Abstract
C4H11ClN2O, monoclinic, P21/n (no. 14), a = 4.8097(2) Å, b = 19.4142(10) Å, c = 7.8686(4) Å, β = 94.399(4)°, V = 732.58(6) Å3, Z = 4, R gt(F) = 0.0438, wR ref(F2) = 0.1308, T = 297 K.
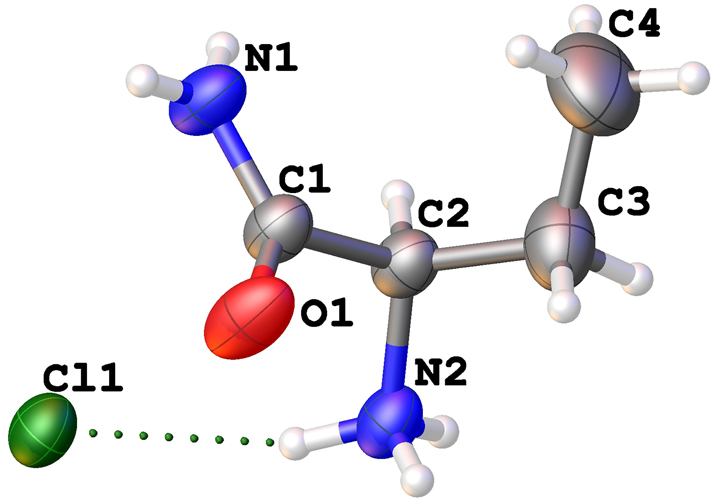
The molecular structure is shown in the figure. Table 1 contains the crystallographic data. The list of the atoms including atomic coordinates and displacement parameters can be found in the cif-file attached to this article.
Data collection and handling.
| Crystal: | Needle, colorless |
| Size: | 0.8 × 0.2 × 0.2 mm |
| Wavelength: | Cu Kα radiation (1.54184 Å) |
| μ: | 3.96 mm−1 |
| Diffractometer, scan mode: | XtaLAB Synergy R, ω-scans |
| θ max, completeness: | 77°, > 99 % |
| N(hkl)measured, N(hkl)unique: | 2743, 2743 |
| Criterion for I obs, N(hkl)gt: | I obs > 2 σ(I obs), 2743 |
| N(param)refined: | 90 |
| Programs: | Rigaku, 1 OLEX2, 2 SHELX 3 , 4 |
1 Source of materials
(R)-2-aminobutanamide hydrochloride and (S)-2-aminobutanamide hydrochloride were bought from Shanghai Titan Technology Co. Ltd. About 10 mg (R)-2-aminobutanamide hydrochloride and (S)-2-aminobutanamide hydrochloride powder were placed into a 10 ml penicillin bottle, followed by addition of 5 ml methanol. After all the powder was completely dissolved. The vial was then removed from the oven and left at room temperature. After 5 days, needle-shaped crystals were harvested.
2 Experimental details
The positions of hydrogen atoms on carbon atoms were calculated geometrically and refined using the riding model. Their coordinates and displacement parameters were constrained to ride on the carrier atom. The hydrogen atom on nitrogen atoms were located from difference Fourier map and refined with U iso(H) = 1.5U eq(N). The distances between the hydrogen atom and the nitrogen atom were refined freely. A DFIX restrain was used to get a better model for the hydrogen atoms. This structure was refined as non-merohedral twin with HKL 5 format, giving the batch scale of 0.5447:0.4553.
3 Comment
The title compound is the racemate of 2-aminobutanamide hydrochloride. 5 The R enantiomer of titled compound can be used for the synthesis of levetiracetam 6 and brivaracetam. 7
The titled compound was crystallized in P21/n space group with one 2-aminobutanamide cation and one chloride anion in its asymmetric unit, forming a 1:1 organic salt. The bond lengths and angles of this molecule are in the expected ranges. 8 Unlike in the structure of the enatiomerically pure R isomer, the four carbons of the racemate do not lie on the same plane, with a torsion angle of −65.8(3)° for C1–C2–C3–C4. The N2 atom forms hydrogen bonds with the three surrounding chloride ions, and the lengths of these hydrogen bonds are 3.1613(18) Å, 3.1740(18) Å, and 3.1594(18) Å, respectively. The N1 atom forms a hydrogen bond with the adjacent amide group, with a bond length of 3.100(3) Å (N1⋯ O11+x,+y,+z). Additionally, the N1 atom forms another hydrogen bond with a chloride ion, and this hydrogen bond has a relatively longer length of 3.392(2) Å (N1⋯Cl1−1/2+x,1/2−y,−1/2+z). This structure extends into a complex three-dimensional network through hydrogen bonding. In general all bond lengths are in the expected ranges. 5 , 8
-
Conflict of interest: The authors declare no conflicts of interest regarding this article.
-
Research funding: Discipline construction of Institute of Chemical Drugs, NIFDC (No.2025HYZX11).
-
Author contribution: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
References
1. Rigaku Oxford Diffraction CrysAlisPRO Software System, version 1.171.42.92a; Rigaku Corporation: Oxford, UK, 2023.Suche in Google Scholar
2. Dolomanov, O. V.; Bourhis, L. J.; Gildea, R. J.; Howard, J. A. K.; Puschmann, H. OLEX2: A Complete Structure Solution, Refinement and Analysis Program. J. Appl. Crystallogr. 2009, 42, 339–341. https://doi.org/10.1107/s0021889808042726.Suche in Google Scholar
3. Sheldrick, G. M. Crystal Structure Refinement with Shelxl. Acta Crystallogr. 2015, C71, 3–8. https://doi.org/10.1107/s2053229614024218.Suche in Google Scholar
4. Sheldrick, G. M. Shelxt – Integrated Space-Group and Crystal-Structure Determination. Acta Crystallogr. 2015, A71, 3–8. https://doi.org/10.1107/s2053273314026370.Suche in Google Scholar PubMed PubMed Central
5. Wang, F.; Luan, L.; Wang, Y.; Hu, X.; Zhang, C.; Liu, Y. The Crystal Structure of (R)-2-Aminobutanamide Hydrochloride, C4H11ClN2O. Z. Kristallogr. N. Cryst. Struct. 2025, 240, 433–434; https://doi.org/10.1515/ncrs-2025-0044.Suche in Google Scholar
6. Domingo Sanchez, J.; Gomez–Carpintero, J.; Gonzalez, J. F.; Menendez, J. C. Twenty-First Century Antiepileptic Drugs. An Overview of their Targets and Synthetic Approaches. Eur. J. Med. Chem. 2024, 272, 116476. https://doi.org/10.1016/j.ejmech.2024.116476.Suche in Google Scholar PubMed
7. Manoj, G.; Hanuman, N.; Gyanchander, E.; Bhosale, R. S.; Yadav, J. S. Synthetic Approaches Toward the Synthesis of Brivaracetam: An Antiepileptic Drug. ACS Omega 2022, 7, 2486–2503. https://doi.org/10.1021/acsomega.1c05378.Suche in Google Scholar PubMed PubMed Central
8. In, Y.; Fujii, M.; Sasada, Y.; Ishida, T. Structural Studies on C-Amidated Amino Acids and Peptides: Structures of Hydrochloride Salts of C-Amidated Ile, Val, Thr, Ser, Met, Trp, Gln and Arg, and Comparison with their C-Unamidated Counterparts. Acta Crystallogr.,Sec. B 2001, 57, 72; https://doi.org/10.1107/S0108768100013975.Suche in Google Scholar
© 2025 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.

