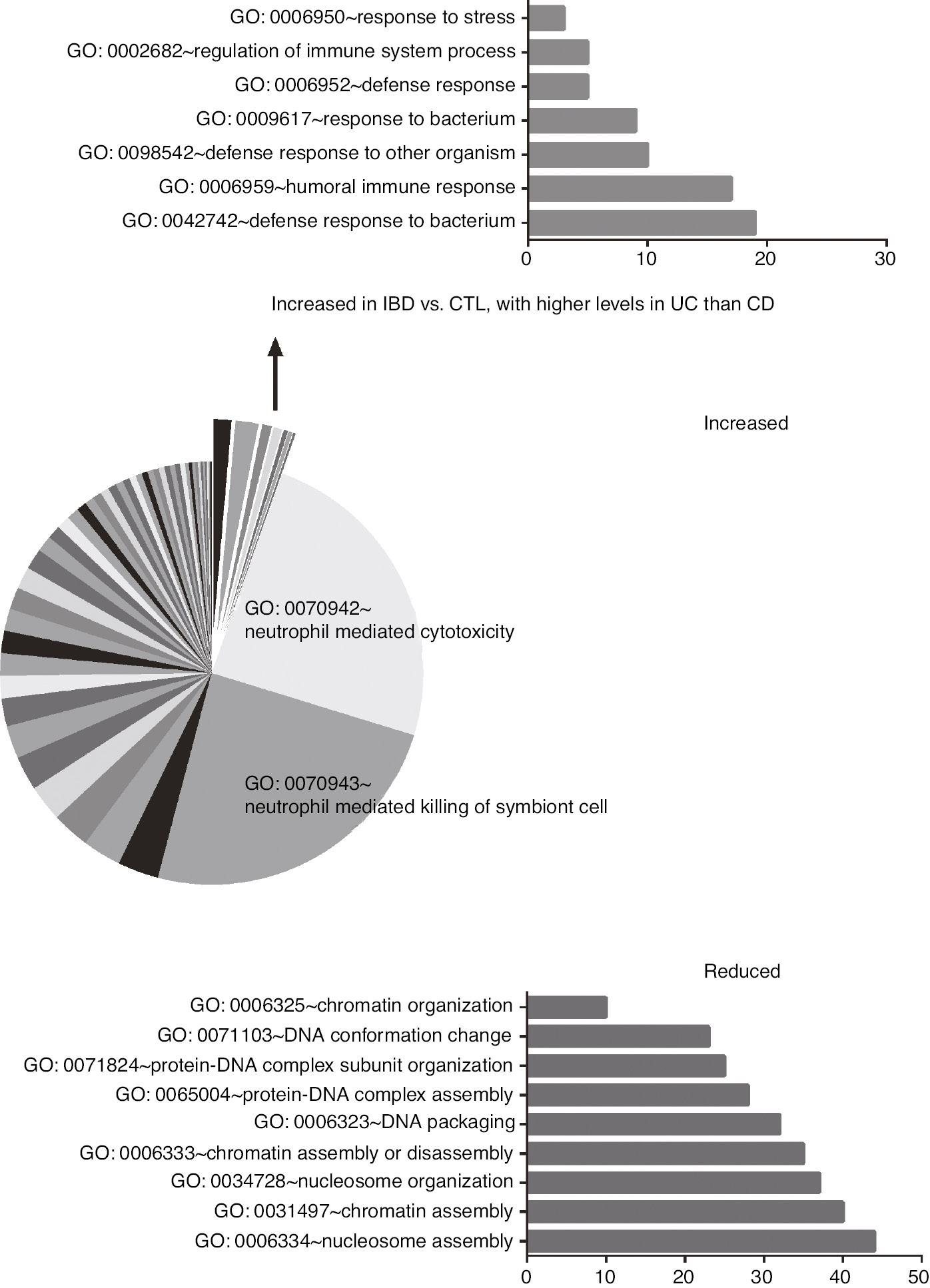
Significant GO terms for biological processes.
The pie chart shows the relative fold enrichment of GO terms found to be significant for stool proteins increased in IBD patients with respect to controls. The sorted slices show the GO terms for proteins with increased expression in IBD patients with respect to controls, with higher levels in UC than in CD. In the bottom, fold enrichment of significant GO terms for proteins with a reduced expression in IBD patients with respect to controls.
Discussion
Serum and faecal inflammatory biomarkers supporting a diagnosis of IBD have poor sensitivities and specificities [7], [8], [9], [10], [11], [13], [17], [18], [29]. The need to discover new biomarkers for diagnosing IBD and stratifying risk for tailored individualised treatment has yet to be met. Stool was chosen as the sample material because it is likely to contain more intestinal biomarkers than blood. Further, we also analysed MALDI-TOF low molecular weight features and proteins on the premise that they might directly reflect disease-associated cellular dysfunction [30]. The MALDI-TOF/MS analysis of stool from the exploratory cohort revealed very few, or no, features in healthy subjects, while stool of IBD patients were highly enriched in features. This finding, suggesting that IBD is associated with increased proteolysis, is in line with data in the literature reporting an increased expression of proteases in the diseased IBD mucosa, including MMP9, which was confirmed to be over-represented in IBD patients’ stool (see Supplementary Table 4) [26]. A higher frequency of proteolysis in IBD than in control samples was further supported by the SDS-PAGE findings exploring faecal proteins in a MW ranging from 200 to 3 kDa, with a MW higher than 45 kDa observed less frequently in IBD than in controls. The overall number of features commonly detected in both controls and patients was limited, while IBD-associated features were very numerous (>350), and each was variably associated with the other features in a very complex manner. In order to decipher and interpret the MALDI-TOF/MS results, each spectrum was classified as control spectrum if flat (no features) or with features (one or more than one) found in controls; MALDI-TOF/MS spectra were classified as IBD spectra if they presented one (or more than one) features found in IBD stool samples only. On applying this classification to the exploratory cohort, 58 control spectra and 109 IBD spectra were defined, this classification allowing a distinction between IBD patients and controls with a sensitivity of 81% and a specificity of 97%. This showed an improvement on fCal, which had a specificity of only 65%. To empower our findings, MALDI-TOF/MS blind analyses and spectra classification were performed using samples from a validation cohort. The obtained results confirmed that the specificity of MALDI-TOF/MS is high (97%), the sensitivity being 55%. In this validation cohort, fCal specificity was even lower (42%) than in the exploratory cohort. To explain why in the validation cohort IBD spectra had a lower sensitivity than in the exploratory cohort, it is important to bear in mind that, in general, validation of new biomarkers in patient cohorts different from those initially studied is almost always associated with reduction in clinical performance. More specifically, a different frequency of active IBD between the two cohorts (37% in the exploratory and 24% in the validation) might have had impact on the results, in view of the fact that positive IBD spectra were correlated with disease activity.
The numerous IBD associated features found prompted investigation of their parent proteins. Peptides were fragmented and analysed by MALDI-TOF/MS-MS, which was successful for 14 features. This small number of identified peptides with respect to the whole IDB associated features is a limitation that depends on the following: (1) MALDI-TOF/MS-MS can be performed on isolated features; (2) for a successful analysis, the features should have a high signal-to-noise ratio; and (3) an uncomplete and unpredictable feature fragmentation prevents the analysis. However, despite these limitations, our results indicated accumulation of immunoglobulins in IBD patients’ stool with respect to controls. This supports the relevance of the adaptive immune response in these inflammatory diseases, in agreement with Lehmann et al. [24], who found increased IgG and decreased IgA in IBD stool samples with respect to healthy controls. Pathways involved in the regulation of gene transcription, in the control of cell cycle and differentiation, and the Wnt pathway also emerged, confirming their involvement in IBD [31], [32]. Of particular interest are our results obtained with the m/z 1810.8 feature being observed frequently in patient stool but rarely in control stool. This matched significantly with the APC2 protein, homologous with the APC protein [33], a polarity regulator and tumour suppressor associated with development of familial adenomatous polyposis and colorectal cancer. To ascertain whether APC expression is altered in IBD mucosa, we performed immunohistochemistry that documented APC overexpression by stromal inflammatory cells which were not limited to the ill mucosa but diffuse throughout all the examined areas of the intestinal mucosa. APC is not only a tumour suppressor but is also involved in tissue damage and repair, especially in IBD, this function being correlated with its expression by vascular endothelial cells as recently demonstrated by Yoshimi et al. in a colitis animal model [34]. Moreover, a close relationship between lymphocyte function and APC expression has been recently reported by Aguera-Gonzalez et al. [35], who demonstrated that APC deficiency reduced NFAT nuclear localisation that determines a reduced Treg differentiation, favouring uncontrolled intestinal inflammation. The APC overexpressing cells infiltrating the IBD intestinal mucosa also showed a different polarisation between CD and UC: while in UC they were mainly polarised towards the intestinal lumen, in CD they were mainly polarised towards the muscularis mucosae. This different polarisation might be one of the mechanisms underlying the differences between the CD and UC intestinal inflammation patterns (transmural in CD, limited to the mucosa in UC).
In order to achieve a comprehensive proteomic analysis to shed further light on the diagnosis of IBD, we also evaluated differently expressed stool proteins by LC-MS. Although most of the identified stool proteins are plasma proteins and their increase in IBD stool might be a consequence of intestinal bleeding, we found also a large series of these plasma proteins that were reduced in IBD patients’ stool with respect to controls. Therefore, IBD associated variations of stool proteins might be the direct expression of disease alterations in the intestinal mucosa and not only of intestinal blood loss. Immunoglobulins were confirmed to be over-expressed in IBD, as were polymorphonuclear derived inflammatory proteins, such as S100A8, partner of S100A9 in the calprotectin heterodimer, neutrophil elastase and defensin. These proteins were found overexpressed in IBD stool also by Lehmann et al. [24]. Overall neutrophil mediated cytotoxicity and neutrophil mediated killing of symbiont cells represented the two main GO biological pathways identified. On the other hand, IBD-associated down expressed proteins were mainly involved in nucleic acid assembly, organisation and conformation, but also proteins related to the adherens junction organisation. Two identified proteins deserve further studies, since their differential expressions indicate not only a potential role for distinguishing between CD and UC but also in understanding why IBD patients are at a higher risk of developing cancer [36], [37]: olfactomedin-4 (OLFM4), absent in CD, and ectonucleotide pyrophosphatase/phosphodiesterase family member 7 (ENPP7), absent in UC. Both proteins, if reduced, might be related to an increased risk of primary cancer and metastases: OLFM4, normally expressed by intestinal crypt base stem cells, was found to be reduced in the subset of poorly differentiated and metastatic colorectal cancer [38], [39], while the lack of ENPP7, involved in sphingomyelin metabolism, significantly enhances colon cancer susceptibility [40].
In conclusion, our study provides evidence of the clinical utility of a new proteomic method for diagnosing IBD and offers insights on the pathogenic role of APC and other newly described proteins that, after validation, might become new tools in the diagnosis of IBD and the assessment of cancer risk in patients with the disease.
Acknowledgments
The authors wish to thank the Cassa di Risparmio di Padova e Rovigo (Cariparo) Holding for funding the acquisition of the LTQ-Orbitrap XL mass spectrometer. The authors thank Mrs. Monica Razetti and Mrs. Cinzia Centobene for their technical support in fCal measurements.
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
Research funding: University of Padova, Department of Medicine – DIMED: BIRD173078/17.
Employment or leadership: None declared.
Honorarium: None declared.
Competing interests: The funding organization(s) played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the report for publication.
References
1. Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012;142:46–54.10.1053/j.gastro.2011.10.001Search in Google Scholar PubMed
2. Kaplan GG, Ng SC. Understanding and preventing the global increase of inflammatory bowel disease. Gastroenterology 2017;152:313–21.10.1053/j.gastro.2016.10.020Search in Google Scholar PubMed
3. Kinnucan J, Binion D, Cross R, Evans E, Harlen K, Matarese L, et al. Inflammatory bowel disease care referral pathway. Gastroenterology 2019;157:242–54.10.1053/j.gastro.2019.03.064Search in Google Scholar PubMed
4. Grainger JR, Konkel JE, Zangerle-Murray T, Shaw TN. Macrophages in gastrointestinal homeostasis and inflammation. Pflugers Arch 2017;469:527–39.10.1007/s00424-017-1958-2Search in Google Scholar PubMed PubMed Central
5. Giuffrida P, Corazza GR, Di Sabatino A. Old and new lymphocyte players in inflammatory bowel disease. Dig Dis Sci 2017;63:277–88.10.1007/s10620-017-4892-4Search in Google Scholar PubMed
6. Pai RK, Geboes K. Disease activity and mucosal healing in inflammatory bowel disease: a new role for histopathology? Virchows Arch 2018;472:99–110.10.1007/s00428-017-2156-5Search in Google Scholar PubMed
7. Menees SB, Powell C, Kurlander J, Goel A, Chey WD. A meta-analysis of the utility of C-reactive protein, erythrocyte sedimentation rate, fecal calprotectin, and fecal lactoferrin to exclude inflammatory bowel disease in adults with IBS. Am J Gastroenterol 2015;110:444–54.10.1038/ajg.2015.6Search in Google Scholar PubMed
8. Waugh N, Cummins E, Royle P, Kandala NB, Shyangdan D, Arasaradnam R, et al. Faecal calprotectin testing for differentiating amongst inflammatory and non-inflammatory bowel diseases: systematic review and economic evaluation. Health Technol Assess 2013;17:xv–xix, 1–211.10.3310/hta17550Search in Google Scholar PubMed PubMed Central
9. Sands BE. Biomarkers of inflammation in inflammatory bowel disease. Gastroenterology 2015;149:1275–85.10.1053/j.gastro.2015.07.003Search in Google Scholar PubMed
10. Lopez RN, Leach ST, Lemberg DA, Duvoisin G, Gearry RB, Day AS. Fecal biomarkers in inflammatory bowel disease. J Gastroenterol Hepatol 2017;32:577–82.10.1111/jgh.13611Search in Google Scholar PubMed
11. Mak WY, Buisson A, Andersen MJ Jr, Lei D, Pekow J, Cohen RD, et al. Fecal calprotectin in assessing endoscopic and histological remission in patients with ulcerative colitis. Dig Dis Sci 2018;63:1294–301.10.1007/s10620-018-4980-0Search in Google Scholar PubMed
12. Panes J, Jairath V, Levesque BG. Advances in use of endoscopy, radiology, and biomarkers to monitor inflammatory bowel diseases. Gastroenterology 2017;152:362–73.10.1053/j.gastro.2016.10.005Search in Google Scholar PubMed
13. Lin JF, Chen JM, Zuo JH, Yu A, Xiao ZJ, Deng FH, et al. Meta-analysis: fecal calprotectin for assessment of inflammatory bowel disease activity. Inflamm Bowel Dis 2014;20:1407–15.10.1097/MIB.0000000000000057Search in Google Scholar PubMed
14. Boschetti G, Garnero P, Moussata D, Cuerq C, Préaudat C, Duclaux-Loras R, et al. Accuracies of serum and fecal S100 proteins (calprotectin and calgranulin C) to predict the response to TNF antagonists in patients with Crohn’s disease. Inflamm Bowel Dis 2015;21:331–6.10.1097/MIB.0000000000000273Search in Google Scholar PubMed
15. Wright EK, Kamm MA, De Cruz P, Hamilton AL, Ritchie KJ, Krejany EO, et al. Measurement of fecal calprotectin improves monitoring and detection of recurrence of Crohn’s disease after surgery. Gastroenterology 2015;148:938–47.10.1053/j.gastro.2015.01.026Search in Google Scholar PubMed
16. Heida A, Park KT, van Rheenen PF. Clinical utility of fecal calprotectin monitoring in asymptomatic patients with inflammatory bowel disease: a systematic review and practical guide. Inflamm Bowel Dis 2017;23:894–902.10.1097/MIB.0000000000001082Search in Google Scholar PubMed PubMed Central
17. Padoan A, D’Incà R, Scapellato ML, De Bastiani R, Caccaro R, Mescoli C, et al. Improving IBD diagnosis and monitoring by understanding preanalytical, analytical and biological fecal calprotectin variability. Clin Chem Lab Med 2018;56:1926–35.10.1515/cclm-2018-0134Search in Google Scholar PubMed
18. D’Angelo F, Felley C, Frossard JL. Calprotectin in daily practice: where do we stand in 2017? Digestion 2017;95:293–301.10.1159/000476062Search in Google Scholar PubMed
19. Reese GE, Constantinides VA, Simillis C, Darzi AW, Orchard TR, Fazio VW, et al. Diagnostic precision of anti-Saccharomyces cerevisiae antibodies and perinuclear antineutrophil cytoplasmic antibodies in inflammatory bowel disease. Am J Gastroenterol 2006;101:2410–22.10.1111/j.1572-0241.2006.00840.xSearch in Google Scholar PubMed
20. Mourad FH, Yau Y, Wasinger VC, Leong RW. Proteomics in inflammatory bowel disease: approach using animal models. Dig Dis Sci 2017;62:2266–76.10.1007/s10620-017-4673-0Search in Google Scholar PubMed
21. Sommer F, Rühlemann MC, Bang C, Höppner M, Rehman A, Kaleta C, et al. Microbiomarkers in inflammatory bowel diseases: caveats come with caviar. Gut 2017;66:1734–8.10.1136/gutjnl-2016-313678Search in Google Scholar PubMed PubMed Central
22. Mohammadi A, Kelly OB, Filice M, Kabakchiev B, Smith MI, Silverberg MS. Differential expression of microRNAs in peripheral blood mononuclear cells identifies autophagy and TGF-beta-related signatures aberrantly expressed in inflammatory bowel disease. J Crohns Colitis 2018;12:568–81.10.1093/ecco-jcc/jjy010Search in Google Scholar PubMed PubMed Central
23. Whitehead SJ, Ford C, Gama RM, Ali A, McKaig B, Waldron JL, et al. Effect of faecal calprotectin assay variability on the management of inflammatory bowel disease and potential role of faecal S100A12. J Clin Pathol 2017;70:1049–56.10.1136/jclinpath-2017-204340Search in Google Scholar PubMed
24. Lehmann T, Schallert K, Vilchez-Vargas R, Benndorf D, Püttker S, Sydor S, et al. Metaproteomics of fecal samples of Crohn’s disease and ulcerative colitis. J Proteomics 2019;201:93–103.10.1016/j.jprot.2019.04.009Search in Google Scholar PubMed
25. Faubion WA Jr, Fletcher JG, O’Byrne S, Feagan BG, de Villiers WJ, Salzberg B, et al. EMerging BiomARKers in inflammatory bowel disease (EMBARK) study identifies fecal calprotectin, serum MMP9, and serum IL-22 as a novel combination of biomarkers for Crohn’s disease activity: role of cross-sectional imaging. Am J Gastroenterol 2013;108:1891–900.10.1038/ajg.2013.354Search in Google Scholar PubMed
26. Farkas K, Saródi Z, Bálint A, Földesi I, Tiszlavicz L, Szűcs M, et al. The diagnostic value of a new fecal marker, matrix metalloprotease-9, in different types of inflammatory bowel diseases. J Crohns Colitis 2015;9:231–7.10.1093/ecco-jcc/jjv005Search in Google Scholar PubMed
27. Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 2006;55:749–53.10.1136/gut.2005.082909Search in Google Scholar PubMed PubMed Central
28. Mechie NC, Mavropoulou E, Ellenrieder V, Petzold G, Kunsch S, Neesse A, et al. Serum vitamin D but not zinc levels are associated with different disease activity status in patients with inflammatory bowel disease. Medicine 2019;98:e15172.10.1097/MD.0000000000015172Search in Google Scholar PubMed PubMed Central
29. Rogler G, Biedermann L. Clinical utility of biomarkers in IBD. Curr Gastroenterol Rep 2015;17:26–9.10.1007/s11894-015-0449-xSearch in Google Scholar PubMed
30. Assadsangabi A, Evans CA, Corfe BM, Lobo A. Application of proteomics to inflammatory bowel disease research: current status and future perspectives. Gastroenterol Res Pract 2019;2019:1426954.10.1155/2019/1426954Search in Google Scholar PubMed PubMed Central
31. Roussel-Gervais A, Naciri I, Kirsh O, Kasprzyk L, Velasco G, Grillo G, et al. Loss of the methyl-CpG-binding protein ZBTB4 alters mitotic checkpoint, increases aneuploidy, and promotes tumorigenesis. Cancer Res 2017;77:62–73.10.1158/0008-5472.CAN-16-1181Search in Google Scholar PubMed
32. Serafino A, Moroni N, Zonfrillo M, Andreola F, Mercuri L, Nicotera G, et al. WNT-pathway components as predictive markers useful for diagnosis, prevention and therapy in inflammatory bowel disease and sporadic colorectal cancer. Oncotarget 2014;5:978–92.10.18632/oncotarget.1571Search in Google Scholar PubMed PubMed Central
33. van Es JH, Kirkpatrick C, van de Wetering M, Molenaar M, Miles A, Kuipers J, et al. Identification of APC2, a homologue of the adenomatous polyposis coli tumour suppressor. Curr Biol 1999;9:105–8.10.1016/S0960-9822(99)80024-4Search in Google Scholar PubMed
34. Yoshimi K, Tanaka T, Serikawa T, Kuramoto T. Tumor suppressor APC protein is essential in mucosal repair from colonic inflammation through angiogenesis. Am J Pathol 2013;182:1263–74.10.1016/j.ajpath.2012.12.005Search in Google Scholar PubMed
35. Agüera-González S, Burton OT, Vázquez-Chávez E, Cuche C, Herit F, Bouchet J, et al. Adenomatous polyposis coli defines Treg differentiation and anti-inflammatory function through microtubule-mediated NFAT localization. Cell Reports 2017;21:181–94.10.1016/j.celrep.2017.09.020Search in Google Scholar PubMed
36. Annese V, Beaugerie L, Egan L, Biancone L, Bolling C, Brandts C, et al. European evidence-based consensus: inflammatory bowel disease and malignancies. J Crohns Colitis. 2015;9:945–65.10.1093/ecco-jcc/jjv141Search in Google Scholar PubMed
37. Choi CR, Bakir IA, Hart AL, Graham TA. Clonal evolution of colorectal cancer in IBD. Nat Rev Gastroenterol Hepatol 2017;14:218–29.10.1038/nrgastro.2017.1Search in Google Scholar PubMed
38. Liu W, Liu Y, Zhu J, Wright E, Ding I, Rodgers GP. Reduced hGC-1 protein expression is associated with malignant progression of colon carcinoma. Clin Cancer Res 2008;14:1041–9.10.1158/1078-0432.CCR-07-4125Search in Google Scholar PubMed
39. van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells.Gastroenterology 2009;137:15–7.10.1053/j.gastro.2009.05.035Search in Google Scholar PubMed
40. Chen Y, Zhang P, Xu SC, Yang L, Voss U, Ekblad E, et al. Enhanced colonic tumorigenesis in alkaline sphingomyelinase (NPP7) knockout mice. Mol Cancer Ther 2015;14:259–67.10.1158/1535-7163.MCT-14-0468-TSearch in Google Scholar PubMed
Supplementary Material
The online version of this article offers supplementary material (https://doi.org/10.1515/cclm-2019-1125).
©2021 Daniela Basso et al., published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Editorial
- Advancements in mass spectrometry as a tool for clinical analysis: part II
- Quantitative protein assessment
- Complexity, cost, and content – three important factors for translation of clinical protein mass spectrometry tests, and the case for apolipoprotein C-III proteoform testing
- Vedolizumab quantitation using high-resolution accurate mass-mass spectrometry middle-up protein subunit: method validation
- Development and evaluation of an element-tagged immunoassay coupled with inductively coupled plasma mass spectrometry detection: can we apply the new assay in the clinical laboratory?
- MALDI-MS for the clinic
- Matrix-assisted laser desorption ionisation (MALDI) mass spectrometry (MS): basics and clinical applications
- Clinical use of mass spectrometry (imaging) for hard tissue analysis in abnormal fracture healing
- Cellular resolution in clinical MALDI mass spectrometry imaging: the latest advancements and current challenges
- Bacterial identification by lipid profiling using liquid atmospheric pressure matrix-assisted laser desorption/ionization mass spectrometry
- Clinical application of ’omics technologies
- Individualized metabolomics: opportunities and challenges
- Diagnostic amyloid proteomics: experience of the UK National Amyloidosis Centre
- The “olfactory fingerprint”: can diagnostics be improved by combining canine and digital noses?
- Peptidomic and proteomic analysis of stool for diagnosing IBD and deciphering disease pathogenesis
- The influence of hypoxia on the prostate cancer proteome
- Laboratory automation and kit-based approaches
- Mass spectrometry and total laboratory automation: opportunities and drawbacks
- The pathway through LC-MS method development: in-house or ready-to-use kit-based methods?
- Evaluation of the 25-hydroxy vitamin D assay on a fully automated liquid chromatography mass spectrometry system, the Thermo Scientific Cascadion SM Clinical Analyzer with the Cascadion 25-hydroxy vitamin D assay in a routine clinical laboratory
Articles in the same Issue
- Frontmatter
- Editorial
- Advancements in mass spectrometry as a tool for clinical analysis: part II
- Quantitative protein assessment
- Complexity, cost, and content – three important factors for translation of clinical protein mass spectrometry tests, and the case for apolipoprotein C-III proteoform testing
- Vedolizumab quantitation using high-resolution accurate mass-mass spectrometry middle-up protein subunit: method validation
- Development and evaluation of an element-tagged immunoassay coupled with inductively coupled plasma mass spectrometry detection: can we apply the new assay in the clinical laboratory?
- MALDI-MS for the clinic
- Matrix-assisted laser desorption ionisation (MALDI) mass spectrometry (MS): basics and clinical applications
- Clinical use of mass spectrometry (imaging) for hard tissue analysis in abnormal fracture healing
- Cellular resolution in clinical MALDI mass spectrometry imaging: the latest advancements and current challenges
- Bacterial identification by lipid profiling using liquid atmospheric pressure matrix-assisted laser desorption/ionization mass spectrometry
- Clinical application of ’omics technologies
- Individualized metabolomics: opportunities and challenges
- Diagnostic amyloid proteomics: experience of the UK National Amyloidosis Centre
- The “olfactory fingerprint”: can diagnostics be improved by combining canine and digital noses?
- Peptidomic and proteomic analysis of stool for diagnosing IBD and deciphering disease pathogenesis
- The influence of hypoxia on the prostate cancer proteome
- Laboratory automation and kit-based approaches
- Mass spectrometry and total laboratory automation: opportunities and drawbacks
- The pathway through LC-MS method development: in-house or ready-to-use kit-based methods?
- Evaluation of the 25-hydroxy vitamin D assay on a fully automated liquid chromatography mass spectrometry system, the Thermo Scientific Cascadion SM Clinical Analyzer with the Cascadion 25-hydroxy vitamin D assay in a routine clinical laboratory


