While conventional statistics is already a closed book for many medical professionals, they often convey a sense of awe or even anxiety when terms such as machine learning (ML) or artificial intelligence (AI) come into play. The idea that a machine can learn something on its own, or can even use mysterious computer structures such as “artificial neurons” to behave intelligently in some way often gives people who are not very familiar with statistics and computer science a feeling of powerlessness.
It is therefore the aim of this special issue of J Lab Med to dissolve the mysticism associated with these terms and provide fact-based knowledge about the fundamentals of data science at the crossroads between conventional statistics and machine learning (Figure 1). While there is considerable overlap between the three fields depicted in the diagram, each has its unique focus and methods. Statistical learning serves as a bridge between statistics as a centuries-old branch of mathematics and machine learning as an upcoming sub-discipline of artificial intelligence.
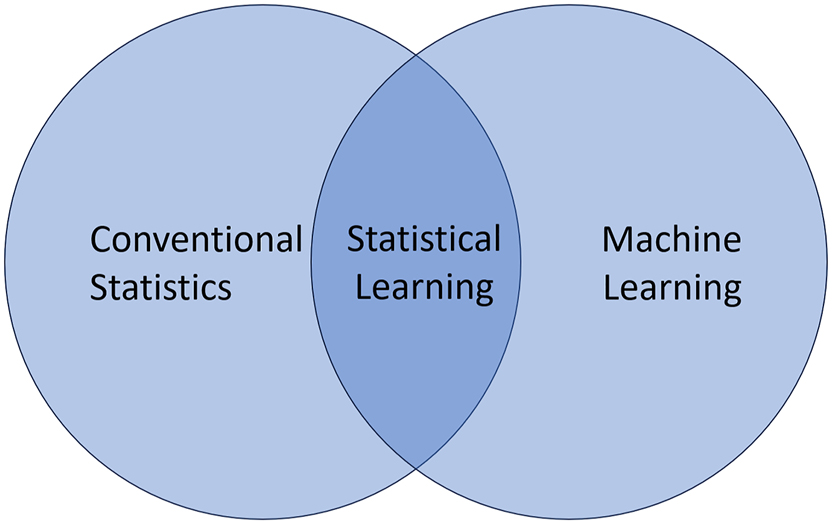
Statistical learning at the crossroads of statistics and machine learning.
To start with the left side of the diagram, a distinction is made between two main fields of statistics: descriptive statistics summarizes specific data (e.g. laboratory results) in the form of key figures (e.g. mean values and standard deviations) or graphics (e.g. histograms and boxplots), whereas inferential statistics applies a mathematical model to this data (e.g. a normal distribution), in order to draw generally valid conclusions about the underlying population (e.g. a reference interval).
To take now a look at the right side of the diagram, machine learning usually aims to make predictions on individual items (e.g. patients) and support decisions based on large amounts of sometimes complex and often less structured data without making strict assumptions on an underlying data model.
Statistical learning uses the principles of both disciplines, trying to detect patterns in the data and make individual predictions based on assumed underlying data models. Its two key concepts are unsupervised and supervised learning. While the former attempts to find hidden structures and relationships in data, the latter is based on data with one or more target attributes (so-called labels) such as ICD codes or effects of a therapy to assign unknown cases to the correct diagnoses or predict risks for specific outcomes.
In this special issue of J Lab Med, our focus is on practical applications of algorithms and tools in laboratory medicine, which should enable readers to analyze their own data and thus gain their own experience. Those who are already familiar with the programming language R (https://r-project.org) will benefit most from this special issue, as the worldwide R community provides countless ready-to-use and free of charge libraries for statistical data analysis and machine learning. But even readers without programming experience can benefit from the practical examples and illustrative figures. And some of them may hopefully be motivated to take an R course in order to use such programs themselves.
Some of the topics were already discussed in the first special issue of the “Applied Biostatistics in Laboratory Medicine” series, which appeared a year ago 1], [2], [3. Due to the current AI hype triggered by the widespread use of ChatGPT and other large language models (LLM), we as editors have decided to dedicate an entire issue now to this crossover area between statistics and artificial intelligence with a focus on methods that are based on statistical models for laboratory results (Table 1).
Overview of articles in this special issue.
| First author | Topic | References |
|---|---|---|
| Annika Meyer | Use of ChatGPT as an educational tool | doi: https://doi.org/10.1515/labmed-2024-0034 |
| Inga Trulson | Machine learning techniques for data exploration and classification | doi: https://doi.org/10.1515/labmed-2024-0100 |
| Amani Al-Mekhlafi | Standardization of laboratory data for machine learning | doi: https://doi.org/10.1515/labmed-2024-0051 |
| Sandra Klawitter | Regression tree model for the estimation of reference intervals | doi: https://doi.org/10.1515/labmed-2024-0083 |
| Tobias Blatter | Reference interval estimation using a statistical optimization algorithm | doi: https://doi.org/10.1515/labmed-2024-0076 |
| Georg Hoffmann | Gaussian mixture modelling for the verification of reference intervals | doi: https://doi.org/10.1515/labmed-2024-0118 |
The first article [4] shows how artificial intelligence can be used to acquire the programming skills in R mentioned above. The second article [5] presents some basic machine learning algorithms that are widely used in clinical studies and can be implemented with little programming effort. In the next article [6], a specific step of this workflow, namely the standardization and scaling of input data, is discussed in more detail.
The remaining three articles 7], [8], [9 deal with the verification of reference intervals in laboratory medicine. This highly topical subject was already dealt with in the last special issue [10, 11], but is presented here from the specific perspective of statistical learning. All three articles show that the borders between conventional statistics and machine learning are rather fluid, and that there is technical skill and medical knowledge rather than mystery behind the proper use of statistical and machine learning algorithms.
References
1. Hoffmann, G, Klawonn, F. Applied biostatistics in laboratory medicine. J Lab Med 2023;47:141–2. https://doi.org/10.1515/labmed-2023-0060.Search in Google Scholar
2. Berns, F, Heilig, N, Stumpe, F, Kirchhoff, J. Medical operational AI: artificial intelligence in routine medical operations. J Lab Med 2023;47:171–9. https://doi.org/10.1515/labmed-2023-0011.Search in Google Scholar
3. Witte, H, Blatter, T, Nagabhushana, P, Schär, D, Ackermann, J, Cadamuro, J, et al.. Statistical learning and big data applications. J Lab Med 2023;47:181–6. https://doi.org/10.1515/labmed-2023-0037.Search in Google Scholar
4. Meyer, A, Ruthard, J, Streichert, T. Dear ChatGPT – can you teach me how to program an app for laboratory medicine? J Lab Med 2024;48:197–201. https://doi.org/10.1515/labmed-2024-0034.Search in Google Scholar
5. Trulson, I, Holdenrieder, S, Hoffmann, G. Using machine learning techniques for exploration and classification of laboratory data. J Lab Med 2024;48:203–14. https://doi.org/10.1515/labmed-2024-0100.Search in Google Scholar
6. Al-Mekhlafi, A, Klawitter, S, Klawonn, F. Standardization with zlog values improves exploratory data analysis and machine learning for laboratory data. J Lab Med 2024;48:215–22. https://doi.org/10.1515/labmed-2024-0051.Search in Google Scholar
7. Klawitter, S, Böhm, J, Tolios, A, Gebauer, J. Automated sex and age partitioning for the estimation of reference intervals using a regression tree model. J Lab Med 2024;48:223–37. https://doi.org/10.1515/labmed-2024-0083.Search in Google Scholar
8. Blatter, T, Nakas, C, Leichtle, A. Direct, age and gender-specific reference intervals: applying a modified M-estimator of the Yeo-Johnson transformation to clinical real-world data. J Lab Med 2024;48:239–50. https://doi.org/10.1515/labmed-2024-0076.Search in Google Scholar
9. Hoffmann, G, Allmeier, N, Kuti, M, Holdenrieder, S, Trulson, I. How Gaussian mixture modelling can help to verify reference intervals from laboratory data with a high proportion of pathological values. J Lab Med 2024;48:251–8. https://doi.org/10.1515/labmed-2024-0118.Search in Google Scholar
10. Meyer, A, Müller, R, Hoffmann, M, Skadberg, Ø, Ladang, A, Dieplinger, B, et al.. Comparison of three indirect methods for verification and validation of reference intervals at eight medical laboratories: a European multicenter study. J Lab Med 2023;47:155–63. https://doi.org/10.1515/labmed-2023-0042.Search in Google Scholar
11. Klawitter, S, Kacprowski, T. A visualization tool for continuous reference intervals based GAMLSS. J Lab Med 2023;47:165–70. https://doi.org/10.1515/labmed-2023-0033.Search in Google Scholar
© 2024 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Editorial
- At the crossroads between statistics and artificial intelligence: statistical learning in laboratory medicine
- Articles
- Dear ChatGPT – can you teach me how to program an app for laboratory medicine?
- Using machine learning techniques for exploration and classification of laboratory data
- Standardization with zlog values improves exploratory data analysis and machine learning for laboratory data
- Automated sex and age partitioning for the estimation of reference intervals using a regression tree model
- Direct, age- and gender-specific reference intervals: applying a modified M-estimator of the Yeo-Johnson transformation to clinical real-world data
- How Gaussian mixture modelling can help to verify reference intervals from laboratory data with a high proportion of pathological values
Articles in the same Issue
- Frontmatter
- Editorial
- At the crossroads between statistics and artificial intelligence: statistical learning in laboratory medicine
- Articles
- Dear ChatGPT – can you teach me how to program an app for laboratory medicine?
- Using machine learning techniques for exploration and classification of laboratory data
- Standardization with zlog values improves exploratory data analysis and machine learning for laboratory data
- Automated sex and age partitioning for the estimation of reference intervals using a regression tree model
- Direct, age- and gender-specific reference intervals: applying a modified M-estimator of the Yeo-Johnson transformation to clinical real-world data
- How Gaussian mixture modelling can help to verify reference intervals from laboratory data with a high proportion of pathological values


