Abstract
Objectives
Most researches have shown that neutrophils are closely related to bladder urothelial carcinoma (BLCA), especially its occurrence and development. Although tumor microenvironment (TME) related genes have an impact on prognosis, the role of neutrophil related genes in BLCA adjuvant therapy is not clear.
Methods
We used sample information from The Cancer Genome Atlas and Gene Expression Omnibus (GEO) databases. And we utilized the CIBERSORT algorithm to obtain the tumor immune microenvironment (TIME) landscape and weighted gene co-expression network analysis (WCGNA) to detect neutrophil-related gene modules. We used univariate Cox regression, multivariate Cox regression, and lasso regression analyses to identify genes that have a relationship with BLCA prognosis. Using the median risk score (RS), we classified the cohort into a high-risk group (HRG) and low-risk group (LRG). External validation of RS was performed via GEO data feeds. Prognostic nomograms were constructed with reference to RS and clinically relevant information and validated using calibration curves. We analyzed the latent connections between RS and tumor mutational burden. Finally, the latent associations between risk markers and chemotherapy prognosis were explored using the pRRophetic algorithm.
Results
In this study, 10 TME-related genes with important prognostic value were screened. Then, by deriving the RS, we constructed a prognostic risk prediction nomogram using parameters such as sex, age, TNM stage, clinical stage, and RS. The area under the receiver operating characteristic curve showed that the predictive accuracy of the constructed nomogram was high. We found that using immunotherapy with new immune checkpoint inhibitors (ICIs) was more beneficial for patients in the LRG. In addition, we can learn from the chemotherapeutic drug model that patients with LRG are more sensitive to cisplatin and imatinib.
Conclusions
In short, the prognostic prediction model based on neutrophil-related genes will help to predict the prognosis and guide the precise treatment of BLCA.
Introduction
Tumors can occur in various parts of the urinary and male reproductive systems, the most common of the tumors being bladder cancer, followed by kidney tumors. Annually, there are approximately 500,000 new cases of bladder cancer and 200,000 resultant deaths worldwide, with more than 80,000 new medical cases of the disease and 17,000 deaths annually in the US alone [1]. Bladder cancer is a malignant tumor originating from the urothelium, mostly from the epithelial tissue, and it is mainly classified into non-muscle-invasive and muscle-invasive types. Advanced age, male sex, and smoking all increase the risk of disease. It is more common among middle-aged and older individuals, and its most common clinical manifestation is painless gross hematuria [2]. The biological characteristics of patients with bladder cancer at different stages and grades are significantly different. For the treatment of non-muscle-invasive bladder cancer, bladder-sparing surgery, such as transurethral resection, can be used. Although postoperative intravesical instillation of specific chemotherapeutic drugs or biological immune preparations can reduce the risk of postoperative recurrence, a considerable number of patients may experience postoperative complications [3]. Patients may experience multiple recurrences after surgery, as well as progression of the tumor, with respect to the stage and grade. For bladder tumors that invade the bladder base, or some recurrent high-grade bladder cancers, radical cystectomy is required, followed by intestinal replacement of the bladder or urinary diversion. At the same time, the quality of life of patients after surgery should be considered. Therefore, it has become the direction of researchers’ efforts to develop new treatment methods on the premise of reducing the trauma of patients and improving the prognosis of patients [4].
Clinical staging and pathological staging are the most valuable parameters to judge the prognosis of bladder cancer. Currently, the 8th version of TNM staging method issued by the United International Cancer Center (UICC) is used clinically. However, different patients with the same TNM stage may also have significant differences in prognosis [5]. In recent years, with the emergence of genetic bioinformatics analysis, more and more bladder cancer related gene markers have been used to evaluate the prognosis, such as epithelial mesenchymal transformation related gene markers and chromatin regulator related gene markers [6].Therefore, we consider screening relevant biomarkers from the perspective of relevant gene signals in tumor microenvironment (TME) to predict individual prognosis and drug response, so as to achieve the results of predicting patients’ prognosis and improving prognosis [7].
In recent years, the close relationship between immune cells in the TIME and the biological behavior of tumors has attracted the attention of researchers. From the point of view of immune cells, the development of new methods for predicting the prognosis of cancer will provide new ideas for the diagnosis and treatment of tumor diseases. The immune microenvironment affects the biological behavior of tumor cells through the secretion of related cytokines and recruitment of immune cells. Neutrophil infiltration plays a major role in this process [8]. According to previous reports, there is an association between tumorigenesis and neutrophils [9]. Although neutrophils are the main immune cells that protect the body from microbial infections and eliminate pathogens, according to recent studies, neutrophils have dual roles as tumor suppressors and tumor promoters. It is hypothesized that neutrophils in normal circulation are of the N1 type that can attack cancer cells. However, neutrophils in cancer cells are of the N2 type, which exerts tumor-promoting functions in the TME, and the switch between the two forms is mainly influenced by cytokines and chemokines [10]. Therefore, the role of neutrophils in tumors has become a hot topic in the field of oncology research. A recent study found that neutrophils contribute to tumor initiation and progression by promoting tumor growth and metastasis, stimulating tumor angiogenesis, and mediating immunosuppressive mechanisms [11, 12]. Till date, the factors and mechanisms that regulate neutrophils to produce either tumor-suppressing or tumor-promoting effects are unclear. In addition, the phenotype and heterogeneity of neutrophils in tumorigenesis and tumor progression have not yet been fully explained. Moreover, the relationship between neutrophils, tumor progression and disease treatment guidance has not been clarified. Therefore, how to apply the mechanism of neutrophils in BLCA tumor to tumor treatment and clinical prognosis evaluation has become the focus of this study.
Materials and methods
Data sorting
In our study, the selected transcriptome data included the TCGA-BLCA and GSE31684 groups. The TCGA-BLCA cohort included 433 BLCA samples divided into 19 normal tissue sequencing samples and 414 tumor samples. After screening, 407 are left. We used somatic mutation data from TCGA database for internal validation. We, then selected the GSE31684 cohort from the Gene Expression Omnibus (GEO) database for external validation.
Landscape of infiltrating immune cells
Through CIBERSORT algorithm [13], we analyzed the sample data of TCGA-BLCA cohort patients to reveal the proportion of 22 tumor infiltrating immune cells (TIICs) in each sample.
Weighted gene co-expression network analysis
The purpose of using WGCNA is to help us build a gene relationship network to identify key genes [14]. We obtained the expression data of sample genes through the TCGA-BLCA cohort, and calculated the correlation coefficient between genes through the Person coefficient. Then, using the weighting function and adjacency function to obtain the parameters β to get the ideal soft threshold. The hierarchical clustering tree constructed by using TOM matrix is divided into 13 candidate modules with different colors by dynamic cutting algorithm, each module contains similar expressed genes. Finally, the relationship graph between 13 candidate modules and 22 kinds of immune infiltrating cells was obtained by correlation analysis between modules and phenotypic traits. Because this study is mainly aimed at neutrophils, we extracted the most relevant modules of neutrophils.
Cox and lasso regression analyses
Since the number of genes that affect prognosis screened out in this study is far greater than the study sample size, in order to avoid model distortion caused by multicollinearity among variables, Lasso+Cox survival analysis mode is used in this study [15]. To facilitate our exploration of the prognostic function of neutrophil-related genes, we screened 4,451 genes in the “grey” module. Firstly, we used univariate regression analysis to determine the genes related to survival. Then, we use lasso regression analysis to avoid over-fitting. Finally, the results were analyzed by multivariate regression to screen out 10 neutrophil-related genes that affect the prognosis of BLCA patients.
Validation of the prognostic neutrophil-related signature
To obtain the RS, we calculated the gene expression associated with prognosis and regression analysis coefficients based on TCGA cohort data. The formula used is as follows:
The median RS value is used for grouping, namely HRG and LRG. Kaplan Meier curve was used to compare the prognosis difference between the two groups. Finally, the value of the prognostic coefficient was proved by the ROC curve.
Establishment and verification of the nomogram
Our nomogram survival plot combined RS and clinical variables to predict patient survival at 1, 3, and 5 years. The validity of the model was verified by using a calibration curve.
Gene set enrichment analysis
We used the c2.cp.kegg.v7.4. symbols collection to study the functional annotation using GSEA software.
Correlation between tumor mutation burden and risk score
Somatic mutation data were acquired from TCGA database. Waterfall charts of the HRG and LRG were drawn with the “maftools” package of R. The survival difference of BLCA patients was analyzed according to median TMB and RS.
Correlation of risk score and tumor immune microenvironment
In order to study the potential relationship between RS and TIICs, we utiliazed seven methods to evaluate the immune infiltration. The proportion of stromal cells and immune cells in BLCA was estimated by ESTIMATE algorithm, and the immune score and matrix score were calculated. Add the two scores to get the ESTIMATE score, which predicts tumor content as well as calculates tumor purity in each tumor sample. The correlation between RS and tumor immune microenvironment was studied by Spearman correlation.
Gene set variation analysis(GSVA)
Though the MSigDB database to conduct relevant pathway analysis [16]. We performed GSVA in the GSVA package for distributional pathway activity estimation, in order to facilitate the assessment of relative pathway activity in a single sample.
Prediction of immunotherapeutic effect
Immune checkpoints refer to programmed death receptors and their ligands, which are the key targets that inhibit the function of immune cells. The immunophenotyping score (IPS) was used to assess the efficacy of immune checkpoint inhibitor therapy and determine tumor immunogenicity. The IPS can calculate scores for four immune phenotypes, including antigen presentation, effector, inhibition, and checkpoint, and the combination of scores of the four immune phenotypes is the IPS z-score. The higher the score, the more immunogenic is the sample.
Prediction of chemotherapeutic effect
We established ridge regression model based on tumor cell line chemosensitivity genomics and TCGA gene expression profile to explore RS and chemosensitivity. The half-maximal inhibitory concentrations (IC50s) of four chemotherapeutic drugs (Imatinid, Cislpain, EHT.1864, and lenalidomide) in patients with BLCA were evaluated using a predictive algorithm.
Statistical analysis
The Wilcoxon test and Kruskal Wallis test [17] were used to compare two groups and more than two groups of variables. Survival analysis was performed using the Kaplan–Meier log-rank test. The Chi-square test was used for analysis of the association between RS and TMB, and Spearman analysis was used to calculate the correlation between coefficients. Two-sided p<0.05 was considered to indicate statistical significance. All statistical analyses were performed using R software (version 4.1.1).
Results
Landscape of TIME in BLCA
We calculated the proportion of TIICs in each sample through CIBERSORT algorithm (Attachment 1: Table S1), revealing the landscape of tumor infiltration microenvironment in BLCA. Then, the relationship between TIME and normal tissues and tumors was discussed and described through comprehensive thermography (Figure 1A). In order to further reveal the potential relationship between these immune cells, we tested the correlation between 22 immune cells in the TIME landscape (Figure 1B). It is noteworthy that neutrophils are positively correlated with activated mast cells (r=0.20; p<0.05) and negatively correlated with regulatory T cells (r=−0.14; p<0.05).

Subpopulation of 22 immune cell subtypes and (A) proportional heat mapof 22 TICs in each BLCA sample. (B) Intrinsic correlation of 22 infiltrating immunecell types in BLCA.
Establishment of the WGCNA network
We calculate the parameters β=9 through the weighting function and adjacency function in WCGNA, and there is a good linear relationship between the validation results of R2 when the visible soft threshold is 9 in Supplementary Figure S1A. Then 13 gene modules with different colors were obtained through the TOM matrix algorithm (Supplementary Figure S1B). Each row in Supplementary Figure S1C has 22 TIC types, and each column has candidate modules of the trait carrier gene. Notably, among the 13 candidate modules, the grey module was associated with neutrophils (cor=−0.23, p=1e−06). Our focus in this study was on neutrophils; therefore, we extracted the genes in the grey module (Additional File 1: Table S2) for further studies.
Development of risk signature
Using TCGA-BLCA data, we obtained expression data and follow-up information to facilitate follow-up studies on the prognostic value of the candidate genes. Using univariate Cox analysis, 4,451 neutrophil-related genes were found to have notable prognostic values (p<0.05, Additional File 1: Table S3). In order to get over fitting results, we conducted Lasso regression analysis on 610 genes screened (Figure 2A and B). Then we further determined 10 neutrophil-related genes (TM4SF1, PCED1B, GJD3, ZNF474, APOL2, PXDNL, BAIAP2, BCL2L14, SIGLEC6, and FAM166B) through multivariate Cox regression analysis. And among these central genes, PCED1B, GJD3, APOL2, FAM166B and BCL2L14 are all useful prognostic indicators (all risk ratios [HRs] are <1, Table S4).

(A) Variation curve of regression coefficient with log (λ) in lasso regression. (B) Ten-fold cross-validation for tuning parameter selection in lasso regression. Vertical lines are drawn from the best data according to the minimum criterion and 1 standard error criterion. (C) Kaplan–Meier curve analysis showing the difference in overall survival between high-risk and low-risk groups in TCGA-BLCA cohort. Kaplan–Meier curve analysis shows the difference in overall survival with respect to APOL2, (D) gene and BAIAP2, (E) gene between the high-expression and low-expression groups. (F) Univariate Cox regression results for overall survival. (G) Multivariate Cox regression results for overall survival.
We calculated the risk score of the 10 central genes in patients with BLCA. Calculation of RS was performed as follows:
Based on the median of RS, we divided the sample into high-risk score group (HRG) and low-risk score group (LRG), so as to facilitate the grouping identification of later research.
Validation of risk prognostic features
From the Kaplan-Meier survival curve, LRG is superior to HRG in survival results (Figure 2C). Taking APOL2 and BAIAP2 as examples, it can be seen in Figure 2D that the survival of APOL2 with high expression is better than that with low expression. In Figure 2E, it can be seen that BAIAP2 survives better under the condition of low expression. According to the results of the two charts, the samples were divided into two groups: high expression group (low risk group) and low expression group (high risk group). From the Kaplan-Meier survival curves in Figure 2D and E, it can be seen that APOL2 and BAIAP2 genes have positive reactions to the prognosis of BLCA patients, suggesting that there is a certain relationship between the screening of neutrophil-related genes and the prognosis of the disease, and its risk score is feasible to be used to construct the prognosis model. We carried out the same verification for the other hub genes (Supplementary Figure S2). Univariate Cox analysis showed that the HR of RS was 1.282 (95 % confidence interval [CI], 1.223−1.344; Figure 2F). Multivariate Cox regression analysis results (HR=1.24295 % CI=1.182−1.306; Figure 2G) showed the effectiveness of RS as an independent prognostic indicator of BLCA. These results show that these 10 hub genes have good clinical prediction ability.
The expression pattern, sample survival status distribution and risk score of 10 genes in TCGA-BLCA cohort are shown in Figure 3A–C. In addition, we used the samples in the GSE31684 queue for external verification (Figure 3D–F). These results all support the above point of view. In the two data samples, we can see that the low risk score has a better survival state.

(A) Confirmation of prognostic risk scores in TCGA cohort. (B) Polygenic model risk score distribution in TCGA cohort. (C) Survival status and duration of patients with BLCA in TCGA cohort. (D) Confirmation of prognostic risk scores in the GEO cohort. (E) Polygenic model risk score distribution in the GEO cohort. (F) Survival status and duration of patients with BLCA in the GEO cohort.
Functional analysis of neutrophil-related genes
We identified the functional enrichment of hub gene expression based on GSEA. Through the Kyoto Encyclopedia of Genesand Genomes(KEGG) pathway enrichment analysis, we found that the main pathways involved in APOL2 are graft vs. host disease, antigen processing and presentation, and Leishmania infection (Figure 4A). We performed the same analysis for the other central genes. KEGG pathway enrichment analysis indicated that the overexpression of BAIAP2 interacted with the signal transduction pathway in small cell lung cancer (Figure 4B). High expression of BCL2L14 and GJD3 was related to the metabolism of xenobiotics through the cytochrome P450 signal transduction pathway (Figure 4C and D). High expression of FAM166B was associated with the systemic lupus erythematosus signal transduction pathway (Figure 4E). High expression of PCED1B, SIGLEC6, and TM4SF1 genes was related to the signal transduction pathway of the intestinal immune network for IgA production (Figure 4F), but high expression of SIGLEC6, and TM4SF1 resulted from their co-action with the signal transduction pathways of viral myocarditis and Leishmania infection, respectively (Figure 4G and H). Overexpression of PXDNL was related to the taste signal transduction pathway (Figure 4I), and high expression of ZNF474 resulted from the interaction between taste transduction and focal adhesion (Figure 4J).

GSEA of samples with high and low expression of two hub genes. (A) Gene set of samples enriched in APOL2 expression collected using KEGG. (B) Gene set of samples enriched in BAIAP2 expression collected using KEGG. (C) Gene set of samples enriched in BCL2L14 expression collected using KEGG. (D) Gene set of samples enriched in GJD3 expression collected using KEGG. (E) Gene set of samples enriched in FAM166B expression collected using KEGG. (F) Gene set of samples enriched in PCED1B expression collected using KEGG. (G) Gene set of samples enriched in SIGLEC6 expression collected using KEGG. (H) Gene set of samples enriched in TM4SF1 expression collected using KEGG. (I) Gene set of samples enriched in PXDNL expression collected using KEGG. (J) Gene set of samples enriched in ZNF474 expression collected using KEGG.
Correlation of risk signature with clinicopathological variables
In Figure 5A, we can clearly observe the distribution difference of clinically relevant variables in HRG and LRG. From Figure 5B–G, we can see that LRG presents a more optimistic result than HRG in terms of disease progression and prognosis.

Clinical significance of prognostic risk characteristics. (A) Heat map showing the distribution of clinical characteristics and corresponding risk scores in each sample. Incidence of subtypes of LRG/HRG clinical variables: (B) sex, (C) tumor grade, (D) clinical stage, (E) T stage, (F) N stage, and (G) M stage.
Construction of prognostic nomogram
The ROC curve shows that the area under the curve (AUC) values of 1 year overall survival rate (OS), 3 year OS and 5 year OS are 0.754, 0.776 and 0.796 respectively. These values are sufficient to explain the high prognostic effectiveness of the features analyzed in this study (Figure 6C). In addition, AUC was used to analyze 1 year (Figure 6D), 3 year (Figure 6E), and 5 year OS (Figure 6F) for a combination of risk characteristics, sex, and clinical stage. Finally, we drew a prognostic nomogram (Figure 6A) so that we could more intuitively evaluate the impact of risk characteristics, sex, TNM stage, and clinical stage on OS in patients with BLCA. The validation results from the calibration curve showed that the nomogram was predictive (Figure 6B).
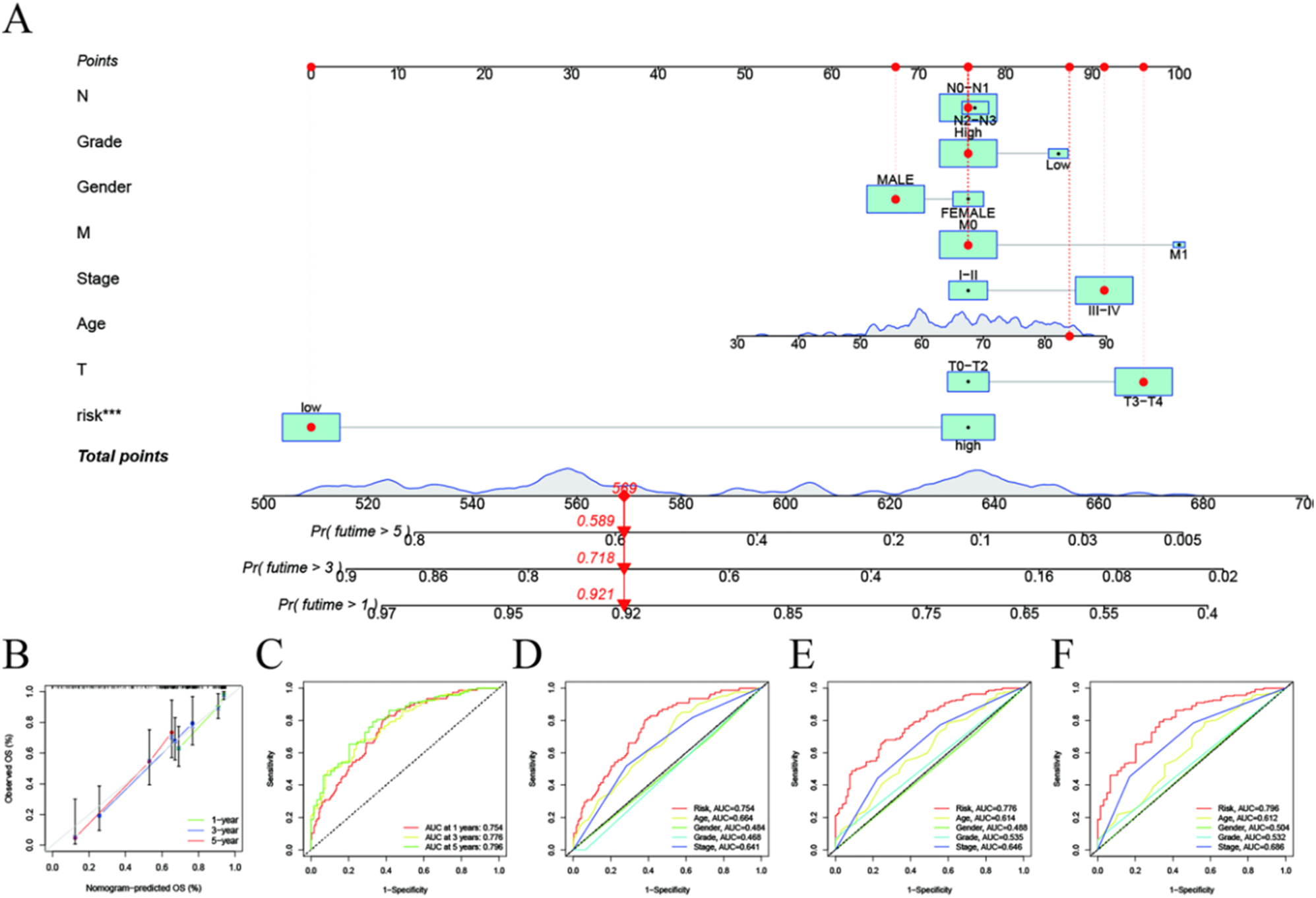
Evaluation of risk signal prediction efficiency. (A) Nomogram used to predict survival in patients with BLCA. (B) 1-, 3-, and 5-year survival nomogram calibration curves. (C) ROC analysis was used to assess the predictive value of prognostic features. The AUC for the risk score was used to predict overall survival at 1, 3, and 5 years and other clinical characteristics (D–F).
Association of risk signature with TMB
Next, we explored the potential associations between TMB and risk characteristics by comparing TMB in samples from the LRG and HRG, and we found that high TMB level is more closely related to LRG (p<0.001, Figure 7A). To facilitate the study of the effect of TMB on the OS of patients with BLCA, we constructed a Kaplan–Meier survival curve, and the grouping of samples from the high- and low-TMB groups in the curve was based on the median TMB value (p<0.001, Figure 7B). Subsequently, we analyzed the correlation between TMB and RS, as shown in Figure 7C, and we found that RS was inversely proportional to TMB. Figure 7D clearly illustrates the interaction between TMB and RS in the prognosis of patients with BLCA.

Correlation between risk score and TMB. (A) Differences in TMB between the HRG and LRG. (B) Scatterplots depicting the positive correlation between risk scores and TMB. (C) Kaplan–Meier curves of the high- and low-TMB groups. (D) Kaplan–Meier curve stratification of patients according to TMB and risk signature. The oncoPrint was constructed using high risk score (E) and low risk score (F). TMB, tumor mutational burden; HRG, high-risk group; LRG, low-risk group.
Subsequently, based on the distribution and type of somatic gene mutations in different RS subpopulations, we created correlation maps of the respective somatic variants of the HRG and LRG (Figure 7E and F). The mutation profiles of significantly mutated genes revealed that TP53 (49 vs. 44 %) had more somatic mutations in the HRG core subtypes, whereas TTN (35 vs. 44 %), KDM6A (19 vs. 25 %), and FGFR3 (7 vs. 21 %) had more somatic mutations in the LRG.
Risk signature in TIME context
Spearman correlations were used to analyze the correlations between the risk signatures of neutrophil-related genes and TIME (Figure 8A, Table S5). According to the analysis results of ESTIMATE score, immune score is higher than stromal score in the LRG. However, the overall ESTIMATE score decreased (p<0.01, Figure 8B). After correlation validation using the four methods—CIBERSORT (Figure 8C), CIBERSORT−ABS (Figure 8D), QUANTISEQ (Figure 8E), and TIMER (Figure 8F)—our analysis was confirmed to be correct.

Estimated abundance of tumor-infiltrating cells. The (A) high-risk group had a stronger correlation with tumor-infiltrating immune cells, as shown by the Spearman correlation analysis. (B) Association between prognostic risk signatures and central immune checkpoint genes. The correlations predicted using the four methods—CIBERSORT (C), CIBERSORT−ABS (D), QUANTISEQ (E), and TIMER (F)—were validated.
Enrichment of signaling pathways in the low/high-risk groups
Through GSVA analysis, we found that RS was negatively correlated with KEGG/PPAR signaling pathway, which indicated that the disease occurrence and development of LRG was related to this pathway. On the contrary, most signal pathways in Figure 9A are in positive proportion to RS.

Enrichment pathways for GSVA. (A) Heat map showing the correlation of representative pathway items in KEGG with risk scores. Predicting immunotherapy response. (B) Association of expression levels of immune checkpoint blockade genes with risk scores. (C–F) IPS distribution map. Estimates of chemotherapeutic effect and risk scores. Sensitivity analysis of (G) EHT.1864 in patients with high and low risk scores. Sensitivity analysis of (H) lenalidomide in patients with high and low risk scores. (I) Sensitivity analysis of cisplatin in patients with high and low risk scores. (J) Sensitivity analysis of Imatini in patients with high and low risk scores.
Prediction of response to immunotherapy
Because of the lack of data on immunotherapy in TCGA-BLCA dataset, we could only use the expression levels of immune checkpoint blockade-related genes to evaluate the prognostic effect of immunotherapy. Moreover, we identified other immune checkpoint blockade-related genes (i.e., TNFRSF8, TNFRSF14, TMIGD2, and CD86) whose expression was significantly proportional to risk characteristics and determined some genes associated with immune checkpoint blockade, such as ICOS, CD27, CD244 and IDO2, whose expression weas inversely associated with risk-related traits (Figure 9B). It can be seen in Figure 9B that GJD3 is negatively correlated with most of the immune checkpoint block-related genes, while FAM166B is positively correlated with most of the immune checkpoint block-related genes, it seems contradictory by comparison, but this is not the case. The influence of neutrophil-related genes on risk score includes the occurrence and development of tumor and the effect of immunotherapy. We can from Figure 9C that the finding suggests that the LRG patients are more inclined to choose new immunotherapy with the ICIs and it can contribute to our understanding of the relationship between immunotherapeutic effects and RS.
Prediction of response to chemotherapy
After data analysis, IC50 value is obtained, which allows us to see the difference of therapeutic sensitivity of four chemotherapy drugs (Cisplatin, Imatini, EHT.1864, and lenalidomide) to HRG and LRG. The HRG was more sensitive to EHT.1864 (Figure 9G) and Lenalidomide (Figure 9H) than the LRG, while the LRG was more sensitive to Cisplatin (Figure 9I) and Imatini (Figure 9J). These results suggest that feasible strategies for treatment can be devised by exploring the relationship between risk characteristics and chemotherapeutic drug sensitivity.
Discussion
Bladder cancer is a common fatal tumor with high incidence and mortality rates. The incidence rate in men is four times higher than that in women, and it is more common in developing countries [18]. At present, the treatment of bladder cancer is mainly surgery and chemotherapy. In general, the prognosis of non muscle invasive bladder cancer is better. For Ta stage bladder cancer, the 10-year survival rate can reach more than 90 %, which means that most patients can survive for a long time [19]. The prognosis of muscle invasive bladder cancer is poor. Because it is easy to metastasize, such patients need platinum based neoadjuvant chemotherapy after surgery. According to previous clinical studies, the overall survival rate improved by the cisplatin based treatment scheme is not obvious, the drug side effects are large, and some patients are also intolerant of cisplatin treatment and other factors, which greatly limit the wide application of this chemotherapy scheme [20]. It can be seen from the immune theory that inhibition of immune checkpoint is one of the reasons for tumor cell immune escape, which has become the focus of current scholars. It is particularly important to find appropriate biomarkers to predict which advanced urothelial carcinoma patients are most suitable for immunocheckpoint block therapy [21, 22].
In this paper, we obtained two different cohorts from a database: TCGA-BLCA cohort for developing risk signatures and GSE31684 cohort for external validation. We investigated the role of neutrophils in the development and clinical prognosis of BLCA, using 409 tumor samples and 16,394 genes. The abundance of 22 TICs subtypes was determined using the CIBERSORT algorithm. We used WGCNA to discover neutrophil-related gene modules (grey), of which 4,451 candidate genes were correlated with neutrophil-related genes.
Obtaining relevant clinical and sequencing data from TCGA-BLCA database helped us to better evaluate the prognostic value of the hub genes for tumor treatment. Thereafter, we performed univariate, lasso, and multivariate Cox analyses to screen for 10 neutrophil-related genes; calculated the Cox HR for each hub gene as a coefficient of RS; and then classified the samples into HRG and LRG groups, using the median RS. We validated the prognostic profile of the risk model using Kaplan–Meier analysis and ROC curves, and we concluded that the risk profile could serve as a qualified independent prognostic predictor in both univariate and multivariate regression analyses. We also validated the data using external datasets (the GSE31684 cohort). From these results, it can be seen that the risk model based on the 10 genes in the study (TM4SF1, PCED1B, GJD3, ZNF474, APOL2, PXDNL, BAIAP2, BCL2L14, SIGLEC6, and FAM166B) can be used alone as a predictor of clinical outcomes in patients with BLCA. To better determine the relationship between the risk status of BLCA and clinical outcomes, we referenced risk characteristics and individual clinical variables to constructed prognostic nomograms to facilitate the evaluation of 1, 3, and 5 year survival rates of patients with BLCA, which were recorded and verified with calibration curves. Furthermore, we used KEGG to determine the enriched signaling pathways, which was facilitated the study of the association between these 10 hub genes and signaling pathways.
Immunotherapy plays a powerful role in the treatment of genetic mutations. We obtained somatic mutation data from TCGA database and classified the samples into high- and low-TMB groups according to the median TMB. We then assessed the risk characteristics and impact of TMB on clinical prognosis in BLCA. To further analyze the relationship between TMB and risk characteristics, we compared the survival of the two groups. It also shows that TMB can be used to assess disease progression and clinical prognosis. Because when somatic mutations occur in tumors, new antigens also appear, which provides new antigen responses for T cells to recognize, making ICB therapy more sensitive. Some studies have demonstrated the correlation between high TMB and anti PD-1 in non-small cell lung cancer and anti CTLA-4 in melanoma. Some studies have also shown that in bladder cancer, the objective response rates (ORRs) of high TMB and ICB treatments are significantly higher than those of low TMB tumors. These results can show the reliability of TMB as a single risk signal predictor.
Currently, platinum-based chemotherapy combined with neoadjuvant chemotherapy and surgical intervention are the mainstay of treatment for muscle-invasive bladder cancer [23]. In this study, the sensitivity of BLCA in the LRG to cisplatin was significantly higher than that in the HRG, and for the LRG, sensitivity to EHT.1864 and lenalidomide was higher than that of the HRG. In summary, RS can be combined in order to make better decisions with respect to the use of chemotherapeutic drugs for BLCA.
In this paper, a prognostic model combining 10 neutrophil related genes (TM4SF1, PCED1B, GJD3, ZNF474, APOL2, PXDNL, BAIAP2, BCL2L14, SIGLEC6, and FAM166B) was constructed, which accurately predicted the survival probability of patients with BLCA. Many studies have been conducted previously based on these genes. Cao et al. proved that the overexpression of TM4SF1 is associated with the progression of muscle invasive bladder cancer, and is proportional to the prognosis of the disease. TM4SF1 is also overexpressed in many tumor diseases, such as colorectal cancer [24], prostate cancer [25] and esophageal cancer [26]. Existing literature show that the remaining genes are also associated with other cancers: for example, low expression of PCED1B is a marker of poor prognosis in multiple myeloma [27]; PXDNL is closely associated with breast cancer progression [28]. SIGLEC6 is a novel target of CAR-T cells in acute myeloid leukemia [29] and BCL2L14 is associated with endometrial and breast cancer [30]. Furthermore, mutations in APOL2 can cause alcohol-use disorders [31]. Rute M.S.M. Pedrosa et al. found that GJD3 is associated with breast cancer metastasis to the brain, and it may have a specific function and potential value as a biomarker for brain metastasis of breast cancer [32]. However, FAM166B and ZNF474 have not yet been clearly associated with cancer. In this study, we elucidated the effects of 10 TME-related genes (TM4SF1, PCED1B, GJD3, ZNF474, APOL2, PXDNL, BAIAP2, BCL2L14, SIGLEC6, and FAM166B) on TIME, TME, and clinical prognosis. It also showed that the expression of these genes was related to the prognosis of BLCA. However, there are still many molecular mechanisms that require further investigation.
Conclusions
This study has shown that neutrophil-based risk scores play important roles in predicting prognosis, TIME heterogeneity, TMB, and treatment response assessment in BLCA. The roles of 10 neutrophil-related genes in BLCA were also explored. However, these findings need to be verified by collecting richer cohort data and conducting related experimental studies.
-
Research funding: This study was supported by National Key R&D Program of China (2021YFA0911600), Shenzhen Science and Technology Program (JCYJ20220818102001002), Municipal Science and Technology Innovation Commission Project in 2020(JCYJ20190806164616292), Sanming Project of Medicine in Shenzhen (No.SZSM202111007), Shenzhen Key Medical Discipline Construction Fund (No.SZXK020) and the Shenzhen High-level Hospital Construction Fund.
-
Author contributions: The authors confirm contribution to the paper as follows: study conception and design: Yanfeng Li,Ying Dong, Yuchen Liu and Hongbing Mei; data collection: Chaojie Xu and Ganglin Su; analysis and interpretation of results: Yanfeng Li, Ying Dong, Chaojie Xu and Ganglin Su ; draft manuscript preparation: Yanfeng Li and Chaojie Xu. All authors reviewed the results and approved the final version of the manuscript.
-
Conflicts of interest: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
-
Ethics approval: Not applicable.
-
Availability of data and materials: The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
References
1. Lenis, AT, Lec, PM, Chamie, K, Mshs, MD. Bladder cancer: a review. JAMA 2020;324:1980–91. https://doi.org/10.1001/jama.2020.17598.Search in Google Scholar PubMed
2. Grayson, M. Bladder cancer. Nature 2017;551:S33. https://doi.org/10.1038/551s33a.Search in Google Scholar PubMed
3. Witjes, JA, Bruins, HM, Cathomas, R, Comperat, EM, Cowan, NC, Gakis, G, et al.. European association of urology guidelines on muscle-invasive and metastatic bladder cancer: summary of the 2020 guidelines. Eur Urol 2021;79:82–104. https://doi.org/10.1016/j.eururo.2020.03.055.Search in Google Scholar PubMed
4. Bin, RI, Khan, AM, Catto, JW, Hussain, SA. Bladder cancer: shedding light on the most promising investigational drugs in clinical trials. Expert Opin Investig Drugs 2021;30:837–55. https://doi.org/10.1080/13543784.2021.1948999.Search in Google Scholar PubMed
5. Ni, L, Tang, C, Wang, Y, Wan, J, Charles, MG, Zhang, Z, et al.. Construction of a miRNA-based nomogram model to predict the prognosis of endometrial cancer. J Personalized Med 2022;12:1154. https://doi.org/10.3390/jpm12071154.Search in Google Scholar PubMed PubMed Central
6. Shi, W, Li, C, Wartmann, T, Kahlert, C, Du, R, Perrakis, A, et al.. Sensory ion channel candidates inform on the clinical course of pancreatic cancer and present potential targets for repurposing of FDA-approved agents. J Personalized Med 2022;12:478. https://doi.org/10.3390/jpm12030478.Search in Google Scholar PubMed PubMed Central
7. Xu, C, Song, L, Peng, H, Yang, Y, Liu, Y, Pei, D, et al.. Clinical eosinophil-associated genes can serve as a reliable predictor of bladder urothelial cancer. Front Mol Biosci 2022;9:963455. https://doi.org/10.3389/fmolb.2022.963455.Search in Google Scholar PubMed PubMed Central
8. Chen, H, Pan, Y, Jin, X, Chen, G. An immune cell infiltration-related gene signature predicts prognosis for bladder cancer. Sci Rep 2021;11:16679. https://doi.org/10.1038/s41598-021-96373-w.Search in Google Scholar PubMed PubMed Central
9. Que, H, Fu, Q, Lan, T, Tian, X, Wei, X. Tumor-associated neutrophils and neutrophil-targeted cancer therapies. Biochim Biophys Acta Rev Cancer. 2022;1877:188762.10.1016/j.bbcan.2022.188762Search in Google Scholar PubMed
10. Che, K, Han, W, Zhang, M, Niu, H. Role of neutrophil gelatinase-associated lipocalin in renal cell carcinoma. Oncol Lett 2021;21:148. https://doi.org/10.3892/ol.2020.12409.Search in Google Scholar PubMed PubMed Central
11. Ibrahim, SA, Kulshrestha, A, Katara, GK, Riehl, V, Sahoo, M, Beaman, KD. Cancer-associated V-ATPase induces delayed apoptosis of protumorigenic neutrophils. Mol Oncol 2020;14:590–610. https://doi.org/10.1002/1878-0261.12630.Search in Google Scholar PubMed PubMed Central
12. Zhou, SL, Zhou, ZJ, Hu, ZQ, Huang, XW, Wang, Z, Chen, EB, et al.. Tumor-associated neutrophils recruit macrophages and T-regulatory cells to promote progression of hepatocellular carcinoma and resistance to sorafenib. Gastroenterology 2016;150:1646–58. https://doi.org/10.1053/j.gastro.2016.02.040.Search in Google Scholar PubMed
13. Chen, B, Khodadoust, MS, Liu, CL, Newman, AM, Alizadeh, AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 2018;1711:243–59. https://doi.org/10.1007/978-1-4939-7493-1_12.Search in Google Scholar PubMed PubMed Central
14. Langfelder, P, Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform 2008;9:559. https://doi.org/10.1186/1471-2105-9-559.Search in Google Scholar PubMed PubMed Central
15. Pak, K, Oh, SO, Goh, TS, Heo, HJ, Han, ME, Jeong, DC, et al.. A user-friendly, web-based integrative tool (ESurv) for survival analysis: development and validation study. J Med Internet Res 2020;22:e16084. https://doi.org/10.2196/16084.Search in Google Scholar PubMed PubMed Central
16. Chan, TA, Yarchoan, M, Jaffee, E, Swanton, C, Quezada, SA, Stenzinger, A, et al.. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol 2019;30:44–56. https://doi.org/10.1093/annonc/mdy495.Search in Google Scholar PubMed PubMed Central
17. Kul, E, Bayindir, F, Gul, P, Yesildal, R, Matori, KA. Effect of coating on the color and surface hardness of the surface of dental ceramics. Dent Res J 2021;18:18. https://doi.org/10.4103/1735-3327.311425.Search in Google Scholar
18. Balar, AV, Kamat, AM, Kulkarni, GS, Uchio, EM, Boormans, JL, Roumiguie, M, et al.. Pembrolizumab monotherapy for the treatment of high-risk non-muscle-invasive bladder cancer unresponsive to BCG (KEYNOTE-057): an open-label, single-arm, multicentre, phase 2 study. Lancet Oncol 2021;22:919–30. https://doi.org/10.1016/s1470-2045(21)00147-9.Search in Google Scholar
19. Magers, MJ, Lopez-Beltran, A, Montironi, R, Williamson, SR, Kaimakliotis, HZ, Cheng, L. Staging of bladder cancer. Histopathology 2019;74:112–34. https://doi.org/10.1111/his.13734.Search in Google Scholar PubMed
20. Patel, VG, Oh, WK, Galsky, MD. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J Clin 2020;70:404–23. https://doi.org/10.3322/caac.21631.Search in Google Scholar PubMed
21. Abbott, M, Ustoyev, Y. Cancer and the immune system: the history and background of immunotherapy. Semin Oncol Nurs 2019;35:150923. https://doi.org/10.1016/j.soncn.2019.08.002.Search in Google Scholar PubMed
22. Zhang, Y, Zhang, Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol 2020;17:807–21. https://doi.org/10.1038/s41423-020-0488-6.Search in Google Scholar PubMed PubMed Central
23. Yin, M, Joshi, M, Meijer, RP, Glantz, M, Holder, S, Harvey, HA, et al.. Neoadjuvant chemotherapy for muscle-invasive bladder cancer: a systematic review and two-step meta-analysis. Oncologist 2016;21:708–15. https://doi.org/10.1634/theoncologist.2015-0440.Search in Google Scholar PubMed PubMed Central
24. Tang, Q, Chen, J, Di, Z, Yuan, W, Zhou, Z, Liu, Z, et al.. TM4SF1 promotes EMT and cancer stemness via the Wnt/beta-catenin/SOX2 pathway in colorectal cancer. J Exp Clin Cancer Res 2020;39:232. https://doi.org/10.1186/s13046-020-01690-z.Search in Google Scholar PubMed PubMed Central
25. Allioli, N, Vincent, S, Vlaeminck-Guillem, V, Decaussin-Petrucci, M, Ragage, F, Ruffion, A, et al.. TM4SF1, a novel primary androgen receptor target gene over-expressed in human prostate cancer and involved in cell migration. Prostate 2011;71:1239–50. https://doi.org/10.1002/pros.21340.Search in Google Scholar PubMed
26. Hou, S, Hao, X, Li, J, Weng, S, Wang, J, Zhao, T, et al.. TM4SF1 promotes esophageal squamous cell carcinoma metastasis by interacting with integrin alpha6. Cell Death Dis 2022;13:609. https://doi.org/10.1038/s41419-022-05067-2.Search in Google Scholar PubMed PubMed Central
27. Kumari, R, Majumder, MM, Lievonen, J, Silvennoinen, R, Anttila, P, Nupponen, NN, et al.. Prognostic significance of esterase gene expression in multiple myeloma. Br J Cancer 2021;124:1428–36. https://doi.org/10.1038/s41416-020-01237-1.Search in Google Scholar PubMed PubMed Central
28. Li, Y, Jiao, Y, Luo, Z, Li, Y, Liu, Y. High peroxidasin-like expression is a potential and independent prognostic biomarker in breast cancer. Medicine 2019;98:e17703. https://doi.org/10.1097/md.0000000000017703.Search in Google Scholar
29. Benmerzoug, S, Chevalier, MF, Verardo, M, Nguyen, S, Cesson, V, Schneider, AK, et al.. Siglec-6 as a new potential immune checkpoint for bladder cancer patients. Eur Urol Focus 2022;8:748–51. https://doi.org/10.1016/j.euf.2021.06.001.Search in Google Scholar PubMed
30. Lee, S, Hu, Y, Loo, SK, Tan, Y, Bhargava, R, Lewis, MT, et al.. Landscape analysis of adjacent gene rearrangements reveals BCL2L14-ETV6 gene fusions in more aggressive triple-negative breast cancer. Proc Natl Acad Sci USA 2020;117:9912–21. https://doi.org/10.1073/pnas.1921333117.Search in Google Scholar PubMed PubMed Central
31. Luo, A, Jung, J, Longley, M, Rosoff, DB, Charlet, K, Muench, C, et al.. Epigenetic aging is accelerated in alcohol use disorder and regulated by genetic variation in APOL2. Neuropsychopharmacol 2020;45:327–36. https://doi.org/10.1038/s41386-019-0500-y.Search in Google Scholar PubMed PubMed Central
32. Pedrosa, R, Wismans, LV, Sinke, R, van der Weiden, M, van Eijck, C, Kros, JM, et al.. Differential expression of BOC, SPOCK2, and GJD3 is associated with brain metastasis of ER-negative breast cancers. Cancers 2021;13:2982. https://doi.org/10.3390/cancers13122982.Search in Google Scholar PubMed PubMed Central
Supplementary Material
This article contains supplementary material (https://doi.org/10.1515/oncologie-2023-0140).
© 2023 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Review Article
- Ethosomes as delivery system for treatment of melanoma: a mini-review
- Research Articles
- Pre-treatment predictors of cardiac dose exposure in left-sided breast cancer radiotherapy patients after breast conserving surgery
- Glycoprofiling of early non-small cell lung cancer using lectin microarray technology
- Overexpression of TRIM28 predicts an unfavorable prognosis and promotes the proliferation and migration of hepatocellular carcinoma
- MiRNA-219a-1-3p inhibits the malignant progression of gastric cancer and is regulated by DNA methylation
- The effect of ubiquitin-specific peptidase 21 on proliferation, migration, and invasion in DU145 cells
- Automatic prediction model of overall survival in prostate cancer patients with bone metastasis using deep neural networks
- Clinical neutrophil-related gene helps treat bladder urothelial carcinoma
- Forkhead Box P4 promotes the proliferation of cells in colorectal adenocarcinoma
- Effect of a CrossMab cotargeting CD20 and HLA-DR in non-Hodgkin lymphoma
- Case Reports
- Endoscopic resection of gastric glomus tumor: a case report and literature review
- Long bone metastases of renal cell carcinoma imaging features: case report and literature review
- The Warthin-like variant of papillary thyroid carcinomas: a clinicopathologic analysis report of two cases
- Corrigendum
- Corrigendum to: Experience of patients with metastatic breast cancer in France: results of the 2021 RÉALITÉS survey and comparison with 2015 results
Articles in the same Issue
- Frontmatter
- Review Article
- Ethosomes as delivery system for treatment of melanoma: a mini-review
- Research Articles
- Pre-treatment predictors of cardiac dose exposure in left-sided breast cancer radiotherapy patients after breast conserving surgery
- Glycoprofiling of early non-small cell lung cancer using lectin microarray technology
- Overexpression of TRIM28 predicts an unfavorable prognosis and promotes the proliferation and migration of hepatocellular carcinoma
- MiRNA-219a-1-3p inhibits the malignant progression of gastric cancer and is regulated by DNA methylation
- The effect of ubiquitin-specific peptidase 21 on proliferation, migration, and invasion in DU145 cells
- Automatic prediction model of overall survival in prostate cancer patients with bone metastasis using deep neural networks
- Clinical neutrophil-related gene helps treat bladder urothelial carcinoma
- Forkhead Box P4 promotes the proliferation of cells in colorectal adenocarcinoma
- Effect of a CrossMab cotargeting CD20 and HLA-DR in non-Hodgkin lymphoma
- Case Reports
- Endoscopic resection of gastric glomus tumor: a case report and literature review
- Long bone metastases of renal cell carcinoma imaging features: case report and literature review
- The Warthin-like variant of papillary thyroid carcinomas: a clinicopathologic analysis report of two cases
- Corrigendum
- Corrigendum to: Experience of patients with metastatic breast cancer in France: results of the 2021 RÉALITÉS survey and comparison with 2015 results

