Abstract
Background and Objectives
Circular RNAs play a vital role in developing triple-negative breast cancer (TNBC). Likewise, the function of circRNAs in TNBC resistance to chemotherapy remains largely unknown. Here, we aimed to investigate whether circPLK1 has a biological efect on anthracycline resistance in TNBC.
Methods
We identified circPLK1—a circRNA—using a circRNA microarray in TNBC cells and paired TNBC samples. We assessed the role of circPLK1 in anthracycline resistance in TNBC. Cytotoxicity assay, colony formation assay, and flow cytometry were performed as functional experiments. Western blot analysis, qRT-PCR, in situ hybridization, and immunohistochemistry were used to evaluate the expression of circPLK1, miR-940, and ETS1. RNA immunoprecipitation and luciferase reporter assay were conducted to evaluate the interaction among circPLK1, miR-940, and ETS1. We evaluated the prognosis value of circPLK1 and ETS1 in 240 TNBC patients.
Results
The upregulation of circPLK1 in non-pCR TNBC patients receiving anthracyclines-based neoadjuvant chemotherapy was significantly associated with aggressive characteristics. Colony formation and doxorubicin resistance of TNBC cells were promoted by circPLK1 overexpression but inhibited by circPLK1 knockdown in vitro. circPLK1 overexpression facilitated doxorubicin resistance of TNBC in the nude mouse xenograft model. We found that circPLK1 could promote ETS1 expression by sponging miR-940. High circPLK1 and ETS1 expression were significantly associated with reduced survival in TNBC.
Conclusion
circPLK1 plays a vital role in the resistance of TNBC to anthracycline and is associated with poor prognosis. The inhibition of circPLK1 may be a practical therapeutic approach to modulate anthracycline resistance in TNBC.
Introduction
Triple-negative breast cancer (TNBC), characterized by negative expression of ER, PR, and ERBB2 receptors, is challenging to treat.[1] In contrast to other subtypes that could be treated with targeted therapies, chemotherapy remains the current treatment option for TNBC patients.[2] Breast cancer is believed to follow the principle by “The clonal evolution theory” and finally to invasive and metastatic stages.[3] The aim of chemotherapy is to reduce the likelihood of cancer invasive and recurrence.[4] However, cytotoxic chemotherapy produces mixed results. Some TNBC patients rapidly develop resistance to chemotherapy, leading to a high incidence of early relapse. Therefore, further research into the mechanisms of TNBC therapeutic resistance is required. It is urgent to identify novel molecular targeted strategies to improve the therapeutic benefit of chemotherapy and the prognosis of TNBC patients.[5]
The circular RNA (circRNAs) family is a class of non-coding RNA with a cyclic structure and exist widely in eukaryotes.[6] Some circRNAs have been identified as a necessary type of competing endogenous RNA (ceRNA) and as essential epigenetic regulators with crucial roles in tumor initiation and malignant progression. They contain multiple binding sites for microRNAs (miRNAs), which sponge miRNAs and modulate the expression of miRNAs’ target genes by participating in a circRNA-miRNA-mRNA regulatory network.[7] MiRNAs are functioning extensively in various cellular processes such as apoptosis and carcinogenesis.[8, 9] Our previous research found that some circular RNAs played a vital role in breast cancer tumorigenesis, development and metastases, such as circEZH2 [10] and circEPSTI1.[11] The role of circRNAs in regulating therapy resistance has attracted increasing attention. For instance, in gastric cancer cells, circAKT3 could target the miR-198/PIK3R1 axis and effectively affect cisplatin sensitivity.[12] CDR1as (hsa_circ_0001946) can affect the sensitivity of non-small cell lung cancer cells to cisplatin.[13] High hsa_circ_100053 expression predicted imatinib resistance in chronic myeloid leukemia.[14] However, the function of circRNAs in TNBC drug resistance remains largely unknown.
To determine the possible roles of circRNAs in TNBC, we performed circRNA microarrays in TNBC patients’ samples and four cell lines (one MCF-10A and three TNBC cell lines) and found 11 upregulated circRNAs in the TNBC tumor. To explore the potential role of circRNAs in chemoresistance, we performed qRT-PCR for 5 upregulated circRNAs in 20 cases of TNBC patients who later received neoadjuvant chemotherapy (anthracyclines-based). We found that hsa_circ_038632, which originates from exons of the Polo-like kinase 1 (PLK1) gene and is termed circPLK1, was significantly upregulated in tumor tissues of non-PCR patients. Our previous study found that circPLK1 regulated tumor progression through the ceRNA mechanism and was a prognostic factor in TNBC.[15] Here we evaluated the functions of circPLK1 in TNBC anthracycline resistance. Understanding of TNBC chemoresistance might provide a new therapeutic approach for TNBC patients.
Materials and methods
CircRNA microarray
This study analyzed three pairs of TNBC patients’ tumors and matched normal breast tissues, normal mammary epithelial cell lines MCF-10A, and three TNBC cell lines (MDA-MB-231, BT549 and MDA-MB-468) (Supplementary Figure S1A and S1B). We used Arraystar Human circRNA Array V2 (Aksomics, Shanghai, China) in this study. All procedures were defined in our previous studies.[11, 15] CircRNAs with a fold change of > 2.0 or < −2.0 (P < 0.05) were screened and presented using hierarchical clustering.
Cell culture and cell transfection
MCF-10A, 184A1, MDA-MB-231, BT20, HCC38, MDA-MB-468, HCC1806, BT549, MCF-7, MDA-MB-415, BT483, BT474, T47D, Skbr-3 and HEK 293T cells were obtained from the ATCC database (Manassas, VA, USA). All cells were cultured according to the ATCC’s protocols (ATCC; https://www.atcc.org/). MDA-MB-231/DOX was a drug resistance cell subline and cultivated using the stepwise increasing method of low-concentration doxorubicin (DOX) (Sigma Aldrich, MO, USA). DOX was added to the culture medium of MDA-MB-231/DOX cells at a concentration of 0.01 g/mL. Phosphorylated deoxyribonucleic acid single strand or plasmid transfection was performed using Lipofectamine 3000.
Patient samples
We collected patient samples who received breast cancer treatment at Sun Yat-Sen University Cancer Centre. We obtained a written informed consent from each participant before study participation and treatment. The response was evaluated using the Response Evaluation Criteria in Solid Tumors guidelines (version 1.1). Defining a pathological complete response (pCR) as the absence of residual invasive breast carcinoma in both breast and axillary nodes. This study was conducted according to local regulations and the Declaration of Helsinki. This study was approved by the Ethics Committee of Sun Yat-Sen University Cancer Center Health Authority (GZR2017-163) and the Ethics Committee of Guangdong Provincial People’s Hospital (2019-040H-1).
Quantitative reverse transcription polymerase chain reaction (qrt-pcr)
Primer information is provided in Supplementary Table S1. The detailed steps are described in our previous study.
RNase R digestion
RNA (2 μg) was incubated with or without RNase R (3 U/μg, Epicenter Biotechnologies, Shanghai, China) for 20 min at 37 °C to confirm the circRNA characteristics.
Fluorescence in situ hybridization (FISH)
Fluorescence in situ hybridization (FISH) was performed using the Fluorescence in situ Hybridization Kit (RiboBio, Guangzhou, China) and a specific probe against circPLK1. After hybridization, the cells were washed using saline sodium citrate solution for 5 min and stained with 4’, 6-diamidino-2-phenylindole dihydrochloride (DAPI) for 10 min without light. The cells were then photographed using a fluorescence microscope.
Oligonucleotide transfection
Transfections were performed using Lipofectamine 3000 (Invitrogen, Cat#L3000015, CA, USA) according to the manufacturer’s instructions. All siRNAs targeting circPLK1 were synthesized by GenePharma (Shanghai, China). The sequences that were used are presented in Supplementary Table S2. GeneCopoeia (Rockville, MD, USA) synthesized miRNA mimics and miRNA inhibitors.
Cell counting kit 8 (CCK8) assay
CCK8 experiments were performed using the CCK8 Kit (Dojindo Laboratories, Japan) according to the manufacturer’s instructions as described previously.[15]
Colony formation assay
At 48 h after transfection, the cells were harvested and seeded in six-well plates at 500 cells per well and cultured for 14 days (treated with or without docetaxel in the first 48 h). The colonies in each well were stained with crystal violet solution 2% ethanol and 0.5% crystal violet), imaged and counted.
Apoptosis assay
The cells were treated with or without DOX (1 μmol/L) for 48 h and harvested. Andy Fluor 488 Annexin V and PI Apoptosis Kit was used to detect cell apoptosis, according to the manufacturer’s protocol (GeneCopoeia, Rockville, MD, USA).
5-ethynyl-2′-deoxyuridine (EdU) assay
Edu assay was performed using the IClick™EdU Andy Fluor™488 Flow Cytometry Assay Kit (GeneCopoeia, Rockville, MD, USA). The specific procedures were conducted according to our previous study.[11]
Dual-luciferase reporter assay
To generate wild-type plasmids (circPLK1 WT and ETS1 3’UTR WT) and mutant-type plasmids (circPLK1 MT and ETS1 3’UTR Mut), the wild-type sequence of circPLK1 or ETS1 containing miR-940 binding sites or the mutant sequences were amplified and inserted into pmirGlO luciferase reporter vector (Promega, WI, USA). The sequence information is provided in Supplementary Figure S2A and S2B. HEK 293T cells, MDA-MB-231 cells, or BT549 cells were subjected to trypsin digestion, followed by centrifugation and resuspension in complete culture medium. The resuspended cells were then seeded into a 96-well plate, which was subsequently placed in a cell incubator for cultivation. The seeding density was adjusted to 50%–60% growth density on the following day. Subsequently, the cells were subjected to serum deprivation by switching to serum-free medium and were starved for 0.5 h. During this period, the transfection mixture was prepared as follows: Initially, 10 μL of Opti-MEM medium (Gibco, Thermo Fisher Scientific, MS, USA). was added to a sterile EP tube. In Solution A, 0.2 μg of wild-type plasmid/mutant plasmid and 5 pmol of miR-940 inhibitors, miR-940 mimics, or NC were thoroughly mixed with Opti-MEM medium. Concurrently, 10 μL of Opti-MEM medium was combined with 0.3 μL of Lipofectamine 3000 (Invitrogen, CA, USA) to form Solution B. After a 5-minute incubation at room temperature, Solutions A and B were mixed thoroughly and allowed to stand at room temperature for an additional 20 min. Following a 6-hour incubation, complete culture medium was added to the cells, and they were continued to be cultured. Cells were harvested 48 h later. Luciferase reporter assay using the Dual-Luciferase reporter assay system (Promega, WI, USA). Relative luciferase activity was normalized to the Renilla luciferase internal control. Triplicate independent experiments were also performed.
Establishment of cell lines with stable circPLK1 overexpression
For constructing of the pLVX-puro plasmid and packaging of the circPLK1 overexpression, we used the empty plasmid as a negative control (Vector). All generated constructs were confirmed by sequencing. CircPLK1 overexpressing cells were named circPLK1/231 and circPLK1/BT549; the corresponding control cells were named Vector/231 and Vector/BT549.
RNA immunoprecipitation assay
In accordance with the manufacturer’s instructions, RIP was performed using a Thermo Scientific RIP kit (Thermo Fisher Scientific, Waltham, MA, USA). The human anti-AGO2 antibody (Millipore, Billerica, MA, USA) or negative control IgG (Abcam, MA, USA) were used for RNA immunoprecipitation assay.
Western blot
Cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer (CWBIO, Cat#CW2333S, Beijing, China) supplemented with a protease inhibitor cocktail (Roche, Cat# 04693116001, Mannheim, Germany) and a phosphatase inhibitor cocktail (Roche, Cat# 04906837001, Mannheim, Germany). Western blot was performed as described previously.[11] Primary antibodies include anti-ETS1 (1∶500, Abcam, Cat# ab186844, MA, USA) and anti-β-actin antibody (1: 1000, Affinity biosciences OH, Cat# AF7018, USA). Prestained Protein Marker II (10–200 kDa, Servicebio, Cat#G2058, Wuhan, China) were used as standard. Protein bands were visualized using ECL reagents (Thermo Fisher Scientific, Cat#34577, Waltham, MA, USA).
Mouse xenograft model
All animal experimental protocols were approved by the Institutional Animal Care and Use Committee of Guangdong Provincial People’s Hospital (2019-040A-1). CircPLK1-overexpressed (circPLK1/231) and control cells (Vector/231) were collected and suspended in 200 μL of phosphate-buffered saline (PBS) (5×106 cells per mL). There are five 4-week-old female BALB/c nude mice in each group. Cells were injected into the mammary fat pads of nude mice. Ten days after cell injection, the mice were injected intraperitoneally with 1 mg/ kg DOX or PBS (once per 3 days) four times. Tumor cell growth every 4 days was monitored using bioluminescence imaging (PerkinElmer, Waltham, MA, USA). Tumor volume was calculated as 0.52×L×W2 (L = length, and W = width). After 28 days of growth, tumors were harvested and samples were collected for further analysis. The mice were sacrificed under anesthesia to minimize discomfort and pain: intraperitoneal injections of sodium pentobarbital (50 mg/ kg) were followed by cervical dislocation.
Immunohistochemistry (IHC) and in situ hybridization (ISH) analysis
Tumor tissues were embedded in paraffin and cut into 4-μm sections. ISH and IHC analyses were performed as described previously. The circPLK1 probe for ISH was Digoxin-5’-TGCAGCTCCAGGAGAGAGAATATTC-3’-Digoxin. The intensity of circPLK1 or ETS1 staining was scored as follows: 1 (no staining), 2 (weak staining), 3 (moderate staining), or 4 (strong staining). The low expression group is defined as IHC or ISH scores of 1 or 2. The high expression group is defined as IHC or ISH scores of 3 or 4. Two pathologists conducted a blinded evaluation of the IHC and ISH scores.
Statistical analysis
The statistical tests indicated in the figure legends were calculated using GraphPad Prism 8.0 software unless specially indicated. Two-tailed Students’ t-tests were used to determine statistical significance. The categorical variables were analyzed using the Chi-square test or Fishers exact test. P < 0.05 was considered statistically significant. Survivals were depicted using Kaplan-Meier plots. Survival differences were assessed using log-rank tests (SPSS version 19.0). The data are presented as mean ± standard error of mean (SEM) unless otherwise indicated.
Results
CircPLK1 is upregulated and associated with non-pCR in TNBC
CircRNA expression in cell lines (MCF-10A, MDA-MB-231, BT549 and MDA-MB-468) and three TNBC tumor and non-cancerous matched breast tissue (NCMT) samples from patients were explored using microarray analysis (Supplementary Figure 1A and 1B). We found some circRNAs with consistent differential expression trends at the cell line level (MCF-10A vs. three TNBC cell lines) and the patient tissue level (TNBC vs. NCMT). The result demonstrated that 11 circular RNAs were upregulated, whereas 3 circular RNAs were downregulated in both TNBC cell lines and TNBC tumor (Figure 1A). Only has_circ_038632 upregulated fold change ≥3.0 in both cell line and patient tissue levels (Figure 1B). Using the human reference genome (GRCh37/hg19), has_circ_038632 was generated from the PLK1 (polo-like kinase) gene, which we termed circPLK1. The predicted PCR products were amplified using divergent primers across the circPLK1 junction and then confirmed using Sanger sequencing (Figure 1C). We confirmed the circular nature of circPLK1 through the RNase R digestion method (Figure 1D). Nuclear and cytoplasmic fractionation assay in MDA-MB-231 and BT549 cells (Figure 1E) and FISH (Figure 1F) in MDA-MB-231 revealed circPLK1 was predominantly localized in the cytoplasm. We tested circPLK1 expression levels in 2 normal mammary epithelial cell lines, six TNBC cell lines and six non-TNBC cell lines (Supplementary Figure 1C). There were 37 pairs of tissues from TNBC and normal breast tissue matched. More than 89% (33/37) of the tumors exhibited increased circPLK1 (P < 0.001) (Supplementary Figure 1D). To explore the biological roles of circPLK1 in anthracycline resistance, we performed qRT-PCR for circPLK1 in 20 cases of TNBC patients who received neoadjuvant chemotherapy (including an anthracycline, 10 pCR cases, and 10 non-pCR cases). The results revealed that circPLK1 expression in the non-pCR group was significantly higher than that in the pCR group (P < 0.001) (Figure 1G).
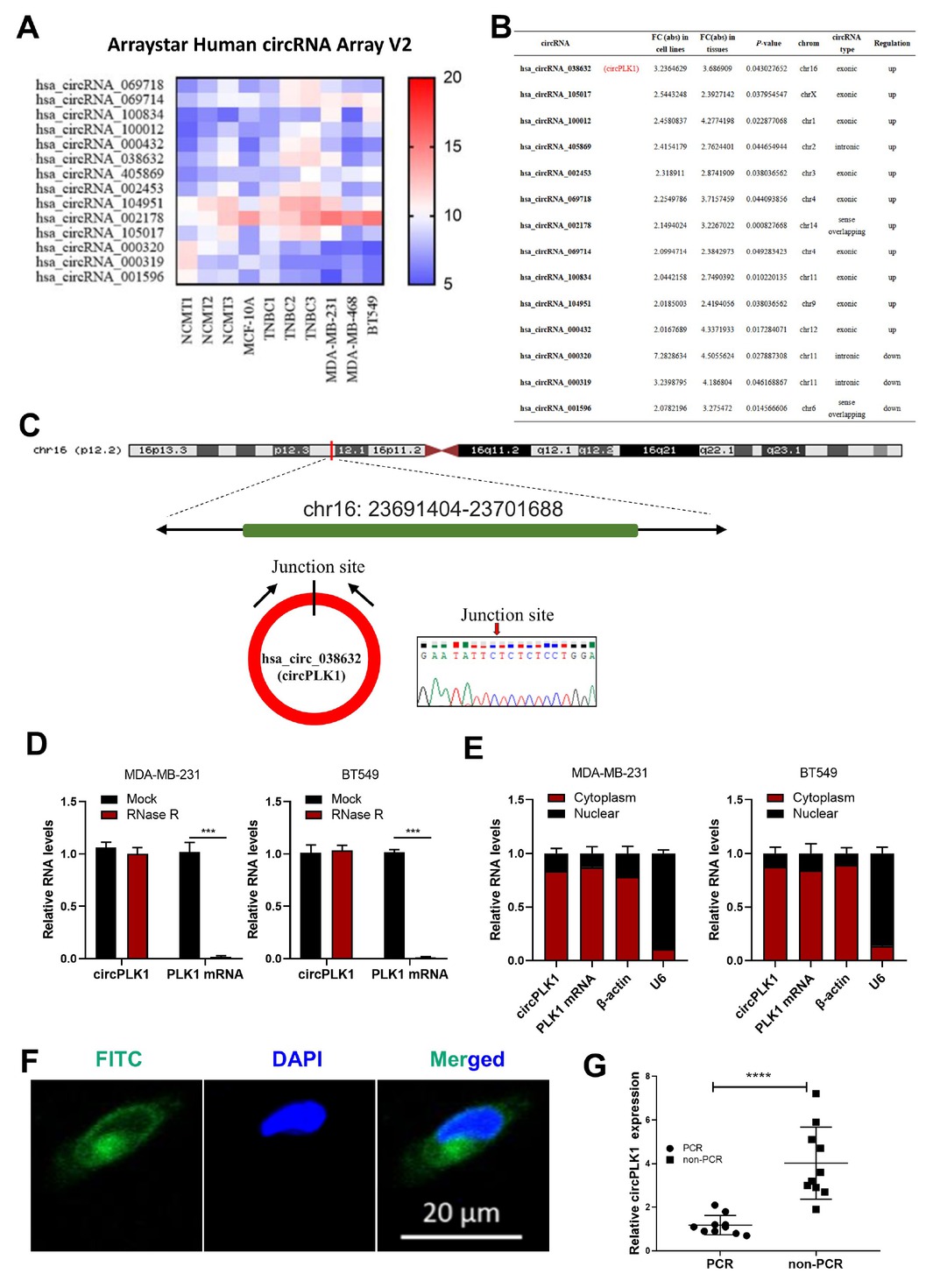
circPLK1 is upregulated expression and associated with non-pCR in TNBC. (A) Heatmap for circRNA expression in cell lines (MCF-10A, MDA-MB-231, BT549 and MDA-MB-468) and three TNBC tumor and non-cancerous matched breast tissue (NCMT) samples from patients. We mixed three biological repetitions of MCF-10A in a pool. (B) has_circ_038632 (circPLK1) upregulated fold change ≥ 3.0 in both TNBC cell line level and TNBC patient tissue level. (C) Schematic representation of the back splicing of circPLK1. The junction of circPLK1 was validated by Sanger sequencing. (D) circPLK1 and PLK1 mRNA levels in MDA-MB-231 (left) and BT549 (right) cells with or without RNase R treatment measured. ***P < 0.001. (E) The abundance of circPLK1 and PLK1 mRNA in either the cytoplasm or nucleus of MDA-MB-231 (left) and BT549 (right) cells. (F) FISH for circPLK1 in MDA-MB-231. Nuclei were stained with DAPI. Scale bar, 20 mm. (G) The expression of circPLK1 in 10 pCR cases and 10 non-pCR cases. ****P < 0.0001.
CircPLK1 enhanced the anthracycline resistance of TNBC cells
Given the inverse relationship between pCR and chemoresistance, we further explored whether circPLK1 affected TNBC anthracycline resistance. Higher expression of circPLK1 was detected in MDA-MB-231/ADR (doxorubicin-resistant cells) compared with MDA-MB-231, indicating it might be related to drug resistance (Figure 2A). Three siRNAs for the back-splice sequence of circPLK1 were designed and verified (Figure 2B). We investigated the effect of circPLK1 on anthracycline resistance in MDA-MB-231/ADR cells. Transfected MDA-MB-231/ ARD cells were treated with different doses of doxorubicin for 48 h. As presented in Figure 2C, the depletion of circPLK1 decreased the cell viability compared with in si-NC transfected cells or cells without transfections. We found that 1 μmol/L doxorubicin treatment had minimal effect on the colony formation of MDA-MB-231/ ADR cells, but when circPLK1 was knocked down, the colony formation decreased sharply (Figure 2D). Based on apoptosis assay results, after doxorubicin treatment, circPLK1 siRNA transfected cells exhibited a higher percentage of apoptotic cells than NC or si-NC transfected cells (Figure 2E). Moreover, the circPLK1 overexpression significantly promoted MDA-MB-231 and BT549 cell viability (Figure 2F), colony formation (Figure 2G), and suppressed doxorubicin-induced cell apoptosis (Figure 2H).

circPLK1 enhanced the anthracycline resistance of TNBC cells. (A) qRT-PCR analysis for the expression of circPLK1 in doxorubicin-resistant cells (MDA-MB-231/ADR) and MDA-MB-231. ***P < 0.001. (B) circPLK1 and PLK1 mRNA expression after treatment with three siRNAs. **P < 0.01, ***P < 0.001. (C) Transfected MDA-MB-231/ADR cells were treated with different doses of doxorubicin for 48 h, and the cell viability was determined using a CCK-8 assay. *P < 0.05, ***P < 0.001. (D) The colony-forming ability of the control (NC), si-NC-transfected and si-circ-1-transfected MDA-MB-231/ADR cells in the absence or presence of doxorubicin (1 μmol/L) for 48 h. (E) The apoptotic rates of MDA-MB-231/ADR cells transfected with si-NC and si-circ-1 in the absence or presence of doxorubicin (1 μmol/L) for 48 h. **P < 0.01. (F) circPLK1/231, Vector/231, circPLK1/BT549 and Vector/BT549 cells were treated with various concentrations of doxorubicin for 48 h, and the cell viability was determined using a CCK-8 assay. *P < 0.05, **P < 0.01. (G) The colony-forming ability of MDA-MB-231 and BT549 cells transfected with circPLK1 or vector after doxorubicin (1 μmol/L) treatment for 48 h. (H) Before apoptosis assay, transfected MDA-MB-231 and BT549 cells with circPLK1 or vector in the presence of doxorubicin (1 μmol/L) for 48 h.
CircPLK1 acted as a miRNA sponge of miR-940
According to the CircInteractome database,[16] circPLK1 served as a molecular sponge for miRNA. In those miRNAs, we found that miR-940 had three potential binding sites within the circPLK1 sequence (Figure 3A). Previous studies have indicated that circRNAs have the potential to function as miRNA sponges if they can form a circRNA-AGO2-miRNA complex by binding to AGO2 and miRNAs.[17, 18] Therefore, to investigate the existence of a circPLK1-AGO2-miR-940 complex, we conducted RIP experiments. The RIP assay on Ago2 revealed a dominant enrichment of circPLK1 and miR-940 to Ago2 in MDA-MB-231 and BT549 cells (Figure 3B). The recruitment of circPLK1 to the Ago2-related RNA-induced silencing complex allows it to interact with miR-940. In luciferase reporter assays, compared with the control group, overexpression of miR-940 reduced the luciferase activity of wild reporter by nearly 50% but had no effect on circPLK1-Mut reporter luciferase activity (Figure 3C). Conversely, an increase in luciferase activity was observed when the wild-type luciferase reporter was co-transfected with a miR-940 inhibitor (Figure 3D). These findings provide compelling evidence for the targeting relationship between circPLK1 and miR-940. Importantly, miR-940 did not significantly alter circPLK1 level, demonstrating that circPLK1 was not digested by miR-940 (Figure 3E). In colony formation assays (Figure 3F) and EdU assays (Figure 3G), the knockdown of circPLK1 in MDA-MB-231 could attenuate the miR-940 knockdown effects.
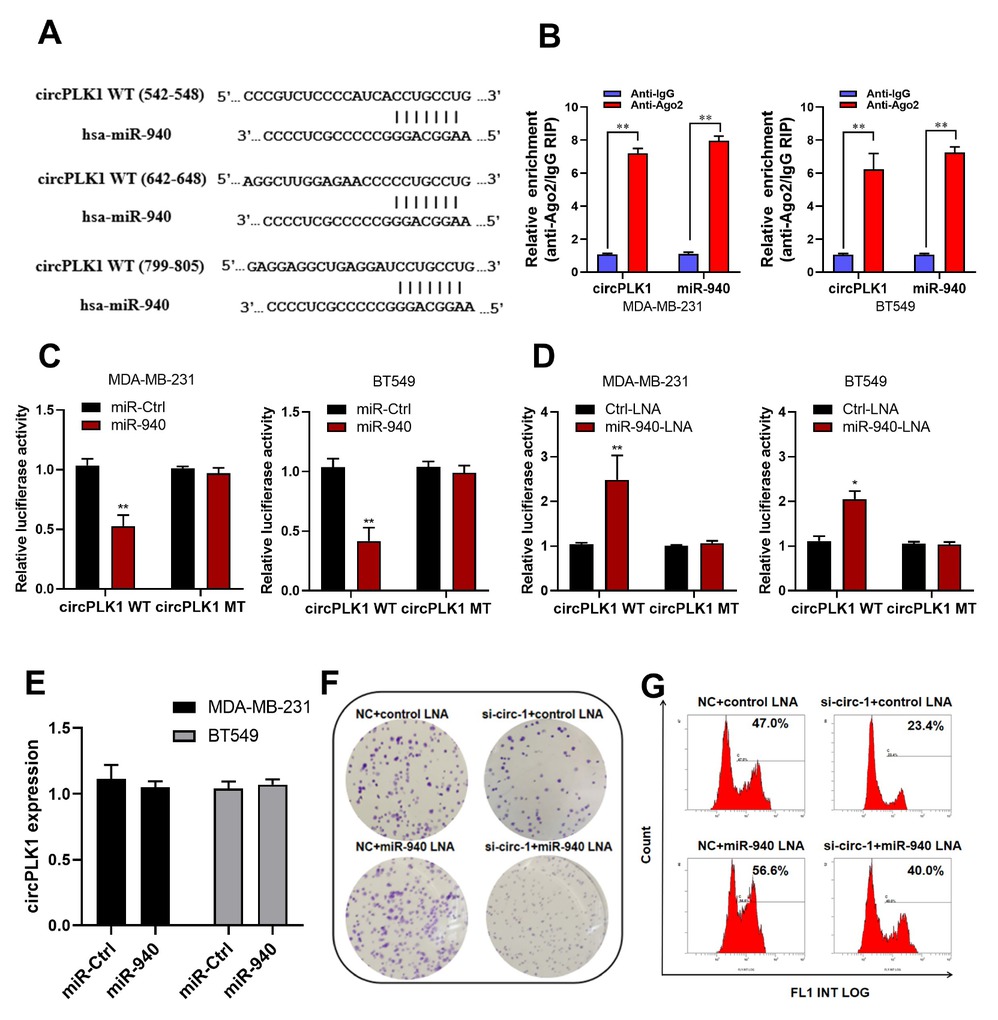
circPLK1 serves as a miRNA sponge for miR-940. (A) Schematic of the predicted miR-940 sites in the circPLK1 on circInteractome (https://circinteractome.nia.nih.gov/). (B) RNA immunoprecipitation assay revealed the enrichment of circPLK1 and miR-940 on Ago2 relative to IgG in MDA-MB-231 and BT549 cells. **P < 0.01. (C) Luciferase assay of MDA-MB-231 and BT549 cells co-transfected with a scrambled control, miR-940 mimics. **P < 0.01. (D) control-LNA, or miR-940-LNA and a luciferase reporter containing circPLK1-WT (circPLK1-WT (542-548), circPLK1-WT (642-648) and circPLK1-WT (799-805) were mixed at equal ratios.) or mutant constructs with mutated miR-940 binding sites (circPLK1-MT: circPLK1-MT (542-548), circPLK1-MT (642-648) and circPLK1-MT (799-805) were mixed at equal ratios.) . *P < 0.05, **P < 0.01. (E) qRT-PCR analysis of circPLK1 after transfection with a scrambled control or miR-940 mimics in MDA-MB-231 and BT549 cells. (F) Colony formation assay and (G) EdU assay were performed in MDA-MB-231 transduced with NC+ control-LNA, si-circ-1+ control-LNA, NC+ miR-940-LNA or si-circ-1+ miR-940-LNA.
ETS1 is a direct target gene of miR-940 and regulated by circPLK1
Using four published algorithms (miRWalk (http://zmf.umm.uni-heidelberg.de/), miRDB,[19] TARGETMINER[20] and TargetScan [21]), a list of predicted miR-940 target genes was generated. Algorithms predicted ETS1 as a target (Figure 4A and 4B). Notably, the accumulation of ETS1 in breast cancer cells promotes tumor growth and multidrug-resistance was reported.[22, 23] Therefore, ETS1 was selected for further observation. We constructed the full-length ETS1 3’-UTR luciferase reporter vectors with or without mutant of miR-940 target sites in the seed region. The results demonstrated that miR-940 mimics led to decreased fluorescence of the wild-type ETS1 3’-UTR but did not affect on the mutant vectors (Figure 4C). qRT-PCR revealed that miR-940 mimics had minimal effects on the ETS1 mRNA level (Figure 4D). Western blots revealed that the level of ETS1 protein was significantly decreased (Figure 4E). We also found that the knockdown of circPLK1 reduced the ETS1 protein levels (Figure 4F). As shown by colony formation and EdU assays, the knockdown of ETS1 also attenuated the effects of miR-940 knockdown on clone formation ability and proliferation (Figure 4G and 4H).

ETS1 is a direct target gene of miR-940 and regulated by circPLK1. (A) Venn diagram (miRWalk, miRDB, TARGETMINER and TargetScan) representing the overlap of co-target genes of miR-940. (B) Schematic of the predicted miR-940 site in the 3'UTR of ETS1 mRNA. (C) HEK 293T cells were co-transfected with wild-type (ETS1-3’UTR-wt) or mutant (ETS1-3'UTR-mut) 3’UTR luciferase reporter constructs as indicated, and either with miR-940 or non-targeting scrambled control. **P < 0.01, ***P < 0.001. (D) qRT-PCR analysis of ETS1 mRNA after transfection with a scrambled control, miR-940 mimics, control-LNA, or miR-940-LNA. (E) Western blot determined the effect of miR-940 mimics or miR-940-LNA on ETS1 protein expression in MDA-MB-231 and BT549 cells. (F) Western blot determined the effect of knockdown of circPLK1 on ETS1 protein expression in MDA-MB-231 and BT549 cells. We used MDA-MB-231/ADR for (G) Colony formation and (H) EdU assays, the knockdown of ETS1 also attenuated the knockdown effects of miR-940 in the presence of doxorubicin (1 μmol/L) for 48 h.
The tumor-promoting roles of circPLK1 overexpression in vivo
To further confirm the aforementioned findings, we used a nude mouse xenograft model was used. circPLK1-overexpressing (circPLK1/231) and control cells (Vector/231) were injected subcutaneously into the mammary fat pads of nude mice, followed by DOX or PBS treatment. The photos of xenograft tumors, tumor volume and tumor weight were shown (Figure 5A–D). Furthermore, we performed immunohistochemistry (Figure 5E) and western blots (Figure 5F) detection on the xenograft tumors. We found that the expression of ETS1 was increased in circPLK1/231 group tumors compared with Vector/231 group tumors.

The tumor-promoting roles of circPLK1 overexpression in vivo. (A) Growth curves of individual tumors are shown. (B) Final tumor weights of xenograft tumors at sacrifice. ***P < 0.001. (C) Tumors from circPLK1/231 and Vector/231 mice treated with PBS or DOX are shown. (D) Representative images of tumors which were detected by luciferase signals. (E) Each tumor was analyzed ETS1 by IHC analysis (Scale bar = 50 μm) and (F) western blot. Representative images for display.
High circPLK1 and high ETS1 expression predict poor prognosis in TNBC
We determined whether circPLK1 affects ETS1 expression levels in 240 TNBC tissue samples. The clinicopathological information of the 240 female TNBC patients were shown in Supplementary Table S3. Representative ISH and IHC images are shown in Figure 6A. As demonstrated by ISH analysis of a tumor microarray (n = 240), 42.9% of the samples (n = 103) exhibited high circPLK1 expression levels. By contrast, 39.2% (n = 94) of the tumor samples exhibited high ETS1 expression. ETS1 expression was higher in the high circPLK1 expression group than in the low circPLK1 expression group (Figure 6B). The clinical prognosis results revealed that patients with high circPLK1 and high ETS1 expression were significantly associated with reduced disease free survival (DFS) (Figure 6C) and overall survival (OS)(Figure 6D). We further detected the expression of ETS1 and circPLK1 in the 20 patients who received neoadjuvant chemotherapy. The results demonstrated that when circPLK1 and ETS1 were both highly expressed, the non-pCR rate of patients was the highest (Figure 6E).

circPLK1 is positively correlated with ETS1 expression and serves as a prognosis factor in TNBC. (A) Representative ISH images of circPLK1 and IHC images of ETS1 expression in two TNBC cases (100×Scale bar = 200 μm; 400×Scale bar = 50 μm. (B) The expression levels of ETS1 in low or high circPLK1 expression group (n = 240). ***P < 0.001. (C) The DFS and (D) OS curves for the four groups are presented (circPLK1 low/ETS1 low n = 112; circPLK1 high/ ETS1 low n = 34; circPLK1 low/ ETS1 high n = 25; circPLK1 high/ETS1 high n = 69). High levels of circPLK1 and ETS1 are correlated with reduced survival. (E) qRT-PCR analysis of circPLK1 and ETS1 in tumor tissues of 20 TNBC patients who received anthracyclines-based neoadjuvant chemotherapy. ***P < 0.001.
Discussion
The improvements in TNBC survival have been associated with the increased use of neoadjuvant or adjuvant chemotherapy. Anthracyclines are cell cycle non-specific chemotherapeutic agents that can induce DNA damage leading to increased toxicity. Anthracyclines serve as cornerstone chemotherapy in the treatment of TNBC patients. However, TNBC is very heterogeneous, as evidenced by genetic and gene expression profiling studies. Congenital or acquired resistance significantly influences anthracycline efficacy in TNBC.[24] The mechanisms of tumor resistance toward anthracycline have not been fully defined. CircPLK1 is a circular RNA generated from the PLK1 gene. Our previous studies have investigated circRNA profiling in breast cancer using a high-throughput circRNA Array and have found abundant differentially expressed circRNAs. We demonstrated the role of some circRNAs, including circPLK1, on breast cancer growth and lung metastasis. circPLK1 also has been proven to act as a carcinogenic driver in osteosarcoma,[25] lung cancer,[26] and malignant pleural mesothelioma.[25, 27] By regulating the miR-4500/IGF1 axis, circPLK1 silence inhibited breast cancer cell growth, migration, and invasion.[28] In osteosarcoma, circ0038632 (circPLK1) sponged miR-186 to upregulate the expression of DNMT3A and to promote tumor progression.[25] Upregulation of exosomal circPLK1 promotes non-small cell lung cancer by interacting with miR-1294 and high mobility group protein A1.[26] By regulating miR-1294/HMGA1, circPLK1 also promotes malignant pleural mesothelioma development.[27] In the present study, we discovered the significance of circPLK1 in the anthracycline resistance of TNBC.
CircRNAs are found in multiple tissues, possessing distinct expression patterns and exerting specific roles during tumor progression. There is growing evidence for the role of circRNAs in gene expression regulation.[29] CircRNAs are abundant, stable, and considered more appropriate tumor biomarkers than other RNAs. Previous investigations have discussed the role of circRNAs in drug resistance. For instance, cESRP1 enhances drug sensitivity by inhibiting TGF-β-mediated EMT in small-cell lung cancer.[30] Downregulation of two circRNAs transcribed from EIF3a reversed cisplatin resistance in lung cancer.[31] In osteosarcoma, the circ_0001258 can regulate GSTM2 levels and was confirmed to contribute to resistance to cisplatin-based chemotherapeutics.[32] By sponging miR-1252, circCELSR1 can upregulate FOXR2 expression to contribute to paclitaxel resistance in ovarian cancer.[33] However, the roles of circRNAs in TNBC anthracycline resistance have not been thoroughly explored.
Here, we demonstrated that circPLK1 upregulates ETS1 via miR-940 suppression to confer anthracycline resistance in TNBC (Figure 7). Higher expression of circPLK1 was detected in non-pCR patients compared with pCR patients in TNBC. Overexpression or knockdown of circPLK1 could affect the viability, colony formation, and apoptosis of doxorubicin-resistant cells. The mechanism of circPLK1 in TNBC anthracycline resistance was explored and verified. CircPLK1 was predominantly localized in the cytoplasm. Numerous studies have confirmed that circRNAs in the cytoplasm could regulate gene expression of genes by acting as miRNA sponges.[34] We screened candidate miRNAs to circPLK1 using bioinformatic approaches and focused on miR-940. MiR-940 was confirmed as a tumor suppressor in TNBC. Liu et al. reported that TNBC with decreased miR-940 and low miR-940 expression could predict poor prognosis.[35] Hou et al. found that miR-940 potentially targeted ZNF24 and inhibited TNBC proliferation and migration.[36] Our study found that the knockdown of circPLK1 promoted miR-940 expression, whereas it was inhibited upon the overexpression of circPLK1 in TNBC cells. We also found a sponging interaction between circPLK1 and miR-940.

A schematic diagram. circPLK1 upregulates ETS1 via miR-940 suppression to confer anthracycline resistance in TNBC.
Furthermore, comprehensive bio-informatics analyses and cell experiments proved that ETS1 was a direct target gene of miR-940 and regulated by circPLK1. ETS1 belongs to the member of the ETS family of transcription factors. It has been implicated as oncogenes and directly regulates several genes involved in invasion and metastasis, such as integrins and matrix metalloproteinase.[22] A major obstacle in tumor therapy is tumor cell resistance to anti-cancer drugs. ETS1 is involved in the drug resistance of pancreatic cancer, ovarian cancer, prostate cancer, hepatocellular carcinoma, bladder cancer and breast cancer.[37, 38, 39, 40] The effect of ETS1 on tumor drug resistance involves different pathways. For instance, in hepatocellular carcinoma, ETS1 regulates mitochondrial ROS pathways to mediate sorafenib resistance.[41] The ETS1/miR-23a-3p/ACSL4 axis contributes to sorafenib resistance by regulating ferroptosis.[42] High expression levels of MDR1 were the cause of cisplatin resistance in T24R2 cisplatin-resistant human bladder cancer cells, and MDR1 promoter activity was dependent on transcription factor ETS1.[43] In breast cancer, ETS1 is primarily expressed in the triple-negative sub-type.[44] Reducing in ETS1 expression enhances chemo-sensitivity to cisplatin and reverses NF-kappaB activation in MDA-MB-231/DDP cells (a cisplatin resistance TNBC cell line) and subcutaneous tumor transplants.[45] Our study found that circPLK1 overexpression increased ETS1 expression and facilitated anthracycline resistance of TNBC in vivo. ETS1 expression was positively correlated with circPLK1 expression in TNBC tissue. In addition, it was confirmed that the prognosis of TNBC with high circPLK1 and ETS1 expression was poor. In this regard, circPLK1 might eventually serve as a pharmacodynamic biomarker, as a prognostic biomarker, or perhaps both in TNBC. Meanwhile, the mechanism of ETS1 affects the sensitivity of TNBC to DOX is worthy of further study.
There are some limitations to our study. We used siRNA technology to knock down circPLK1. The siRNA technique has a risk of off-target effects, which might result in false-negative or false-positive results. Besides TNBC, it remains unclear whether circPLK1 confers anthracycline resistance in the other breast cancer subtypes and whether additional regulatory mechanisms are involved. The effects of circPLK1 on the sensitivity of other drugs remain to be clarified.
Conclusion
In summary, circPLK1 plays a vital role in the resistance of TNBC to anthracycline and is associated with poor prognosis. circPLK1 offers potential utility as a pharmacodynamic and prognostic biomarker. The inhibition of circPLK1 may be a practical therapeutic approach to modulate anthracycline resistance in TNBC.
Supplementary Information
Supplementary materials are only available at the official site of the journal (www.intern-med.com).
Funding statement: This work was supported by funds from the National Natural Science Foundation of China (81902828, 82272998, Bo Chen), the Science and Technology Program of Guangzhou (202201011427, Bo Chen), the Guangdong Basic and Applied Basic Research Foundation (2022A1515011599, Bo Chen), the Excellent Young Talent Program of Guangdong Provincial People’s Hospital (KY012021190, Bo Chen) and the Natural Science Foundation of Guangdong (2022A1515012213, Yunxian Mo).
Acknowledgements
None.
-
Author Contributions
Dai DN: Conceptualization, Data curation, Investigation, Methodology, Validation, Visualization, Writing-original draft. Mo YX: Data curation, Investigation, Methodology, Validation. Wu S: Conceptualization, Visualization, Software, Supervision. Zhuang ZH: Visualization, Software, Supervision. Song CL: Data curation, Investigation, Methodology. Liu LR: Methodology. Chen ZS: Conceptualization, Project administration, Supervision. Tang HL: Conceptualization, Methodology, Project administration, Supervision, Writing-review & editing. Chen B: Conceptualization, Methodology, Project administration, Supervision, Funding acquisition, Writing-review & editing. All authors contributed to this manuscript read and approved the final version.
-
Ethical Approval
The procedure and protocol have been reviewed and approved by the Ethics Committee of Sun Yat-Sen University Cancer Center Health Authority (GZR2017-163) and the Ethics Committee of Guangdong Provincial People’s Hospital (2019-040H-1). All animal experiments have been approved by the Institutional Animal Care and Use Committee of Guangdong Provincial People’s Hospital and conducted according to the guidance (2019-040A-1).
-
Informed Consent
All patients have consented to the study and written informed consent before study participation and treatment.
-
Conflict of Interest
The authors declare no competing interests.
-
Use of Large Language Models, AI and Machine Learning Tools
None declared.
-
Data Availability Statement
The datasets used in the study are available from the corresponding author on reasonable request.
References
1 Bianchini G, De Angelis C, Licata L, Gianni L. Treatment landscape of triple-negative breast cancer - expanded options, evolving needs. Nat Rev Clin Oncol. 2022;19:91–113.10.1038/s41571-021-00565-2Search in Google Scholar PubMed
2 Waks AG, Winer EP. Breast cancer treatment: A review. JAMA. 2019;321:288–300.10.1001/jama.2018.19323Search in Google Scholar PubMed
3 Man YG, Mannion C, Stojadinovic A, Peoples GE, Cho WC, Fu SW, et al. The most likely but largely ignored triggering factor for breast (or all) cancer invasion. J Cancer. 2023;14:573–590.10.7150/jca.82291Search in Google Scholar PubMed PubMed Central
4 Ye F, Dewanjee S, Li Y, Jha NK, Chen ZS, Kumar A, et al. Advancements in clinical aspects of targeted therapy and immunotherapy in breast cancer. Mol Cancer. 2023;22:105.10.1186/s12943-023-01805-ySearch in Google Scholar PubMed PubMed Central
5 Garrido-Castro AC, Lin NU, Polyak K. Insights into molecular classifications of triple-negative breast cancer: Improving patient selection for treatment. Cancer Discov. 2019;9:176–198.10.1158/2159-8290.CD-18-1177Search in Google Scholar PubMed PubMed Central
6 Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 2020;21:475-490.10.1038/s41580-020-0243-ySearch in Google Scholar PubMed
7 Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691.10.1038/s41576-019-0158-7Search in Google Scholar PubMed
8 Gao Z, Jiang J, Hou L, Zhang B. Dysregulation of MiR- 144–5p/RNF187 Axis contributes to the progression of colorectal cancer. J Transl Int Med. 2022;10:65–75.10.2478/jtim-2021-0043Search in Google Scholar PubMed PubMed Central
9 Chen B, Tang H, Liu X, Liu P, Yang L, Xie X, et al. miR-22 as a prognostic factor targets glucose transporter protein type 1 in breast cancer. Cancer Lett. 2015;356:410–417.10.1016/j.canlet.2014.09.028Search in Google Scholar PubMed
10 Liu P, Wang Z, Ou X, Wu P, Zhang Y, Wu S, et al. The FUS/circEZH2/ KLF5/ feedback loop contributes to CXCR4-induced liver metastasis of breast cancer by enhancing epithelial-mesenchymal transition. Mol Cancer. 2022;21:198.10.1186/s12943-022-01653-2Search in Google Scholar PubMed PubMed Central
11 Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, et al. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8:4003–4015.10.7150/thno.24106Search in Google Scholar PubMed PubMed Central
12 Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18:71.10.1186/s12943-019-0969-3Search in Google Scholar PubMed PubMed Central
13 Huang MS, Liu JY, Xia XB, Liu YZ, Li X, Yin JY, et al. Hsa_circ_0001946 inhibits lung cancer progression and mediates cisplatin sensitivity in non-small cell lung cancer via the nucleotide excision repair signaling pathway. Front Oncol. 2019;9:508.10.3389/fonc.2019.00508Search in Google Scholar PubMed PubMed Central
14 Lei P, Chen JJ, Liao CS, Liu GH, Zhou M. High circ_100053 predicts a poor outcome for chronic myeloid leukemia and is involved in imatinib resistance. Oncol Res 2019. Online ahead of print.Search in Google Scholar
15 Kong Y, Yang L, Wei W, Lyu N, Zou Y, Gao G, et al. CircPLK1 sponges miR-296–5p to facilitate triple-negative breast cancer progression. Epigenomics. 2019;11:1163–1176.10.2217/epi-2019-0093Search in Google Scholar PubMed
16 Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42.10.1080/15476286.2015.1128065Search in Google Scholar PubMed PubMed Central
17 Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W, et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer. 2020;19:65.10.1186/s12943-020-01152-2Search in Google Scholar PubMed PubMed Central
18 Zeng Y, Zou Y, Gao G, Zheng S, Wu S, Xie X, et al. The biogenesis, function and clinical significance of circular RNAs in breast cancer. Cancer Biol Med. 2021;19:14–29.10.20892/j.issn.2095-3941.2020.0485Search in Google Scholar PubMed PubMed Central
19 Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43: D 146–D152.10.1093/nar/gku1104Search in Google Scholar PubMed PubMed Central
20 Bandyopadhyay S, Mitra R. TargetMiner: microRNA target prediction with systematic identification of tissue-specific negative examples. Bioinformatics. 2009;25:2625–2631.10.1093/bioinformatics/btp503Search in Google Scholar PubMed
21 Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798.10.1016/S0092-8674(03)01018-3Search in Google Scholar PubMed
22 Lu G, Zhang Q, Huang Y, Song J, Tomaino R, Ehrenberger T, et al. Phosphorylation of ETS1 by Src family kinases prevents its recognition by the COP1 tumor suppressor. Cancer Cell. 2014;26:222–234.10.1016/j.ccr.2014.06.026Search in Google Scholar PubMed PubMed Central
23 Kars MD, Iseri OD, Gunduz U. Drug resistant breast cancer cells over-express ETS1 gene. Biomed Pharmacother. 2010;64:458–462.10.1016/j.biopha.2010.01.008Search in Google Scholar PubMed
24 Szekely B, Silber AL, Pusztai L. New therapeutic strategies for triple-negative breast cancer. Oncology (Williston Park). 2017;31:130–137.Search in Google Scholar
25 Tan X, Zeng C, Li H, Tan Y, Zhu H. Circ0038632 modulates MiR-186/ DNMT3A axis to promote proliferation and metastasis in osteosarcoma. Front Oncol. 2022;12:939994.10.3389/fonc.2022.939994Search in Google Scholar PubMed PubMed Central
26 Li C, Wang G, Ma X, Tao T, Li Q, Yang Y, et al. Upregulation of exosomal circPLK1 promotes the development of non-small cell lung cancer through the miR-1294/ high mobility group protein A1 axis. Bioengineered. 2022;13:4185–4200.10.1080/21655979.2022.2026727Search in Google Scholar PubMed PubMed Central
27 Zhang Q, Wang Z, Cai H, Guo D, Xu W, Bu S, et al. CircPLK1 Acts as a carcinogenic driver to promote the development of malignant pleural mesothelioma by governing the miR-1294/HMGA1 pathway. Biochem Genet. 2022;60:1527–1546.10.1007/s10528-022-10186-8Search in Google Scholar PubMed
28 Lin G, Wang S, Zhang X, Wang D. Circular RNA circPLK1 promotes breast cancer cell proliferation, migration and invasion by regulating miR-4500/IGF1 axis. Cancer Cell Int. 2020;20:593.10.1186/s12935-020-01694-xSearch in Google Scholar PubMed PubMed Central
29 Liu CX, Chen LL. Circular RNAs: Characterization, cellular roles, and applications. Cell. 2022;185:2016–2034.10.1016/j.cell.2022.04.021Search in Google Scholar PubMed
30 Huang W, Yang Y, Wu J, Niu Y, Yao Y, Zhang J, et al. Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93–5p to inhibit TGF-beta signalling. Cell Death Differ. 2020;27:1709–1727.10.1038/s41418-019-0455-xSearch in Google Scholar PubMed PubMed Central
31 Huang MS, Yuan FQ, Gao Y, Liu JY, Chen YX, Wang CJ, et al. Circular RNA screening from EIF3a in lung cancer. Cancer Med. 2019;8:4159–4168.10.1002/cam4.2338Search in Google Scholar PubMed PubMed Central
32 Zhu KP, Zhang CL, Ma XL, Hu JP, Cai T, Zhang L. Analyzing the Interactions of mRNAs and ncRNAs to predict competing endogenous RNA networks in osteosarcoma chemo-resistance. Mol Ther. 2019;27:518–530.10.1016/j.ymthe.2019.01.001Search in Google Scholar PubMed PubMed Central
33 Zhang S, Cheng J, Quan C, Wen H, Feng Z, Hu Q, et al. circCELSR1 (hsa_circ_0063809) Contributes to paclitaxel resistance of ovarian cancer cells by regulating FOXR2 expression via miR-1252. Mol Ther Nucleic Acids. 2020;19:718–730.10.1016/j.omtn.2019.12.005Search in Google Scholar PubMed PubMed Central
34 Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188–206.10.1038/s41571-021-00585-ySearch in Google Scholar PubMed
35 Liu W, Xu Y, Guan H, Meng H. Clinical potential of miR-940 as a diagnostic and prognostic biomarker in breast cancer patients. Cancer Biomark. 2018;22:487–493.10.3233/CBM-171124Search in Google Scholar PubMed
36 Hou L, Chen M, Yang H, Xing T, Li J, Li G, et al. MiR-940 inhibited cell growth and migration in triple-negative breast cancer. Med Sci Monit. 2016;22:3666–3672.10.12659/MSM.897731Search in Google Scholar
37 Khanna A, Mahalingam K, Chakrabarti D, Periyasamy G. Ets-1 expression and gemcitabine chemoresistance in pancreatic cancer cells. Cell Mol Biol Lett. 2011;16:101–113.10.2478/s11658-010-0043-zSearch in Google Scholar PubMed PubMed Central
38 Rahim S, Uren A. Emergence of ETS transcription factors as diagnostic tools and therapeutic targets in prostate cancer. Am J Transl Res. 2013;5:254–268.Search in Google Scholar
39 Wilson LA, Yamamoto H, Singh G. Role of the transcription factor Ets-1 in cisplatin resistance. Mol Cancer Ther. 2004;3:823–832.10.1158/1535-7163.823.3.7Search in Google Scholar
40 Kalet BT, Anglin SR, Handschy A, O’Donoghue LE, Halsey C, Chubb L, et al. Transcription factor Ets1 cooperates with estrogen receptor alpha to stimulate estradiol-dependent growth in breast cancer cells and tumors. PLoS One. 2013;8: e68815.10.1371/journal.pone.0068815Search in Google Scholar PubMed PubMed Central
41 Vishnoi K, Ke R, Viswakarma N, Srivastava P, Kumar S, Das S, et al. Ets1 mediates sorafenib resistance by regulating mitochondrial ROS pathway in hepatocellular carcinoma. Cell Death Dis. 2022;13:581.10.1038/s41419-022-05022-1Search in Google Scholar PubMed PubMed Central
42 Lu Y, Chan YT, Tan HY, Zhang C, Guo W, Xu Y, et al. Epigenetic regulation of ferroptosis via ETS1/miR-23a-3p/ACSL4 axis mediates sorafenib resistance in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2022;41:3.10.1186/s13046-021-02208-xSearch in Google Scholar PubMed PubMed Central
43 Oh SS, Lee KW, Madhi H, Jeong JW, Park S, Kim M, et al. Cordycepin resensitizes T24R2 cisplatin-resistant human bladder cancer cells to Cisplatin by inactivating Ets-1 dependent MDR1 transcription. Int J Mol Sci. 2020;21:171010.3390/ijms21051710Search in Google Scholar PubMed PubMed Central
44 Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol. 2015;35:20–38.10.1016/j.semcancer.2015.09.010Search in Google Scholar PubMed
45 Zhang Y, Wu J, Ye M, Wang B, Sheng J, Shi B, et al. ETS1 is associated with cisplatin resistance through IKKalpha/NF-kappaB pathway in cell line MDA-MB-231. Cancer Cell Int. 2018;18:86.10.1186/s12935-018-0581-4Search in Google Scholar PubMed PubMed Central
© 2025 Danian Dai, Jinhui Zhang, Yunxian Mo, Cailu Song, Lingrui Liu, Zhe-Sheng Chen, Hailin Tang, Bo Chen, published by De Gruyter on behalf of the SMP
This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Articles in the same Issue
- Perspective
- Advances in the diagnosis and treatment of gastrointestinal tumors under the concept of super minimally invasive surgery
- Review Article
- Harnessing the potential of small extracellular vesicle biomarkers for cancer diagnosis and prognosis with advanced analytical technologies
- Current understanding and controversy on brain access of GLP-1 and GLP-1 receptor agonists
- Mitochondrial quality control as a therapeutic target in cardiovascular disease: Mechanistic insights and future directions
- Original Article
- Causal association between gut microbiota composition and the risk of atrial fibrillation
- Artificial intelligence-based predictive model for relapse in acute myeloid leukemia patients following haploidentical hematopoietic cell transplantation
- CircPLK1 upregulates ETS1 to confer anthracycline resistance in triple-negative breast cancer
- N-acetylglucosaminyltransferase V attenuates myocardial infarction by mediating the insulin-like growth factor 1 receptor signaling pathway
- A similar effect of fibrinogen on efficacy and safety of tenecteplase versus alteplase in acute ischemic cerebrovascular events (TRACE II) trial
Articles in the same Issue
- Perspective
- Advances in the diagnosis and treatment of gastrointestinal tumors under the concept of super minimally invasive surgery
- Review Article
- Harnessing the potential of small extracellular vesicle biomarkers for cancer diagnosis and prognosis with advanced analytical technologies
- Current understanding and controversy on brain access of GLP-1 and GLP-1 receptor agonists
- Mitochondrial quality control as a therapeutic target in cardiovascular disease: Mechanistic insights and future directions
- Original Article
- Causal association between gut microbiota composition and the risk of atrial fibrillation
- Artificial intelligence-based predictive model for relapse in acute myeloid leukemia patients following haploidentical hematopoietic cell transplantation
- CircPLK1 upregulates ETS1 to confer anthracycline resistance in triple-negative breast cancer
- N-acetylglucosaminyltransferase V attenuates myocardial infarction by mediating the insulin-like growth factor 1 receptor signaling pathway
- A similar effect of fibrinogen on efficacy and safety of tenecteplase versus alteplase in acute ischemic cerebrovascular events (TRACE II) trial

