Abstract
Objectives
Kaposi’s sarcoma (KS) develops from the lining cells of blood or lymphatic vessels and may appear as red, purple, brown, or black lesions. E-cadherin, CDH1, is a cell adhesion molecule located on the surface of epithelial cells. CDH1 gene expression is downregulated in several cancers and is considered a tumor suppressor gene involved in epithelial-mesenchymal transition in carcinomas. Loss of CDH1 gene expression is observed in many carcinomas, mainly diffuse gastric carcinomas and lobular breast carcinomas, as well as skin tumors. This study investigates the CDH1 germline mutations in HIV-negative (Human Immunodeficiency) Virus KS patients in the Turkish population.
Methods
The study examined 25 peripheral blood mononuclear cells from KS patients using the Sanger sequencing technique.
Results
Sixteen exons of the CDH1 gene were sequenced, and a pathogenic functional germline mutation, HET c.2245C > T, p.(Arg749Trp) rs776975632, NM _004360.5, was identified in a patient with a family history of gastric and breast cancer with a high number of lesions compared to other KS patients. Discussion: KS patients with a family history of cancer could be screened for CDH1 gene and cancer-related-gen variants in the future.
Conclusions
KS is a rare malignancy, and genetic analysis will benefit KS patients. Further studies are needed to describe better the variations detected in a large number of KS patients in this study.
Introduction
Kaposi’s sarcoma (KS) arises from the cells lining the blood and lymph vessels. The tumors (lesions) are usually found on the skin, but can also occur in the mouth, gastrointestinal tract, and respiratory tract. The skin lesion appears as red, purple, brown, and black spots [1].
In 1872, Moritz Kaposi, a Hungarian dermatologist, described KS as a skin lesion with red, purple, brown, and black spots. Human herpesvirus-8 (HHV-8), commonly known as Kaposi’s sarcoma-associated herpesvirus (KSHV), has been shown to cause KS [2]. Unlike other KS variants, Non-HIV (Human Immunodeficiency Virus) related Kaposi’s Sarcoma (NHKS) is a rare disease that usually progresses slowly and is not associated with HIV [3]. KS lesions are most commonly found on the mucosa but can affect any organ or anatomic region [4]. The malignancy of KS starts s1lowly and progresses steadily, worsening abruptly and leading to death [5]. Viruses are responsible for 12% of all human malignancies. The expression of viral oncogenes causes viral oncogenesis in most cases [6]. KS is caused by KSHV infection, as shown in several studies [2]. Because the immune system regulates KSHV infection, it does not usually cause symptoms in healthy individuals. On the other hand, KSHV can cause KS in people with weakened immune systems. In the Turkish population, the prevalence of KSHV is low, and KS usually occurs in HIV-negative people [7]. Currently, KSHV infection is thought to be a necessary but not sufficient condition for the development of KS, whereas genetic, immunologic, and environmental factors are also required for carcinogenesis. One of the most important processes in tumorigenesis and development is the regulation of cell adhesion and motility. In addition to genes involved in cell proliferation and survival, these genes contribute to malignancy [8]. Cadherins are a class of calcium-dependent adhesion proteins belonging to transmembrane group type one and play an important role in cell adhesion.
Classical cadherins, especially E- and N-cadherins, play a crucial role in tissue development during gastrulation, neurulation, and organogenesis [9]. In epithelial tissues, E-cadherin (CDH1) is one of the most important molecules for cell-cell adhesion. It is located on the surfaces of epithelial cells at the adherens junctions, which are areas of cell-cell interaction [10]. Loss of function of the CDH1 gene is associated with cancer metastasis. CDH1 gene down-regulation reduces tissue’s cellular adhesion strength, resulting in increased cellular motility. As a result, cancer cells spread to the surrounding tissues by passing through the basement membrane. CDH1 is considered a tumor suppressor gene due to its property [11, 12]. Various mechanisms such as somatic mutation, downregulation of gene expression, promoter methylation, and transcriptional repression inactivate the activity of CDH1 during tumor progression [13]. Many carcinomas show loss of CDH1 expression as cancer progresses. Hypermethylation of the CDH1 gene promoter has been associated with invasive behaviour in many malignancies, including diffuse gastric, breast, oesophagal, prostate, bladder, kidney, cervical, endometrial, lung, skin, and hepatocellular carcinomas [14]. In Turkey, there are a few studies on KS, and these studies are related to the clinical features of KS. Almost all patients with KS in our country are HIV-negative based only on clinical observations that have not yet been published in the scientific literature. While 23 (23/25) of the 25 Turkish KS patients included in the study had Classic Kaposi Sarcoma (CKS), 2 (2/25) patients had Iatrogenic Kaposi’s sarcoma. All patients were HIV-negative, and no African endemic Kaposi’s sarcoma patients were included in the study. In this study, both viral etiologic factors and CDH1 gene mutations, identified in some skin cancers, are investigated in cases with KS in Turkey. In this study, the prevalence of KSHV infection in KS in Turkey and CDH1 gene mutations, which have not been previously studied in this disease group, are investigated using the Sanger sequencing technique.
Materials and methods
Patient collection
The study was conducted in accordance with the Declaration of Helsinki [15]. The Ethics Committee of Istanbul University accepted the study (Approval No. 1490, September 22, 2011). CDH1 gene mutations were investigated in 25 patients diagnosed with KS who presented to the Oncology Institute of Istanbul University clinics between 2016 and 2020. DNA samples were isolated from the patients’ peripheral blood mononuclear cells, and each exon was amplified with primers specific for the CDH1 gene. Patients’ records were examined for viral etiologic factors, Human immunodeficiency virus (HIV), Kaposi’s sarcoma-associated herpesvirus (KSHV), Hepatitis B surface antigen (HbsAg), hepatitis B core antibody (ANTI-HBc), anticytomegalovirus (ANTI-CMV), Epstein-Barr virus capsid antigen (EBV VCA), human herpesvirus 1 (HHV-1), human herpesvirus 2 (HHV-2), and Varicella-zoster virus (ANTI-VARI ZOS) are listed in Table 1. After the Dye Terminator Cycle Sequencing (DTCS) reaction, the gene regions loaded into the sequence analyzer were analyzed and scored.
The distribution of viral agents in the KS patients.
| Tests | Patients (n:25) | Positivity, % | Healty controls (n:15) | Positivity, % |
|---|---|---|---|---|
| HIV (+) | 0/25 | 0% | 0/15 | 0% |
| KSHV (+) | 13/25 | 52% | 0/15 | 0% |
| HBsAg (+) | 15/25 | 60% | 2/15 | 13.3% |
| ANTI-HBc (+) | 9/25 | 36% | 0/15 | 0% |
| ANTI-CMV (+) | 24/25 | 96% | 8/15 | 53.3% |
| EBV VCA (+) | 22/25 | 88% | 4/15 | 26.6% |
| HHV-1 (+) | 20/25 | 80% | 6/15 | 40% |
| HHV-2 (+) | 2/25 | 8% | 0/15 | 0% |
| ANTI-VARI ZOS (+) | 25/25 | 100% | 9/15 | 60% |
-
HIV, human immunodeficiency virus; KSHV, Kaposi’s sarcoma-associated herpesvirus; HbsAg, hepatitis B surface antigen; ANTI-HBc, hepatitis B core antibody; ANTI-CMV, anticytomegalovirus; EBV VCA, Epstein–Barr virus viral-capsid antigen; HHV-1, human herpesvirus 1; HHV-2, human herpesvirus 2; ANTI-VARI ZOS, varicella zoster virus.
The median age of patients diagnosed with KS was 64 years (46–76 years). 28% (7/25) of patients are female, and 72% (18/25) are male. While 44% (11/25) of the patients smoke, 32% (8/25) use alcohol. 24% (6/25) of patients have diabetes mellitus, hypertension, and chronic heart failure. 8% (2/25) of patients have an iatrogenic subtype, and 92% (23/25) have a classic subtype of KS. One extremity involvement was observed in 48% (12/25) of the patients, 2–5 in 36% (9/25), >5 16% (4/25).
5A family history of cancer was noted in 52% (13/25) of patients. Recurrence was observed in 76% (19/25) of patients. 56% of the patients have patches, 36% have plaque, and 8% have nodule histopathological subtype.
In addition, benign variants were detected in 84% (21/25) of patients, Variant of uncertain significance (VUS) in 12% (3/25), and pathogenic variants in 4% (1/25) of KS patient.
Leukocytes isolation
First, 20 mL of the patients’ peripheral blood mononuclear cells were collected and placed in a tube containing 3 mL of Ficoll solution (Sigma-Aldrich, Darmstadt, Germany). The peripheral blood mononuclear cells were diluted with phosphate-buffered saline (AppliChem, Darmstadt, Germany). Peripheral blood mononuclear cells diluted with phosphate-buffered saline were centrifuged at 1,910 rpm for 30 min at room temperature. Cells were frozen at −80 °C (Sanyo, Moriguchi, Japan) for 24 h before removal and storage in a tank of liquid nitrogen (Thermo Fisher Scientific, Gothenburg, Sweden) for long-term storage.
DNA isolation
DNA isolation was performed according to the protocol of the Quick DNA Kit (Zymo Research, CA, USA). First, lysis buffer (AppliChem, Darmstadt, Germany) was added to leukocytes isolated from peripheral blood. The lysed leukocytes isolated from peripheral blood were transferred to a DNA filter and centrifuged to obtain the DNA adhering to the filter. The DNA filter was washed with wash solutions and centrifuged. A separation solution was added to dissolve the filter adhering to the DNA and washed. Eppendorf tubes were placed under the DNA filters for the final centrifugation. The DNA of peripheral blood mononuclear cells from KS patients was then transferred to tubes and prepared for quality control and further testing.
Agarose gel electrophoresis
1 µl of DNA samples from the patient’s peripheral blood mononuclear cells were mixed with 5 µl of bromophenol blue (Sigma-Aldrich, Darmstadt, Germany) loading buffer and loaded onto a 1.5% agarose gel (Invitrogen, MA, USA). The current was set at 150 V, and the gel was allowed to run for 25 min. The DNA of the isolated peripheral blood mononuclear cells was visualized under UV light (Vilber Lourmat, Marne-la-Vallée Cedex 3, France).
RT-PCR reaction and purification
DNA from peripheral blood mononuclear cells of patients were isolated in a 0.2 mL tube containing 0.5 μLTaq polymerase enzyme (Thermo Fisher Scientific, CA, USA), 0.5 μL dNTP (Thermo Fisher Scientific, CA, USA), MgCl (Thermo Fisher Scientific, CA, USA), 0.5 μL forward and reverse primer (Oligomer, Istanbul, Turkey), 39.5 μL dH2O, 5 μL PCR buffer using a thermal cycler (BioRad T100TM Thermal Cycler, WI, USA) according to the appropriate conditions. The PCR product was stored at −20 °C (Bosch, Turkey) for later use in DTCS. Tubes were placed in a thermal cycler. The following program was set at 98 °C for initial denaturation for 30 s, at 60 °C for denaturing for 30 s, at 72 °C for annealing for 15 s, at 72 °C for extension for 10 min, and holded at 4 °C.
DTCS reaction and purification
The High Pure PCR Product Purification Kit (Sigma-Aldrich, Darmstadt, Germany) was used according to the company’s instructions. The purified PCR products were run on an agarose gel (Sigma Aldrich, Darmstadt, Germany) in the column after purification.
The DTCS reaction was performed with the purified PCR product for 25 ng per kilobase. 8.0 µL of DTCS Quick Start Kit master mix, 2.0 µL of Sequencing Primer 1.6 pmol/μL, and 8.3 µL of dH2O were added as PCR Reaction Content.
PCR product purification and Sanger sequencing
After the DTCS reaction, a second purification was performed. After this purification, agarose gel electrophoresis was performed to verify the presence of PCR products and their suitability for sequencing. PCR products from the isolated peripheral blood mononuclear cells of KS patients were then prepared for sequencing using a Beckman Coulter CEQ 8000 (Beckman Coulter CEQ-8000 Analyzer, CA, USA). For sequence analysis, PCR products were stopped immediately after the DTCS reaction with a 1:2:2 glycogen-sodium-EDTA-sodium acetate mixture (AppliChem, Darmstadt, Germany). Subsequently, 60 µL of 95% cold ethanol (Sigma Aldrich, Darmstadt, Germany) was added to the PCR products and centrifuged for 15 min at 4 °C and 14,000 rpm (VWR MICRO-STAR 17/17R Leuven, Belgium). The supernatant was removed, and 200 µL of 70% cold ethanol (Sigma Aldrich, Darmstadt, Germany) was added to the pellet and centrifuged at 14,000 rpm for 5 min. The alcohol was collected, and the pellet remaining at the bottom of the tube was dried using a vacuum concentrator (CentriVap Benchtop Vacuum Concentrators, Kansas City, USA) to evaporate the alcohol. 40 µL of sample loading solution (SLS) buffer was used to dissolve the dry pellet. The thawed material was added to the instrument on the plate for Beckman Coulter 8000 (GenomeLab™ GeXP, CA, USA) sequence analysis. Beckman Coulter CEQ 8000 Analysis Software was used to analyze the data. The results were evaluated after comparing the analyzed samples with the reference gene sequence.
This study used the genome sequence of Homo sapiens (human) GRCh37 (hg19) from the Genome Reference Consortium as a control.
Reading of sequencing results
The presence of PCR products was detected by agarose gel electrophoresis, and the purified PCR products were analyzed by direct Sanger sequencing. Sanger reads were manually evaluated for all queried variants to emulate techniques typically used in a clinical setting. The Genome Analyzer (Beckman Coulter CEQ-8000 Analyzer, CA, USA) was used to examine the sequence analysis data generated by the capillary electrophoresis instrument and the electropherogram image was validated by comparison with the reference gene sequences. This study used the H. sapiens (human) GRCh37 (hg19) genome assembly from the Genome Reference Consortium as a control. The ALAMUT [16], dbSNP [17], SIFT [18], and ClinVar [19] databases, as well as in silico analyses, were used to investigate the functional effects of variants and to predict the effects of amino acid substitution variations on the human genome in the study.
Statistical analysis
All statistical analyses were performed using the SPSS program (SPSS version 27; SPSS Science, Chicago, IL, USA), with a p-value of 0.05 considered statistically significant. The chi-square, ANOVA, and Fisher-exact tests were used to evaluate statistical significance. Kaplan-Meier analysis was used to determine the survival of KS patients according to mutation subtype.
Results
We performed the Sanger sequencing technique on peripheral blood mononuclear cells from 25 Turkish KS patients. In this study, the distribution of the viral agents was also investigated. HIV, KSHV, HbsAg, ANTI–HBc, ANTI-CMV, EBV VCA, HHV-1, HHV-2, and ANTI-VARI ZOS were obtained from the records of KS patients. Viral agent infection rates of patients are shown in Table 1.
In this study, all patients diagnosed with KS were HIV-negative (there were no AIDS-linked [epidemic] KS patients). The study group consisted of 23 patients with classic KS and two patients with iatrogenic KS; no African endemic KS patients were included in the study. The evaluation of the variant frequencies of the classic and iatrogenic KS patients is shown in Table 2.
Variant frequencies in classical KS patients, and iatrogenic KS patients.
| EX | Sequence variations | Total number of affected classical KS patients, n (%) | Total number of affected iatrogenic KS patients, n (%) | Total number of patients, n |
|---|---|---|---|---|
| Intron 4 | HET c.531 + 10G > C | 1 (4%) | 1 (4%) | 25 |
| Intron 5 | HET c.688-20C > A | 1 (4%) | 0 (0%) | 25 |
| Intron 7 | HET c.1008 + 12G > T | 1 (4%) | 0 | 25 |
| Intron 7 | HET c.1009-16G > A | 0 | 1 (4%) | 25 |
| Intron 8 | HOM c.1137 + 144_ 1137 + 145dupT | 1 (4%) | 0 | 25 |
| Intron 9 | HOM c.1320 + 39C > T | 1 (4%) | 0 | 25 |
| Exon 12 | HET c.1849G > A | 0 | 1 (4%) | 25 |
| Intron 12 | HET c.1937-13T > C | 4 (16%) | 1 (4%) | 25 |
| Intron 13 | HET c.2164 + 17dupA | 2 (8%) | 1 (4%) | 25 |
| Exon 14 | HET c.2245C > T | 1 (4%) | 0 | 25 |
| Exon 14 | HET c.2268T > C | 0 | 1 (4%) | 25 |
| Exon 14 | HET c.2287G > C | 1 (4%) | 0 | 25 |
| Intron 14 | HET c.2296-14T > C | 0 | 1 (4%) | 25 |
| Intron 15 | HET c.2439 + 19A > G | 1 (4%) | 0 | 25 |
| Intron 15 | HET c.2439 + 161dupT | 1 (4%) | 1 (4%) | 25 |
| Exon 16 | HET c.2471C > T | 0 | 1 (4%) | 25 |
Although there are not many studies linking genetic alterations to the pathogenesis of KS, loss of function or inactivation of the CDH1 gene is thought to play a role in tumor metastasis. There is no information in the literature about the efficacy of the CDH1 gene in Turkish KS patients. In this project, we aimed to investigate CDH1 gene mutations in HIV-negative KS cases and explore the importance of CDH1 in the etiology and prognosis of KS. We performed the Sanger sequencing technique on the peripheral blood mononuclear cells of 25 Turkish KS HIV-negative patients.
Sixteen exons of the CDH1 gene were sequenced using the Sanger sequencing technique, and one functional mutation was detected, whereas 15 polymorphisms were detected in 10 exons (Table 3). In these patients, 16 polymorphisms were found as germline variants, including eight benign, four likely benign, and four variants of uncertain significance (Table 3). Of the 16 genotypes, the frequencies of 11 novel intronic, Single Nucleotide Polymorphism (SNP) regions and five exonic variants were detected in KS patients were shown in Table 3. While the role of the CDH1 gene in KS was clarified in this study, the cases of KS in Turkey were also evaluated in terms of viral etiologic variables providing insight into the situation in Turkey. In other societies, 20–100% of people with KS have HIV and HHV-2 viral infections, these viral infections are uncommon in Turkish patients. In Turkish society, EBV virüs infection is found in 88% of KS patients, while KSHV infection is compatible with European society with a rate of 52%. According to the results of our study, the average age at which KS occurs in our society is 64 (46–76) years, while according to Globocan 2018 data, the average age of onset in other societies is 54 years. EBV virus infection and KSHV is considered more efficient in the Turkish population’s etiology of KS.
The variations analysis of CDH1 gene.
| Number of the patients | EX | cDNA | Protein | dbSNP | G1000 | ExAC | GnomAD | Clinical significance | HGMD | PolyPhen-2 | SIFT | Mutation taster |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 patients | Intron 4 | HET c.531 + 10G > C | p.? | rs33963999 | 0.0180 | 0.038282 | 0.039074 | Benign | N.R. | N.A. | N.A. | N.A. |
| 1 patient | Intron 5 | HET c.688-20C > A | p.? | rs1064795881 | N.A. | N.A. | N.A. | Uncertain-significance | N.R. | N.A. | N.A. | N.A. |
| 1 patient | Intron 7 | HET c.1008 + 12G > T | p.? | rs756154578 | N.A. | 0.000017 | 0.000007 | Likely-benign | N.R. | N.A. | N.A. | N.A. |
| 2 patients | Intron 7 | HET c.1009-16G > A | p.? | rs1057521282 | N.A. | N.A. | 0.000021 | Likely-benign | N.R. | N.A. | N.A. | N.A. |
| 1 patient | Intron 8 | HOM c.1137 + 144_ 1137 + 145dupT | p.? | N.A. | N.A. | N.A. | Benign | N.R. | N.A. | N.A. | N.A. | |
| 1 patient | Intron 9 | HOM c.1320 + 39C > T | p.? | N.A. | N.A. | N.A. | Benign | N.R. | N.A. | N.A. | N.A. | |
| 1 patient | Exon 12 | HET c.1849G > A | p. (Ala617Thr) | rs33935154 | 0.0144 | 0.004225 | 0.014120 | Uncertain-significance | DM? | 0.21 | 0.82 | 0.71 |
| Tolerated | Tolerated | Might be affected | ||||||||||
| 5 patients | Intron 12 | HET c.1937-13T > C | p.? | rs2276330 | 0.0579 | 0.104767 | 0.096345 | Benign | Altered splicing | N.A. | N.A. | N.A. |
| 3 patients | Intron 13 | HET c.2164 + 17dupA | p.? | rs1370519975 | 0.0459 | 0.045231 | 0.028983 | Benign | N.R. | N.A. | N.A. | N.A. |
| 1 patient | Exon 14 | HET c.2245C > T | p. (Arg749Trp) | rs776975632 | N.A. | 0.000008 | N.A. | Uncertain-significance | DM | N.A. | N.A. | 0.97 |
| likely pathogenic | ||||||||||||
| 1 patient | Exon 14 | HET c.2268T > C | p. (Asp756=) | rs1567515480 | N.A. | N.A. | 0.000007 | Likely benign | N.R. | N.A. | N.A. | 0.76 |
| Might be affected | ||||||||||||
| 1 patient | Exon 14 | HET c.2287G > C | p. (Glu763Gln) | N.A. | N.A. | N.A. | Benign | N.R. | N.A. | N.A. | 0.58 | |
| Might be affected | ||||||||||||
| 1 patient | Intron 14 | HET c.2296-14T > C | p.? | rs755950355 | N.A. | N.A. | N.A. | Likely-benign | N.R. | N.A. | N.A. | N.A. |
| 1 patient | Intron 15 | HET c.2439 + 19A > G | p.? | rs1567516245 | N.A. | N.A. | N.A. | Benign | N.R. | N.A. | N.A. | N.A. |
| 2 patients | Intron 15 | HET c.2439 + 161dupT | p.? | rs67262350 | N.A. | N.A. | N.A. | Benign | N.R. | N.A. | N.A. | N.A. |
| 1 patient | Exon 16 | HET c.2471C > T | p. (Ala824Val) | rs1450920019 | N.A. | N.A. | 0.000004 | Uncertain-significance | N.R. | N.A. | N.A. | 0.68 |
| Might be affected |
-
EX, exon; G1000, 1000 Genomes Project; ExAC, The Exome Aggregation Consortium; Gnomad, Genome Aggregation Database; HGMD, The Human Gene Mutation Database; PolyPhen-2, Polymorphism Phenotyping v2; SIFT, The Sorting Intolerant from Tolerant; N.A., not applied; N.R., not reported; DM, disease mutation.
Of 16 polymorphisms, two homozygous variants, HOM c.1137 + 144_ 1137 + 145dupT and HOM c.1320 + 39C > T were detected in two CKS patients. The demographic, clinical, and pathologic characteristics of KS patients are shown in Table 4. Statistical evaluation of clinical parameters and mutation types revealed no significant difference between age, alcohol, subtype, recurrence, extremities involvement, and a number of nodules. According to the F- ANOVA test performed with a 95% confidence interval, statistical significance was found between benign and VUS variants depending on gender (p=0.031). A higher level of significance was found between benign and VUS variants compared to smokers (p<0.001). Benign and pathogenic variant’s statistical significance was determined using additional disease status (p=0.014). A high significance level was found between benign and VUS according to family history (p<0.001). Significance between benign and pathogenic was determined based on histopathological stage (p=0.006). According to the Kaplan-Meier survival analysis, the expected survival was 5 years in the benign group, 4 years in VUS, and the survival of patient with pathogenic variant was 1 year shown in Figure 1.
Demographic, clinical, and pathologic characteristics of KS patients.
| n=25 | n (%) | p-Value |
|---|---|---|
| Gender | p=0.031 | |
| Female | 7 (28%) | |
| Male | 18 (72%) | |
| Age | 64 (46–76) years | p>0.005 |
| Duration of follow-up, years | 8 years | p>0.005 |
| Smoking | 11 (44%) | p<0.001 |
| Alcohol use | 8 (32%) | p>0.005 |
| Additional systemic disease | p=0.014 | |
| Diabetes mellitus | 5 (20%) | |
| Hypertension | 9 (36) | |
| Chronic heart failure | 2 (8%) | |
| Number of involved extremities | p>0.005 | |
| 1 | 12 (48%) | |
| 2–5 | 9 (36%) | |
| >5 | 4 (16%) | |
| Histopathological stage | p=0.006 | |
| Patch | 14 (56%) | |
| Plaque | 9 (36%) | |
| Nodule | 2 (8%) | |
| Number of lesions | p>0.005 | |
| 1 | 6 (24%) | |
| 2–10 | 14 (56%) | |
| >10 | 5 (20%) | |
| Family history | 13 (52%) | p<0.001 |
| Yes/No | 11 (44%) | |
| Secondary malignancy | 2 (8%) | p>0.005 |
| Yes/No | 23 (92%) |
-
n, number of the patients.
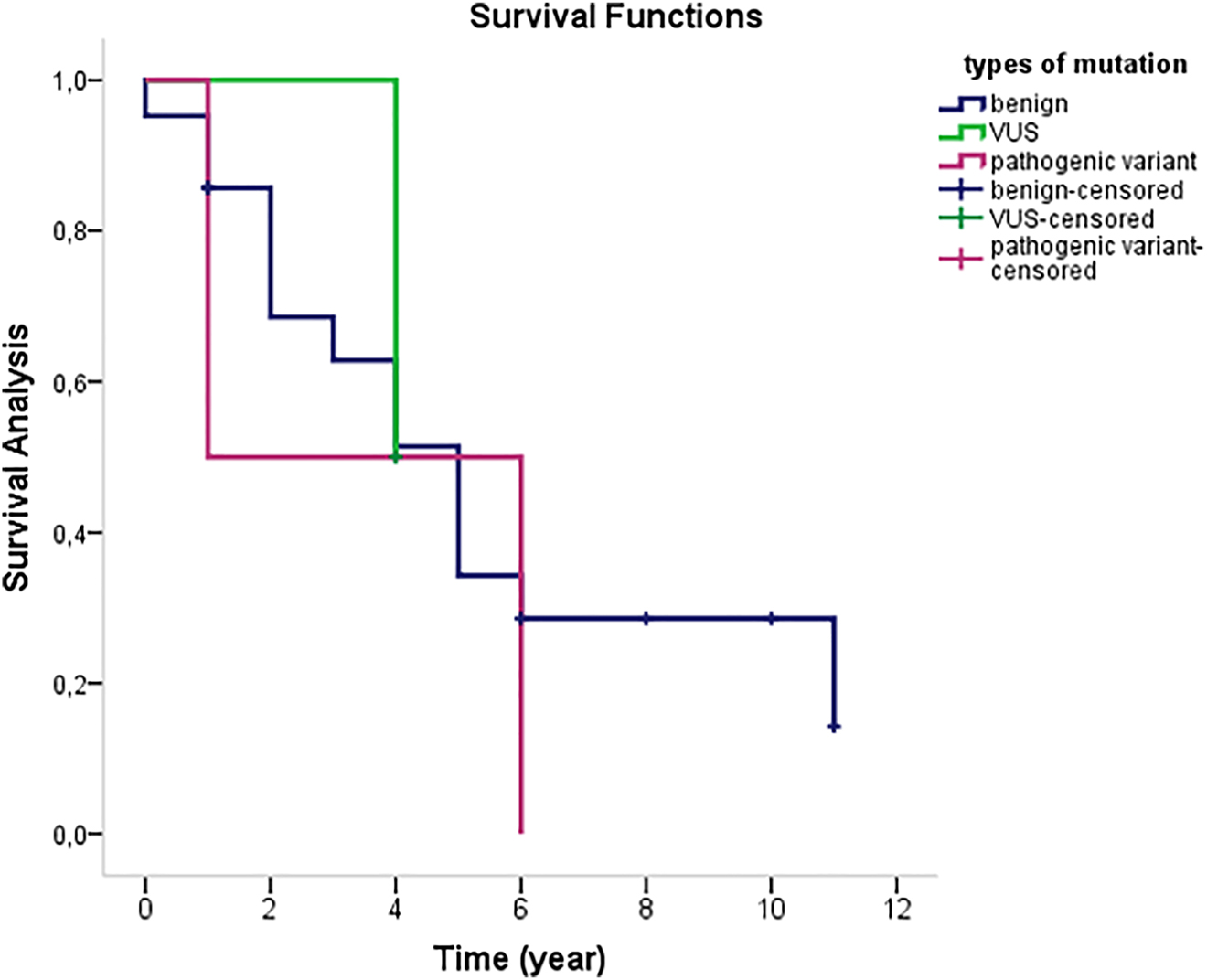
Kaplan-Meier survival analysis for the expected survival according to types of mutation.
Discussion
KS is very rare but common worldwide, with 34,270 cases and 15,086 deaths in 2020 [20] Currently, there are four clinical subtypes of the disease, Classic KS,Endemic KS, Iatrogenic KS, and Epidemic KS (AIDS-related) [15]. The variant frequencies from peripheral blood mononuclear cells of the 25 Turkish patients with classic KS and iatrogenic KS were analyzed for the first time by Sanger sequencing for the CDH1 gene. The CDH1 gene encodes the cell-cell adhesion protein, E-cadherin, whose intracellular domain binds various catenins and functions as a tumor suppressor [16]. CDH1 deficiency accelerates cancer progression through increased proliferation, invasion, and/or metastasis and expression of CDH1 is frequently inactivated or absent in a variety of epithelial tumors [17]. During tumor metastasis, CDH1 gene function is inactivated by several mechanisms, including somatic mutation, downregulation of gene expression, promoter methylation, and transcriptional suppression [13]. Although the loss of CDH1 gene expression is seen in many carcinomas, inactivating mutations of the gene are commonly observed in diffuse gastric carcinomas and lobular breast carcinomas [18]. Hypermethylation of the promoter of the CDH1 gene is associated with invasive behaviour in certain subtypes of skin cancer. While CDH1 germline mutations have previously been associated with certain forms of epithelial carcinomas, such as diffuse gastric cancer [19] and invasive lobular breast cancer [21] screening for pathogenic CDH1 variations may be beneficial in KS patients.
In silico analyses of the missense variants
A total of 16 variants of the CDH1 gene were identified: HET c.531 + 10G > C, HET c.688-20C > A, HET c.1008 + 12G > T, HOM c.100916G > A, HET c.1137 + 144_1137 + 145dupT, HOM c.1320 + 39C > T, HET c.1849G > A, HET c.193713T > C, HET c.2164 + 17dupA, HET c.2245C > T, HET c.2268T > C, HET c.2287G > C, HET c.2296-14T > C, HET c.2439 + 19A > G, HET c.2439 + 161dupT, HET c.2471C > T. In silico analyses of these variants using five different prediction tools have shown conflicting results. We searched the Human Genome Mutation Database (HGMD), ALAMUT, Single Nucleotide Polymorphism Database (dbSNP), Sorting Intolerant From Tolerant (SIFT), and Clinically observed variants (ClinVar) databases, which classified variants as benign, likely benign, or uncertain significance [1], [2], [3], [4, 7]. The pathogenic mutation HET c.2245C > T, p.(Arg749Trp) rs776975632 NM _004360.5 was found in a KS patient with a family history of gastric and breast cancer (patient ID: 25). HGMD considered HET c.2245C > T, p.(Arg749Trp) rs776975632 NM _004360.5 as a potential disease-causing mutation. In a previous study by Kaurah and colleagues, the missense mutation HET c.2245C > T, p.(Arg749Trp) rs776975632 NM _004360.5 was predicted to be a pathogenic variant with the prediction software SIFT [3]. Moreover, and they proposed this variant as a novel pathogenic variant with a family history of gastric cancer in a Colombian family [22]. Aggregation and invasion analyses were tested in CHO cells to validate the pathogenicity of the novel missense mutation HET c.2245C > T, p.(Arg749Trp) rs776975632 NM _004360.5 by Kaurah and colleagues as these analyses are common approaches to compare the function of epithelial cadherin missense variants with wild-type epithelial cadherin, which increases cell aggregation and prevents cell invasion [23, 24]. In addition, Kaurah et al. described HET c.2245C > T, p.(Arg749Trp) rs776975632 NM _004360.5 as a known CDH1 mutation [22]. In our research group, we also found the CDH1 HET c.2245C > T, p.(Arg749Trp) rs776975632 NM _004360.5 variant in KS patients with a family history of gastric cancer and, and it may have an impact on the initiation and metastasis of KS tumors with a family history of gastric and breast cancer.
In addition, the patient in whom the pathogenic variant HET c.2245C > T, p.(Arg749Trp) rs776975632 NM _004360.5 was detected had five affected extremities and a high number of lesions compared with other KS patients. Because KS is an epithelial lesion resembling gastric cancer and the CDH1 gene has been associated with gastric malignancies, the CDH1 gene was investigated in KS patients. Consistent with the literature, pathogenic variant (HET c.2245C > T, p.(Arg749Trp) rs776975632 NM _004360.5) was detected in one of the 25 KS patients with a family history of cancer in the study [22]. Since patient with this pathogenic mutation in the CDH1 gene had more active lesions and a more aggressive disease course KS patients should have germline test of CDH1.
Previously, the missense mutation HET c.1849G > A, p. (Ala617Thr) rs33935154 was discovered as a somatic pathogenic variation in endometrial cancer and identified as a pathogenic germline mutation in a patient with diffuse gastric cancer [25, 26]. In our study, the variation HET c.1849G > A, p. (Ala617Thr) rs33935154 was detected in one patient. This variation affects a conserved region encoding one of the calcium-binding motifs in the extracellular part of CDH1. Since the presence of calcium ions stabilizes the active form of the protein, these calcium-binding motifs are functionally important. Because of their position, this mutation has been associated with developing an unstable intercellular protein complex. Suriano et al. discovered this germline mutation in two African American female patients with diffuse gastric cancer who were 43 years old at the time [24]. In a study by Valente and colleagues, the identical germline variant was found in 6% (10/165) of African American patients with ductal or mixed breast cancer [27]. Here in this study, the variant HET c.1849G > A p. (Ala617Thr) rs33935154 was detected in a patient with a family history cancer, and this variant may be investigated in terms of pathogenicity in the etiology of KS in future studies of KS patients.
On the other hand, immunosuppression, HHV-8, advanced age, diabetes, and corticosteroid treatment have been reported as the most important causes of KS. In this study, a KS patient with a variant of HET c.1849G > A, rs33935154, was also found to have diabetes and hypertension in his late 60 s.
In the present study, five exonic variants (HET c.1849G > A rs33935154, HET c.2245C > T rs776975632, HET c.2268T > C rs1567515480, HET c.2287G > C, HET c.2471C > T rs1450920019) were detected in KS patients. These patients, in whom exonic variants were detected, had more plaque lesions, which are a sign of early-stage KS [25]. Patients with the exonic variant HET c.2245C > T, p.(Arg749Trp) rs776975632 had a higher number of affected extremities and increased number of lesions compared with patients with intronic variants.
NHKS is a multifocal vascular endothelial carcinoma of KS that is quite rare and not associated with HIV, as in all patients studied in this study. We reported the CDH1 variants in the etiology and prognosis of KS in only 25 HIV-negative KS cases. Blood DNA testing has its limitations. However, basal transcription of the genome is known to occur at low levels, and blood has been found to contain approximately 80% of all human coding sequences. In addition, this study was modest in scope and relied on secondary data from clinical diagnostic laboratories. Further studies are needed to better characterise the variants detected in this study in many patients with KS.
Secondary primary malignancies (SPMs) are also a severe problem for people with KS. SPMs can occur in KS patients up to 15 years after the diagnosis of CKS. However, the guidelines have not yet established the mechanism for detecting SPMs [26]. Maruyama et al. reported that more than 280 patients with KS had solid malignancy related to head and neck, esophagus, stomach, duodenum, or colorectal cancer. As SPMs are crucial complications in patients with KS, these malignancies should be detected as early as possible [27]. We believe that life-threatening SPM in patients with CKS should be screened by genetic analysis at early stage.
It has been shown that in patients who are considered immunocompromised, inherited genetic mutations may also be the cause of elderly diseases, such as KS. Since genetic disorders may cause disease in immunosuppressed patients similar to KS, patients should be screened in this direction. More comprehensive genetic screening is recommended for individuals with HIV-independent KS with a family history of cancer. In addition, further large-scale studies, and particularly multicenter cohort studies, will provide further insight into the role of CDH1 in carcinogenesis and in KS patients with a family history of cancer.
-
Research ethics: The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Committee of Istanbul Medical Faculty at Istanbul University (Ethical Approval No:1490, dated September 22, 2011).
-
Informed consent: Informed consent was obtained from all individuals included in this study.
-
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
-
Competing interests: Authors state no conflict of interest.
-
Research funding: None declared.
-
Data availability: The data sets generated and/or analyzed in the current study are available on request from the corresponding author.
References
1. Antman, K, Chang, Y. Kaposi’s sarcoma. N Engl J Med 2000;342:1027–38, https://doi.org/10.1056/nejm200004063421407.Search in Google Scholar
2. Chang, Y, Cesarman, E, Pessin, MS, Lee, F, Culpepper, J, Knowles, DM, et al.. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994;266:1865–9, https://doi.org/10.1126/science.7997879.Search in Google Scholar PubMed
3. Tappero, JW, Conant, MA, Wolfe, SF, Berger, TG. Kaposi’s sarcoma. Epidemiology, pathogenesis, histology, clinical spectrum, staging criteria and therapy. J Am Acad Dermatol 1993;28:371–95, https://doi.org/10.1016/0190-9622(93)70057-z.Search in Google Scholar PubMed
4. Radu, O, Pantanowitz, L. Kaposi sarcoma. Arch Pathol Lab Med 2013;137:289–94, https://doi.org/10.5858/arpa.2012-0101-rs.Search in Google Scholar PubMed
5. Dezube, BJ. Clinical presentation and natural history of AIDS--related Kaposi’s sarcoma. Hematol Oncol Clin N Am 1996;10:1023–9, https://doi.org/10.1016/s0889-8588(05)70382-8.Search in Google Scholar PubMed
6. Morrison, K, Manzano, M, Chung, K, Schipma, MJ, Bartom, ET, Gottwein, E. The oncogenic Kaposi’s sarcoma-associated herpesvirus encodes a mimic of the tumor-suppressive miR-15/16 miRNA family. Cell Rep 2019;29:2961–9, https://doi.org/10.1016/j.celrep.2019.11.005.Search in Google Scholar PubMed PubMed Central
7. Sen, F, Tambas, M, Ciftci, R, Toz, B, Kilic, L, Bozbey, HU, et al.. Factors affecting progression-free survival in non-HIV-related Kaposi sarcoma. J Dermatol Treat 2016;27:275–7, https://doi.org/10.3109/09546634.2015.1094177.Search in Google Scholar PubMed
8. Pecina-Slaus, N. Tumor suppressor gene E-cadherin and its role in normal and malignant cells. Cancer Cell Int 2003;3:17.10.1186/1475-2867-3-17Search in Google Scholar PubMed PubMed Central
9. Barth, AI, Nathke, IS, Nelson, WJ. Cadherins, catenins and APC protein: interplay between cytoskeletal complexes and signaling pathways. Curr Opin Cell Biol 1997;9:683–90, https://doi.org/10.1016/s0955-0674(97)80122-6.Search in Google Scholar PubMed
10. Gumbiner, BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 1996;84:345–57, https://doi.org/10.1016/s0092-8674(00)81279-9.Search in Google Scholar PubMed
11. Thiery, JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol 2003;15:740–6, https://doi.org/10.1016/j.ceb.2003.10.006.Search in Google Scholar
12. Birchmeier, W, Behrens, J. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim Biophys Acta 1994;1198:11–26, https://doi.org/10.1016/0304-419x(94)90003-5.Search in Google Scholar
13. Peinado, H, Portillo, F, Cano, A. Transcriptional regulation of cadherins during development and carcinogenesis. Int J Dev Biol 2004;48:365–75, https://doi.org/10.1387/ijdb.041794hp.Search in Google Scholar
14. Chiles, MC, Ai, L, Zuo, C, Fan, CY, Smoller, BR. E-cadherin promoter hypermethylation in preneoplastic and neoplastic skin lesions. Mod Pathol 2003;16:1014–8, https://doi.org/10.1097/01.mp.0000089779.35435.9d.Search in Google Scholar
15. Patrikidou, A, Vahtsevanos, K, Charalambidou, M, Valeri, RM, Xirou, P, Antoniades, K. Non-AIDS Kaposi’s sarcoma in the head and neck area. Head Neck 2009;31:260–8, https://doi.org/10.1002/hed.20945.Search in Google Scholar
16. Mendonsa, AM, Na, TY, Gumbiner, BM. E-cadherin in contact inhibition and cancer. Oncogene 2018;37:4769–80, https://doi.org/10.1038/s41388-018-0304-2.Search in Google Scholar
17. Zeng, W, Zhu, J, Shan, L, Han, Z, Aerxiding, P, Quhai, A, et al.. The clinicopathological significance of CDH1 in gastric cancer: a meta-analysis and systematic review. Drug Des Dev Ther 2015;9:2149–57, https://doi.org/10.2147/dddt.s75429.Search in Google Scholar
18. Berx, G, Becker, KF, Hofler, H, Van Roy, F. Mutations of the human E-cadherin (CDH1) gene. Hum Mutat 1998;12:226–37, https://doi.org/10.1002/(sici)1098-1004(1998)12:4<226::aid-humu2>3.0.co;2-d.10.1002/(SICI)1098-1004(1998)12:4<226::AID-HUMU2>3.0.CO;2-DSearch in Google Scholar
19. Guilford, P, Hopkins, J, Harraway, J, Mcleod, M, Mcleod, N, Harawira, P, et al.. E-cadherin germline mutations in familial gastric cancer. Nature 1998;392:402–5, https://doi.org/10.1038/32918.Search in Google Scholar
20. Sung, H, Ferlay, J, Siegel, RL, Laversanne, M, Soerjomataram, I, Jemal, A, et al.. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca – Cancer J Clin 2021;71:209–49, https://doi.org/10.3322/caac.21660.Search in Google Scholar
21. Xie, ZM, Li, LS, Laquet, C, Penault-Llorca, F, Uhrhammer, N, Xie, XM, et al.. Germline mutations of the E-cadherin gene in families with inherited invasive lobular breast carcinoma but no diffuse gastric cancer. Cancer 2011;117:3112–7, https://doi.org/10.1002/cncr.25876.Search in Google Scholar PubMed
22. Kaurah, P, Macmillan, A, Boyd, N, Senz, J, De Luca, A, Chun, N, et al.. Founder and recurrent CDH1 mutations in families with hereditary diffuse gastric cancer. JAMA 2007;297:2360–72, https://doi.org/10.1001/jama.297.21.2360.Search in Google Scholar PubMed
23. Suriano, G, Oliveira, MJ, Huntsman, D, Mateus, AR, Ferreira, P, Casares, F, et al.. E-cadherin germline missense mutations and cell phenotype: evidence for the independence of cell invasion on the motile capabilities of the cells. Hum Mol Genet 2003;12:3007–16, https://doi.org/10.1093/hmg/ddg316.Search in Google Scholar PubMed
24. Suriano, G, Oliveira, C, Ferreira, P, Machado, JC, Bordin, MC, De Wever, O, et al.. Identification of CDH1 germline missense mutations associated with functional inactivation of the E-cadherin protein in young gastric cancer probands. Hum Mol Genet 2003;12:575–82, https://doi.org/10.1093/hmg/ddg048.Search in Google Scholar PubMed
25. Niedt, GW, Myskowski, PL, Urmacher, C, Niedzwiecki, D, Chapman, D, Krown, SE, et al.. Histologic predictors of survival in acquired immunodeficiency syndrome-associated Kaposi’s sarcoma. Hum Pathol 1992;23:1419–26, https://doi.org/10.1016/0046-8177(92)90063-9.Search in Google Scholar PubMed
26. Mukhtar, F, Ilozumba, M, Utuama, O, Cimenler, O. Change in pattern of secondary cancers after Kaposi sarcoma in the era of antiretroviral therapy. JAMA Oncol 2018;4:48–53, https://doi.org/10.1001/jamaoncol.2017.2395.Search in Google Scholar PubMed PubMed Central
27. Maruyama, N, Okubo, Y, Umikawa, M, Matsuzaki, A, Hokama, A, Hirano, F, et al.. Quadruple multiple primary malignancies: early detection of second primary malignancy by esophagogastroduodenoscopy/colonoscopy is crucial for patients with classic Kaposi’s sarcoma. Diagnostics 2020;10:218, https://doi.org/10.3390/diagnostics10040218.Search in Google Scholar PubMed PubMed Central
© 2022 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Editorial
- A brief history of hematology analyzers and recent advancements: the available testing wealth
- Research Articles
- Correlation between serum 1,25-dihydroxyvitamin D and 25-hydroxyvitamin D in response to analytical procedures; a systematic review and meta-analysis
- Investigation of CDH1 germline mutations in Turkish patients with Kaposi’s sarcoma
- Frequency and pattern of test utilization rate in clinical biochemistry laboratory: two different large hospital examples
- High serum angiopoietin-like protein-4 levels are associated with gestational hypertension and preeclampsia: a case-control study
- Comparison of efficacy and reliability of six commercial COVID-19 diagnostic PCR kits
- Does COVID-19 infection alter serum biochemical and hematological biomarkers in deceased dementia patients?
- Activity of protein C, protein S and antithrombin 3 in COVID-19 patients treated with different modalities of oxygen supplementation
- Effectiveness after immunization with BNT162b2 and Gam-COVID-Vac for SARS-CoV-2 and neutralizing antibody titers in health care workers
- Relationship between methylation pattern of the SYN2 gene and schizophrenia
- Revealing distinct DNA methylation patterns in hepatic carcinoma through high-throughput sequencing
- 4-h mean lactate clearance as a good predictor of adverse outcome in acute cardiogenic pulmonary edema: a pilot study
- Elucidating the role of ZRF1 in monocyte-to-macrophage differentiation, cell proliferation and cell cycle in THP-1 cells
- Inflammatory factors secreted from endothelial cells induced by high glucose impair human retinal pigment epithelial cells
- Influence of TLR4 signaling on cannabidiol’s antitumor effectiveness in lung adenocarcinoma cells
- Renoprotective effect of diacerein in rats with partial unilateral ureteral obstruction model
- Cytotoxic and apoptotic effectiveness of Cypriot honeybee (Apis mellifera cypria) venom on various cancer cells
- Resveratrol modulates signalling to inhibit vascular smooth muscle cell proliferation induced by angiotensin II and high glucose
- Development and production of antibodies against gamma inactivated pathogenic bacterial spores
Articles in the same Issue
- Frontmatter
- Editorial
- A brief history of hematology analyzers and recent advancements: the available testing wealth
- Research Articles
- Correlation between serum 1,25-dihydroxyvitamin D and 25-hydroxyvitamin D in response to analytical procedures; a systematic review and meta-analysis
- Investigation of CDH1 germline mutations in Turkish patients with Kaposi’s sarcoma
- Frequency and pattern of test utilization rate in clinical biochemistry laboratory: two different large hospital examples
- High serum angiopoietin-like protein-4 levels are associated with gestational hypertension and preeclampsia: a case-control study
- Comparison of efficacy and reliability of six commercial COVID-19 diagnostic PCR kits
- Does COVID-19 infection alter serum biochemical and hematological biomarkers in deceased dementia patients?
- Activity of protein C, protein S and antithrombin 3 in COVID-19 patients treated with different modalities of oxygen supplementation
- Effectiveness after immunization with BNT162b2 and Gam-COVID-Vac for SARS-CoV-2 and neutralizing antibody titers in health care workers
- Relationship between methylation pattern of the SYN2 gene and schizophrenia
- Revealing distinct DNA methylation patterns in hepatic carcinoma through high-throughput sequencing
- 4-h mean lactate clearance as a good predictor of adverse outcome in acute cardiogenic pulmonary edema: a pilot study
- Elucidating the role of ZRF1 in monocyte-to-macrophage differentiation, cell proliferation and cell cycle in THP-1 cells
- Inflammatory factors secreted from endothelial cells induced by high glucose impair human retinal pigment epithelial cells
- Influence of TLR4 signaling on cannabidiol’s antitumor effectiveness in lung adenocarcinoma cells
- Renoprotective effect of diacerein in rats with partial unilateral ureteral obstruction model
- Cytotoxic and apoptotic effectiveness of Cypriot honeybee (Apis mellifera cypria) venom on various cancer cells
- Resveratrol modulates signalling to inhibit vascular smooth muscle cell proliferation induced by angiotensin II and high glucose
- Development and production of antibodies against gamma inactivated pathogenic bacterial spores

